Significance
Our work reveals the genome-wide prevalence of G4 structural variations (G4Vs) associated with single-nucleotide variations (SNVs) in the human genome and how such G4Vs interact with gene-regulatory elements and subsequently affect the activities of genes accommodating the G4Vs. The G4Vs are enriched in genes around transcription start sites, implying their potential involvement in gene expression. Because G4 structures participate in essential physiological and pathological processes, the G4Vs should make crucial contributions to the genotype and phenotype connection and serve as potential therapeutic targets for personalized remedies and medicine.
Keywords: G-quadruplexes, single nucleotide variations, genetic variations
Abstract
G-quadruplexes (G4s) formed by guanine-rich nucleic acids play a role in essential biological processes such as transcription and replication. Besides the >1.5 million putative G-4–forming sequences (PQSs), the human genome features >640 million single-nucleotide variations (SNVs), the most common type of genetic variation among people or populations. An SNV may alter a G4 structure when it falls within a PQS motif. To date, genome-wide PQS–SNV interactions and their impact have not been investigated. Herein, we present a study on the PQS–SNV interactions and the impact they can bring to G4 structures and, subsequently, gene expressions. Based on build 154 of the Single Nucleotide Polymorphism Database (dbSNP), we identified 5 million gains/losses or structural conversions of G4s that can be caused by the SNVs. Of these G4 variations (G4Vs), 3.4 million are within genes, resulting in an average load of >120 G4Vs per gene, preferentially enriched near the transcription start site. Moreover, >80% of the G4Vs overlap with transcription factor–binding sites and >14% with enhancers, giving an average load of 3 and 7.5 for the two regulatory elements, respectively. Our experiments show that such G4Vs can significantly influence the expression of their host genes. These results reveal genome-wide G4Vs and their impact on gene activity, emphasizing an understanding of genetic variation, from a structural perspective, of their physiological function and pathological implications. The G4Vs may also provide a unique category of drug targets for individualized therapeutics, health risk assessment, and drug development.
G-quadruplexes (G4s) are four-stranded secondary structures formed by guanine-rich nucleic acids via a union of four guanine tracts (G-tracts). G4s play a role in essential cellular processes such as transcription, replication, genome instability, carcinogenesis, and other diseases (for recent reviews, see refs. 1 and 2). In the human genome, there exist >1.5 million putative G-4 sequences (PQSs) with a preferential enrichment near transcription start sites (TSSs) (3–6), implying a role of G4s in transcription regulation. PQSs can be classified into four major subtypes according to the variation in G-tracts or loops. The most studied canonical PQS motifs are defined by a consensus of G≥3(N1–7G≥3)≥3 (N denotes any of the four nucleotides) (4, 7), which can form a G4 of three or more guanine-tetrad (G-tetrad) layers, named 4G in this work (Fig. 1). Other well-characterized noncanonical G4s include those with a long loop of 8 to 15 nucleotides (4GL15) (8), a G-vacancy in one of the G-tetrad (GVBQ) (5), or a bulge of one non-G nucleotide in one of the G-tracts (Bulge) (9) (Fig. 1). These subtypes of G4s have been recently detected in the genome of living human cells (10).
Fig. 1.
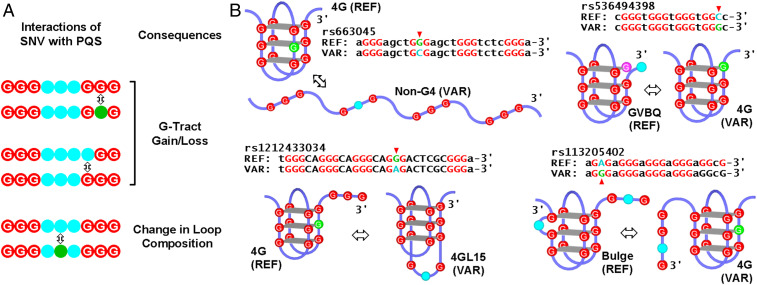
PQS–SNV interactions and examples of structural changes of G4 they may induce. (A) Interactions of an SNV with a PQS (showing only two G-tracts). (B) Representative structural changes (⇔) that may be caused by SNV at the indicated guanine (green to cyan). REF, reference; VAR, variant.
Genomic DNA also features single-nucleotide variations (SNVs), a type of genetic variation at single-nucleotide positions. SNVs can influence a variety of functions of a genome. Many SNVs are associated with diseases or disease susceptibility, response to drugs, treatments, and vaccines (for review, see refs. 11 and 12). An SNV within a PQS may affect the assembly of a G4 (Fig. 1). It has been reported that the change of a single nucleotide in a PQS could cause a drastic change in G4 structure (13, 14) and change gene expression, for example, by threefold of the c-Myc oncogene (14). A few studies showed that SNVs in the G4-L1 DNA PQS in humans are subject to evolutional selection (15), and those in the PQSs at the 5′ untranslated region (UTR) of human RNA are of physiological relevance (16, 17).
An SNV falling within a PQS either causes a gain/loss of G-tract or changes the composition of a loop (Fig. 1A), which can further result in complex changes in the physical properties of the G4 due to the extreme structural polymorphism of G4s arising from different folding pathways and base interactions (18). In particular, an SNV disrupting a G-tract is expected to have a more profound impact than that in a loop and may prevent G4 formation or cause a structural conversion from one subtype to another (Fig. 1B, arrowheads). Such structural changes can affect G4s in multiple aspects such as the following: 1) folding topology and/or conformation (13, 14); 2) folding/unfolding kinetics and stability (19); 3) equilibrium between G4 and duplex DNA (20); 4) local G4-protein interactions (21); 5) translocation of motor proteins on DNA (22); and 6) transmission of negative supercoiling wave in DNA, which can modulate G4 formation at a distal region (21, 23). Therefore, G4-interacting SNVs not only bring variations to sequences but also lead to diverse changes in secondary structures in genomic DNA and, perhaps, the corresponding RNA products, eventually leading to physiological consequences (2, 24, 25).
The number of SNVs disclosed increased significantly in recent years. According to build 154 of the Single Nucleotide Polymorphism Database (dbSNP) released in 2020 by the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/), over 640 million SNV entries have been achieved for humans. Since the size of the human genome is ∼3 billion nucleotides, this gives an average of one SNV in every five nucleotides. At such a high frequency, every PQS motif may interact with three or more SNVs on average, since a minimal canonical PQS motif is 15 nucleotides (i.e., G3NG3NG3NG3). For this reason, it is becoming ever more important to ask how genome-wide PQS–SNV interactions result in structural variations in G4s and consequently affect the functionality of genomes—a question that has not yet been addressed.
