Significance
Across the Tree of Life, life’s phenotypic diversity has been accompanied by a massive expansion of the protein universe. Compared with simple prokaryotes that harbor thousands of proteins, plants and animals harbor hundreds of thousands of proteins that are also longer, multidomain, and comprise a variety of folds and fold combinations, repeated segments, and beta-rich architectures that make them prone to misfolding and aggregation. Surprisingly, the relative representation of core chaperones, those dedicated to maintaining the folding quality of these increasingly complex proteomes, did not change from prokaryotic to mammalian genomes. To reconcile the expanding proteomes, core chaperones have rather increased in cellular abundance and evolved to function cooperatively as a network, combined with their supporting workforce, the cochaperones.
Keywords: Tree of Life, expansion of proteomes, core chaperones, cochaperones, chaperone network
Abstract
Across the Tree of Life (ToL), the complexity of proteomes varies widely. Our systematic analysis depicts that from the simplest archaea to mammals, the total number of proteins per proteome expanded ∼200-fold. Individual proteins also became larger, and multidomain proteins expanded ∼50-fold. Apart from duplication and divergence of existing proteins, completely new proteins were born. Along the ToL, the number of different folds expanded ∼5-fold and fold combinations ∼20-fold. Proteins prone to misfolding and aggregation, such as repeat and beta-rich proteins, proliferated ∼600-fold and, accordingly, proteins predicted as aggregation-prone became 6-fold more frequent in mammalian compared with bacterial proteomes. To control the quality of these expanding proteomes, core chaperones, ranging from heat shock proteins 20 (HSP20s) that prevent aggregation to HSP60, HSP70, HSP90, and HSP100 acting as adenosine triphosphate (ATP)-fueled unfolding and refolding machines, also evolved. However, these core chaperones were already available in prokaryotes, and they comprise ∼0.3% of all genes from archaea to mammals. This challenge—roughly the same number of core chaperones supporting a massive expansion of proteomes—was met by 1) elevation of messenger RNA (mRNA) and protein abundances of the ancient generalist core chaperones in the cell, and 2) continuous emergence of new substrate-binding and nucleotide-exchange factor cochaperones that function cooperatively with core chaperones as a network.
All cellular life is thought to have stemmed from the last universal common ancestor (LUCA) (1, 2), that emerged more than 3.6 billion y ago. Two major kingdoms of life diverged from LUCA: bacteria and archaea, which about 2 billion y later merged into the eukaryotes (3). Since the beginning of biological evolution, life’s volume has increased on a grand scale: The average size of individual cells has increased ∼100-fold from prokaryotes to eukaryotes (4), the number of cell types has increased ∼200-fold from unicellular eukaryotes to humans (5), and average body size has increased ∼5,000-fold from the simplest sponges to blue whales (6).
This expansion in organismal complexity and variability was accompanied by an expansion in life’s molecular workforce, proteomes in particular, which in turn presented a challenge of reaching and maintaining properly folded and functional proteomes. Most proteins must fold to their native structure in order to function, and their folding is largely imprinted in their primary amino acid sequence (7–9). However, many proteins, and especially large multidomain polypeptides, or certain protein types such as all-beta or repeat proteins, tend to misfold and aggregate into inactive species that may also be toxic (10). Life met this challenge by evolving molecular chaperones that can minimize protein misfolding and aggregation, even under stressful out-of-equilibrium conditions favoring aggregation (11, 12). Chaperones can be broadly divided into core and cochaperones. Core chaperones can function on their own, and include ATPases heat shock protein 60 (HSP60), HSP70, HSP100, and HSP90 and the adenosine triphosphate (ATP)-independent HSP20. The basal protein holding, unfolding, and refolding activities of the core chaperones are facilitated and modulated by a range of cochaperones such as J-domain proteins (13–15).
Starting from LUCA, as proteomes expanded, so did the core chaperones and their respective cochaperones. Indeed, chaperones have been shown to facilitate the acquisition of destabilizing mutations and thereby accelerate protein evolution (16–18). However, the coexpansion of proteomes and of chaperones, underscoring a critical balance between evolutionary innovation and foldability, remains largely unexplored. We thus embarked on a systematic bioinformatics analysis that explores the evolution of both proteomes and chaperones, and of both core and their auxiliary cochaperones, along the Tree of Life.
Results
A Tree of Life Analysis of the Expansion of Proteomes and Chaperones.
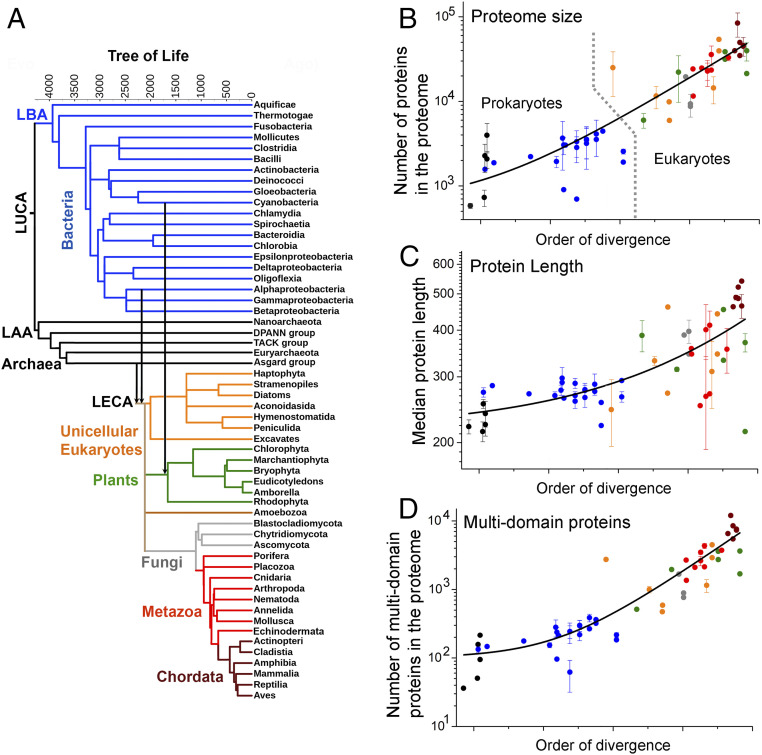
We aimed to explore, systematically, across the Tree of Life (ToL) the expansion of proteomes and compare it with the chaperone composition and level. To this end, we collected proteome sequences from representative organisms belonging to all the major bacterial, archaeal, and eukaryotic clades and constructed a ToL (Dataset S1). The overall topology of our tree was borrowed from TimeTree (19) and we also adhered to their order of divergence, which is based on molecular dating and geological records. TimeTree also provides putative dates of emergence and these are provided as branch lengths, yet because our analysis is primarily comparative, branch lengths were only used here as a graphical aid (Fig. 1A).
