Significance
Evolutionary transitions between species with separate sexes and species in which individuals have both sex functions have wide-ranging biological implications. It is largely unknown how such transitions occur in systems with haploid male- and female-determining chromosomes in algae and bryophytes. We investigated such a transition in the algal genus Volvox by making whole-genome sequences of two closely related species, one of which is heterothallic (with distinct males and females) and the other homothallic (with only bisexual, self-compatible individuals). The heterothallic species harbors a sex-determining region (SDR), while the homothallic species retains a nearly intact female-derived SDR-like region and separate regions containing key male genes. Thus, an ancestral female has probably become homothallic by acquiring genes that confer male functions.
Keywords: evolution, sex, Volvox, heterothallism, homothallism
Abstract
Transitions between separate sexes (dioecy) and other mating systems are common across eukaryotes. Here, we study a change in a haploid dioecious green algal species with male- and female-determining chromosomes (U and V). The genus Volvox is an oogamous (with large, immotile female gametes and small, motile male gametes) and includes both heterothallic species (with distinct male and female genotypes, associated with a mating-type system that prevents fusion of gametes of the same sex) and homothallic species (bisexual, with the ability to self-fertilize). We date the origin of an expanded sex-determining region (SDR) in Volvox to at least 75 Mya, suggesting that homothallism represents a breakdown of dioecy (heterothallism). We investigated the involvement of the SDR of the U and V chromosomes in this transition. Using de novo whole-genome sequences, we identified a heteromorphic SDR of ca 1 Mbp in male and female genotypes of the heterothallic species Volvox reticuliferus and a homologous region (SDLR) in the closely related homothallic species Volvox africanus, which retained several different hallmark features of an SDR. The V. africanus SDLR includes a large region resembling the female SDR of the presumptive heterothallic ancestor, whereas most genes from the male SDR are absent. However, we found a multicopy array of the male-determining gene, MID, in a different genomic location from the SDLR. Thus, in V. africanus, an ancestrally female genotype may have acquired MID and thereby gained male traits.
As first noted by Darwin when studying plant sexuality, self-fertilization can lead to inbreeding depression, though this potential disadvantage relative to outcrossing can be offset by higher probability of fertilization success (1, 2). Thus, transitions between inbreeding and outbreeding mating systems attract the attention of evolutionary biologists and have been documented in sexual systems across a broad range of taxa including animals, land plants, algae (Fig. 1), protists, and fungi (3–9). Hermaphroditic mating systems in diploid animals and plants are fairly common, and the evolution of hermaphroditism in animals has been extensively studied (10–12). The molecular genetic bases of hermaphroditism have also been recently studied in several flowering plants (13–15).
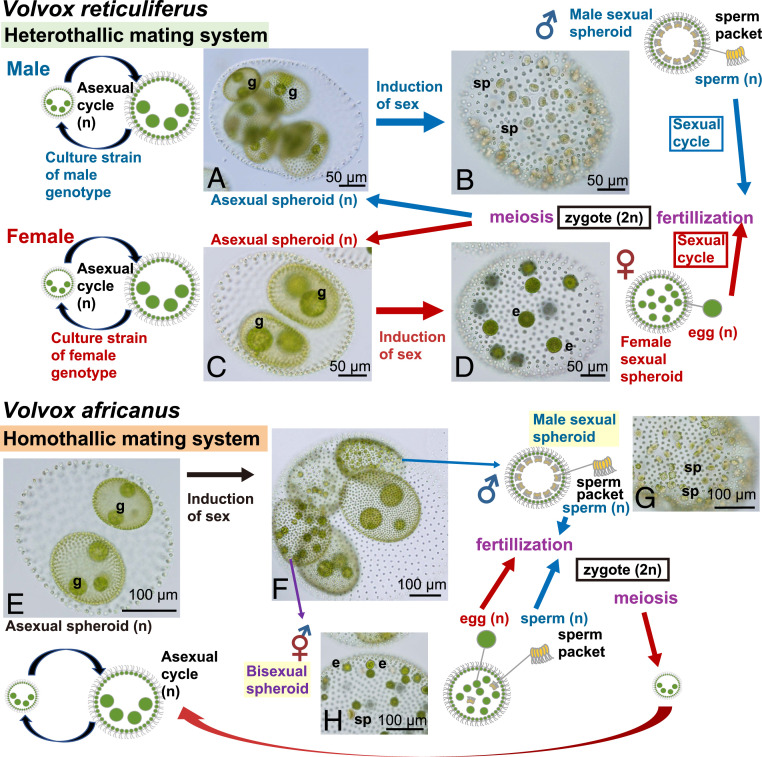
Fig. 1.
Schematic representation of life cycles of two closely related species of Volvox sect. Merrillosphaera (SI Appendix, Fig. S1) showing different sexual systems (heterothallism and homothallism), based on Nozaki et al. (29). Note gonidia (g) in asexual spheroids and sperm packets (sp) and eggs (e) in sexual spheroids. (A) NIES-3781. (B) NIES-3783. (C and D) NIES-3782. (E and H) NIES-3784. (F and G) NIES-3780. For materials and methods of light micrographs, refer to Nozaki et al. (29). All photographs are original.
In haploid organisms, two basic types of mating systems are recognized: heterothallic (with two or more self-incompatible mating types in isogamous species or with males and females in anisogamous/oogamous species) and homothallic (self-compatible with isogamy or bisexual with anisogamy/oogamy) (SI Appendix, Table S1). In heterothallic species, gamete compatibility in isogamy or maleness versus femaleness is usually determined by a single complex locus on a chromosome (9). The process by which this genetic system breaks down to allow a single genotype to acquire functions of both sexes and mating-types in the homothallic species is poorly understood outside of fungi (8).
Organisms with a heterothallic haploid generation such as algae and early diverging land plants like mosses and liverworts have sex-determining regions (SDRs) that are differentiated between the two sexes and located on male and female determining (U and V) chromosomes (9). SDRs exhibit suppressed recombination and harbor fully sex-linked genes including sex-specific genes (found in only one of the two sexes) and gametologs (genes with alleles in both haplotypes of the SDR) (9). A recent study has characterized genes involved in a change from sexual reproduction to parthenogenesis in a heterothallic brown alga with UV chromosome (16). However, genomics of transitions between heterothallism and homothallism have not been previously studied in the context of haploid SDRs, and the fates of sex-determining and sex-related genes present on ancestral SDRs in UV chromosomes after transitions to homothallism are unknown.