Therefore, we surveyed genome-wide PQS–SNV interactions in the human genome and explored their impact on G4 assembly and gene activity. Based on dbSNP build 154, we identified 5 million gains/losses or structural conversions of G4 variations (G4Vs) that may be caused by SNVs. Of them, 3.4 million are associated with genes with a profound enrichment near TSSs; 80% overlap with transcription factor–binding sites (TFBSs) and 14% with enhancers, implying they can impact transcription by affecting DNA-transcription factor/enhancer interactions. Interestingly, we found G4Vs displayed significantly higher occurrence in oncogenes and tumor-suppressor genes than in bulky genes. Our experiments show such G4Vs can significantly influence DNA processivity and gene expression at both RNA and protein levels. Collectively, our work reveals a prevalence of G4Vs and their impact on the functionality of the human genome. The structure–functionality relationship associated with the G4Vs should be a vital factor in determining the consequences of genetic variations.
Results
SNVs and PQSs in the Human Genome.
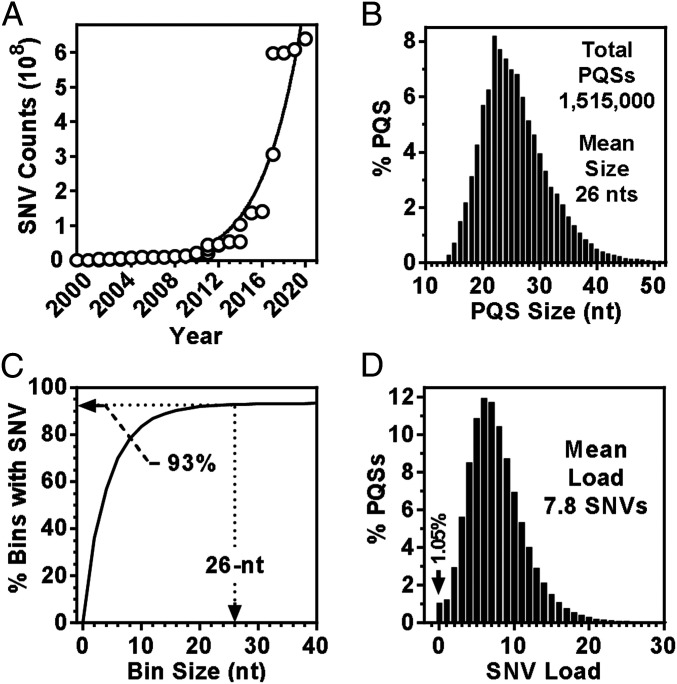
In dbSNP build 154, the total entries of SNVs, excluding the indels, have reached >640 million (Fig. 2A). The probability of a genomic region having an SNV is determined by the size of the region and the frequency of SNV occurrence. For convenience, we surveyed the size distribution of all the four subtypes of PQSs that have recently been shown to form G4 in living animal cells (10), which yielded an average of 26 nucleotides (nts) based on the consensus used (Fig. 2B). It should be mentioned here that the value depends on the definition of PQS, particularly the maximum length of loops. To evaluate the frequency of SNVs, we divided the human chromosomes into contiguous bins of defined sizes and calculated the percentage of bins that contained at least one SNV. This result gives the probability for a genomic DNA region of defined size to have one or more SNVs. As shown in Fig. 2C, if an SNV and PQS are random to each other, 93% of the PQSs would be SNV positive. In reality, however, nearly 99% of the PQSs were found SNV positive when we mapped the SNVs to the PQSs (Fig. 2D), giving an average load of 7.8 SNVs per PQS.
Fig. 2.
SNVs and PQSs in the human genome. (A) Accumulation of SNVs in the NCBI dbSNP database over the last decade. (B) The size distribution of PQS motifs. (C) SNV coverage across intervals of chromosomes. Arrow indicates the probability of finding at least one SNV in a 26-nt-sequence motif. (D) Distribution of SNV loads in PQSs.
A Preferential Occurrence of SNV in PQS.
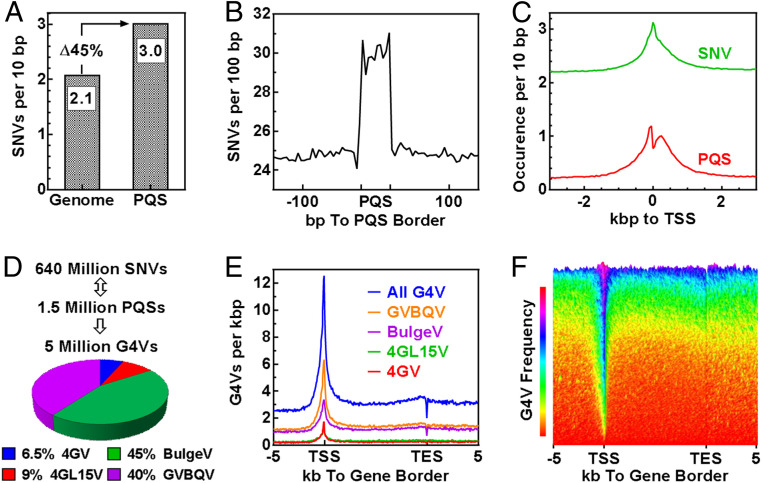
The higher percentage of SNV-positive PQSs than predicted suggested that the SNVs did not occur randomly but with a preference toward PQSs, which is supported by an observation that G4-forming sequences are more mutagenic (26). By comparing their density in the entire genome and the PQS regions, we found that the frequency of SNV was 45% greater in the latter (Fig. 3A). In agreement with this, the distribution of SNV across the PQS regions also showed an increased occurrence in the PQSs compared with the flanking regions (Fig. 3B). PQSs enrich at TSSs. Because of the elevated occurrence of SNVs at PQSs, the SNVs also peaked at the TSSs (Fig. 3C). If the SNVs occurred randomly, the SNV distribution would be a flat line across the TSSs.
Fig. 3.
The occurrence of SNVs in human genomic regions. (A) Preference of SNV occurrence in PQS region versus in the entire genome. (B) Enrichment of SNVs in PQS regions. (C) The overlap frequency of SNV and PQS over genome intervals around TSS. (D) Total G4Vs caused by SNVs in the human genome. (E) The occurrence of G4V across human Refseq genes. (F) Three-dimensional presentation of G4V occurrence across human Refseq genes sorted in descending order by the mean of G4V frequency.
