Significance
Secondary metabolites (SMs) produced by fungi mediate ecological interactions, define fungal niches, and are of profound pharmacological importance to humans. Most work on SMs has focused on a small number of individuals from each species, not fully reflecting the importance of intraspecific diversity. We demonstrate that even in one of the best-studied model fungi, the carcinogen-producing Aspergillus flavus, more than 25% of SM-producing biosynthetic gene clusters (BGCs) are novel and/or show population-specific variants. These results support the finding that the organization of BGC diversity into population-specific patterns may sometimes result from ecologically important interactions and may inform evolutionary and etiological inferences of SM capacities within a species. Importantly, our work also presents a vision of sources of potential pharmaceuticals.
Keywords: secondary metabolism, population genomics, allopatric speciation, eukaryotic pangenome, comparative genomics
Abstract
Fungi produce a wealth of pharmacologically bioactive secondary metabolites (SMs) from biosynthetic gene clusters (BGCs). It is common practice for drug discovery efforts to treat species’ secondary metabolomes as being well represented by a single or a small number of representative genomes. However, this approach misses the possibility that intraspecific population dynamics, such as adaptation to environmental conditions or local microbiomes, may harbor novel BGCs that contribute to the overall niche breadth of species. Using 94 isolates of Aspergillus flavus, a cosmopolitan model fungus, sampled from seven states in the United States, we dereplicate 7,821 BGCs into 92 unique BGCs. We find that more than 25% of pangenomic BGCs show population-specific patterns of presence/absence or protein divergence. Population-specific BGCs make up most of the accessory-genome BGCs, suggesting that different ecological forces that maintain accessory genomes may be partially mediated by population-specific differences in secondary metabolism. We use ultra-high-performance high-resolution mass spectrometry to confirm that these genetic differences in BGCs also result in chemotypic differences in SM production in different populations, which could mediate ecological interactions and be acted on by selection. Thus, our results suggest a paradigm shift that previously unrealized population-level reservoirs of SM diversity may be of significant evolutionary, ecological, and pharmacological importance. Last, we find that several population-specific BGCs from A. flavus are present in Aspergillus parasiticus and Aspergillus minisclerotigenes and discuss how the microevolutionary patterns we uncover inform macroevolutionary inferences and help to align fungal secondary metabolism with existing evolutionary theory.
Understanding how ecologically important traits are organized among locally adapted, genetically distinct populations allows for ecological and evolutionary inferences that cannot be approached when treating species as monomorphic groups. In fungi, biosynthetic gene clusters (BGCs) that produce secondary metabolites (SMs) can define niches (1), provide selective advantages in specific ecological conditions (2–4), and affect the host range of pathogens (5, 6). BGCs are characterized by the presence of a gene encoding a core-biosynthetic enzyme (often called a backbone gene) that creates a core chemical structure and by the presence of additional genes encoding enzymes, transporters, and transcription factors that together determine the structure and localization of resulting SMs. In contrast to primary metabolites, SMs are often assumed to be retained only when specific ecological interactions select for them. Given the ecological and economic importance of SMs (e.g., penicillin and lovastatin), their distribution in fungal species has been the subject of much research. While it has been widely held that species differ in BGC content (7), relatively few genomic studies have examined intraspecific variation. Large-scale genomic studies of intraspecific diversity have focused on strain-level differences in medically important species that are assumed to be panmictic (i.e., without population structure) (8, 9). However, many phylogenetically and ecologically diverse fungi are known to comprise genetically distinct populations (i.e., have population structure) (10–13), with recent evidence that even some species previously considered panmictic have population structure (14). Studies assuming panmixis have drawn macroevolutionary conclusions treating BGC diversity as reflecting a single ecology. While these studies are of clear value to our understanding of the evolution of secondary metabolism in fungi, alternative hypotheses that are based on macroevolutionary ideas rooted in the importance of ecological differentiation at the population level are not well represented in the current literature (8, 15). With this basis, a growing number of prominent comparative genomics papers have drawn ecological and evolutionary inferences between species using a small number of (or often single) genomes to represent species. Such inferences reflect a narrow view of species ecology and may miss the overall niche breadth or even the most common ecology of species that may sometimes manifest in different populations.
Population structure allows BGCs to evolve not only within specific environmental conditions encountered by a distinct population but also within the context of other genes (including other BGCs) that may be rare in a species as a whole. There is considerable evidence that different combinations of BGCs present in the same genome provide important genomic context in fungi. For example, the synthesis of several SMs is known to require two physically distinct BGCs (16, 17). Some SMs have ecologically synergistic effects with other SMs (18) or associations with other ecologically important traits (19); such SMs may be of limited ecological value when not found in the same genome, potentially resulting in fitness costs (i.e., recombination load). The evolutionary and ecological significance of such genetic interactions is particularly evident in populations that occur at the edge of species ranges (i.e., peripheral populations). Such populations often exhibit decreased recombination rates, as mixing of gene combinations that are selected for at the edge of a species’ range may result in maladapted offspring when combined with genes that are selected for in the general population (i.e., recombination load) (20, 21). Importantly, most genes are highly conserved within species and are thus of little significance for population-level differentiation. When treating species as panmictic, ecologically important coevolved combinations of genes that are at high frequency within small locally adapted peripheral populations are likely to be interpreted as rare anomalies that explain little of the overall species’ ecology. A better understanding of how SM diversity is distributed between populations will present opportunities to study ecological and evolutionary processes that are founded upon population-specific differentiation of some ecologically important genes.
Few SMs have received as much attention as aflatoxin, a mycotoxin produced by the fungus Aspergillus flavus and several closely related species in Aspergillus section Flavi. Aflatoxin is a notoriously potent hepatotoxin that causes acute toxicosis, cancer, immune suppression, and stunted growth in children (22–24), chronically impacting an estimated 4.5 billion people (25). However, only 40 to 60% of A. flavus isolates produce aflatoxin. While the ecological significance of BGCs are often unknown, aflatoxin production is thought to be favored in the presence of insects (2) but costly in their absence (26). Recently, Drott et al. (27) demonstrated that three distinct populations of A. flavus in the United States differ quantitatively in aflatoxin production. Interestingly, a small population at the northern edge of the species’ range (28) that is closely related to the domesticated soy sauce–producing fungus Aspergillus oryzae was also the least aflatoxigenic population. Some have suggested (29, 30) that the lower prevalence of insects at higher latitudes favors nonaflatoxigenic isolates. These findings suggest that population-specific ecologies of A. flavus populations may favor differentiation of aflatoxin-producing ability. However, emblematic of the secondary metabolism literature, it is unclear if such population-specific patterns are common in the secondary metabolome or represent rare differentiation of specific BGCs. Recently, Uka et al. (31) demonstrated high levels of chemotypic diversity in A. flavus using 50 metabolites (including 11 precursors and derivatives of aflatoxin). However, their use of only a small number of molecular markers to identify population structure led them to assume that populations, as defined by geographic location, were panmictic. As mentioned above, such assumptions preclude the interpretation of intraspecific diversity as reflecting local adaptation of populations. We suggest that inferences of population-specific variation in the secondary metabolome of A. flavus will clarify the potential for these ecologically important genes to drive population-level ecological and evolutionary processes.