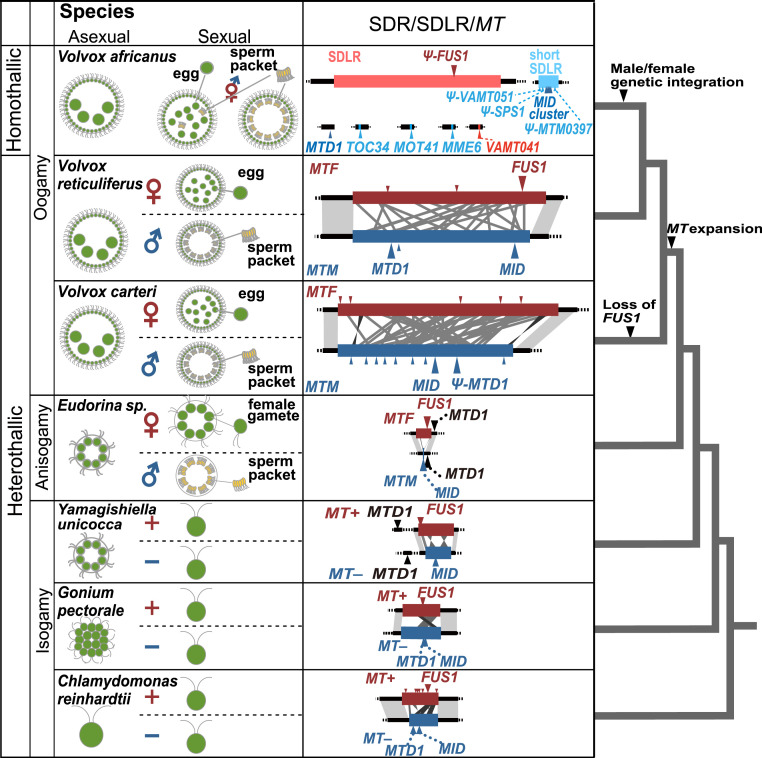
The volvocine green algal lineage is an especially well-studied evolutionary model for investigating the origins of sexes and transitions in sexuality, because it includes extant organisms with a graded range of sexual or mating phenotypes from unicellular isogamous Chlamydomonas through genera with increasing degrees of sexual dimorphism, such as multicellular, isogamous Gonium, and oogamous Volvox (Fig. 2) (17–22). In heterothallic species of the isogamous volvocine genera such as Chlamydomonas, there are two mating-types, plus and minus, both of which are needed for sexual reproduction (9). Ferris and Goodenough (23) characterized the first mating-type locus (MT) in Chlamydomonas reinhardtii, which was discovered to have features similar to those of a typical SDR, including large size and suppressed recombination. Subsequent studies of heterothallic species of multicellular volvocine algae revealed conservation of orthologs of the mating-type determining gene MID in the mating type minus genotype of the isogamous genera Gonium and Yamagishiella and in the SDR of males of anisogamous Eudorina and the oogamous Volvox (19, 21, 24). This indicates that the evolution of anisogamy has involved genetic changes that are closely associated with the MT, presumably because this ensures the ability of male and female gametes to fuse with each other as proposed on theoretical grounds (25). The MT or SDR haplotypes in heterothallic volvocine algae range from 7 kbp to 1 Mbp and are structurally heteromorphic with chromosome rearrangements that distinguish the haplotypes and various degrees of genetic differentiation between them (21) (Fig. 2). The ca 1 Mbp, highly differentiated Volvox carteri SDR has expanded in size about fivefold relative to that in Chlamydomonas (19), and this was likely to have occurred after the transition from isogamy to anisogamy/oogamy (21). However, the timing of the SDR expansion in the genus Volvox and its significance with respect to the evolution of sexual dimorphism remain unknown.
Fig. 2.
Volvocine green algal phylogeny and SDR or MT evolution. Asexual or vegetative phase, sexual phase, MTM and MTF, or SDLR and phylogenetic positions of V. reticuliferus and V. africanus are illustrated with those of five other species previously studied [C. reinhardtii, Gonium pectorale, Yamagishiella unicocca, Eudorina sp., and V. carteri (19, 21, 23, 24)]. Note that all three of these Volvox species belongs to the section Merrillosphaera (SI Appendix, Fig. S1).
To date, genomic studies of volvocine green algae have focused on heterothallic species (Fig. 2), but most genera within this lineage have members that also underwent transitions from heterothallism to homothallism (7). These algae are found in freshwater habitats and form a dormant diploid zygote with a resistant wall that allows it to survive under unfavorable conditions; diploidy in the zygote could be of selective value for survival when chromosomal damage and mutation may be occurring at increased frequencies (26). Thus, transitions from heterothallism to homothallism in the volvocine algae may be favored for production of the resistant diploid cells (zygotes) by self-fertilization. We previously noted the presence of a MID gene in two homothallic species of the genus Volvox (27), and an artificial homothallic phenotype was shown in the male genotype of V. carteri (28). However, it remains unknown how a naturally occurring homothallic mating system could arise from an ancestral SDR in UV chromosome system (or vice versa).
In most members of the genus Volvox, the transition from vegetative to sexual reproduction involves modified embryogenesis and the production of morphologically distinct male and/or female sexual spheroids (7). Recently, we characterized two species of Volvox that are closely related but have different mating systems: heterothallic Volvox reticuliferus, which upon sexual induction makes differentiated male or female, and homothallic Volvox africanus, which produces both male spheroids and bisexual spheroids from single clonal cultures (27, 29) (Figs. 1 and 2 and SI Appendix, Table S1). These two species diverged ca 11 MYA; together with the more distantly related V. carteri, they represent a monophyletic group, the “section Merrillosphaera,” which originated ca 75 MYA and is an infrageneric taxon of the polyphyletic genus Volvox (SI Appendix, Fig. S1). While ancestral heterothallism in Merrillosphaera is more likely than homothallism, statistical support for this inference is not strong, and the directionality of transitions between these two types of sexuality within this clade remains somewhat inconclusive (7).