G4Vs in the Genome.
To find out how the SNVs might affect G4 structures, we extracted the immediate 50 nts from both sides of each SNV and searched the 101-nt region for PQSs in the reference (REF) and variant (VAR) sequences, respectively. Previous studies showed that a substitution in a G-tract has a more profound impact on the physical properties of a G4 than that in a loop. For example, the changes in the thermal melting temperature of G4s in the former case could be up to several tens of degrees (27–29) while being several degrees in the latter (30, 31). Therefore, we focused only on those G-tract–affecting SNVs afterward. By comparing the changes in the number of G4s in the two sequences, we identified 5 million of such G4Vs (Fig. 3D).
Formation of G4s can be induced by transcription at both the upstream (21, 32) and downstream (7, 33–37) side of a TSS, which in return regulates transcription (32, 36, 37). To explore the physiological relevance of G4Vs concerning gene regulation, we surveyed their occurrence across genes and found they were significantly enriched near the TSSs (Fig. 3E). This enrichment resulted from two features combined: an enrichment of PQSs at TSS (6, 38–41) and a higher occurrence of SNVs at the PQSs (Fig. 3C). The frequency of G4V in the ∼100,000 reference sequence (Refseq) genes is highly variable. Some genes are abundant, and others are low in G4V load, but a G4V enrichment at TSS was seen in most genes (Fig. 3F). Since TSSs are where transcriptions initiate and fire, the G4Vs near TSSs may have a greater impact than those in other regions on gene activity.
G4Vs in Genes.
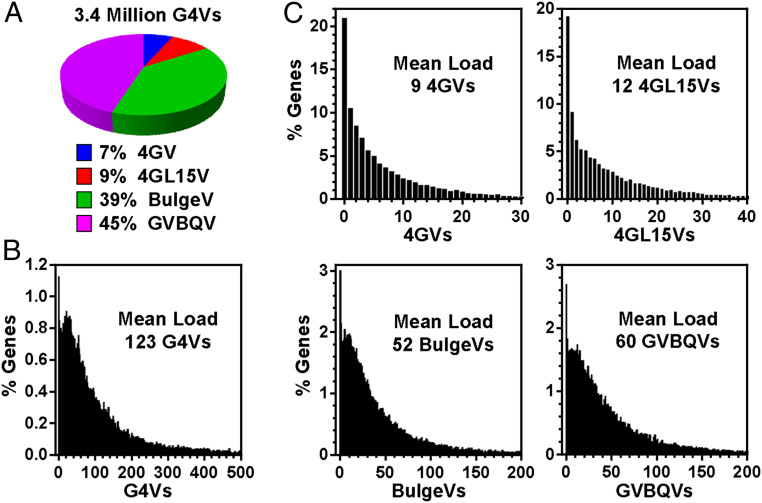
We then focused more on the G4Vs in genes (i.e., gene bodies plus the 2-kbp flanking regions), in which we found 3.4 million G4Vs (Fig. 4A). G4Vs were present in almost every gene when SNVs disrupting any of the four subtypes of PQSs were counted, giving an average load of 123 per gene (Fig. 4B). For each subtype of G4s, the average load is provided in Fig. 4C, which showed that the G4Vs involving noncanonical PQSs were more dominant.
Fig. 4.
Distribution of the number of G4Vs assigned to human genes. (A) Total G4Vs and their subtypes. (B) G4V loads in genes. (C) G4V loads in genes classified by subtypes.
G4Vs in Gene-Regulatory Elements.
Genomic DNA functions through DNA–protein interactions. A structural change in DNA caused by a G4V at a protein binding site directly affects DNA–protein interaction (21) and, expectedly, the relevant function it mediates. A single nucleotide mutation affecting a G4 upstream of the P1 promoter of the c-MYC gene could change the promoter activity by several folds (14). Transcription regulation is mainly mediated by bindings of transcription factors (TFs) to short sections of DNA known as TFBSs. Such binding should be affected if a G4V occurs within or near a TFBS.
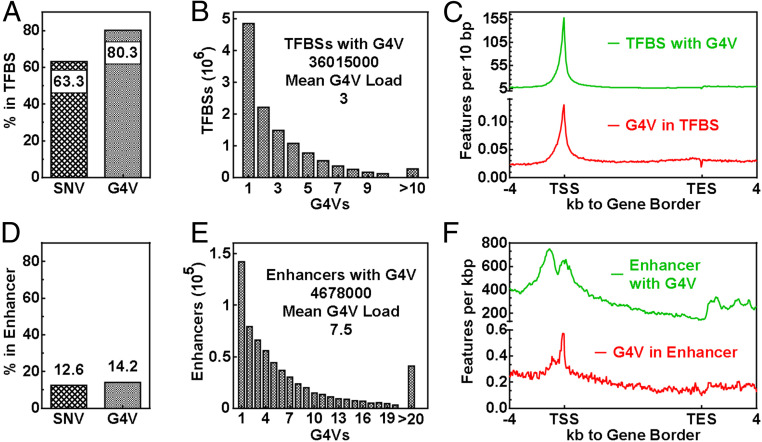
To seek more insight, we surveyed the interaction between SNV/G4Vs and TFBSs using the data from the Gene Transcription Regulation Database (GTRD, gtrd.biouml.org/) containing TFBSs identified from multiple chromatin immunoprecipitation sequencing (ChIP-Seq) experiments (42). We found that >60% of the SNVs and >80% of the G4Vs overlapped with the TFBS (Fig. 5A). On the other hand, a total of 36 million TFBS entries interacted with G4V with an average load of 3 (Fig. 5B). These interactions were both enriched around TSSs (Fig. 5C), implying rich G4V–TFBS interactions in the initiation of transcription.
Fig. 5.
Interaction of G4V with gene-regulatory elements. (A–C) TFBSs. (D–F) Enhancers. (A) Percent features within TFBSs. (B) G4V loads in G4V-positive TFBSs. (C) Frequency of G4V–TFBS interactions across Refseq genes. (D) Percent features within enhancers. (E) G4V loads in G4V-positive enhancers. (F) Frequency of G4V–enhancer interactions across Refseq genes.