Modern macroevolutionary theory infers ecological and evolutionary processes that occur at the population level. However, a dearth of information about how SM gene clusters are organized on this scale limits our understanding of SM evolution. The overall objective of this study was to determine how variation in BGCs and resulting SMs are distributed across the three A. flavus populations found in the United States (27). Because of the ecological relevance of many SMs, ecological differences are often inferred solely from differences in BGC content of genomes. However, it is phenotypes (chemotypes), not genotypes, that mediate ecological interactions and are subject to selection. We thus sought to understand the ecological and evolutionary significance of intraspecific variation in secondary metabolism by testing the following hypotheses: 1) variation in some BGCs show population-specific patterns in A. flavus; 2) population-specific BGCs that are missing from some populations are present in closely related species, illuminating ecological similarities and varied evolutionary histories of genes; and 3) population-specific differences in BGCs result in chemotypic differences that could be subject to selection.
Results
Pangenomic BGC Display Three Patterns of Population-Specific Conservation.
Using genome sequences with the BGCs removed, we identified the same populations (A, B, and C) as with the whole-genome data set (SI Appendix, Fig. S1), confirming that A. flavus population structure reflects differentiation across the genome, not just of BGCs. Network analysis of BGCs identified by antiSMASH (32) and by manual curation of characterized clusters dereplicated 7,821 BGCs across all isolates into 92 unique BGCs (individual isolates had between 78 and 85 BGCs with a median of 82 BGCs) that were shared by two or more individuals in the A. flavus pangenome (SI Appendix, Fig. S1 and Table S1). Of the pangenomic BGCs, 83 were found in the genome of the reference isolate obtained from Northern Regional Research Library (NRRL), NRRL3357 (BGCs and associated genes are listed in SI Appendix, Table S2). Three BGCs unique to single A. flavus isolates were removed from further analysis. While this approach determined the presence/absence of a BGC based on total gene content, given the magnitude of the dataset, we focused gene-level analyses on the 9,782 core-biosynthetic backbone genes (key genes that define the central structure of resulting SMs, e.g., polyketide synthase [PKS], dimethylallyl tryptophan synthase [DMATS], terpene synthases/cyclase, and nonribosomal peptide synthetase [NRPS]) that were identified within clusters. Consistent with previous assessments of intraspecific diversity (8, 9), we find isolate-specific variation in the form of deletions, single nucleotide polymorphisms (SNPs), and differences in gene content. For example, BGC 43 is only missing in isolate 41mAF (SI Appendix, Fig. S2). However, isolate-specific differences explained relatively little of the variation in BGCs when compared with population-specific variation (Fig. 1 compared with SI Appendix, Fig. S2).
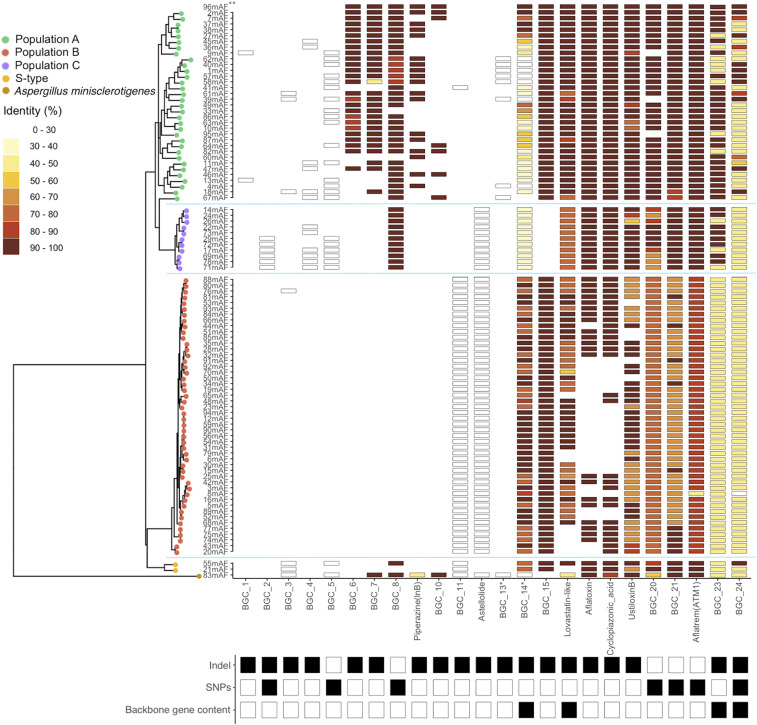
Fig. 1.
Heatmap showing BGCs with confirmed population-specific patterns among three genetically isolated L-type A. flavus populations, A (green), B (orange), and C (purple) identified previously (27) and confirmed in this study (SI Appendix, Fig. S1). In addition to 92 L-type A. flavus isolates, two S-strains (55mAF and 21mAF, Bottom, yellow) and one Aspergillus minisclerotigenes isolate (83mAF, Bottom, brown—formerly identified as Aspergillus texensis [Materials and Methods]) were included for context. The presence of a BGC, as indicated with a black-bordered box, was determined through network analysis of antiSMASH results that uses total gene content of BGCs or by previous studies (Materials and Methods). Absence of a BGC is indicated by blank spaces. When a BGC is not present in the reference genome, all corresponding boxes are white (BGCs 1 to 5 and 11 to 13). The protein sequence identity of backbone genes (core biosynthetic genes) was determined relative to the NRRL3357 reference genome (**a high-quality assembly [96mAF] was used as the reference and was compared with an assembly [2mAF—which is located immediately below 96mAF in the figure] that resulted from methodology used on all other isolates [Materials and Methods]; the high-quality assembly was not included in phylogeny). Genetic relationships among isolates are depicted on the left side using a maximum likelihood tree based on genome-wide SNPs that were thinned to be at least 3 kb apart. Population structure was determined using discriminant analysis of principle components. Population-specific variation was categorized below the heatmap as being caused by insertion/deletion (indel) or differences in protein sequence identity of backbone genes (SNPs). A third type of variation was identified when the previous types of variation occurred differentially across multiple backbone genes in a single cluster (backbone gene content). The protein sequence identity and number of backbone genes, including those found by reciprocal best-hit BLAST but not incorporated into a BGC by antiSMASH, are indicated for all 92 BGCs analyzed in SI Appendix, Fig. S2 (reference SI Appendix, SI Text for validation and incorporation of reciprocal best-hit genes). *These BGCs are physically overlapping but were distinguished by antiSMASH based on differences in gene content (see main text).
Population-specific patterns of variation were evident in 24 BGCs (Fig. 1). Population-specific differences fell into three major categories (Fig. 1): The first two categories describe BGCs with a single backbone gene, where population-specific differences manifest as differences in protein sequence identity caused, in the first category, by the accumulation of SNPs or, in the second category, by partial or complete deletion. Differences in the third category of BGCs arose by the aforementioned mechanisms but in BGCs where there were multiple backbone genes that showed different patterns of variation from each other, for example, a backbone gene was deleted in some isolates while another backbone gene in the same BGC varied in identity. If all backbone genes within a BGC showed the same type of variation, they were categorized as one of the previous two types of variation.