Here, we performed de novo whole-genome sequencing of male and female genotypes of heterothallic V. reticuliferus and of homothallic V. africanus in order to characterize their SDR and SD-like regions (SDLRs), respectively. We used this sequence information to infer the likely ancestral state of heterothallism in this clade and to reconstruct the SDR changes which occurred during the evolution of homothallism in V. africanus, including tracking the fates of male- and female-specific genes and male versus female gametologs derived from the putative heterothallic ancestral species.
Results and Discussion
Whole-Genome Assembly.
We constructed three de novo nuclear genomes of Volvox by assembling a combination of long and short sequencing reads into contigs (Materials and Methods). Each of the three genomes was composed of 200 to 448 gap-free contigs (N50 = 1.36 to 1.91 Mbp) that yielded nuclear genome assemblies of around 130 Mbp, similar to that of V. carteri and other volvocine species (SI Appendix, Tables S2 and S3). Protein coding gene predictions were performed with the assistance of transcriptome data to estimate ca 13,000 to 14,000 expressed genes in each genome (SI Appendix, Table S2). Assembly quality of the three genomes was high based on the presence of the vast majority of benchmarking universal single-copy orthologs (BUSCO) reference genes (30) (94.9 to 98.1% complete genes) (SI Appendix, Table S2). In addition, genome sizes estimated based on k-mer frequencies (SI Appendix, Fig. S2) using GenomeScope (31) were consistent with the total genome assembly sizes (SI Appendix, Table S2), suggesting high coverage rates of the assembled contigs for the whole genome of each culture strain or genotype.
An Expanded SDR in Heterothallic V. reticuliferus.
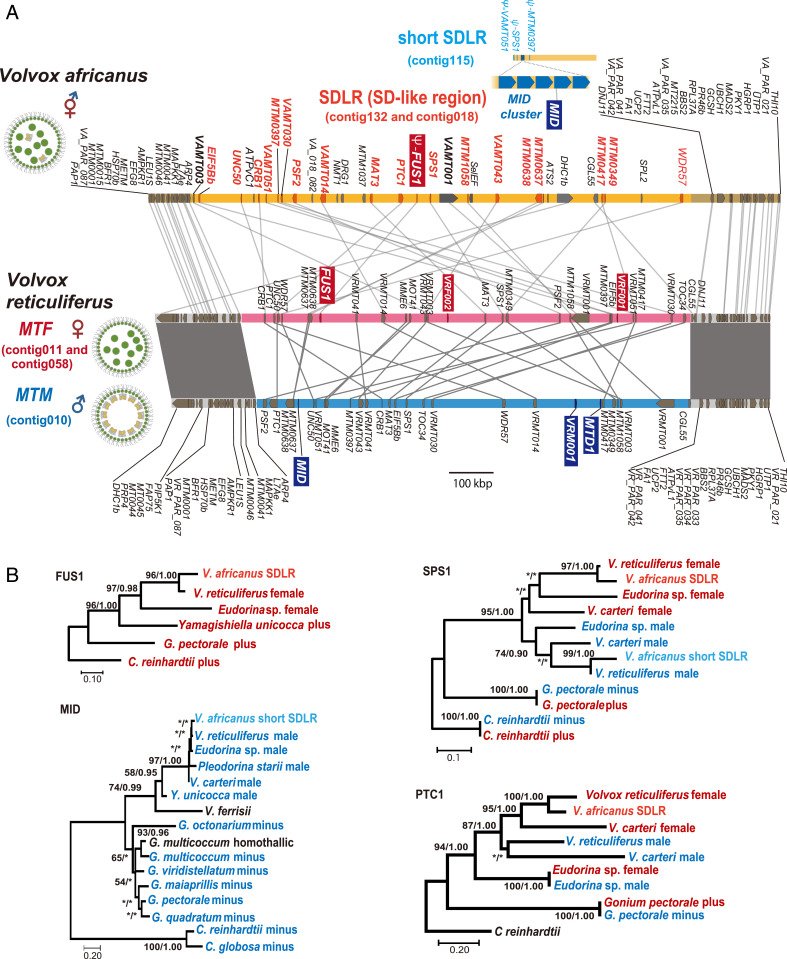
Here, we focused on identifying and analyzing the V. reticuliferus SDRs, with some additional whole-genome analyses. Based on comparison of whole-genome sequences between male and female genotypes of V. reticuliferus and directed searching for candidate homologs of MID and other fully sex-linked genes [i.e., male or female SDR sex-specific genes plus gametologs (9) {“shared genes” (19)}] in various volvocine SDRs (Materials and Methods), we identified a single large (ca 1 Mbp) heteromorphic and highly rearranged MT region (SDR) that differed between male and female in gene ordering and composition (Figs. 2 and 3 and SI Appendix, Figs. S3 and S4). The male SDR (MTM) was 0.98 Mbp long and contained three predicted male-specific genes (MID, MTD1, and VRM001) whereas the female SDR (MTF) was 1.02 Mbp long with three predicted female-specific genes (FUS1, VRF001, and VRF002) (Figs. 2 and 3 and SI Appendix, Fig. S4). The male and female SDRs also harbored 24 gametologs that were highly differentiated as in V. carteri (SI Appendix, Fig. S5). Similar to the V. carteri SDR, the V. reticuliferus SDR haplotypes had very different gene ordering without conserved syntenic gene blocs (19, 21) (Fig. 3 and SI Appendix, Fig. S3). Also similar to the V. carteri SDR, the V. reticuliferus SDR had low gene density (28 Mbp versus 104 to 105 Mbp genome average) and was strongly enriched for repeats (70% compared with 27% genome average) (SI Appendix, Table S2), features typical for heteromorphic SDRs and other nonrecombining genomic regions (13, 19).
Fig. 3.