Fig. 1.
Expansion of proteome size across the Tree of Life. (A) The ToL used in this study. Leaves represent extant phylogenetic clades, while internal nodes represent their presumed ancestors. Branch lengths are in million years, as available from TimeTree (19), and they refer to the relative order of divergence of the corresponding clades rather than the absolute dates of their emergence. The major phylogenetic groups in the tree (Bacteria, Archaea, unicellular eukaryotes, plants, Fungi, Metazoa, and Chordata) are highlighted in different colors. Vertical arrows highlight the two major endosymbiosis events: the Alphaproteobacterial origin of mitochondria and the cyanobacterial origin of plastids. LAA, last archaeal ancestor; LBA, last bacterial ancestor. (B–D) The average per-clade values for various proteome size parameters (y axis, in log scale) are plotted against their order of divergence (along the x axis in linear scale). These parameters include proteome size (B), median protein length (C), and multidomain proteins in the proteome (D). In these scatterplots, the colors of data points represent their major phylogenetic group in the tree (A). Error bars represent the clade SD (no error bars relate to clades comprising only one representative organism). The lines were derived by a fit to an exponential equation, and are provided merely as visual guides. Prokaryotic and eukaryotic organisms are separated by a dashed line in B.
The Tree of Life begins with LUCA at the root (Fig. 1A). The edges of the ToL represent the extant three kingdoms—archaea, plotted throughout in black; bacteria, plotted in blue; and eukaryotes. The latter emerged by endosymbiosis of an Alphaproteobacterium and an Asgard-like archaeon (20, 21). The emergence of green algae and subsequently of plants occurred with a secondary endosymbiosis of a Cyanobacterium into a nonphotosynthetic eukaryote (22). The major eukaryotic clades therefore comprised unicellular, early-diverging eukaryotes (in orange), fungi (gray), plants (green), Metazoa (invertebrate animals; red), and Chordata (vertebrate animals; wine). Overall, our analysis was based on comparing the proteomes of 188 representative organisms, covering 56 major clades of bacteria, archaea, and eukaryote (Dataset S1). The various proteome parameters analyzed below were initially derived for each representative organism in the core tree. The representative organisms of each clade were then pulled together to calculate the clade average and the SD for this average. The clade average values were subsequently plotted using the order of divergence for the x axis. Accordingly, these plots also broadly divide into prokaryotes (the left part) and eukaryotes (the right part), and the latter’s right edge comprises Chordata including Mammalia (Fig. 1B and the following figures).
The Expansion of Proteome Size.
Initially, we scrutinized the expansion of proteome size by examining 1) the total number of proteins per proteome in a given clade; 2) the median protein length; and 3) the number of multidomain proteins in the proteome. The clade average values of these three parameters are plotted in Fig. 1 B‒D, with colors of points matching the branch colors in Fig. 1A.
The total number of proteins per proteome expanded ∼200-fold.
Proteomes that comprise a larger number of proteins unavoidably present a greater challenge for their protein quality control chaperone machinery. To examine the expansion in the number of proteins per proteome, proteome sequences of the 188 representative organisms were obtained. Across the ToL, the number of proteins per proteome expanded roughly 200-fold (Fig. 1B) from ∼700 proteins in the simplest free-living DPANN (23) archaea to ∼120,000 proteins in humans (Dataset S2). Proteome size is similar across prokaryotes, on the order of 3,000 proteins, although the smallest proteomes belong to the earliest-diverging free-living DPANN archaea and Aquificae bacteria (23, 24) (not counting parasites and symbionts). Eukaryote proteomes are substantially larger and, considering free-living organisms only, the smallest eukaryotic proteomes harboring ∼10,000 proteins belong to Amoebozoa, one of the earliest-diverging eukaryotes (25, 26). Land plants and metazoans comprise hundreds of thousands of proteins per proteome. However, as described later, this dramatic increase in the number of proteins in eukaryotes occurred not only by duplication of preexisting proteins but also by the emergence of completely new domains and folds.
The median protein length increased ∼2-fold.
The longer the polypeptides are the more prone they are to misfold and aggregate instead of readily reaching their native functional state (27, 28). Analyzing the lengths of all proteins in each representative proteome (SI Appendix), we found that compared with ∼250 residues across prokaryotes, median protein length increased about 2-fold in multicellular eukaryotes (Fig. 1C), with ∼400 residues in plants and ∼500 residues in Chordata (Dataset S2). Longer proteins were found primarily in multicellular eukaryotes (average lengths of the top 10% largest proteins were roughly 1,300 residues in plants, ∼1,500 residues in metazoans, and ∼2,150 residues in mammals). The longest polypeptides in mammals are predominantly muscle proteins, including different variants of titin (>34,000 residues) or adhesins (>5,000 residues).
There are different ways by which proteins can increase in size. First, the domains themselves can grow larger by decorating an ancestral core domain with additional segments. Second, the fusion of multiple domains can result in a larger multidomain protein. Third, domain-flanking regions (C- and N-terminal segments, and interdomain linkers), that are typically disordered, can expand. A systematic analysis of 38 distinct folds that are conserved across the ToL (including parasites and symbionts) showed that lengths of individual domains increased mildly, nearly 1.5-fold, across the ToL (SI Appendix, Fig. S1A). The expansion of domain-flanking segments was also modest, nearly 3-fold, from prokaryotes to multicellular eukaryotes (SI Appendix, Fig. S1B). Indeed, as elaborated below, length expansion primarily stemmed from the increase in the fraction of multidomain proteins.
Multidomain proteins expanded ∼50-fold.
Multidomain proteins are inherently more prone to misfolding and aggregation than single-domain proteins, and may therefore demand more chaperone holding–unfolding–refolding action (29–31). To examine their expansion, domain annotations of all proteins in the 188 representative organisms were obtained from Pfam (32). Across the ToL, multidomain proteins comprising ≥3 Pfam-annotated domains have expanded ∼50-fold (Fig. 1D), from ∼100 proteins per proteome in prokaryotes to ∼5,000 in plants and animals (Dataset S2). Further, multidomain proteins have expanded beyond the expansion of proteome size, to become nearly 3-fold more frequent in eukaryotic proteomes compared with prokaryotes (SI Appendix, Fig. S1C) with the expected corresponding shrinkage of proteins comprising one or two domains (SI Appendix, Fig. S1D). As described later, this expansion occurred not only by duplication of preexisting multidomain proteins but foremost by the emergence of new domain combinations.