We performed extensive validations using a variety of alignment- and assembly-based methods (SI Appendix, SI Text) that extend substantially beyond validation typically reported in similar studies. We only interpret population-specific differences in BGCs where we could identify associated changes in protein domains (as identified using the National Center for Biotechnology Information's [NCBI's] conserved domain tool). We speculate that ecological forces may select for and maintain differentiation of population-specific patterns among BGCs. Estimates of selection derived from the site-frequency spectrum (e.g., comparing the number of nonsynonymous [dN] and synonymous [dS] SNPs) were originally developed for deeply divergent species and are insensitive to selective pressures when estimated within species, with dN/dS ratios deviating substantially from classic interpretations applied between species (reviewed by ref. 33). We find that the vast majority of population-specific BGCs have dN/dS ratios that indicate purifying selection (< 1.0) (SI Appendix, Table S3). This result is consistent with our interpretation that BGCs with population-specific variation are being retained by selection within respective populations. However, as ancient signatures of purifying selection can be very strong, new selective forces acting between recently diverged lineages may not be evident over ancient signals. Given problems associated with dN/dS estimates between closely related groups, we emphasize that selection acts on phenotypes and thus focus our efforts on identifying how population-specific variation is associated with differences in corresponding SM production. Furthermore, our focus on protein domains avoids issues where dN/dS ratios may not capture the outsized importance that individual SNPs can have (e.g., those that may impact the sequences associated with protein domains). Differences in proteins that result from relatively small differences in nucleotide and/or amino acid sequences are difficult to differentiate from assembly and annotation errors with complete confidence (notably, patterns of indels described in the Pattern 1 section are easier to interpret than differences that arise from SNPs). Despite having validated population-specific differences using NCBI’s conserved domain tool, the importance of these domains to the biosynthetic abilities of a protein are not always clear. We outline the details of the population-specific differences we found in the Pattern 1, Pattern 2, and Pattern 3 sections to emphasize that a large portion of the diversity in BGCs can be explained by population-specific differences.
Pattern 1: Indels Result in Population-Specific Differences in BGC Gene Content and Presence/Absence.
We found that ∼15% of BGCs show clear population-specific patterns in presence/absence caused by deletions (BGCs 1, 3, 4, 6, 7, 10, 11, 13, 15, astellolide, aflatoxin, cyclopiazonic acid [CPA], and piperazine [lnB]) (Fig. 1). Most of these BGCs, with the exception of 1 and 3, were found at relatively high frequency in at least one population but were completely missing in others. Interestingly, the backbone gene of BGC 13 is present in all isolates but was usually incorporated into BGC 14 (discussed in the Pattern 3 section). BGC 13 was only considered a distinct cluster by antiSMASH in a subset of isolates where three additional cytochrome P450 enzymes and a transporter were also present. Production of astellolide was previously unknown in A. flavus but has been described in several other Aspergillus spp (34), as is discussed in the Population-Specific BGCs Are Found in Isoaltes from Closely Related Species section. This BGC was found in all isolates of populations B and C but in no isolates of population A, which contains the NRRL3357 A. flavus reference strain. Piperazine synthesis is mediated by two unlinked BGCs, lnA and lnB (16). While lnA is present in all isolates, lnB is missing from all isolates in populations B and C, raising questions about the role of this cluster without its biosynthetic counterpart. Deletions in the aflatoxin and CPA BGCs as well as the lovastatin-like (also discussed in the Pattern 3 section) and ustiloxin B BGCs were only found in isolates of population B. Similar, though not identical, sets of isolates with deletions observed in the aflatoxin and CPA BGCs may be explained by the physical proximity of these BGCs (SI Appendix, Fig. S3).
Pattern 2: Population-Specific SNPs Result in Protein-Domain Shifts.
In contrast to the highly conserved nature (>95% protein sequence identity) of most BGC backbone genes, we identified seven BGCs where backbone genes have diverged through the accumulation of SNPs to extents that result in protein-domain differences among populations (Fig. 1). For example, the identity of the backbone gene of the aflatrem (ATM1) BGC relative to the reference genome is lower for all isolates in population B (∼89%) compared with population A (∼97%), a difference that was associated with the presence of putative protein domains in population A isolates that were missing in population B isolates. Similarly, the terpene backbone gene of BGC 21 is truncated in most population B isolates, resulting in the loss of one of two putative terpene cyclase domains. Similar patterns of full domain or partial domain loss that result from the accumulation of SNPs were also observed in BGC 20. While these BGCs were still found by antiSMASH, some BGCs with similar patterns of divergence were not. A terpene backbone gene of BGC 5 was not found in any isolate of population B and was also missing in several isolates from population A despite >98% nucleotide identity in the associated region. While the domains in the backbone gene of BGC 8 appear to be retained, a less recognizable (lower e-value) DMAT domain may explain why antiSMASH did not identify these clusters in population B, despite being identified by reciprocal best-hit Basic Local Alignment Search Tool (BLAST) (SI Appendix, Fig. S2). Most isolates from populations A and B have the backbone gene of BGC 2 completely deleted; however, 15 isolates have variants where many of the NRPS domains, including a pp-binding and an AMP-binding domain, are missing, which we interpret as pseudogenized variants.
Pattern 3: BGCs Where Variable Backbone Gene Content Shows Population-Specific Evolutionary Patterns.
Because most of the BGCs we identified here are not fully characterized, it is unclear if differences in backbone gene content of BGCs with multiple backbone genes may modify or stop production of a resulting metabolite. In BGC 24, a terpene synthase backbone gene was deleted from eight isolates in population A and the Aspergillus minisclerotigenes isolate (Fig. 1). A full version of this backbone protein was only found in 13 population A isolates and one S-type isolate, suggesting pseudogenization of this gene in isolates with no apparent deletion. In both the lovastatin-like BGC and BGC 23, deletions of variable sizes were observed in only a subset of backbone genes found in each BGC. BGCs 13 and 14 (discussed above in the Pattern 1 section) are physically overlapping clusters and together are part of a highly variable region where several different combinations of SM backbone genes exist. Some of the variation at this locus had previously been identified in comparisons with A. oryzae (35). In Aspergillus fumigatus, a similar set of BGC alleles was dubbed an “idiomorphic” BGC (8).
Population-Specific BGCs Are Found in Isolates from Closely Related Species.
The two S-type isolates of A. flavus have together retained all of the BGCs that were present in all isolates of the three L-type populations (SI Appendix, Fig. S2), as well as five BGCs that are otherwise only found in specific populations (Fig. 1 and SI Appendix, Fig. S2). While some S-type isolates are still considered to be part of A. flavus (36, 37), we also observed a similar pattern in the BGCs of the A. minisclerotigenes isolate. A. minisclerotigenes is a very closely related species to A. flavus that was used to determine if population-specific BGCs could be identified in other species. We found that nine population-specific BGCs were present in the A. minisclerotigenes isolate, for example, BGCs 23 and 24 (Fig. 1), as well as an additional 10 BGCs that were not found in any A. flavus isolates. Furthermore, the astellolide BGC, which has previously been described only in A. oryzae and Aspergillus parasiticus (34), is present in all population B and C isolates of A. flavus. Because A. oryzae is thought to be the domesticated form of A. flavus, we only interpret the presence of this BGC by A. parasiticus. These results emphasize the importance of population-level assessments of Aspergillus species, via pangenomic sequencing of populations, to better understand how population-specific BGCs may impact ecological inferences of comparative genomics and may inform varied evolutionary histories of some BGCs.
Genetic Differences in BGCs Can Result in Population-Specific Patterns of SM Production.