SDR or MT of heterothallic V. reticuliferus (male and female) and two SD-like regions (SDLR and short SDLR) in homothallic V. africanus and phylogeny of four genes in SDR and two SDLRs. (A) Comparison of homothallic V. africanus SDLRs (accession LC586641 through LC58662) and V. reticuliferus male and female SDR (MTM and MTF, respectively; accession LC586643 through LC586644). Note male- and female-specific genes (with blue or red backgrounds, respectively) and gametologs of V. reticuliferus and their homologs in SDLRs of V. africanus. Red and blue regions represent MTF and MTM, respectively. Yellow regions represent SDLR and short SDLR. Gray shading indicates a syntenic bloc of pseudo autosomal regions. Red and blue homologs in SDLR and short SDLR represent MTF- and MTM-origin, respectively (deduced from phylogenetic analyses; B and SI Appendix, Figs. S6–S8). (B) Maximum likelihood (ML) phylogeny of homologs of female-specific FUS1, male-specific MID, and two gametologs (PTC1 and SPS1). Red and blue represent homologs of fully sex-linked genes from female and male MT (SDLR and short SDLR), respectively. Note that V. africanus has male-related and female-related SPS1 homologs in short SDLR and SDLR, respectively. Numbers in left and right sides at branches indicate bootstrap values of ML analysis and posterior probabilities of Bayesian inference, respectively. For phylogenetic analyses of homologs of male-specific MTD1 and other gametologs, refer to SI Appendix, Figs. S6–S8 and S11.
As expected, the male (or minus)-specific gene MID was found in the V. reticuliferus male SDR, as well as an intact MTD1 gene (Fig. 3 and SI Appendix, Fig. S4), which is mating-type minus–specific in C. reinhardtii and Gonium pectorale and widely conserved in the volvocine lineage [though it became a pseudogene in the V. carteri lineage (19)] (Fig. 2). Also as expected, the V. reticuliferus female SDR carried the female-specific gene FUS1 (Fig. 3 and SI Appendix, Fig. S4), which is conserved in Eudorina sp. female SDR and mating-type plus MTs of all isogamous volvocine species (21), though also lost recently in the V. carteri lineage (Fig. 2). The remaining three sex-specific genes in the V. reticuliferus SDRs (male-specific VRM001 and female-specific VRF001 and VRF002) had no predicted functional domains and no homologs in other species or elsewhere in the V. reticuliferus genome (Fig. 3 and SI Appendix, Fig. S4). Whether any of these recently evolved sex-specific genes play roles in male or female sexual cycles remains to be determined.
In total, 17 of the 24 gametologs in the V. reticuliferus SDR had homologs in the V. carteri SDR, suggesting they were present in the ancestral SDR of Merrillosphaera (SI Appendix, Figs. S6–S8), though the physical order of these 17 gametologs was not conserved between homologous haplotypes (SI Appendix, Fig. S9). Additionally, 13 other gametologs from the V. carteri SDR had homologs distributed in pseudo autosomal regions of V. reticuliferus, further supporting the idea that the SDRs of the two species share common ancestry (SI Appendix, Fig. S9). Overall, our data indicate that an expanded and highly differentiated SDR was present at least as far back as the emergence of the Volvox sect. Merrillosphaera lineage originating around 75 MYA (Fig. 2 and SI Appendix, Fig. S1) and has persisted in extant taxa. Further evidence for shared common ancestry of Volvox SDRs based on gametolog molecular evolution data is described below.
An Expanded SDLR in Homothallic V. africanus.
Using V. reticuliferus male and female contigs including SDR sequences (Fig. 2 and SI Appendix, Fig. S3), we searched for and identified homologous sequences in V. africanus, including a ca 1 Mbp genomic region in V. africanus that we designated “SDLR” (SI Appendix, Fig. S10). The SDLR was flanked by sequences corresponding to the pseudo autosomal regions flanking the V. reticuliferus SDR (Fig. 3 and SI Appendix, Fig. S10), indicating a syntenic chromosomal location for the SDLR and SDR of these two species.
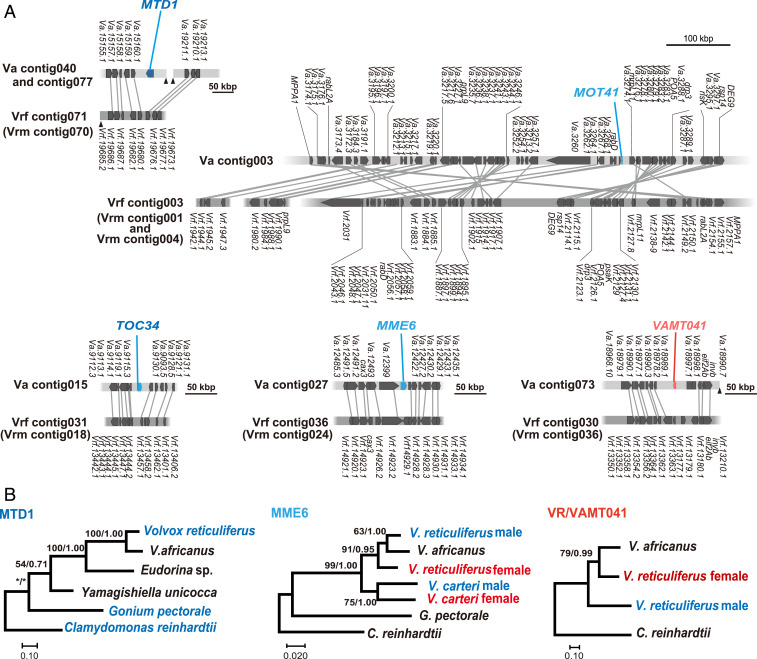
The SDLR contained homologs for 20 of 24 V. reticuliferus gametologs and a FUS1-related pseudogene (Fig. 3 and SI Appendix, Fig. S4) but no homologs of three male- and two other female-specific genes from V. reticuliferus. Based on the phylogenetic analyses of these homologs in the V. africanus SDLR (see below), it was inferred to have originated from an ancestral female SDR. In addition, the change to homothallism in the ancestor of V. africanus may have been recent, as the SDLR retained SDR-like properties including low gene density (29 Mbp versus 100 Mbp genome average) and high repeat content (75% compared with 26% genome average) (SI Appendix, Table S2) which are expected to be purged if normal meiotic recombination were restored to this region. Homologs of two male-specific genes (MID and MTD1) and four remaining gametologs from V. reticuliferus were found in separate, discontinuous genomic locations (see below) (Fig. 4), but homologs of the three remaining sex-specific genes of V. reticuliferus (male-specific VRM001 and female-specific VRF001 and VRF002) were not found in V. africanus whole-genome data.