Proteome Expansion by Innovation.
Most proteins emerge by duplication and divergence of a preexisting protein. The outcome is paralogous proteins with the same overall fold and domain arrangement (for multidomain proteins). Thus, duplication and “local” divergence (point mutations and short insertions or deletions) certainly increase proteome size (the total number of proteins) but do not dramatically change proteome composition or complexity. The latter relates primarily to the birth of completely new proteins possessing new folds, and to the emergence of multidomain proteins with new fold combinations. Additions of new folds and fold combinations likely impose an additional burden on the chaperone machinery. We thus analyzed additional proteome parameters that represent expansion by innovation, rather than by mere duplication and divergence, as detailed below.
Fold types expanded ∼5-fold.
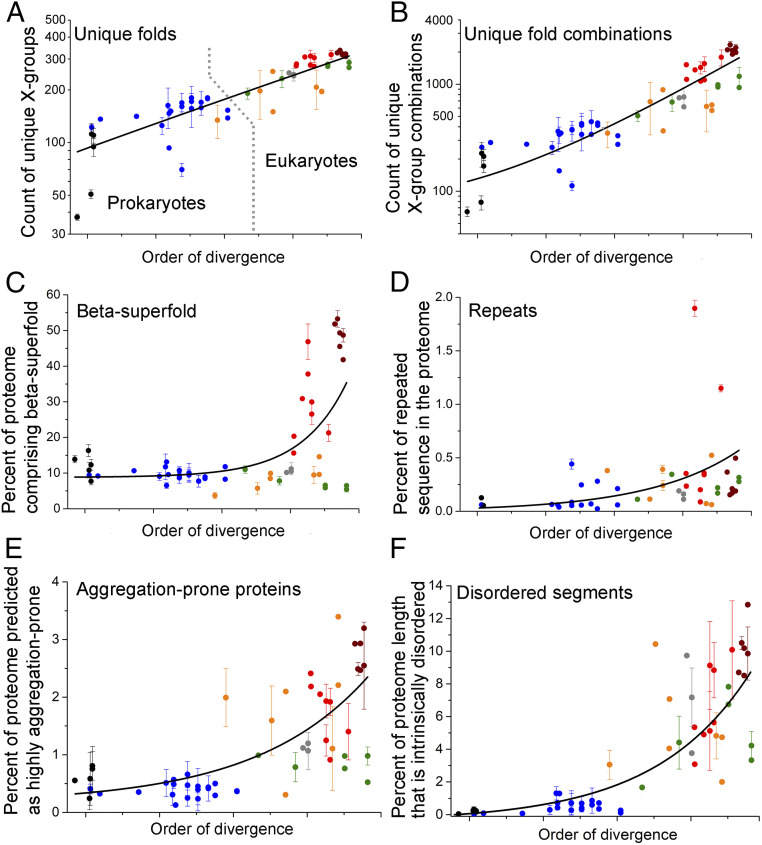
To assess the emergence of new folds, we used evolutionary classification of domains (ECOD)—a hierarchical classification of protein folds that uses both sequence and structural similarities and clusters all domains with known structures into independently evolved lineages, termed “X-groups” (33). For each representative organism, the Pfam-annotated domains were mapped to ECOD X-groups (SI Appendix). We found that from prokaryotes to eukaryotes, the number of unique folds (i.e., unique ECOD X-groups) per proteome expanded about 5-fold (Fig. 2A), from ∼150 in prokaryotes to ∼450 in metazoans (Dataset S3).
Fig. 2.
Expansion of proteomes by innovations. Figure features follow those of Fig. 1. (A) Shown on the y axis (log scale) is the average number of unique folds [ECOD X-groups (33)] in each phylogenetic clade. The lines were derived by a fit to an exponential equation, and are provided merely as visual guides. Prokaryotic (black and blue dots) and eukaryotic organisms (orange, gray, green, red, and wine dots) are largely separated by a dashed line in A. (B) Same as A, for the count of unique fold combinations. (C) Same as A, for the percentage of proteins in the proteome comprising at least one beta-superfold domain (note the linear scale). (D) Same as A, for the percentage of total proteome length that is repeated (also on a linear scale). (E) Same as A, for the percentage of proteins in the proteome predicted to have ≥20 aggregation hotspots per proteome (also on a linear scale; see also Dataset S10). (F) Same as A, for the percentage of proteome length that is intrinsically disordered (also on a linear scale).
New fold combinations expanded ∼20-fold.
As shown above, multidomain proteins expanded nearly 3-fold (their fraction out to the total number of proteins) alongside a parallel shrinkage of proteins comprising one or two domains (SI Appendix, Fig. S1 C and D). This expansion of multidomain proteins occurred not only by duplication, namely by amplifying preexisting multidomain proteins, but also via the emergence of new combinations. To assess the latter, we examined the number of unique combinations of domains per proteome, with domains being assigned by ECOD X-groups. It appeared that new domain combinations arose throughout evolution and, from prokaryotes to eukaryotes, the number of unique combinations per proteome increased ∼20-fold (Fig. 2B), from ∼100 combinations in bacteria to ∼2,000 combinations in Chordata (Datasets S4 and S5).
All-beta and beta-rich folds expanded up to ∼600-fold.
Proteins that are beta-rich are known to be prone to misfolding and aggregation (34). In the simplest free-living bacteria and archaea, proteins comprising the ancient all-alpha and alpha-beta architectures are the most frequent. Remarkably, upon the emergence of eukaryotes, and in metazoans especially, all-beta or beta-rich architectures (beta superfold) expanded massively, nearly 600-fold (Fig. 2C). Beta-rich proteins, in proportion to the total number of proteins, became nearly 6-fold more frequent in mammalian proteomes, as compared with bacteria and archaea (SI Appendix, Fig. S2 and Dataset S6). The immunoglobulin fold had a major contribution to this expansion, owing to its diverse roles in immunity, multicellularity, and signaling (35).
Repeat sequences expanded ∼700-fold.