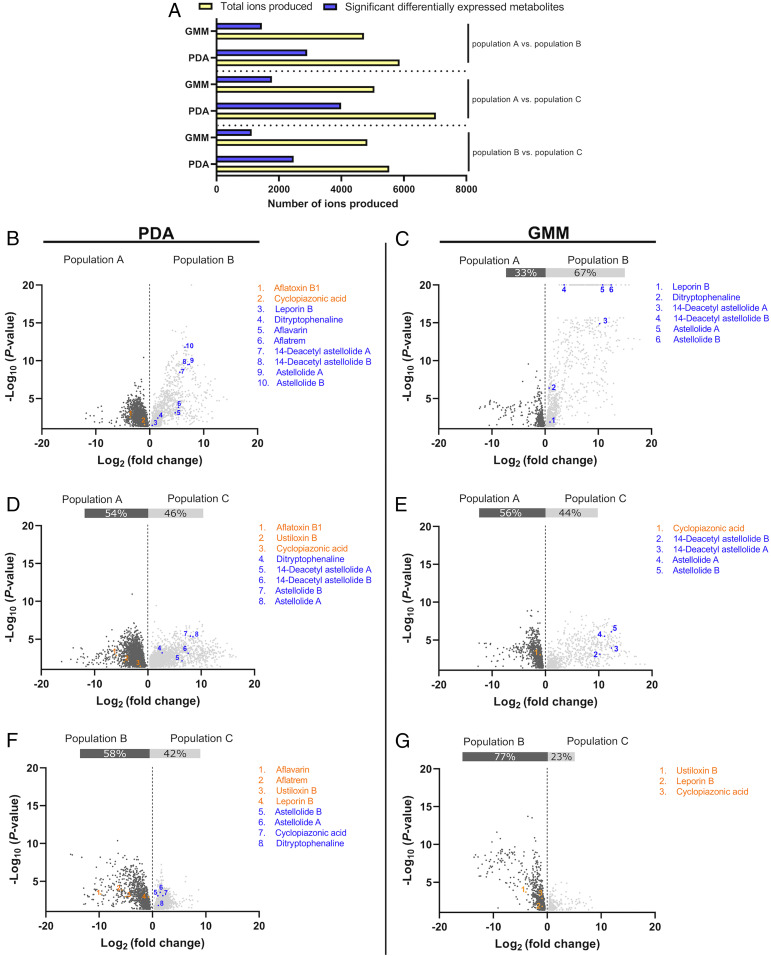
For selection to act upon population-specific differences in BGCs, it is necessary that genetic differences result in associated phenotypes. We established that population-specific differences in BGC content resulted in chemotypic differences between isolates by identifying population-specific patterns in 13 characterized BGCs with known products (SI Appendix, Table S3 and Materials and Methods). We found that aflatoxin, astellolide, CPA, ustiloxin B, aflatrem, and the piperazine BGCs showed population-specific genetic variation (Fig. 1). Complementarily, we identified differential production of most of these compounds, but piperazine [specifically, piperazine compound 7 identified by Forseth et al. (16)] was not detected in this analysis (Fig. 2 B–G). While sclerotia are produced by some isolates of A. flavus grown on both glucose-minimal medium (GMM) and potato dextrose agar (PDA) media, piperazine expression has previously been measured using growth media that greatly increases sclerotial production (16, 19), which we did not use, perhaps explaining the absence of these metabolites in our study. In general, populations where some isolates were missing associated BGCs had lower mean production of the associated SMs (Fig. 2).
Fig. 2.
Production of metabolites by A. flavus isolates of populations A, B, and C [identified previously (27)] grown on PDA and GMM as detected by ultra-high-performance liquid chromatography–high-resolution mass spectrometry in positive mode. Ions showing significant differences (P < 0.05) in pairwise comparison of populations were assumed to be metabolites and are referred to as significantly differentially expressed metabolites (SDEMs) (A). The magnitude and significance of SDEMs for each pairwise comparison are depicted in volcano plots for PDA (B, D, and F) and GMM (C, E, and G). Putative SMs are colored blue or orange to differentiate from unidentified metabolites, which are colored gray or dark gray. Points within each volcano plot were colored orange or dark gray if they were produced in higher abundance by the first-named population and blue or gray if produced in higher abundance by the second-named population. The identity of SMs was confirmed by fragmentation analysis (SI Appendix, SI Text).
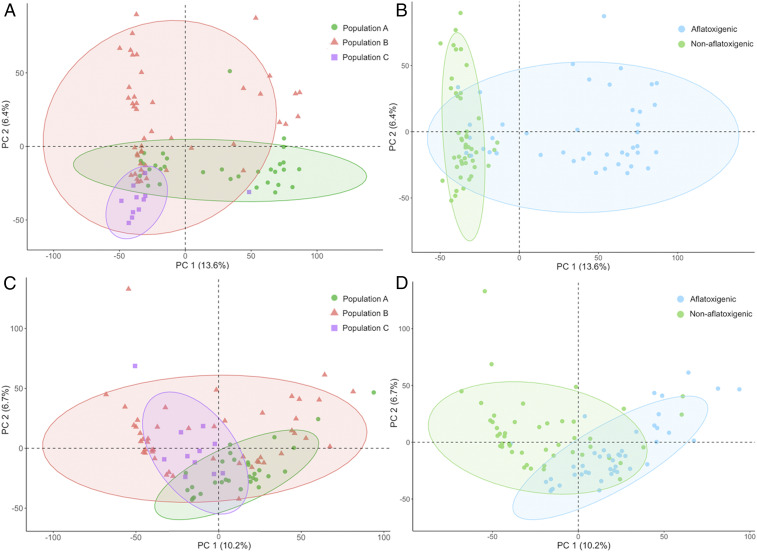
In addition to differential production of compounds corresponding to population-specific patterns in BGCs, we also observed two SMs, aflavarin and leporin B, as differing significantly between populations despite having BGCs that were indistinguishable with our analyses. This finding may be explained by regulatory differences not within the scope of this study. Consistent with previous work on fungal SMs, we find that the magnitude of these differences varies based on media type (Fig. 2). Metabolic differentiation of populations was more evident in principle component analysis (PCA) plots stemming from growth on PDA medium than from growth on GMM (Fig. 3), likely reflecting a larger number or concentration of ions detected on PDA (Fig. 2A). Populations largely overlapped in metabolic space, although populations A and B both occupied some unique metabolic space (Fig. 3). Interestingly, the aflatoxin-producing ability of isolates differentiated some of the overall metabolome, with little overlap in metabolic space between nonaflatoxigenic and aflatoxigenic isolates (Fig. 3). This differentiation was particularly apparent along dimension 1, where aflatoxin explained more variation than 65% of other ions. However, aflatoxin’s overall contribution to this axis was less than 1% for both PDA and GMM profiles.
Fig. 3.
PCA of all metabolites produced by isolates of A. flavus populations A, B, and C [identified previously (27)] on PDA (A and B) and GMM (C and D). Isolates were assigned to populations in this study using discriminant analysis of principle components on variant data that does not include BGCs. Aflatoxin-producing ability was determined previously on several different media types including PDA (27, 28) but not GMM as aflatoxin production on the latter is typically low or absent. Ellipses represent 95% CI. While metabolic space of populations largely overlapped (A and B), aflatoxin-producing ability (C and D) was observed to differentiate metabolic space.
Discussion
Our finding that ∼25% of BGCs present within the A. flavus pangenome show population-specific differences (Fig. 1) has profound implications for inferences of ecological and macroevolutionary processes. Approximately 15% of BGCs were found in only a subset of populations, with population-specific differences in protein sequence identity accounting for another ∼10% of BGCs (Fig. 1). Because selection acts on phenotypes, we emphasize the potential ecological and evolutionary importance of population differentiation of BGCs by demonstrating that, in several cases, these genomic differences result in different chemotypes (Fig. 2). Aflatoxin-producing ability differentiated a large portion of overall metabolic space (Fig. 3), suggesting that the metabolic consequences of differences in a single BGC can be wide-ranging. While differentiation of metabolic space based on aflatoxin-producing ability was independent of population structure, this finding raises the question whether similar patterns may also exist for population-specific differences in BGCs identified here. In addition to microevolutionary inferences, we show that several BGCs that are found in only a subset of populations, but not in the A. flavus reference genome, are present in closely related species, providing important context for macroevolutionary inferences and comparative genomic studies (as discussed below in this section). These results better align our understanding of fungal secondary metabolism with existing evolutionary theories that predict population-specific processes and have important implications for interpretation of past work and design of future studies.