Fig. 4.
V. africanus autosome-like regions inserted by homologs of the male-specific gene MTD1 and four gametologs of V. reticuliferus (Fig. 3A). (A) Comparison between autosome-like regions inserted by SDR-related gene homologs in homothallic V. africanus (Va, upper bars) and their homologous autosomal regions of heterothallic V. reticuliferus female (Vrf, lower bars). Contigs harboring the corresponding autosomal regions of V. reticuliferus male (Vrm) are described within parentheses and have the same gene arrangements as in Vrf autosomal regions shown here. Blue and red color of the gene names of V. africanus represent homologs of male or female gametologs of V. reticuliferus, respectively, based on the phylogenetic analyses (B; SI Appendix, Fig. S8). (B) Maximum likelihood (ML) phylogeny of homologs of MTD1 and two gametologs (MME-6 and VR/VAMT041). Red and blue gene names represent fully sex-linked genes from female SDR (MTF) and male SDR (MTM), respectively. Numbers in left and right sides at branches indicate bootstrap values of ML analysis and posterior probabilities of Bayesian inference, respectively. For MOT41 and TOC34, refer to SI Appendix, Fig. S8.
Fates of SDR Genes after Transition to Homothallism.
Interestingly, V. africanus MID was found in a short contig (0.20 Mbp) as a tandem five-copy array flanked by three pseudogenes (VAMT051, SPS1, and MTM0397) (Fig. 3A). These pseudogenes were apparently male related because each had the closest homolog among gametologs of the male SDR of V. reticuliferus (Fig. 3 and SI Appendix, Fig. S7). Interestingly, their female-related counterparts were all intact and localized in SDLR (Fig. 3 and SI Appendix, Fig. S7). We designated this short MID-containing contig “short SDLR” because, like the SDLR, it has low gene density (20 Mbp versus 100 Mbp genome average) and is strongly enriched for repeats (85% compared with 26% genome average) (SI Appendix, Table S2) also suggesting a recent origin from a heteromorphic male SDR. The short SDLR was apparently separated from SDLR because SDLR itself was continuous without gaps and flanked by sequences corresponding to the pseudo autosomal regions of V. reticuliferus (Fig. 3 and SI Appendix, Fig. S10). Continuity or discontinuity of the short SDLR to the contig harboring MTD1 (contig040) was not resolved in the present genome analyses. However, possibility of such continuity is low because V. africanus MTD1 was inserted in the genomic region homologous to the autosomal region of V. reticuliferus (Fig. 4A).
Phylogenies of SDR, SDLR, and short SDLR genes from V. reticuliferus, V. africanus, and V. carteri were used to infer their origins and relationships to one another and by extension to understand the origins of the SDLR and short SDLR in V. africanus (Fig. 3). As expected, the phylogenies of FUS1, MID, and MTD1 were consistent with vertical inheritance in Volvox and other volvocine species, with independent losses or pseudogenization of FUS1 in V. carteri and V. africanus (Figs. 2 and 3B). Of the 20 genes in the V. africanus SDLR that it shares with V. reticuliferus gametologs, 18 descend from a female SDR ancestor, and in five cases, these genes date back to a common ancestor with a V. carteri female SDR homolog (Fig. 3 and SI Appendix, Fig. S6); in the case of SPS1, they go as far back as the SDR from Eudorina sp. (Figs. 2 and 3B). In contrast, none of the SDLR-predicted proteins grouped with male SDR proteins. In two cases, VAMT001 and VAMT003, the phylogenetic relationships of the V. africanus proteins suggest that their corresponding genes might have entered the SDR/SDLR after the V. reticuliferus and V. africanus lineages split (SI Appendix, Fig. S6). There was a single gene likely belonging to the ancestral female SDR, VAMT041, that was not present in the V. africanus SDLR but instead was found inserted into an autosome-like region (Figs. 3 and 4). These results unequivocally support the ancestral state of the Merrillosphaera clade as being heterothallic with large nonrecombining SDRs and the transition to homothallism in V. africanus as being a derived feature.
If V. africanus descended from an ancestor with UV chromosomes, then what were the fates of the male SDR genes from that ancestor? As described above, the sex-related genes MID and MTD1 are present elsewhere in the V. africanus genome. Strikingly, only three male SDR–derived ancestral gametologs (MME6, MOT41, and TOC34) remained intact in V. africanus, and all three were found inserted into different discontinuous autosomal locations (Figs. 3 and 4 and SI Appendix, Fig. S11). Notably, female-derived homologs of these three genes were absent from the SDLR and other autosomal regions, suggesting the ancestral female alleles were lost after the transition to homothallism. Three pseudogenes in the short SDLR (VAMT051, SPS1, and MTM0397) are male derived and had intact female-derived paralogs in the SDLR (Fig. 3 and SI Appendix, Fig. S7). In no case, did we find both intact male-derived and female-derived copies of ancestral gametologs in V. africanus.
Taken together, our data allow us to draw several conclusions regarding the transition from heterothallism to homothallism in the V. africanus algae. First, we conclude that maleness is mainly conferred by the conserved genes MID and MTD1 retained from a heterothallic ancestor. In volvocine algae female (or mating-type plus), gamete production is the default state when MID is absent (28). However, it is likely that the female-derived SDLR has retained some sex-specific functions. This inference of sex-specific functions encoded in the female SDLR is supported by the transgenetic study of V. carteri (28) (see below). The loss of the only conserved female gene, FUS1, from the SDLR in V. africanus suggests that its function became partly or wholly dispensable for female gamete function or was replaced by something else. In Chlamydomonas, the FUS1 protein localizes to the site of membrane fusion during fertilization and is essential to complete fertilization (32). Precedent for the dispensability of FUS1 comes from heterothallic V. carteri, which also lost FUS1 but which still can produce fertilizable eggs in a chromosomally male SDR background (19, 28), implying that other sex-linked genes in the female SDR did not replace FUS1 function in this species. The other losses from the ancestrally female SDLR in V. africanus were gametologs that each has a male-derived allele inserted into an autosomal location (Figs. 3 and 4 and SI Appendix, Fig. S11). This finding suggests that retention of two copies of each gametolog was unstable and raises the question of why the ancestral female gametolog copy was not retained for all of the SDLR genes? While we cannot rule out that the loss or retention of male versus female gametologs was largely stochastic, we speculate that in the cases in which a male-derived gametolog copy was retained (MME6, MOT41, and TOC34), the male gametolog may have had a history of sexually antagonistic selection in the heterothallic ancestor, resulting in a differential fitness penalty for retaining male versus female allele (see below). Interestingly, one of the three male SDR–derived gametologs in V. africanus, MOT41, encodes a homolog of IFT43, an intraflagellar transport protein that plays an essential role in flagellar biogenesis (33). Although functional flagella must be produced in somatic cells of both sexes of Volvox, it is possible that the ancestrally male allele of MOT41 was needed for some critical aspect of sperm motility, and this function could not be adequately fulfilled by the female allele. The potential for sexually antagonistic functions of MME6, a subunit of the NADP malic enzyme (34), and TOC34, a chloroplast outer membrane transit peptide receptor (35), are less clear, but they could be involved in specialized aspects of sperm cell energy metabolism and chloroplast biogenesis, respectively.