Proteins comprising tandem repeats of nearly identical sequences emerge readily yet are prone to misfolding and aggregation. We identified proteins with repeated sequences of the size of a single “foldon” unit, ∼20 amino acids (aa) (9), with ≥90% sequence similarity (Dataset S7). Most of the early-diverging archaea and bacteria do not possess repeat proteins. Indeed, repeat proteins appear in more recently diverged prokaryotes and foremost in eukaryotes (a similar trend was described in ref. 36). Indeed, metazoan proteomes contain large proteins with long repeated segments, for example, Drosophila Ank2p (21 Ankyrin repeats, in total 836 residues) or human Dmbt1p (11 cysteine-rich repeats of a total 1,419-residue length). The cumulative length of repeat sequences in metazoan proteomes can be up to 100,000 residues. Overall, from prokaryotes to eukaryotes, a 700-fold expansion (Fig. 2D) of repeated sequences was observed along the ToL (SI Appendix, Fig. S3). Repeated sequences also expanded beyond the expansion of proteome size—the percentage of total proteome length that comprises repeats increased nearly 7-fold from prokaryotes to metazoans (Fig. 2D).
Proteins predicted as aggregation-prone became ∼6-fold more frequent in the proteome.
To further examine the expansion of aggregation-prone proteins, for each representative organism, we identified how many proteins in the proteome are predicted to have an unusually high number of “aggregation hotspots” (defined as a poorly soluble protein segment of ≥5 aa in length, with solubility predicted from the sequence). The threshold for comparison was set at ≥20 hotspots per protein (at this threshold, ≤3% of proteins are aggregation-prone; the same trend was observed with lower thresholds; Dataset S8). We then calculated what percentage of the entire proteome these aggregation-prone proteins represent. This prediction is restricted by the fact that some of the predicted segments actually reside in the hydrophobic cores of stably folded proteins and hence do not comprise aggregation hotspots (37). However, such segments are likely to be as frequent in prokaryote and eukaryote proteomes (or even less frequent in the latter, where disordered proteins are abundant). Overall, our results indicate that compared with prokaryotes, aggregation-prone proteins have become nearly 6-fold more frequent in eukaryotes (Fig. 2E), with the highest frequency seen in Chordata proteomes (Fig. 2E).
Intrinsically disordered regions became ∼20-fold more frequent in the proteome.
From prokaryotes to eukaryotes, did all the changes in proteome composition demand more chaperone action in the latter? The expansion of intrinsically disordered regions, that in principle would not demand increased chaperone action, could be an exception. Though explored before (38, 39), to have this expansion on the same scale and set of organisms used for analyzing all other proteome factors, each protein of the representative proteomes was scanned to infer disordered segments ≥100 aa long (Dataset S9). As plotted in Fig. 2F, from prokaryotes to eukaryotes, the percentage of proteome length that is disordered has expanded nearly 20-fold.
Overall, it appears that although gene and whole-genome duplications dominate, and in particular along with the evolution of eukaryotes (40), dramatic changes in proteome composition have occurred owing to bona fide innovations. Specifically, concerning the burden on the chaperone machinery, proteome compositions have changed massively with respect to new folds, beta superfolds, repeat proteins, fold combinations, and aggregation propensity.
The Evolutionary History of Chaperones.
In parallel to estimating the expansion of proteome size and composition, we investigated the evolutionary history of chaperones, aiming to date their emergence and their expansion along the ToL. To that end, absence or presence, and copy numbers, of the core chaperones (HSP20, HSP60, HSP70, HSP100, and HSP90) was determined for the representative proteomes. Subsequently, protein trees were generated and compared with the ToL, to account for gene loss and horizontal transfer events. Protein sequences of all core-chaperone families were extracted from the representative proteomes (Dataset S10). These sequences were aligned and used to generate maximum-likelihood, midpoint-rooted protein trees which were then compared with the ToL (SI Appendix, Table S1).
The core chaperones emerged in early-diverging prokaryotes.
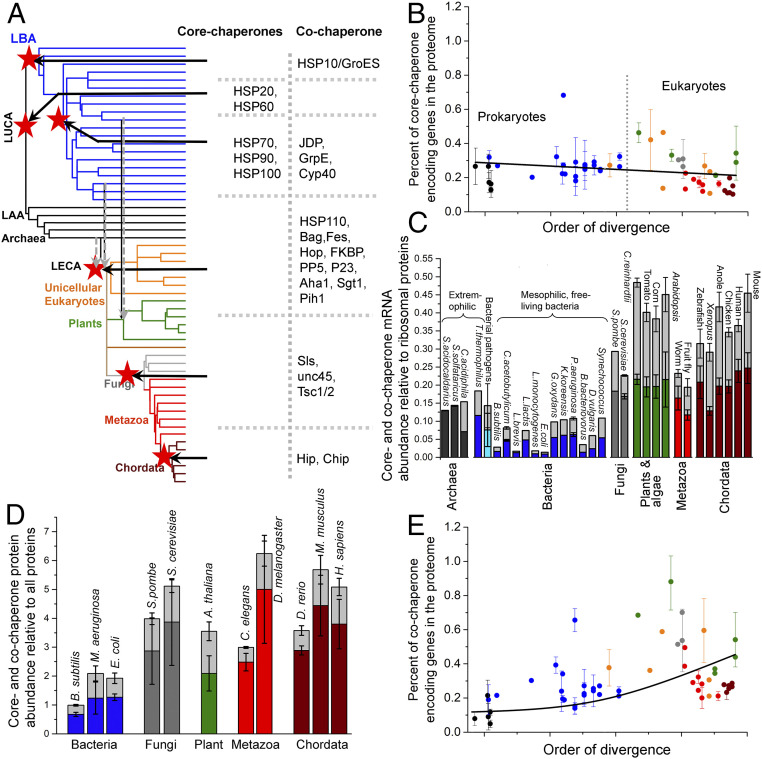
Our analysis traced the origin of all five core-chaperone families in early-diverging prokaryotes. The phylogenetic tree of a single protein, typically of a few hundred amino acids in length, often lacks the resolution required to reliably date the emergence, especially when horizontal transfer events are frequent. Dating emergence to LUCA is particularly challenging. We followed the recommendations of Berkemer and McGlynn (41) and demanded that for a chaperone family to be assigned to LUCA, there must be a single split between bacterial and archaeal sequences at the root of the protein tree, with strong bootstrap support for this split, and that interkingdom branches would be longer than the intrakingdom branches. These criteria assigned the emergence of only one core chaperone, HSP60—a cage-like ATP-fueled unfoldase (11, 42), to LUCA (Fig. 3A). The protein tree of HSP60 further indicated an ancient horizontal gene transfer (HGT) from archaea to Firmicutes, as previously noted (43).
Fig. 3.