Enhancers are sequences that can be bound by TFs and can act at regions more distal to TSSs to activate gene promoters (43). We also examined SNV/G4V-enhancer interactions using the data from the Human Active Enhancers to Interpret Regulatory Variants (HACER, bioinfo.vanderbilt.edu/AE/HACER/index.html) database (44). Our results show a much smaller fraction of the SNV/G4Vs overlapped with the enhancers (Fig. 5D) in comparison with the TFBSs (Fig. 5A). A total of 4.7 million enhancer entries overlapped with an average of 7.5 G4Vs (Fig. 5E). Similar to the G4V–TFBS interaction, the enhancers with G4V and G4Vs in enhancers both peaked at the TSSs (Fig. 5F). However, the enhancers with G4Vs showed a much broader distribution near the TSSs.
Effect of PQS–SNV Interaction on G4 Formation.
All our above analyses pointed to a large scale of PQS–SNV interaction at TFBSs and enhancers near TSSs, revealing a mechanism by which SNVs may affect G4 formation and, consequently, transcription. In the following sections, we demonstrate how such SNVs could affect G4 structures and, as a result, impact gene activities.
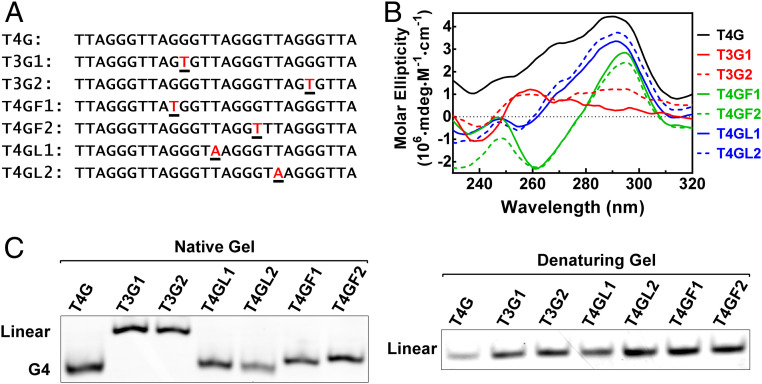
Regarding the alteration in G4 structure, we first tested several single point mutations in the telomeric DNA sequence (Fig. 6A) to mimic SNVs that fall into the three regions in Fig. 1A, respectively. Among the sequences, the wild type (T4G) readily formed a canonical antiparallel G4 as judged from the positive peak at 290 nm in its circular dichroism (CD) spectrum (Fig. 6B) and faster migration in native gel electrophoresis (Fig. 6 C, Left). The formation of G4 was prevented by a mutation in the middle of a G-tract (T3G1, T3G2), resulting in a flat CD spectrum (Fig. 6B) and slower migration in the native gel than the more compact G4s (Fig. 6C). The other mutations in the loop or at the G-tract–loop interface did not abolish G4 formation as expected, suggesting a G-tract–disrupting SNV is more effective in affecting a G4.
Fig. 6.
The structural change caused by a single point mutation in single-stranded telomeric DNA revealed by CD spectroscopy and gel electrophoresis. (A) DNA sequences used. Letters in red indicate mutation. (B) CD spectroscopy. (C) Gel electrophoresis. G4 migrates faster than an equivalent linear DNA. The DNAs showed identical migration when denatured.
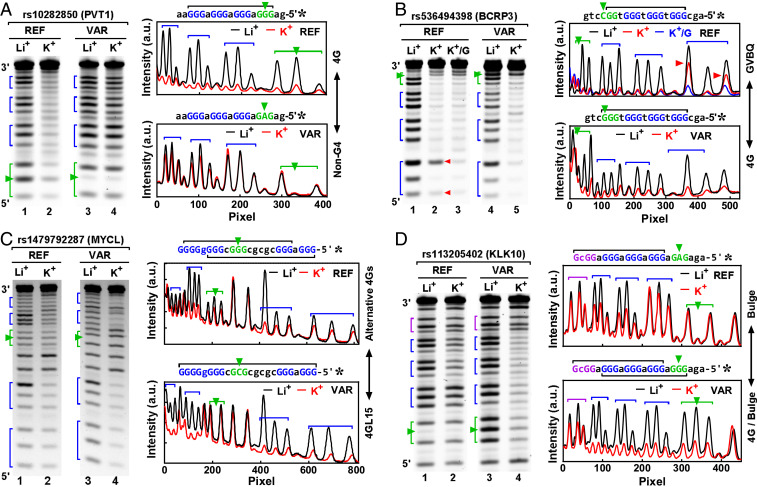
We next tested seven native SNVs that induced conversions between representative G4 structures (Fig. 1) by a more informative technique: dimethyl sulfate (DMS) footprinting. The rs10282850, an intron variant in the PVT1 gene, is an A/G substitution that turns the GGG(aG3)3 motif in the REF into GaG(aG3)3 in the VAR. With four intact G3 tracts, the REF readily formed a canonical G4 according to the protection of the four G3 tracts against chemical cleavage. In the VAR, the disruption of the G3 tract at the 5′ side completely prevented G4 formation, leaving no protection for the G-tracts (Fig. 7A). For the rs536494398, the protection to the one G2 and three G3 tracts in the REF suggested a formation of G4 (Fig. 7B). The hyper-cleavage at the two Gs (red arrowheads) in the first G3 tract from the 5′ end and its return to a fully protected status in the presence of guanosine (K+/G) indicated a formation of GVBQ in the REF (5, 45, 46). The C/G substitution led to a formation of a canonical G4 with three intact G-tetrad layers in the VAR. For the rs1479792287 (Fig. 7C), a G4 can form in the REF at two alternative positions using the four G-tracts from either the 5′ or 3′ end (black bracket). The G/C substitution in the VAR caused a loss of a G-tract in the middle, resulting in a formation of a long-loop 4GL15 G4. As to the rs113205402 (Fig. 7D), the GGcG tract at the 3′ side of the PQS joined the three G3 tracts at its 5′ side to form a bulge G4 in the REF. In the VAR, the GaG tract at the 5′ end was turned into a G3 that joined the other three neighboring G3 tracts at its 3′ side to form a more stable canonical G4 as indicated by the greater protection of the G-tracts. Further examples of more subtle changes in G4 structure can be found in SI Appendix, Figs. S1–S3.
Fig. 7.
Examples of G4V caused by SNV: (A) rs10282850, (B) rs536494398, (C) rs1479792287, and (D) rs113205402. G4 formation in single-stranded DNAs was detected by the protection of the G-tracts in DMS footprinting (gels at the left side and digitization at the right side). SNV IDs are at the top of the gels, followed by the names of their host genes in parentheses. The G4 was stabilized by K+, but not by Li+. G-tracts are indicated by brackets and SNVs by arrowheads. Structural change was indicated at the right side of the digitization panel.