Transition from Heterothallism to Homothallism in a UV Chromosome System.
In heterothallic fungi, mating-type–specific idiomorphs (nonhomologous mating-type proteins) govern mating compatibility, whereas in homothallic fungi, both mating-type–specific idiomorphs are generally present or are fused (8). In contrast, the transition to homothallism in Volvox resolved in the present study is not so simple. The presence of a long female-derived SDLR in homothallic V. africanus indicates that this locus remained largely intact during the transition from heterothallism to homothallism (SI Appendix, Fig. S11). Starting with this observation, we can begin to infer how homothallism arose in V. africanus. Our hypotheses are based on the assumptions that MID (and probably MTD1) is essential for male gametogenesis and that none of the fully sex-linked genes in the female SDR of the heterothallic ancestor were absolutely essential for female gametogenesis as suggested by transgenic studies of V. carteri (28); an artificial homothallic phenotype can be created by suppressing MID expression in a V. carteri male genotype (28). However, some ancestrally female gametologs probably did play a role in female reproductive fitness. The MID gene promotes male gametogenesis and suppresses female gametogenesis and would need to be present in the homothallic species but have its expression modulated to prevent complete dominance of male gametogenesis over female (Fig. 1). This type of modulation is consistent with the previous results (27, 28) in which lowering MID expression allowed for hermaphroditic development. In V. carteri, developmental and fertility defects were observed in a culture strain in which a chromosomally male genotype was converted to an egg-producing pseudofemale by suppression of MID expression (28). These fertility defects were likely due to the absence of female SDR genes in this genotype. Therefore, functional homothallism in Volvox could be initiated simply by a MID-containing fragment of an ancestrally male SDR being retained in a chromosomally female genotype. The subsequent genomic modifications that led to present-day V. africanus could have included 1) serial amplification of MID and loss or degeneration of male genes linked to MID; 2) integration of MTD1, three male gametologs, and one female gametolog into autosomes; and 3) loss of the corresponding female or male allele for the autosomally integrated SDR genes to maintain them as single copies (SI Appendix, Fig. S11).
Conclusion
The molecular genetic basis for the evolution of hermaphrodism (bisexuality) from an ancestral male–female mating system have been little studied except in nematodes of the genus Caenorhabditis. Here, hermaphroditism has evolved three times independently from an ancestral male–female mating system, and sex determination is based on an XX/XO sex chromosome system (XX females or hermaphrodites and XO males) (12). It has been suggested that the ancestral XX females became XX hermaphrodites by acquiring the male traits via the co-option of preexisting genes (12). The evolution of hermaphrodites from dioecy is not well understood in other groups of animals. There are numerous examples of transitions from hermaphroditism to dioecy in plants, probably involving the spread of male and female sterility mutation (1, 2, 5) as well as transitions in the opposite direction that involve a restoration of the missing sex function by a simple genetic change (36). Thus, the evolutionary transition of the mating system in the green algal genus Volvox studied here is unique in that an ancestral female genotype has probably acquired male genes to become hermaphrodite (bisexual).
The ancestral female genotype of homothallic V. africanus had to receive sperm from the male genotype to produce heavy-walled resistant zygotes, which can survive cold and/or dry seasons in freshwater habitats. However, the homothallic V. africanus is able to produces zygotes by self-fertilization between eggs and sperm within the same bisexual spheroid (Fig. 1), which may provide reproductive assurance when population density is low, as seen in flowering plants (1, 2, 37). Furthermore, V. africanus also performs “outcrossing” between eggs of a bisexual spheroid and sperm from the separate male spheroid in order to avoid inbreeding depression. By using these two fertilization strategies, V. africanus might have invaded various freshwater habitats from small ponds to large lakes (29).
The remnant female SDLR in V. africanus with distinct properties of SDRs (low gene density and high repeat content) suggests that this transition may have been recent. It is likely that some of the gametologs in the SDR of the heterothallic ancestor of V. africanus had undergone masculinization or feminization and contributed to sex-specific developmental patterning and other sex-specific functions. The V. africanus SDLR harbors at least 18 female-derived genes that have common ancestors with V. reticuliferus female gametologs, and five of the 18 also have a common ancestor with the V. carteri female SDR, making them ancestrally female with respect to the section Merrillosphaera (Fig. 3 and SI Appendix, Fig. S6). The V. africanus female-like SDLR genes may therefore play a role in the development of female functions in this species (Figs. 1 and 2) (29). Similarly, the three male-derived ancestral gametologs in V. africanus may have been retained due to their promotion of male functions. The impacts on reproductive fitness of losing either a male or female gametologs and sex-limited genes (e.g., FUS1) during the transition to homothallism in V. africanus remain to be determined and are an important area of future work.
Materials and Methods
Algal Culture Strains.
V. reticuliferus male and female culture strains and a homothallic V. africanus culture strain (NIES-3786, 3785 and 3780, respectively; Microbial Culture Collection at the National Institute for Environmental Studies) (38) were used in the present study.
Genomic DNA Sequencing and de Novo Whole-Genome Assembly.