Evolutionary history of core and cochaperones. (A) The de novo emergence of core- and cochaperone families is summarized on the ToL. The ToL is the same as in Fig. 1A; clade names are omitted for clarity. Ancestral nodes in which a chaperone family emerged are marked with red stars and the core- and cochaperone families emerged in that node are listed. The dashed gray arrows reflect the endosymbiotic integration of archaeal and bacterial chaperone systems in LECA and eukaryotic and cyanobacterial chaperone systems in photosynthetic algae. (B) The percentage of core-chaperone genes per proteome (shown is the average percentage for each phylogenetic clade). Figure features follow Fig. 1. The line was derived by a fit to a linear equation and is provided merely as a visual guide. Prokaryotic (black and blue dots) and eukaryotic organisms (orange, gray, green, red, and wine dots) are largely separated by a dashed line. (C) Core- and cochaperone gene expression, relative to ribosomal proteins, in cells. Plotted are model organisms for which sufficient, processed, and reliable expression data were available. The columns include core chaperones (colors represent the phylogenetic clades in Fig. 1A) and cochaperones (light gray color). The error bars represent the SD among different nonredundant abundance datasets. (D) Same as C, for the relative basal abundance of core and cochaperones in cells. (E) Same as B, for cochaperone genes per proteome. The line was derived by a fit to an exponential equation and is provided merely as a visual guide.
The protein tree of HSP20—an antiaggregation “holding” chaperone—depicted a clear single split of bacterial and archaeal domains at the root, albeit with weak bootstrap support, and interkingdom branch lengths were shorter than intrakingdom branch lengths (SI Appendix, Table S1). Similar uncertainties were noted for the majority of protein families formerly assigned to LUCA (41). Previous studies assigned HSP20 to LUCA (44, 45), which, despite the above uncertainties, we concur with (Fig. 3A), although later emergence and HGT is an alternative. Indeed, in accordance with a previous study (46), the HSP20 protein tree suggests multiple HGT events, though given the weak bootstrap support it was difficult to distinguish between phylogenetic uncertainty and actual HGT events, let alone to assign donor and acceptor clades.
The remaining core-chaperone families, HSP70, HSP90, and HSP100, appear to have emerged in bacteria, though phylogenetic uncertainties, and probably extensive horizontal transfer between different bacterial clades and between bacteria and archaea, prevent the reliable assignment of their points of origin (Fig. 3A). The ATP-dependent core chaperone HSP70—that controls protein unfolding, disaggregation, and degradation (47, 48)—was detected in the earliest-diverging bacterial clades Aquificae and Thermotogae. However, the protein tree clustered their HSP70 sequences with those from the later-diverging bacterial lineages Deltaproteobacteria, Clostridia, and Bacilli, with bootstrap values being too low to distinguish between phylogenetic uncertainty and true HGT events (SI Appendix, Table S1). Thus, HSP70 appears to have a bacterial origin, around or after the emergence of terrestrial bacteria, that is, around the divergence of Fusobacteria (49) (Fig. 3A). Following its emergence, HSP70 was likely horizontally transferred to archaea, but the current analysis could not reliably assign donor and acceptor clades.
The protein trees of both HSP90 and HSP100 depict a similar scenario. Both seem to have emerged in bacteria around or after terrestrial bacteria emerged (Fig. 3A). Although a reliable point of origin could not be assigned for either of these two chaperones, biochemical assays show that whereas the activity of HSP90 or HSP100 strictly depends on the presence of HSP70, HSP70 itself can act independently. Further, across the ToL, every organism that harbors genes for HSP90 and/or HSP100 also harbors genes for HSP70, but not vice versa. Thus, it is likely that HSP90 and HSP100 have both emerged after HSP70. Similar to HSP70, HSP90 and HSP100 were likely horizontally transferred to archaea. While our protein trees do indicate such trends, the bootstrap values are low.
The archaeal and bacterial core chaperones were integrated into the last eukaryotic common ancestor (LECA), and no new core chaperones emerged with the birth of eukaryotes (Fig. 3A). Chaperones of archaeal origin mostly continued to function in their original compartment, the cytosol. Although most Alphaproteobacterial endosymbiont genes were transferred to the nucleus, most of the chaperones of bacterial origin evolved to translocate back to the compartment from which they originated, namely to the mitochondria (50–53). Chaperone evolution in eukaryotes involved gene loss as well; for example, cytosolic and mitochondrial HSP100s have been lost in metazoans (53).
The expansion of core chaperones.
Whereas no new core-chaperone family emerged in eukaryotes, gene copy numbers of the existing families did increase via gene duplication, to support expanding proteomes, for condition-specific expression, and also to cater for the emergence of multiple subcellular compartments. Bacteria and archaea typically harbor the same five core-chaperone families as eukaryotes. In any bacterial or archaeal genome, gene copy numbers of individual chaperone families range between 1 and 4, summing up to an average of 8 core-chaperone genes per proteome (Dataset S10). In comparison, the number of core-chaperone genes in higher plants, which are among the most complex eukaryotes, increased ∼30-fold for HSP20, ∼50-fold for HSP60, ∼40-fold for HSP70, ∼20-fold for HSP90, and ∼10-fold for HSP100 (SI Appendix, Fig. S4). Parasitic microbes, such as Mycoplasma pneumoniae, Plasmodium falciparum, and Entamoeba histolytica, and photosynthetic bacteria, algae, and plants often harbor unusually high chaperone gene copy numbers, likely to counter the immune response of the host (54, 55) and the oxidative stress (56, 57). However, when proteome size is accounted for, it is evident that the expansion of core chaperones largely coincides with the overall expansion of proteome size. In fact, core chaperones comprise ∼0.3%, that is, 3 out of 1,000 proteins, in all proteomes, from the simplest free-living prokaryotes to mammals (Fig. 3B). Further, the expansion of core chaperones occurred by gene duplication only, with no bona fide innovation, as all five core-chaperone families seem to have preexisted in prokaryotes.
The cellular abundance of core chaperones increased ∼6-fold.
As described above, the relative representation of core-chaperone genes is roughly the same in the genomes of prokaryotes and eukaryotes. However, gene expression levels could vary, and higher cellular levels of chaperones could support the increasingly complex eukaryotic proteomes. To assess expression levels, the messenger RNA (mRNA) and protein abundance of core or cochaperones was compared in prokaryotic and eukaryotic cells/tissues not subjected to stress or genetic modifications. Curated expression data could be obtained for 30 free-living model organisms spanning 14 major clades along the ToL (Dataset S11). The mRNA abundance of core-chaperone genes, relative to that of ribosomal proteins (as a proxy for the overall level of protein synthesis), has elevated ∼6-fold in chordates compared with mesophilic bacteria (Fig. 3C). Protein abundance data available for 11 model organisms along the ToL (Dataset S12) indicate the very same trend (Fig. 3D). Further, chaperone levels in eukaryotes are >3-fold compared with extremophilic prokaryotes and pathogenic bacteria that in turn show >2-fold higher levels than those of mesophilic nonpathogenic bacteria.