Effect of G4V on DNA Processing.
Genomic DNA is processed by various proteins to function. We then used the exonuclease I hydrolysis assay (22, 47) to evaluate how a G4V would affect DNA processing. In these assays, two DNAs with a PQS from the REF and VAR of Fig. 7 and SI Appendix, Figs. S1–S3, respectively, were mixed and subjected to cleavage from the 3′ end by the exonuclease. The formation of G4 in the DNAs prevented them from being cleaved depending mainly on the stability and folding/unfolding kinetics of the G4s, since the enzyme only cleaves relaxed DNA. Therefore, the extent of DNA cleavage assesses how the G4s would affect the exonuclease-catalyzed DNA processing. We can see that all the G4Vs identified in Fig. 7 and SI Appendix, Figs. S1–S3 remarkably affected the susceptibility of the DNAs to the exonuclease (SI Appendix, Table S1), illustrating an impact of G4V on this and potentially other DNA-processing events.
Effect of G4V on Gene Expression.
Many previous studies have shown that G4-interacting chemical ligands can disturb G4s in many ways similar to SNVs by mediating structural changes affecting G4 kinetics, stability, and G4-protein interaction (48). Treating cells with such ligands could cause a change in the transcriptome (SI Appendix, Fig. S4), demonstrating a structure–function connection for G4s. The enrichment of G4V near TSSs implied that the structural changes are likely to affect gene expression. To gain more insight, we constructed luciferase-expressing plasmids in which a native promoter sequence amplified from the human genome containing an REF or VAR of an SNV was used to control the expression of the luciferase gene. These plasmids were transfected into cultured HEK293T cells to examine the effect of the corresponding G4V.
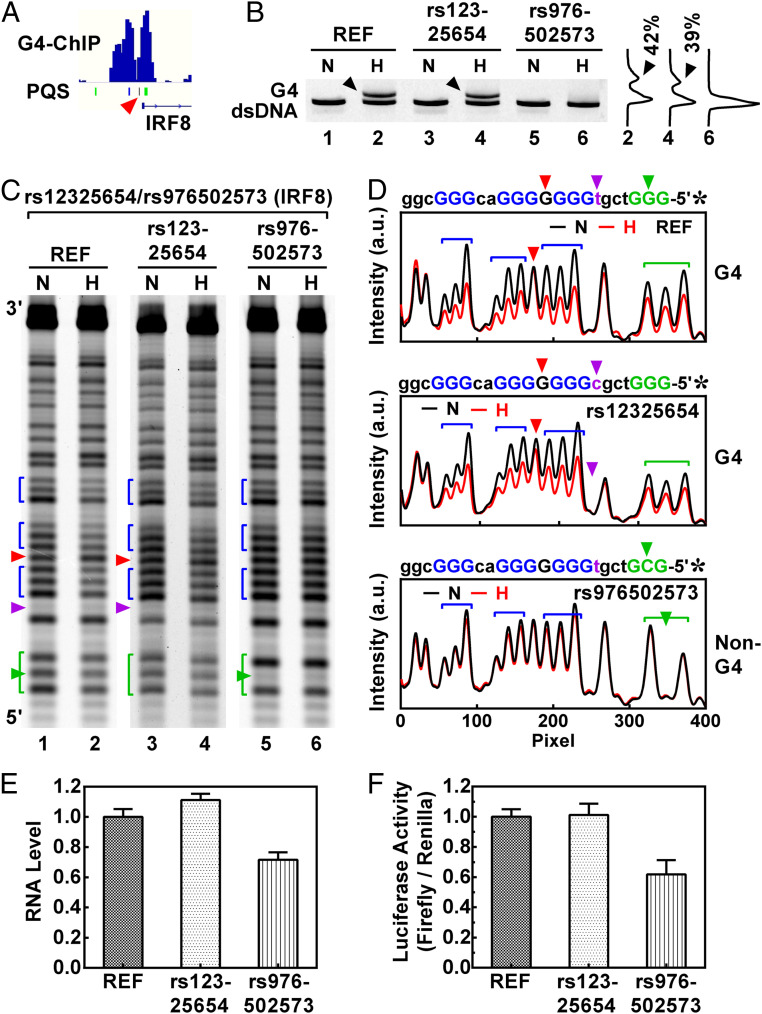
In Fig. 8, two SNVs in the promoter region of the IRF8 gene, rs12325654 and rs976502573, were tested. Our recent work (10) has shown a formation of G4s at the SNV sites (Fig. 8A). The two SNVs both occurred in a G3tcgtG7acG3 motif that formed a canonical G4 in a DNA duplex in the REF as indicated by an extra band in native gel electrophoresis (Fig. 8B, lane 2, arrowhead). The DMS footprinting (Fig. 8 C and D) revealed that the guanine residual in the middle of the G7 tract (red arrowhead) was not protected and therefore served as a 1-nt loop in the G4. Because the rs12325654 is a T/C substitution in a loop, it only caused a tiny reduction in the formation of the G4 (Fig. 8B, lane 4, arrowhead). In contrast, the G-tract–disrupting rs976502573 completely prevented G4 formation by changing the first G3 tract into GcG (Fig. 8B, lane 6).
Fig. 8.
Effect of SNV-mediated loop change and G-tract disruption on IRF8 gene expression. (A) Formation of G4s near the TSS of the IRF8 gene detected by G4-ChIP in living human HEK293T cells. The red arrowhead indicates the PQS bearing the SNVs. (B) Formation of G4s in the REF and VAR duplex DNAs detected by native gel electrophoreses in the PQS indicated by an arrowhead in A. The DNA was heated (H) to generate G4 or not heated (N) to remain fully annealed. (C) Same as B, except that G4 formation was detected by DMS footprinting. (D) Digitization of C. (E) RNA and (F) protein expression of the luciferase reporter downstream of the IRF8 promoter in a pGL3-basic plasmid transfected into HEK293T cells.
We then amplified the promoter and 5′ UTR region of the IRF8 gene and inserted the amplicon with or without the SNV into a pGL3-basic plasmid upstream of a luciferase reporter. Gene activity assay showed that the rs12325654, which occurred in a loop and slightly reduced the formation of G4 (Fig. 8B, lane 4), did not show a meaningful effect on IRF8 expression (Fig. 8 E and F). In contrast, the G4-disrupting rs976502573 significantly reduced IRF8 expression at both the RNA (Fig. 8E) and enzyme (Fig. 8F) level, indicating a strong effect of the G4V on IRF8 expression.