The defining role of SMs on fungal niches is emphasized by a large number of recent studies that have used BGCs to infer ecological and evolutionary differences between species (38–44). However, many of these studies use single reference genomes to represent species. Without using ecological groupings, for example, pathogenic versus nonpathogenic species, whereby reference genomes become replicates within an ecotype (5, 45, 46), studies may effectively be unreplicated. Previous studies that looked for intraspecific variation in fungal secondary metabolism compared the mutations and extent of variation with that found between species but did not infer or discuss ecological distinctions within species or how such intraspecific variation might impact evolutionary inferences drawn between species (8, 40). Using population genomics, we demonstrated that a large portion of the intraspecific variation in BGCs in A. flavus is explained by differences between discrete populations. We speculate that population-specific variation reflects local adaptation associated with different BGCs or combinations of BGCs as mediated through chemotypes. Although we found that dN/dS ratios suggested evidence of purifying selection in most BGCs with population-specific variation (dN/dS < 1 as detailed in SI Appendix, Table S3), methods for inferring selection from dN/dS ratios were originally developed to describe evolution of genes in deeply diverged species and may be relatively insensitive and/or deviate from classical interpretations when applied between closely related groups as we have done here (reviewed by ref. 33). Because selection acts on phenotypes, we have focused our analysis on demonstrating that the patterns we identify are associated with chemotypic differences that could mediate ecological and evolutionary processes. Furthermore, we emphasize that even nonadaptive differentiation (e.g., as can occur through genetic drift) that occurs between populations may still be important for some ecological and evolutionary interactions if associated traits retain functionality (as we show using domain structures and chemotypes). Environmental change that is often associated with punctuated equilibrium and/or some epidemics (discussed further below in this section) can result in novel selective pressures that favor traits that were not previously under selection.
The potential ecological relevance of this diversity raises concerns about studies that do not account for population structure, as they may mischaracterize ecologically important differences between species. For example, comparing the A. flavus NRRL3357 reference genome with A. minisclerotigenes would incorrectly suggest that five clusters present in A. minisclerotigenes are not present in A. flavus. Similarly, we find the astellolide (also known as parasiticolide) BGC, which is also present in A. parasiticus (34), in two of the three A. flavus populations but not in population A, of which NRRL3357 is a member. As ecological functions are often (and increasingly) being assigned to SMs, it is important to interpret comparisons between species in the context of such intraspecific variation. Indeed, ∼15% of A. flavus BGCs are specific to a single or subset of population(s), a number of BGCs similar to that found in some comparisons between closely related species (40). While delineations of populations from species are not always clear, the genetic distances we observe between the three A. flavus populations are much less than between A. flavus and the closely related species A. minisclerotigenes (as is evident in the phylogeny of Fig. 1); therefore, we interpret these as populations not species. Furthermore, we suggest that bio-prospecting efforts that have focused on intraspecific variation (47) may benefit from intentionally sampling populations, an effort that may be facilitated in some species by existing population genetic data.
Two recent analyses of the pangenomes of model fungi have concluded that differences in BGCs between species can be explained by strain-specific differences (8, 9). However, we find in A. flavus that by incorporating analysis of population structure, most strain-specific differences are organized into population-specific differences. While some fungi may have panmictic population structure, precluding similar patterns, we emphasize that many ecologically and taxonomically diverse fungi do have population structure (10–13), and recent large-scale population studies have even identified population structure in well-studied fungi that were previously inferred to be panmictic (14). The distinction between strain-specific and population- (or lineage-) specific differences is of great importance as the latter represents varied ecologies that arise from local adaptation while the former interprets differences between strains as representing variation around a single mean ecology. The strain-specific interpretation lends itself to ecological comparisons between species using a small number of isolates (discussed above), as interspecific variation is assumed to be the result of a gradual accretion of mutations in a species as a whole (e.g., phyletic gradualism). However, Gould (48) referred to such generalization of microevolutionary processes to explain macroevolution as “extrapolationism.” He suggested that this approach precludes inference of important ecological and evolutionary processes by not considering alternative hypotheses that are rooted in ecological differentiation of populations within species (as discussed in the introduction). While we do not expect all fungi to show population structure, we suggest that patterns like those we find in A. flavus may be common across fungi and that differences in underlying population structure may allow for comparison of how secondary metabolism may evolve differently in structured versus unstructured species.
In bacteria, accessory genomes (comprising genes present in only a subset of isolates) are thought to be maintained through selection for ecological conditions that are relatively rare across the entire range of a species whereas core genomes (comprising genes present in all isolates) are thought to reflect a core ecology of species (49–51). While there have been rare suggestions that specific populations of fungi may contain unique BGCs (52) or unique alleles of BGCs (53), our findings that much of the accessory secondary metabolome is explained by population-specific patterns in BGCs (Fig. 1) suggests that, at least in some species, SMs may be of profound importance for local adaptation and overall niche breadth of species. Furthermore, our results show how inferences of population structure raise questions about the ecology of coevolved combinations of genes. For example, the two piperazine BGCs (lnA and lnB) that are present in the A. flavus reference genome are known to have important regulatory cross talk (16). We find that while lnA is part of the core genome (SI Appendix, Fig. S2), lnB is part of the accessory genome, only present in a subset of isolates from population A (Fig. 1). While we were unable to detect piperazine production in this study, we speculate that cross talk between the accessory-genome BGC (lnB) may have important ecological implications for the function of the core-genome BGC (lnA).
The ecological and evolutionary implications of variation in BGCs is complemented by inferences of corresponding SM profiles (chemotypes)—this is an important addition to an evolutionary genomics study as selection acts upon phenotypic variation. While comparative genomic studies of fungal secondary metabolism have revealed interesting differences between species (38–44), inferences on adaptive significance in BGCs cannot be appreciated without determining SM profiles. Perhaps the best evidence of population-specific BGCs and subsequent SM production in fungi comes from Fusarium graminearum, where slight genetic variation in the trichothecene BGC between populations also results in different chemotypes (53–55). However, because of the very close relationship between the resulting SMs, there is controversy about the ecological significance of these differences (53), with some suggesting that inferences of population-specific patterns may sometimes result from methodological approaches (56). However, evidence that such variation in the trichothecene BGC is retained in an ancient transspecies polymorphism (57) does suggest differential ecological function of these BGC variants. In contrast to results on a single SM, our results with A. flavus clarify that population-specific patterns can affect a large portion of the pansecondary metabolome. We demonstrate clear differences in fungal chemotypes that result from population-specific patterns in the presence/absence of multiple BGCs (e.g., astellolide [Figs. 1 and 2]) and significant differences in metabolite expression that were not immediately explained by genetic differentiation (e.g., the aflavarin and leporin B BGCs were genetically indistinguishable using our methods [SI Appendix, Fig. S2]). Furthermore, PCA analysis of the entire metabolome differentiated aflatoxin-producing and non–aflatoxin-producing strains (Fig. 3), emphasizing that differences in a single metabolite may be associated with large-scale differences in fungal metabolism. While we have not explicitly tied these differences to ecological differentiation of populations, we suggest that differences in the geographic distribution of some populations assayed here (27) are consistent with potential variation in niche (58). In contrast to primary metabolites, SMs are often assumed to be maintained through niche-specific ecological interactions. Differences in BGC content (59) and in gene expression of some SMs may underlie differences between some biotrophic and necrotrophic life strategies (60). Similar differences in SM production are thought to be associated with niche adaptation of peripheral populations of some plant species (61). While we assume that conservation of SM production between populations reflects ecological importance, future studies are needed to elucidate the specific ecological functions of these BGCs (1, 27, 58).