Cochaperones expanded ∼9-fold.
In eukaryotes, as the gene copy numbers of core chaperones expanded, and their protein abundance also increased, what happened to their auxiliary workforce, the cochaperones? The number of unique cochaperone families per proteome expanded from ∼3 in prokaryotes to ∼20 in humans. Most cochaperones are eukaryote-specific (Dataset S10), and therefore have likely emerged relatively recently, and only a few cochaperones are found in all three domains of life. To date their emergence, protein trees were generated. These suggest that, as expected, these cochaperones emerged after the core chaperone they work with. HSP60 is assigned to LUCA, while its bacterial cochaperone HSP10/GroES appears to have emerged later along with the emergence of bacteria (Fig. 3A). HSP70’s cochaperones, the J-domain proteins (JDPs) and GrpE, and HSP90’s cochaperone, Cyp40, appear to have emerged at the same node as their respective core chaperones. Given the phylogenetic uncertainties, possible extensive horizontal transfers, and the resolution that protein trees allow, a more precise dating could not be performed. It appears, however, that JDPs, GrpE, and Cyp40 have all emerged after the emergence of terrestrial bacteria (Fig. 3A), and therefore likely emerged after their respective core chaperones. Several cochaperone families emerged in eukaryotes (Fig. 3A), including HSP110 that diverged by duplication of HSP70 (58), and Pih1, Aha1, and Chip that harbor eukaryote-specific folds and hence likely emerged de novo. Overall, it is evident that cochaperones tail core chaperones, and not vice versa. With the birth of several cochaperones in eukaryotes, the percentage of genes encoding for cochaperones in the proteome expanded ∼5-fold from prokaryotes to eukaryotes (Fig. 3E). Notably, the JDPs, cochaperones of HSP70, are the major contributor to this copy-number expansion (17). Further, alongside the increase in copy numbers and the emergence of new families, the protein expression levels of cochaperones also increased across the ToL (cochaperone protein abundance is ∼2-fold higher in chordates compared with mesophilic bacteria and mRNA abundance is ∼4-fold higher; Fig. 3 C and D).
Our analysis, therefore, suggests that evolutionary innovation occurred primarily at the level of cochaperones that facilitated the basal core-chaperone activity, thus expanding the chaperone network to meet the challenges of newly emerging protein folds and increasingly complex proteomes.
Discussion
How did the proteomes expand across the ToL, and how did chaperones evolve to support this expansion? To address this question, we compiled data from multiple sources and analyzed them under one roof, thus allowing a systematic, quantitative comparison, as summarized in Fig. 4. From the simplest free-living prokaryotes to plants and animals, proteomes have continuously expanded by both duplications and innovations. It is primarily due to the latter that proteome “complexity” has continuously increased in various ways that demand increased chaperone action. Across the ToL and especially when comparing prokaryotes with eukaryotes, we see a larger number of proteins per proteome (Fig. 4A) as well as larger proteins. The latter relates to multidomain proteins being increasingly represented. The number of different folds increases as well as of unique combinations of folds in multidomain proteins. Proteomes also contain a larger fraction of protein types that are prone to misfolding, such as repeat proteins and proteins comprising beta sheets, and those that are predicted to be highly aggregation-prone. The birth of a new fold, or of a new domain combination, likely results in poor foldability. With time, mutation and selection would improve foldability (59) and could ultimately render a newly born chaperone-independent protein. Nonetheless, the cumulative impact of newly evolved proteins, and of certain protein types (repeat or beta-rich proteins), likely demands increased chaperone capacity.
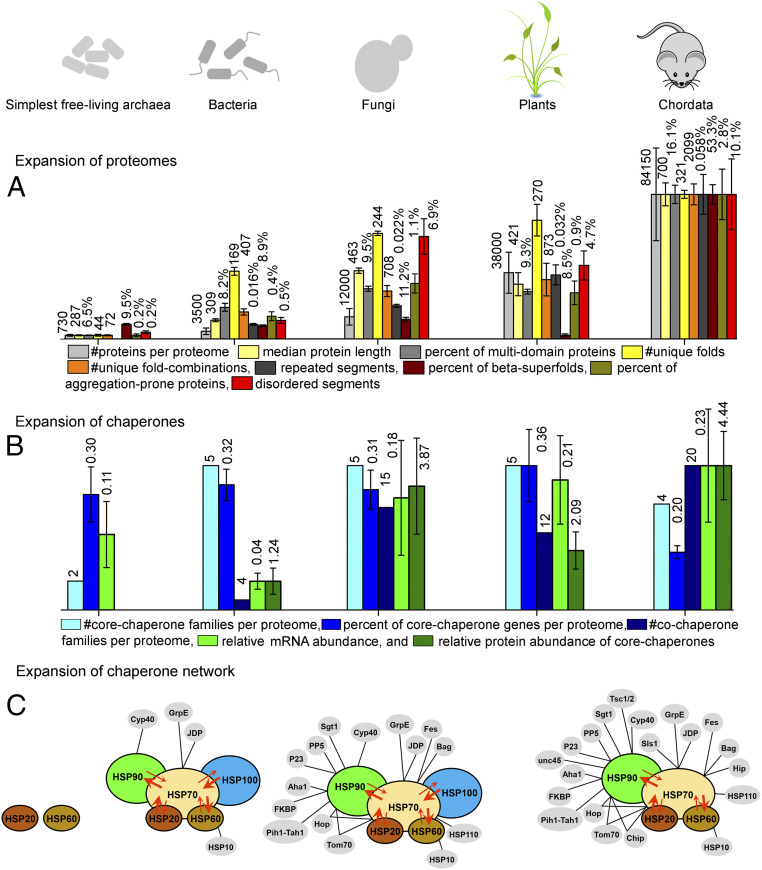
Fig. 4.