Next, we examined the rs757592196, a G/A substitution in a five–G-tract PQS with the VEGFA gene, which turned the middle G5 into G3aG. G4 formation was detected in the neighborhood of this SNV in living human cells (10) (SI Appendix, Fig. S5A). This SNV did not abolish the formation of G4 (SI Appendix, Fig. S5B) but shortened the G-tract in the middle and enlarged the loop beside it. According to the literature, these two alterations both destabilize the G4 (49, 50). As a result, the formation of G4 reduced from 66% in the REF to 37% in the VAR (SI Appendix, Fig. S5B), accompanied by a reduction of protection to the G-tracts in DMS footprinting (SI Appendix, Fig. S5 C and D). The DMS footprinting also suggested a change in folding topology as judged from the altered participation of guanines in the G-tracts. Altogether, these changes led to a reduced VEGFA expression (SI Appendix, Fig. S5 E and F).
We also carried out the same set of assays with six more SNVs that could cause different structural changes, including 4G to alternative 4Gs (SI Appendix, Fig. S6), 4G to Bulge (SI Appendix, Fig. S7), G4 disruption (SI Appendix, Fig. S8), less 4GL15 to more 4G/4GL15 (SI Appendix, Fig. S9), GVBQ to 2-tetrad 4G (SI Appendix, Fig. S10), and Bulge to 2-tetrad 4G (SI Appendix, Fig. S11). The expressions of the corresponding genes all changed in association with these G4Vs. Of them, rs17335710 is of particular interest. With four G-tracts, a G4 could form in the REF (SI Appendix, Fig. S6). A T/G substitution in the VAR only had a marginal effect on the extent of G4 formation (SI Appendix, Fig. S6B) but added an extra G3 tract at the 3′ side of the original PQS (SI Appendix, Fig. S6 C and D, green arrowheads). The protection to the five G-tracts indicated that a G4 could alternatively arise with four G-tracts either from the 5′ or 3′ side of the PQS in the VAR, creating a “spare tire” that has been hypothesized to aid in the repair process when such sequences are damaged (51). This alternative G4 formation may cause different folding topologies and change the way the sequence responds to the environment that might contribute to the reduced EGFR expression. It is also noted that rs976502573 (Fig. 8) and rs151333 (SI Appendix, Fig. S8) both caused disruption of G4 but led to opposite effects on gene expression. This observation implies that each SNV may have unique effects depending on a combination of multiple factors. Collectively, the results of the nine SNVs in this section revealed that a G4V could dramatically affect the activity of a host gene.
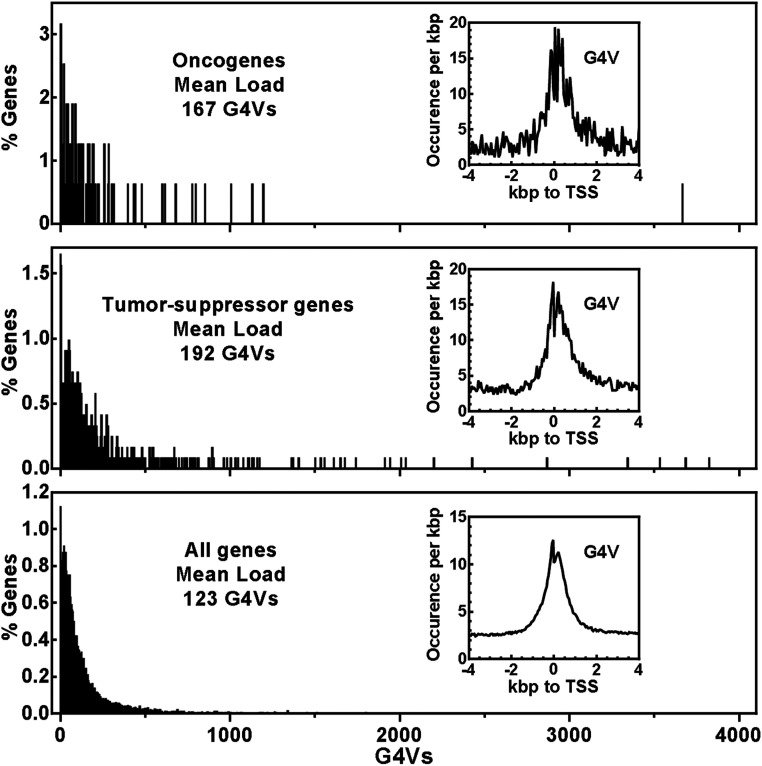
G4Vs in Oncogenes and Tumor-Suppressor Genes.
The association of SNVs with diseases has been the most explored genotype–phenotype connection. The impact of G4V on gene activity strongly suggests their biological and medical implications. The association of G4Vs with diseases and abnormal phenotypes is also of particular interest because G4 structures are therapeutic targets (52). A G4V may imply a gain or loss of a potential drug target in a specific group of people. Information on G4Vs may, therefore, promote developing therapeutic approaches for personalized medicine. We examined the occurrence of G4Vs in oncogenes (53) and tumor-suppressor genes (54) to exemplify their potential involvement in diseases (Fig. 9). On average, 167 and 192 G4Vs were found per oncogene and tumor-suppressor gene, respectively, which are both enriched at TSSs and significantly greater than the average load of 123 G4Vs for all genes. This suggests G4Vs may have particular importance in carcinogenesis that can be induced by abnormal expression of oncogenes and tumor-suppressor genes.
Fig. 9.
Distribution of G4Vs in 803 oncogenes, 1,217 tumor-suppressor genes, and 104,986 Refseq genes and their enrichment around TSSs (Insets).
Discussion
In this work, we disclosed the existence of millions of G4Vs associated with SNVs in the human genome (Figs. 2 and 3) and surveyed their presence in genes (Figs. 4 and 9) and regulatory elements (Fig. 5). We further illustrated that they could efficiently affect G4 structure (Figs. 6 and 7 and SI Appendix, Figs. S1–S3) and, consequently, influence DNA processing (SI Appendix, Table S1) and gene activity (Fig. 8 and SI Appendix, Figs. S5–S11). Because an SNV influences a G4 in diverse aspects, the ultimate consequence of a G4V on gene expression results from a combination of many factors.