In addition to evolutionary and ecological implications, population-specific patterns may also be important for applied efforts to control fungal disease outbreaks. The southern-corn-leaf-blight epidemic of 1970 was caused by a previously undetected lineage (race T) of Cochliobolus heterostrophus that harbored the T-toxin BGC. Corn (maize) varieties particularly susceptible to this toxin were planted widely across the United States, resulting in race T’s decimation of corn production in 1970. Since that outbreak, questions have remained about why race T had not been previously observed. Recently, Condon et al. (62) showed that the T-toxin BGC is ancient in C. heterostrophus, ruling out the possibility of a horizontal gene transfer near the time of the outbreak. The prevalence of population-specific SM demonstrated here raises questions of whether BGCs that are maintained in genetically isolated populations could explain past epidemics or could result in future epidemics when coupled with environmental change. The potential of locally adapted BGCs to drive novel epidemics may be of great importance as it has recently been suggested that climate change may be an important factor in favoring human pathogens (63). Consistent with ideas of punctuated equilibrium, environmental change may allow some populations to undergo range expansion. In new geographic areas, BGCs evolved in one ecological context may find novel functions (i.e., exaptation). Our results demonstrate the importance of intraspecific diversity for secondary metabolism that aligns this field with existing evolutionary theory in a way that offers opportunities to understand the ecology, evolution, and epidemiology of fungi.
Materials and Methods
Isolates of A. flavus.
We performed all analyses on existing genomic data from 92 A. flavus L-type isolates, two A. flavus S-type isolates, and a single isolate of the closely related S-type species A. minisclerotigenes [which was formerly identified as Aspergillus texensis, but whose taxonomy has since been further resolved (64) (SI Appendix, Table S1)]. In this manuscript, we refer to S-type isolates of A. flavus (37) as “S-type” while we refer to isolates from other species with similar morphology by their species name. Species-level identifications were previously confirmed (27) based on comparison of house-keeping gene sequences (SI Appendix, Fig. S4). These isolates comprise a stratified random sample from corn field soils in seven states spanning the eastern and central United States as described by Drott et al. (27). While isolate numbers are identical to those used by Drott et al. (27), we have amended isolate numbers with “mAF” to avoid confusion with similarly numbered BGCs. We used a high-quality assembly of NRRL3357 as a reference isolate (96mAF) but confirmed the consistency of our methods on lower-quality genome assemblies by also incorporating a genome assembly of NRRL3357 that was generated using identical methodologies to all other isolates (2mAF) (27). When we refer to the reference genome sequence, we are referring to the high-quality assembly of 96mAF from Drott et al. (27).
Determination of Population Structure.
In order to confirm that previous inference of population structure (27) was not the result of between-subpopulation differences in BGCs, we removed SNPs from the Drott et al. (27) dataset that fell into regions containing BGCs (identified below in the Identification of BGCs section) using VCFtools (65). The remaining SNPs were analyzed using the non–model-based multivariate discriminant analysis of principle components from “adegenet” v2.1.1 (66) implemented in R v3.5.2 (67) according to procedures outlined in the “adegenet” tutorial (68).
Phylogenetic relationships between isolates was determined from a maximum likelihood analysis executed in Mega X (69) on a data set of whole-genome SNPs thinned to a minimum distance of 3,000 bp. This data set identically reflected isolate relationships evident in neighbor-net created using whole-genome data (27) but was more computationally feasible for this analysis. The resulting tree was graphed using “treeio” (70), “ggtree” (71), and “ggplot2” (72) in R.
Identification of BGCs.
BGCs were identified from genome assemblies from Drott et al. (27) using antiSMASH v4.1.0 (32). We manually curated the borders of 13 characterized SM BGCs that have been associated with the production of specific SMs including the following: two aflatrem BGCs (ATM1 and ATM2) (17), aflatoxin (73), aflavarin (74), asparasone A (75), aspergillic acid (76), aspergillicins (77), CPA (78), ditryptophenaline (79), imizoquin (80), leporin B (81), and two piperazine BGCs (lnA and lnB) (16). Additionally, we added the kojic acid (82) and ustiloxin B (83) BGCs, which were not found by antiSMASH. In a small number of instances where manual curation of characterized BGCs did not include nearby backbone genes (key genes that define the central structure of resulting SMs, e.g., polyketide and NRPS) that were incorporated into a BGC by antiSMASH, we separated out the genes unique to the antiSMASH call and analyzed them separately from the curated BGC.
BGCs were dereplicated by creating networks with Biosynthetic Gene Similarity Clustering and Prospecting Engine (BiG-SCAPE) (84), which were visualized with Cytoscape v3.7.1 (85). Extensive validation of inferred BGC distributions and site-frequency estimates of selection (dN/dS) acting on these BGCs are detailed in SI Appendix, SI Text.
Secondary Metabolite Extraction and Quantification.
Because selection acts upon phenotypes, we determined if differences in BGC content resulted in different chemotypes. SM production was assessed by growing all isolates on both PDA and on GMM at 30 °C for 14 d. Metabolites were extracted in ethyl acetate from three 1.2-cm plugs from each plate similar to methods described previously (86). Metabolic profiles were determined from these extracts using ultra-high-performance liquid chromatography with high-resolution mass spectrometry (UHPLC-HRMS) on a Thermo Scientific-Vanquish UHPLC system connected to a Thermo Scientific Q Exactive Orbitrap mass spectrometer in ES+ mode between 200 and 1,000 m/z to identify metabolites as described previously (86). Acquisition and processing of UHPLC-MS data were done using the open-source software program, Maven version 2011.6.17 and the Thermo Scientific Xcalibur software version 4.3. The identification of putative SMs was further confirmed using the fragmentation patterns produced in tandem mass spectra (MS/MS) to annotate major identifiable peaks (SI Appendix, Table S3 and Figs. S5–S29). Additional details of metabolite identification and chemical analyses can be found in SI Appendix, SI Text.
Supplementary Material
Acknowledgments
This project was supported by US Department of Agriculture, National Institute of Food and Agriculture Postdoctoral Fellowship Award 2019-67012-29662 to M.T.D. This research was performed using the computational resources and assistance of the University of Wisconsin–Madison Center for High Throughput Computing in the Department of Computer Sciences. Work performed by J.M.S. and N.L.G. was supported by a grant from the Innovative Genomics Institute, University of California Berkeley. This work has been performed in collaboration with J.L.L., T.A.R., R.J.G., and P.E.A., supported by the Genomic Science Program, US Department of Energy, Office of Science, Biological and Environmental Research as part of the Plant Microbe Interfaces Scientific Focus Area (https://pmi.ornl.gov/). Oak Ridge National Laboratory is managed by UT-Battelle LLC, for the US Department of Energy under Contract DE-AC05-00OR22725.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021683118/-/DCSupplemental.