Summary figure describing the parallel expansion of proteomes and chaperones. Bar heights (y axis) were scaled such that the highest value per parameter assumed the same height (the absolute values are listed above the bars). (A) Bar graphs describing the expansion of proteomes in a nutshell. For the simplest free-living archaea, bacteria, fungi, plants, and chordates, plotted are the number of proteins per proteome (light gray), median protein length (light yellow), number of unique folds (gray), number of unique fold combinations per proteome (yellow), percentage of multidomain proteins (out of all proteins in the proteome; orange), percentage of proteome length that corresponds to repeat proteins (calculated by residue length; dark gray), percentage of proteins that have the beta-superfold architecture (wine), percentage of proteins predicted as highly aggregation-prone (dark yellow), and percentage of proteome length predicted as intrinsically disordered (red). (B) Same as A, for the expansion of chaperones. Plotted are the number of core- (cyan) and cochaperone families per proteome (navy), percentage of core-chaperone genes in the proteome (blue), relative mRNA abundance of core chaperones compared with ribosomal proteins (green), and relative protein abundance of core chaperones compared with all other proteins (dark green). (C) A schematic description of the expansion of the integrated chaperone network. Core chaperones are shown in various colors and with black outlines, while cochaperones are in gray with no outline. Cochaperones of HSP60, HSP70, and HSP90 are connected to their respective core chaperone by black lines. Cooperativity between core chaperones is represented by overlaps between circles, and substrate sharing between different core chaperones is shown by red arrows. Arrow direction and width represent the direction and magnitude of substrate sharing. Note that the network is shown for the simplest free-living archaea, bacteria, fungi, and chordates.
This dramatic increase in proteome complexity, and hence the demand for chaperone action, has not been met by the emergence of new core chaperones. Eukaryotes possess the same five core-chaperone families as prokaryotes, and metazoans and chordates have in fact lost HSP100 (Fig. 4B). Further, the relative representation of core-chaperone genes does not vary between prokaryotes and eukaryotes. Rather, the need for increased chaperone action was met in two ways: first, by an increased cellular abundance of core chaperones, and second, though, by the emergence of new cochaperones—while in bacteria ∼4 cochaperone families are found, in eukaryotes their number increased to 15 or even 20 in Mammalia.
Most of the expanding proteome features are likely the outcome of adaptive evolution (e.g., the emergence of new folds and domain combinations). However, expansion may also occur by drift, that is, by fixation of genetic changes by chance, due to population bottlenecks. Indeed, the effective population size (Ne) has dropped from prokaryotes (typically >108) to unicellular eukaryotes (∼107), invertebrates and land plants (∼106), and chordates (∼105) (60). Consequently, neutral, or even mildly deleterious mutations that would be purged in prokaryotes, might readily fix in multicellular eukaryotes. For example, drift may have driven the accumulation of hydrophobic residues on protein surfaces regardless of their protein–protein interaction potential, thus leading to lower protein stability and oligomerization but also increased aggregation propensity in eukaryotic proteins (61). Similarly, insertions fixed by drift could elongate disordered segments (62) and repeat proteins. The higher chaperone levels in eukaryotes (Fig. 3 C and D) may relate to the mitigation of the deleterious effects of such accumulating mutations (63). Pathogenic bacteria often experience severe population bottlenecks, and their chaperone expression levels are comparable to those of extremophiles (Fig. 3C and Dataset S11). Overall, the impact of drift on proteome and chaperone evolution merits further investigation.
The above trends highlight two features that comprise hallmarks of the chaperone machinery: the generalist nature of core chaperones, and their ability to act in a cooperative mode alongside cochaperones as an integrated network. HSP60, HSP70, HSP90, and HSP100 are core chaperones acting as generalist unfolding–refolding machineries that work on a broad range of differently misfolded and aggregated protein substrates, largely regardless of size [except HSP60 (64)], structure, and function (11, 31). While core chaperones can exert high-affinity binding to few specific substrates at their native folded state, they generally tend to bind misfolded and aggregated polypeptides that abnormally expose hydrophobic surfaces (31, 65, 66). The main driving force for duplication and specialization is a functional tradeoff—optimization of one function comes at the expense of other functions (67). However, given a “generalist” mode of function, the quality control of increasingly large and complex proteomes could be achieved by an elevated abundance of existing core chaperones, rather than by the emergence of new core-chaperone families. Indeed, although gene copy numbers of core chaperones have indeed increased by gene duplication, their relative representation compared with proteome size remained constant (Fig. 3B) and the resulting paralogous copies have mostly relocalized to different subcellular compartments or are expressed under different stress conditions (68). In parasitic microbes and photosynthetic organisms, duplicates of HSP70 and HSP90 have subspecialized to resist host immune responses and oxidative stress (54–57). However, consistent with their generalist nature, the challenge of maintaining large, complex proteomes (Fig. 4A) has primarily been met by increased abundances of preexisting core chaperones rather than by the de novo emergence of new ones.
In healthy cells, an integrated chaperone network, comprising both core and cochaperones, controls protein quality (69–71). In this network (Fig. 4C), the highly abundant core chaperones operate cooperatively, namely they not only share, and exchange incompletely processed misfolded or unfolded protein substrates, but also trigger the activities of one another. HSP70 plays a critical role in this network by mediating cooperative communications between the other core chaperones. For example, HSP70 triggers the disaggregase activity of HSP100, and jointly they disaggregate aggregated proteins and promote their subsequent refolding (72–74). In another example, HSP20 can transfer misfolded substrates to HSP70 for ATP-driven unfolding, from which they can be further transferred to HSP60 for final refolding to the native state (75). Likewise, HSP90 can promote the maturation of incompletely processed HSP70 substrates (76, 77). Cooperativity and substrate sharing between core chaperones are schematically represented in Fig. 4C. Together, these generalist, cooperative core chaperones constitute the core of an integrated chaperone network that has emerged from a simple two-component system in LUCA (Fig. 4C).
Alongside the expansion of proteome complexity, the chaperone network has also expanded—primarily by the emergence of cochaperones (Fig. 4C). This expanding array of cochaperones augmented the ability of core chaperones to efficiently share substrates and to function cooperatively. In contrast to the generalist core chaperones, cochaperones are more diverse and accordingly seem to subspecialize in specific roles, including cochaperones that handle specific proteins. Examples include UNC45, a cochaperone that emerged in Fungi, and facilitates HSP90-mediated maintenance of myosin in metazoan skeletal and cardiac muscles (78). Another Fungi-born cochaperone, the Tsc1/2 heteromer, specializes in recruiting kinase and some nonkinase substrates to HSP90 (79). Other cochaperones mediate protein transport; examples include Tom70 and P23 that facilitate protein trafficking through Golgi and mitochondrial membranes (80–82). The specialist mode of function of cochaperones coincides with how they expanded, namely by duplication and divergence of ancient prokaryote-born cochaperones but also via bona fide innovations, namely by the emergence of completely new specialized cochaperones in eukaryotes. As shown here, the emergence of new cochaperones coincides with the emergence of new proteins (i.e., by de novo emergence rather than by duplication of preexisting proteins). However, co-occurrence does not mean coevolution—indeed, we know very little about the latter. Did certain cochaperones emerge to support the de novo emergence of a specific protein or protein class? If so, does chaperone dependency persist, hence making codependency a “selfish” irreversible trait? Alternatively, as some newly emerged proteins evolved further, their foldability improved, allowing them to become chaperone-independent.