G4s are involved in a variety of biological processes by acting at the transcription and translation layers of gene expression (1). The enrichment of G4V near TSSs creates structural changes in these key regulatory elements in transcription. Our results illustrated that a G4V can affect gene activity by directing the interaction between DNA and transcription factors, activators, and silencers. Examples of such G4 structures and their participation in gene expression have been found in the promoters of many genes such as HRAS (55), KRAS (56), WT1 (57), MET (58), BCL2 (59), and C-MYC (14, 60, 61), to mention a few examples. Moreover, a gain/loss or change in a G4 in an RNA transcript originated from a G4V in DNA can affect translation (62, 63). On the other hand, a G4 may serve as a regulatory element in a more specific manner by specific interactions with proteins and other molecules (2, 25).
Our current work focused only on the SNVs that change a G-tract. Besides, those in loops or flanking sequences may also affect G4 formation to various degrees. In some particular cases, for example, in which G4 forms along with or in competition against a hairpin in a loop or flanking sequence (64–66), an SNV would interfere with the cooperation or competition to bring sophisticated changes in structures. Therefore, the actual scope of G4Vs should go further beyond what we disclosed here. On the other hand, new rules governing the formation of G4s are being discovered, which can be explored by various prediction algorithms (for a recent review, see ref. 67). The identification of new G4s will further increase the diversity of both G4s and G4Vs as part of the genetic variations. At present, knowledge regarding the contribution of G4Vs to the functional diversity of the human genome is almost absent and awaits systematic investigation. The structural aspect of the SNVs should be considered in inferring their biological function and pathological implication. The G4V-function relationship should enrich our understanding of the health issues associated with SNVs in individualized therapeutics, health risk assessment, and drug development. For example, a disease-causing G4V suggests that the corresponding G4 structure may be relevant to and serve as a unique target for the disease (52). This information may help develop therapeutic approaches to such diseases in a specific population. A PQS–SNV interaction that destabilizes a G4 and changes gene expression may be corrected by stabilization of the G4 with chemical ligands.
Materials and Methods
Details are in SI Appendix, SI Materials and Methods, including the following: the source of genome data files; identification of PQSs and G4V-inducing SNVs; coverage of SNVs in genome; profiling of SNVs in genomic regions; DNA and plasmids; CD spectroscopy; DMS footprinting; native gel electrophoresis; exonuclease I hydrolysis; in vitro transcription of dsDNA; transfection; reverse transcription; qPCR; and determination of reporter activity.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 21977094 and 22037004); the National Science and Technology Major Project of China (Grant 2018ZX09J18111); and the Program for the 13th Five-Year Plan of China (Grant 2017ZX10202101004).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013230118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Hänsel-Hertsch R., Di Antonio M., Balasubramanian S., DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 18, 279–284 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Rhodes D., Lipps H. J., G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 43, 8627–8637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huppert J. L., Balasubramanian S., Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todd A. K., Johnston M., Neidle S., Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 33, 2901–2907 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X. M., et al., Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives. Proc. Natl. Acad. Sci. U.S.A. 112, 14581–14586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao S., Zhang J. Y., Zheng K. W., Hao Y. H., Tan Z., Bioinformatic analysis reveals an evolutional selection for DNA:RNA hybrid G-quadruplex structures as putative transcription regulatory elements in warm-blooded animals. Nucleic Acids Res. 41, 10379–10390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao S., et al., Formation of DNA:RNA hybrid G-quadruplexes of two G-quartet layers in transcription: Expansion of the prevalence and diversity of G-quadruplexes in genomes. Angew. Chem. Int. Ed. Engl. 53, 13110–13114 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Guédin A., Gros J., Alberti P., Mergny J. L., How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 38, 7858–7868 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukundan V. T., Phan A. T., Bulges in G-quadruplexes: Broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 135, 5017–5028 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Zheng K. W., et al., Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res. 48, 11706–11720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan S., Clements J. A., Batra J., Single nucleotide polymorphisms in clinics: Fantasy or reality for cancer? Crit. Rev. Clin. Lab. Sci. 53, 29–39 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Frazer K. A., Murray S. S., Schork N. J., Topol E. J., Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10, 241–251 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Miyoshi D., Karimata H., Sugimoto N., Drastic effect of a single base difference between human and tetrahymena telomere sequences on their structures under molecular crowding conditions. Angew. Chem. Int. Ed. Engl. 44, 3740–3744 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H., Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puig Lombardi E., et al., Thermodynamically stable and genetically unstable G-quadruplexes are depleted in genomes across species. Nucleic Acids Res. 47, 6098–6113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee D. S. M., Ghanem L. R., Barash Y., Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat. Commun. 11, 527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaudoin J. D., Perreault J. P., 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 38, 7022–7036 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lightfoot H. L., Hagen T., Tatum N. J., Hall J., The diverse structural landscape of quadruplexes. FEBS Lett. 593, 2083–2102 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Gros J., et al., Guanines are a quartet’s best friend: Impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 35, 3064–3075 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhary S., Kaushik M., Ahmed S., Kukreti R., Kukreti S., Structural switch from hairpin to duplex/antiparallel G-quadruplex at single-nucleotide polymorphism (SNP) site of human apolipoprotein E (APOE) gene coding region. ACS Omega 3, 3173–3182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C., Liu H. H., Zheng K. W., Hao Y. H., Tan Z., DNA G-quadruplex formation in response to remote downstream transcription activity: Long-range sensing and signal transducing in DNA double helix. Nucleic Acids Res. 41, 7144–7152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J. Q., Chen C. Y., Xue Y., Hao Y. H., Tan Z., G-quadruplex hinders translocation of BLM helicase on DNA: A real-time fluorescence spectroscopic unwinding study and comparison with duplex substrates. J. Am. Chem. Soc. 132, 10521–10527 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Xia Y., et al., Transmission of dynamic supercoiling in linear and multi-way branched DNAs and its regulation revealed by a fluorescent G-quadruplex torsion sensor. Nucleic Acids Res. 46, 7418–7424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim N., The interplay between G-quadruplex and transcription. Curr. Med. Chem. 26, 2898–2917 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bochman M. L., Paeschke K., Zakian V. A., DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruisselbrink E., et al., Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr. Biol. 18, 900–905 