Data Availability
Raw reads used to construct assemblies in this study were previously made available by Drott et al. (27, 87). A curated list of genes found in BGCs in the reference genome are available in SI Appendix, Table S2. Liquid chromatography with tandem mass spectrometry (LC-MS) data are available through the MassIVE repository (ID no. MSV000087134, https://doi.org/doi:10.25345/C54226) (88). All isolates are available upon request. Additionally, three isolates from each population and the three S-type isolates used in this study have been submitted to NRRL (searchable at https://nrrl.ncaur.usda.gov/cgi-bin/usda/index.html): 18mAF (NRRL66969), 33mAF (NRRL66970), 60mAF (NRRL66971), 68mAF (NRRL66972), 20mAF (NRRL66973), 85mAF (NRRL66974), 29mAF (NRRL66975), 71mAF (NRRL66976), 24mAF (NRRL66977), 12mAF (NRRL66978), 55mAF (NRRL66979), and 83mAF (NRRL66980) (as is also indicated in SI Appendix, Table S1).
References
- 1.Schimek C., “13 Evolution of special metabolism in fungi: Concepts, mechanisms, and pathways” in Evolution of Fungi and Fungal-like Organisms, Pöggler S., Wöstmeyer J., Eds. (Springer, 2011), pp. 293–329. [Google Scholar]
- 2.Drott M. T., Lazzaro B. P., Brown D. L., Carbone I., Milgroom M. G., Balancing selection for aflatoxin in Aspergillus flavus is maintained through interference competition with, and fungivory by insects. Proc. Biol. Sci. 284, 20172408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khudhair M., et al., Fusaristatin A production negatively affects the growth and aggressiveness of the wheat pathogen Fusarium pseudograminearum. Fungal Genet. Biol. 136, 103314 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Pfliegler W. P., Pócsi I., Győri Z., Pusztahelyi T., The Aspergilli and their mycotoxins: Metabolic interactions with plants and the soil biota. Front. Microbiol. 10, 2921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu X., et al., Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc. Natl. Acad. Sci. U.S.A. 111, 16796–16801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan P., et al., Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 197, 1236–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Keller N. P., Turner G., Bennett J. W., Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Lind A. L., et al., Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol. 15, e2003583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy C. G. P., Fitzpatrick D. A., Pan-genome analyses of model fungal species. Microb. Genom. 5, e000243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H.-X., Brewer M. T., Spatial genetic structure and population dynamics of gummy stem blight fungi within and among watermelon fields in the southeastern United States. Phytopathology 106, 900–908 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Ahmadpour A., et al., Population structure, genetic diversity, and sexual state of the rice brown spot pathogen Bipolaris oryzae from three Asian countries. Plant Pathol. 67, 181–192 (2018). [Google Scholar]
- 12.Nakamura N., Tanaka C., Takeuchi-Kaneko Y., Recombination and local population structure of the root endophytic fungus Glutinomyces brunneus based on microsatellite analyses. Fungal Ecol. 41, 56–64 (2019). [Google Scholar]
- 13.Steenwyk J. L., Soghigian J. S., Perfect J. R., Gibbons J. G., Copy number variation contributes to cryptic genetic variation in outbreak lineages of Cryptococcus gattii from the North American Pacific Northwest. BMC Genomics 17, 700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashu E. E., Hagen F., Chowdhary A., Meis J. F., Xu J., Global population genetic analysis of Aspergillus fumigatus. mSphere 2, e00019-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drott M. T., et al., Diversity of secondary metabolism in Aspergillus nidulans clinical isolates. mSphere 5, e00156-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forseth R. R., et al., Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew. Chem. Int. Ed. Engl. 52, 1590–1594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson M. J., et al., Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 75, 7469–7481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowd P. F., Synergism of aflatoxin B1 toxicity with the co-occurring fungal metabolite kojic acid to two caterpillars. Entomol. Exp. Appl. 47, 69–71 (1988). [Google Scholar]
- 19.Georgianna D. R., et al., Beyond aflatoxin: Four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 11, 213–226 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otto S. P., Lenormand T., Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3, 252–261 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Whitlock A. O. B., Azevedo R. B. R., Burch C. L., Population structure promotes the evolution of costly sex in artificial gene networks. Evolution 73, 1089–1100 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Williams J. H., et al., Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 80, 1106–1122 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., Wu F., Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 118, 818–824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wild C. P., Gong Y. Y., Mycotoxins and human disease: A largely ignored global health issue. Carcinogenesis 31, 71–82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CDC, Health Studies Branch . 2012. Understanding chemical exposures: aflatoxin. Atlanta, GA. https://www.cdc.gov/nceh/hsb/chemicals/aflatoxin.htm. Accessed 1 July 2020.
- 26.Drott M. T., Debenport T., Higgins S. A., Buckley D. H., Milgroom M. G., Fitness cost of aflatoxin production in Aspergillus flavus when competing with soil microbes could maintain balancing selection. mBio 10, e02782–e02718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drott M. T., et al., The frequency of sex: Population genomics reveals differences in recombination and population structure of the aflatoxin-producing fungus Aspergillus flavus. mBio 11, e00963-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drott M. T., Fessler L. M., Milgroom M. G., Population subdivision and the frequency of aflatoxigenic isolates in Aspergillus flavus in the United States. Phytopathology 109, 878–886 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Horn B. W., Biodiversity of Aspergillus section Flavi in the United States: A review. Food Addit. Contam. 24, 1088–1101 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Wicklow D. T., Dowd P. F., Gloer J. B., “Antiinsectan effects of Aspergillus metabolites” in The Genus Aspergillus, Powell K. A., Renwick A., Peberdy J. F., Eds. (Plenum Press, New York, 1994), pp. 93–109. [Google Scholar]
- 31.Uka V., et al., Secondary metabolite dereplication and phylogenetic analysis identify various emerging mycotoxins and reveal the high intra-species diversity in Aspergillus flavus. Front. Microbiol. 10, 667 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blin K., et al., antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kryazhimskiy S., Plotkin J. B., The population genetics of dN/dS. PLoS Genet. 4, e1000304 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frisvad J. C., et al., Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 93, 1–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbons J. G., et al., The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr. Biol. 22, 1403–1409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh P., Orbach M. J., Cotty P. J., Aspergillus texensis: A novel aflatoxin producer with S morphology from the United States. Toxins (Basel) 10, 513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohkura M., Cotty P. J., Orbach M. J., Comparative genomics of Aspergillus flavus S and L morphotypes yield insights into niche adaptation. G3 (Bethesda) 8, 3915–3930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kjærbølling I., et al., Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc. Natl. Acad. Sci. U.S.A. 115, E753–E761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kjærbølling I., et al., A comparative genomics study of 23 Aspergillus species from section Flavi. Nat. Commun. 11, 1106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vesth T. C., et al., Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat. Genet. 50, 1688–1695 (2018). [DOI] [PubMed] [Google Scholar]
- 41.de Vries R. P., et al., Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 18, 28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao S., Nandineni M. R., Genome sequencing and comparative genomics reveal a repertoire of putative pathogenicity genes in chilli anthracnose fungus Colletotrichum truncatum. PLoS One 12, e0183567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pi B., et al., A genomics based discovery of secondary metabolite biosynthetic gene clusters in Aspergillus ustus. PLoS One 10, e0116089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tralamazza S. M., Rocha L. O., Oggenfuss U., Corrêa B., Croll D., Complex evolutionary origins of specialized metabolite gene cluster diversity among the plant pathogenic fungi of the Fusarium graminearum species complex. Genome Biol. Evol. 11, 3106–3122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martino E., et al., Comparative genomics and transcriptomics depict ericoid mycorrhizal fungi as versatile saprotrophs and plant mutualists. New Phytol. 217, 1213–1229 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Schuelke T. A., et al., Comparative genomics of pathogenic and nonpathogenic beetle-vectored fungi in the genus Geosmithia. Genome Biol. Evol. 9, 3312–3327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greco C., Keller N. P., Rokas A., Unearthing fungal chemodiversity and prospects for drug discovery. Curr. Opin. Microbiol. 51, 22–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gould S. J., The Structure of Evolutionary Theory (Harvard University Press, 2002). [Google Scholar]