Genomic DNAs were prepared according to the method of Miller et al. (39) Whole-genome sequencings of male and female genotypes of V. reticuliferus and a homothallic genotype of V. africanus (SI Appendix, Table S2) were performed using PacBio and Illumina technologies as described previously (20). Genomic DNA was sheared using a DNA shearing tube, g-TUBE (Covaris). A 30 kb library for each of male and female of V. reticuliferus and a 20 kb library for V. africanus were constructed and sequenced on single molecule, real-time (SMRT) cells in PacBio Sequel (Pacific Biosciences). These reactions generated 1.61 M, 2.10M, and 2.08 M sub reads (total bases: 19.0 Gb, 26.4 Gb, and 19.1 Gb, respectively), respectively, for male, female, and homothallic genotypes of Volvox, respectively. Sequencing coverage was about 142x, 198x, and 148x based on the estimated genome size, respectively. In each of the two genotypes of V. reticuliferus, the PacBio reads were assembled de novo with Falcon/Falcon_Unzip v0.9.0 assembler and genome assembly for V. africanus was performed using HGAP4 assembler (Pacific Biosciences). Furthermore, genomic DNA was fragmented with a DNA Shearing System, S2 Focused-ultrasonicator (Covaris Inc.). Illumina paired-end libraries (average insert sizes with 658 bp and 573 bp for male and female genotypes, respectively, of V. reticuliferus and 667 bp for homothallic genotype of V. africanus) were constructed with a TruSeq DNA PCR-Free Sample Prep Kit (Illumina) according to the manufacturer’s instructions and were size selected on an agarose gel using a Zymoclean Large Fragment DNA Recovery Kit (Zymo Research). These libraries were sequenced on the Illumina HiSeq 2500 sequencers (72.9 M, 91.0 M, and 75.4 M reads with 250 bp read length for male, female, and homothallic genotypes, respectively). Total bases and sequencing coverage were 18.2 Gb (136x), 22.8 Gb (133x), and 18.9 Gb (129x), respectively. The Illumina data were then used to improve the assembly sequence with Pilon v.1.22 software tool, ultimately giving a set of nuclear genome sequence (SI Appendix, Table S2).
SDR Identification.
Candidate contigs for the entire SDR or MT (V. reticuliferus male: contig010; V. reticuliferus female: contig011/058 [SI Appendix, Supplementary Information Text 1]) were screened as major significant matching subjects with more than three nonoverlapping protein hits (cutoff maximum E-value: 1 × 10−10) by TBLASTN (National Center for Biotechnology Information) on de novo assemblies of V. reticuliferus with 80 proteins on V. carteri female SDR (MTF) (GU784915) as queries and then dotplot analyzed between haplotypes of same species using YASS (https://bioinfo.lifl.fr/yass/index.php) (40) to detect the rearranged genomic regions or SDR. By using the V. reticuliferus SDR sequences, a long SDLR was determined in V. africanus contig018/132 (SI Appendix, Supplementary Information Text 2).
Gene Identification.
We performed TBLASTN searches against the genome assembly databases of V. reticuliferus male and female genotypes and the homothallic V. africanus genotype with the volvocine fully sex-linked gene proteins (SI Appendix, Table S3) as the queries, retrieved sequences with the highest similarity. For FUS1 homolog of V. africanus, coding regions were determined by using RT-PCR with specific primers (SI Appendix, Supplementary Information Text 3). Other gene models on SDR and SDLR/short SDLR contigs and on autosomal/autosome-like regions were constructed manually after predicted by AUGUSTUS (41) with the C. reinhardtii parameter and by RNA sequencing mapping (SI Appendix, Supplementary Information Text 4).
Molecular Phylogenetic Analyses.
Homologs of fully sex-linked genes in V. reticuliferus and V. africanus were analyzed as described in SI Appendix, Supplementary Information Text 5. The alignments used for the analyses are available in TreeBASE (https://www.treebase.org/treebase-web/home.html; Study ID S26767).
Molecular Evolutionary Analysis.
Divergence scores of synonymous and nonsynonymous substitutions between gametologs were computed using yn00 of the phylogenetic analysis by maximum likelihood (PAML) 4 package (42); nonsynonymous and synonymous site divergence of aligned coding sequences of gametologs was calculated based on Yang and Nielsen (43) with equal weighting between pathways and the same codon frequency for all pairs (24).
Supplementary Material
Acknowledgments
We thank the staff of the Comparative Genomics Laboratory at the National Institute of Genetics (NIG) for supporting genome sequencing. Computations were partially performed on the NIG supercomputer at Research Organization of Information and Systems (ROIS) NIG. This work was supported by a Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Research Fellow (Grant No. 18J11391 to K.Y.), the Scientific Research on Innovative Areas “Platform for Advanced Genome Science” (Grant No. 16H06279 to A.T.), Scientific Research (A) (Grant No. 16H02518 to H. Nozaki), Scientific Research (B) (Grant No. 20H03299 to H. Nozaki), Scientific Research (C) (Grant No. 17K07510 to H.K.-T.), a Grant-in-Aid for Scientific Research on Innovative Areas (Grant No. 17H05840 to T.H. and Y.N.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/JSPS KAKENHI, and the NSF (Grant IOS 1755430 to J.G.U.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100712118/-/DCSupplemental.
Data Availability
Raw reads of genome and RNA-seq data and genome assemblies with annotations data have been deposited in DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank (DDBJ Sequence Read Archive [DRA]: DRA010672; whole-genome assemblies with annotations: BNCO01000001 through BNCO01000448 [V. africanus], BNCP01000001 through BNCP01000200 [V. reticuliferus female genotype], and BNCQ01000001 through BNCQ01000230 [V. reticuliferus male genotype]). SDR genome sequences harboring rearranged domains with fully sex-linked genes in V. reticuliferus male and female genotypes (MTM and MTF, respectively), SDLR, and short SDLR sequences in V. africanus are available under accession numbers LC586641 through LC586644. All other study data are included in the article and/or SI Appendix.