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Tomasko M., Vorlícková M., Sagi J., Substitution of adenine for guanine in the quadruplex-forming human telomere DNA sequence G(3)(T(2)AG(3))(3). Biochimie 91, 171–179 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Agarwala P., Kumar S., Pandey S., Maiti S., Human telomeric RNA G-quadruplex response to point mutation in the G-quartets. J. Phys. Chem. B 119, 4617–4627 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Marušič M., Plavec J., Towards understanding of polymorphism of the G-rich region of human papillomavirus type 52. Molecules 24, 1294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guédin A., Alberti P., Mergny J. L., Stability of intramolecular quadruplexes: Sequence effects in the central loop. Nucleic Acids Res. 37, 5559–5567 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatzakis E., Okamoto K., Yang D., Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: Effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry 49, 9152–9160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng K. W., et al., Superhelicity constrains a localized and R-loop-dependent formation of G-quadruplexes at the upstream region of transcription. ACS Chem. Biol. 12, 2609–2618 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Liu J. Q., Xiao S., Hao Y. H., Tan Z., Strand-biased formation of G-quadruplexes in DNA duplexes transcribed with T7 RNA polymerase. Angew. Chem. Int. Ed. Engl. 54, 8992–8996 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Zheng K. W., et al., A competitive formation of DNA:RNA hybrid G-quadruplex is responsible to the mitochondrial transcription termination at the DNA replication priming site. Nucleic Acids Res. 42, 10832–10844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J. Y., Zheng K. W., Xiao S., Hao Y. H., Tan Z., Mechanism and manipulation of DNA:RNA hybrid G-quadruplex formation in transcription of G-rich DNA. J. Am. Chem. Soc. 136, 1381–1390 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Zheng K. W., et al., Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res. 41, 5533–5541 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu R. Y., Zheng K. W., Zhang J. Y., Hao Y. H., Tan Z., Formation of DNA:RNA hybrid G-quadruplex in bacterial cells and its dominance over the intramolecular DNA G-quadruplex in mediating transcription termination. Angew. Chem. Int. Ed. Engl. 54, 2447–2451 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y., Du Z., Li N., Extensive selection for the enrichment of G4 DNA motifs in transcriptional regulatory regions of warm blooded animals. FEBS Lett. 581, 1951–1956 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Eddy J., et al., G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 39, 4975–4983 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huppert J. L., Balasubramanian S., G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huppert J. L., Bugaut A., Kumari S., Balasubramanian S., G-quadruplexes: The beginning and end of UTRs. Nucleic Acids Res. 36, 6260–6268 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yevshin I., Sharipov R., Kolmykov S., Kondrakhin Y., Kolpakov F., GTRD: A database on gene transcription regulation-2019 update. Nucleic Acids Res. 47, D100–D105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterjee S., Ahituv N., Gene regulatory elements, major drivers of human disease. Annu. Rev. Genomics Hum. Genet. 18, 45–63 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Wang J., et al., HACER: An atlas of human active enhancers to interpret regulatory variants. Nucleic Acids Res. 47, D106–D112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Y. D., et al., Selective targeting of guanine-vacancy-bearing G-quadruplexes by G-quartet complementation and stabilization with a guanine-peptide conjugate. J. Am. Chem. Soc. 142, 11394–11403 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Li X. M., Zheng K. W., Hao Y. H., Tan Z., Exceptionally selective and tunable sensing of guanine derivatives and analogues by structural complementation in a G-quadruplex. Angew. Chem. Int. Ed. Engl. 55, 13759–13764 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Yao Y., Wang Q., Hao Y. H., Tan Z., An exonuclease I hydrolysis assay for evaluating G-quadruplex stabilization by small molecules. Nucleic Acids Res. 35, e68 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Hagan M. P., Morales J. C., Galan M. C., Binding and beyond: What else can G-quadruplex ligands do? Eur. J. Org. Chem. 2019, 4995–5017 (2019). [Google Scholar]
- 49.Rachwal P. A., Brown T., Fox K. R., Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry 46, 3036–3044 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Risitano A., Fox K. R., Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 32, 2598–2606 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fleming A. M., Zhou J., Wallace S. S., Burrows C. J., A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent. Sci. 1, 226–233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian T., Xiao H., Zhou X., A review: G-Quadruplex’s applications in biological target detection and drug delivery. Curr. Top. Med. Chem. 15, 1988–2001 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Liu Y., Sun J., Zhao M., ONGene: A literature-based database for human oncogenes. J. Genet. Genomics 44, 119–121 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Zhao M., Kim P., Mitra R., Zhao J., Zhao Z., TSGene 2.0: An updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 44, D1023–D1031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cogoi S., Shchekotikhin A. E., Xodo L. E., HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 42, 8379–8388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaiser C. E., et al., Insight into the complexity of the i-Motif and G-quadruplex DNA structures formed in the KRAS promoter and subsequent drug-induced gene repression. J. Am. Chem. Soc. 139, 8522–8536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zidanloo S. G., Hosseinzadeh Colagar A., Ayatollahi H., Raoof J. B., Downregulation of the WT1 gene expression via TMPyP4 stabilization of promoter G-quadruplexes in leukemia cells. Tumour Biol. 37, 9967–9977 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Yan J., Zhao X., Liu B., Yuan Y., Guan Y., An intramolecular G-quadruplex structure formed in the human MET promoter region and its biological relevance. Mol. Carcinog. 55, 897–909 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Onel B., et al., A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J. Am. Chem. Soc. 138, 2563–2570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hurley L. H., Von Hoff D. D., Siddiqui-Jain A., Yang D., Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element. Semin. Oncol. 33, 498–512 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Yang D., Hurley L. H., Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides Nucleic Acids 25, 951–968 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Endoh T., Sugimoto N., Mechanical insights into ribosomal progression overcoming RNA G-quadruplex from periodical translation suppression in cells. Sci. Rep. 6, 22719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Endoh T., Kawasaki Y., Sugimoto N., Translational halt during elongation caused by G-quadruplex formed by mRNA. Methods 64, 73–78 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Yu Z., et al., Tertiary DNA structure in the single-stranded hTERT promoter fragment unfolds and refolds by parallel pathways via cooperative or sequential events. J. Am. Chem. Soc. 134, 5157–5164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuo M. H., et al., Conformational transition of a hairpin structure to G-quadruplex within the WNT1 gene promoter. J. Am. Chem. Soc. 137, 210–218 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Bugaut A., Murat P., Balasubramanian S., An RNA hairpin to G-quadruplex conformational transition. J. Am. Chem. Soc. 134, 19953–19956 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lombardi E. P., Londoño-Vallejo A., A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 48, 1603 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.