- 49.Belbahri L., et al., Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 8, 1438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livingstone P. G., Morphew R. M., Whitworth D. E., Genome sequencing and pan-genome analysis of 23 Corallococcus spp. strains reveal unexpected diversity, with particular plasticity of predatory gene sets. Front. Microbiol. 9, 3187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brockhurst M. A., et al., The ecology and evolution of pangenomes. Curr. Biol. 29, R1094–R1103 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Niehaus E.-M., et al., Comparative genomics of geographically distant Fusarium fujikuroi isolates revealed two distinct pathotypes correlating with secondary metabolite profiles. PLoS Pathog. 13, e1006670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly A. C., Ward T. J., Population genomics of Fusarium graminearum reveals signatures of divergent evolution within a major cereal pathogen. PLoS One 13, e0194616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulcher M. R., Winans J. B., Quan M., Oladipo E. D., Bergstrom G. C., Population genetics of Fusarium graminearum at the interface of wheat and wild grass communities in New York. Phytopathology 109, 2124–2131 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Kelly A., et al., The geographic distribution and complex evolutionary history of the NX-2 trichothecene chemotype from Fusarium graminearum. Fungal Genet. Biol. 95, 39–48 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Crippin T., Renaud J. B., Sumarah M. W., Miller J. D., Comparing genotype and chemotype of Fusarium graminearum from cereals in Ontario, Canada. PLoS One 14, e0216735 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward T. J., Bielawski J. P., Kistler H. C., Sullivan E., O’Donnell K., Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. U.S.A. 99, 9278–9283 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levins R., Evolution in Changing Environments: Some Theoretical Explorations (Princeton University Press, 1968). [Google Scholar]
- 59.Collemare J., Lebrun M. H., “Fungal secondary metabolites: Ancient toxins and novel effectors in plant–microbe interactions” in Effectors in Plant–Microbe Interactions, Martin F., Sophien K., Eds. (Wiley-Blackwell, 2011), pp. 377–400. [Google Scholar]
- 60.Collemare J., et al., Secondary metabolism and biotrophic lifestyle in the tomato pathogen Cladosporium fulvum. PLoS One 9, e85877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demasi S., et al., Latitude and altitude influence secondary metabolite production in peripheral alpine populations of the Mediterranean species Lavandula angustifolia Mill. Front. Plant Sci. 9, 983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Condon B. J., et al., Clues to an evolutionary mystery: The genes for T-Toxin, enabler of the devastating 1970 Southern corn leaf blight epidemic, are present in ancestral species, suggesting an ancient origin. Mol. Plant Microbe Interact. 31, 1154–1165 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Casadevall A., Kontoyiannis D. P., Robert V., On the emergence of Candida auris: Climate change, azoles, swamps, and birds. mBio 10, e01397-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houbraken J., et al., Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 95, 5–169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danecek P.et al.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jombart T., adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008). [DOI] [PubMed] [Google Scholar]
- 67.R Core Team , R: A Language and Environment for Statistical Computing (Version 3.5.2, R Foundation for Statistical Computing, 2018).
- 68.Jombart T. (2012). An introduction to adegenet 2.0.0. http://adegenet.r-forge.r-project.org/files/tutorial-basics.pdf. Accessed 8 August 2020.
- 69.Tamura K., Dudley J., Nei M., Kumar S., MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Wang L.-G., et al., treeio: An R package for phylogenetic tree input and output with richly annotated and associated data. Mol. Biol. Evol. 37, 599–603 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T. Y., ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 72.Wickham H., Ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016). [Google Scholar]
- 73.Cary J. W., Linz J. E., Bhatnagar D., Microbial Foodborne Diseases: Mechanisms of Pathogenesis and Toxin Synthesis (CRC Press, 1999). [Google Scholar]
- 74.Cary J. W., et al., Transcriptome analysis of Aspergillus flavus reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot. Cell 14, 983–997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cary J. W., et al., Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genet. Biol. 64, 25–35 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Lebar M. D., et al., Identification and functional analysis of the aspergillic acid gene cluster in Aspergillus flavus. Fungal Genet. Biol. 116, 14–23 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Greco C., Pfannenstiel B. T., Liu J. C., Keller N. P., Depsipeptide aspergillicins revealed by chromatin reader protein deletion. ACS Chem. Biol. 14, 1121–1128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang P. K., Horn B. W., Dorner J. W., Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet. Biol. 46, 176–182 (2009). [DOI] [PubMed] [Google Scholar]
- 79.Saruwatari T., et al., Cytochrome P450 as dimerization catalyst in diketopiperazine alkaloid biosynthesis. ChemBioChem 15, 656–659 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Khalid S., et al., NRPS-derived isoquinolines and lipopetides mediate antagonism between plant pathogenic fungi and bacteria. ACS Chem. Biol. 13, 171–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cary J. W., et al., An Aspergillus flavus secondary metabolic gene cluster containing a hybrid PKS-NRPS is necessary for synthesis of the 2-pyridones, leporins. Fungal Genet. Biol. 81, 88–97 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Terabayashi Y., et al., Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 47, 953–961 (2010). [DOI] [PubMed] [Google Scholar]
- 83.Umemura M., et al., Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet. Biol. 68, 23–30 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Navarro-Muñoz J. C.et al., A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16, 60–68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shannon P., et al., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfannenstiel B. T., Greco C., Sukowaty A. T., Keller N. P., The epigenetic reader SntB regulates secondary metabolism, development and global histone modifications in Aspergillus flavus. Fungal Genet. Biol. 120, 9–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Drott M., et al., Raw genomic reads from “The frequency of sex: population genomics reveals differences in recombination and population structure of the aflatoxin-producing fungus Aspergillus flavus.” National Center for Biotechnology Information (NCBI). https://www.ncbi.nlm.nih.gov/bioproject/PRJNA639008. Deposited 12 June 2020. [DOI] [PMC free article] [PubMed]
- 88.Drott M. T., et al., Liquid chromatography with mass spectrometry data for “Microevolution in the pan-secondary metabolome of Aspergillus flavus and its potential macroevolutionary implications for filamentous fungi”. MassIVE. 10.25345/C54226. Deposited 30 April 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads used to construct assemblies in this study were previously made available by Drott et al. (27, 87). A curated list of genes found in BGCs in the reference genome are available in SI Appendix, Table S2. Liquid chromatography with tandem mass spectrometry (LC-MS) data are available through the MassIVE repository (ID no. MSV000087134, https://doi.org/doi:10.25345/C54226) (88). All isolates are available upon request. Additionally, three isolates from each population and the three S-type isolates used in this study have been submitted to NRRL (searchable at https://nrrl.ncaur.usda.gov/cgi-bin/usda/index.html): 18mAF (NRRL66969), 33mAF (NRRL66970), 60mAF (NRRL66971), 68mAF (NRRL66972), 20mAF (NRRL66973), 85mAF (NRRL66974), 29mAF (NRRL66975), 71mAF (NRRL66976), 24mAF (NRRL66977), 12mAF (NRRL66978), 55mAF (NRRL66979), and 83mAF (NRRL66980) (as is also indicated in SI Appendix, Table S1).