References
- 1.Darwin C., The Correspondence of Charles Darwin. Vol. 9, Burkhardt F., Browne J., Porter D. M., Richmond M., Eds. (Cambridge University Press, Cambridge/New York, 1994), 9: 1861, pp. 1–609. [Google Scholar]
- 2.Barrett S. C. H., The evolution of plant sexual diversity. Nat. Rev. Genet. 3, 274–284 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Policansky D., Sex change in plants and animals. Annu. Rev. Ecol. Syst. 13, 471–495 (1982). [Google Scholar]
- 4.Jarne P., Auld J. R., Animals mix it up too: The distribution of self-fertilization among hermaphroditic animals. Evolution 60, 1816–1824 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Renner S. S., The relative and absolute frequencies of angiosperm sexual systems: Dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 101, 1588–1596 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Wong J. L., Wolfner M. F., Is gender just a category? The two-plus sex advantage. Mol. Reprod. Dev. 84, 89–90 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Hanschen E. R., Herron M. D., Wiens J. J., Nozaki H., Michod R. E., Repeated evolution and reversibility of self-fertilization in the volvocine green algae. Evolution 72, 386–398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni M., Feretzaki M., Sun S., Wang X., Heitman J., Sex in fungi. Annu. Rev. Genet. 45, 405–430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coelho S. M., Gueno J., Lipinska A. P., Cock J. M., Umen J. G., UV chromosomes and haploid sexual systems. Trends Plant Sci. 23, 794–807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghiselin M. T., The evolution of hermaphroditism among animals. Q. Rev. Biol. 44, 189–208 (1969). [DOI] [PubMed] [Google Scholar]
- 11.Reinboth R., Ed., Intersexuality in the Animal Kingdom, (Springer, 1975), pp. 1–449. [Google Scholar]
- 12.Ellis R. E., Lin S. Y., The evolutionary origins and consequences of self-fertility in nematodes. F1000Prime Rep. 6, 62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlesworth D., Plant sex chromosomes. Annu. Rev. Plant Biol. 67, 397–420 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Massonnet M., et al., The genetic basis of sex determination in grapes. Nat. Commun. 11, 2902 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badouin H., et al., The wild grape genome sequence provides insights into the transition from dioecy to hermaphroditism during grape domestication. Genome Biol. 21, 223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mignerot L., et al., A key role for sex chromosomes in the regulation of parthenogenesis in the brown alga Ectocarpus. PLoS Genet. 15, e1008211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirk D. L., A twelve-step program for evolving multicellularity and a division of labor. BioEssays 27, 299–310 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Nozaki H., Mori T., Misumi O., Matsunaga S., Kuroiwa T., Males evolved from the dominant isogametic mating type. Curr. Biol. 16, R1018–R1020 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Ferris P., et al., Evolution of an expanded sex-determining locus in Volvox. Science 328, 351–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanschen E. R., et al., The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat. Commun. 7, 11370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamaji T., et al., Anisogamy evolved with a reduced sex-determining region in volvocine green algae. Commun. Biol. 1, 17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umen J., Coelho S., Algal sex determination and the evolution of anisogamy. Annu. Rev. Microbiol. 73, 267–291 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Ferris P. J., Goodenough U. W., The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell 76, 1135–1145 (1994). [DOI] [PubMed] [Google Scholar]
- 24.Hamaji T., et al., Sequence of the Gonium pectorale mating locus reveals a complex and dynamic history of changes in volvocine algal mating haplotypes. G3 (Bethesda) 6, 1179–1189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlesworth B., The population genetics of anisogamy. J. Theor. Biol. 73, 347–357 (1978). [DOI] [PubMed] [Google Scholar]
- 26.van den Ende H., VanWinkle-Swift K. P., Mating-type differentiation and mate selection in the homothallic Chlamydomonas monoica. Curr. Genet. 25, 209–216 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto K., et al., Molecular evolutionary analysis of a gender-limited MID ortholog from the homothallic species Volvox africanus with male and monoecious spheroids. PLoS One 12, e0180313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geng S., De Hoff P., Umen J. G., Evolution of sexes from an ancestral mating-type specification pathway. PLoS Biol. 12, e1001904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nozaki H., Matsuzaki R., Yamamoto K., Kawachi M., Takahashi F., Delineating a new heterothallic species of Volvox (Volvocaceae, Chlorophyceae) using new strains of ‘Volvox africanus’. PLoS One 10, e0142632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Vurture G. W., et al., GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferris P. J., Woessner J. P., Goodenough U. W., A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7, 1235–1248 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu B., et al., Functional exploration of the IFT-A complex in intraflagellar transport and ciliogenesis. PLoS Genet. 13, e1006627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubini A., Mus F., Seibert M., Grossman A. R., Posewitz M. C., Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J. Biol. Chem. 284, 7201–7213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalanon M., McFadden G. I., The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: A bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179, 95–112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crossman A., Charlesworth D., Breakdown of dioecy: Models where males acquire cosexual functions. Evolution 68, 426–440 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Pannell J. R., The evolution and maintenance of androdioecy. Annu. Rev. Ecol. Syst. 33, 397–425 (2002). [Google Scholar]
- 38.Kawachi M., et al., MCC-NIES list of strains, 9th Edition, Microbial Culture Collection at National Institute for Environmental Studies, Tsukuba, Japan. https://mcc.nies.go.jp/download/list9th_e.pdf. Accessed 1 April 2021.
- 39.Miller S. M., Schmitt R., Kirk D. L., Jordan, an active Volvox transposable element similar to higher plant transposons. Plant Cell 5, 1125–1138 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noé L., Kucherov G., YASS: Enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 33, W540–W543 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoff K. J., Stanke M., WebAUGUSTUS–A Web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 41, W123–W128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Yang Z., Nielsen R., Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17, 32–43 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads of genome and RNA-seq data and genome assemblies with annotations data have been deposited in DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank (DDBJ Sequence Read Archive [DRA]: DRA010672; whole-genome assemblies with annotations: BNCO01000001 through BNCO01000448 [V. africanus], BNCP01000001 through BNCP01000200 [V. reticuliferus female genotype], and BNCQ01000001 through BNCQ01000230 [V. reticuliferus male genotype]). SDR genome sequences harboring rearranged domains with fully sex-linked genes in V. reticuliferus male and female genotypes (MTM and MTF, respectively), SDLR, and short SDLR sequences in V. africanus are available under accession numbers LC586641 through LC586644. All other study data are included in the article and/or SI Appendix.