Thus, across the Tree of Life, proteomes have massively expanded, not just by duplication of preexisting proteins but also by the emergence of completely new ones. Eukaryotic proteomes became particularly large and specifically richer in repeat, beta-rich, and aggregation-prone proteins whose folding is inherently challenging. These changes in proteome size and composition intensified the demand for chaperone action. Curiously, however, no new core chaperones emerged in response to this increased demand. Instead, they increased in abundance relative to all other proteins in the cell. Foremost, an entire network of cochaperones had evolved that facilitate the basal core-chaperone activity.
Materials and Methods
For details, see SI Appendix, Methods.
Proteome Size and Median Protein Length.
A nonredundant set of 188 prokaryotic and eukaryotic organisms was collected from the TimeTree database (19) (listed in Dataset S1) and their proteome sequences (sequences of all proteins including splice variants, if relevant) were obtained from the National Center for Biotechnology Information genome database (83). For each organism, the total numbers of proteins in the respective proteome, and their lengths, were computed.
Multidomain Proteins.
For each protein, domain annotations were collected from Pfam (32). The number of proteins comprising <3 and those comprising ≥3 domains were then counted per each proteome.
Number of Unique Folds and Fold Combinations.
Proteins were clustered into independently evolved lineages using the ECOD database (which considers both sequence and structural similarities) where independently evolved lineages are termed X-groups (33). For each representative organism, the Pfam-assigned domains were mapped to their corresponding ECOD X-groups. The numbers of unique X-groups identified in a given proteome were considered as a measure of the number of unique folds. Similarly, we counted the X-group combinations, considering also their order along the polypeptide chain (AB ≠ BA) present in each protein. The total number of X-group combinations identified in a given proteome was considered as a measure of the total number of fold combinations.
Beta Superfolds in the Proteome.
We counted how many proteins in the proteome contain at least one domain annotated as all-beta fold (annotated in ECOD’s top hierarchy groups: beta barrel, beta meander, beta sandwich, beta duplicates or obligate multimers, and beta-complex topology). By normalizing this number by the total number of proteins in the proteome, we derived the fraction of beta-superfold proteins per proteome.
Repeated Sequences.
Each protein in the representative proteomes was scanned by the T-REKS repeat-identifier program (84) to detect repeats that are ≥20 aa long and exhibit ≥90% sequence similarity. The percentage of proteome length that is repeated was subsequently derived (the sum of the number of residues of all repeated segments multiplied by 100, divided by the total length of all proteins).
Aggregation-Prone Proteins.
An aggregation hotspot was defined as a “poorly soluble” protein segment of ≥5 aa in length, with solubility predicted from the protein’s sequence using CamSol v2.1 (85). For each representative organism, we computed the percentage of proteins in the proteome that contain ≥20 aggregation hotspots.
Intrinsically Disordered Regions.
Intrinsically disordered segments were identified by scanning all proteins in the representative proteomes by IuPred2A (86). Disordered segments ≥100 residues were considered. The percentage of proteome length that is disordered was subsequently derived (the total number of residues assigned to disordered segments multiplied by 100, divided by the sum of the length of all proteins).
Evolutionary History of Chaperones.
To determine the evolutionary appearance and expansion of the core-chaperone and cochaperone families, we identified their occurrences in the 188 representative organisms, using two complementary methods. The first method involved manual curation of annotated chaperones in model organisms that were subsequently used as queries to find orthologous sequences in the other organisms by protein–protein BLAST (87). The second method involved identifying the Pfam-assigned domain combinations of the various known chaperones in model organisms. Subsequently, any protein in the representative proteomes comprising these domain combinations was assigned as a member of the corresponding chaperone family. The orthologous and paralogous sequences for each chaperone family were aligned using MUSCLE v3.8.31 (88). Maximum-likelihood phylogenetic trees were generated by MEGA X (89). To date the emergence of individual chaperone families, the protein trees were manually compared with the ToL to assign the node of emergence and possible HGT events.
Chaperone Abundance Analysis.
To quantify the variation in chaperone mRNA abundance, RNA sequencing (RNA-seq)-based expression data for various model organisms were collected from available resources. Only processed RNA-seq data were considered where the normalized abundances of mRNA transcripts are provided as transcripts per million (TPMs). For each experiment, the sum of the TPM values of the core and the cochaperones was divided by the sum of TPMs of all ribosomal proteins. The average and SD over all experiments per given organism were computed.
To quantify protein abundance, mass spectrometry-based protein abundance data were collected for various model organisms from PaxDb (90). For each dataset, the sum of abundance values of all core chaperones and cochaperones was normalized by the sum of abundance values of all other proteins. The average and SD over all experiments per given organism were computed.
Supplementary Material
Acknowledgments
We thank Jagoda Jablonska for helping to establish the representative ToL, and Ita Gruic-Sovulj for proposing the chaperone abundance analysis. We thank Rina Rosenzweig, Harm H. Kampinga, Matthias P. Mayer, Sudip Kundu, Marc Robinson-Rechavi, and Agnieszka Klosowska for valuable suggestions regarding the manuscript. This work was supported by a Minerva Foundation grant (to D.S.T.) and Grant 31003A_175453 from the Swiss National Fund (to P.G. and M.E.R.). S.M. was supported by a postdoctoral fellowship, provided by Israel’s Council for Higher Education Planning and Budgeting Committee. P.G. is an associate professor in the Department of Plant Molecular Biology at the University of Lausanne. D.S.T. is the Nella and Leon Benoziyo Professor of Biochemistry at The Weizmann Institute of Science.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020885118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Woese C., The universal ancestor. Proc. Natl. Acad. Sci. U.S.A. 95, 6854–6859 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doolittle W. F., Phylogenetic classification and the universal tree. Science 284, 2124–2129 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Eme L., Spang A., Lombard J., Stairs C. W., Ettema T. J. G., Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Cooper G. M., The Cell: A Molecular Approach (Sinauer Associates, 2000). [Google Scholar]
- 5.Arendt D., The evolution of cell types in animals: Emerging principles from molecular studies. Nat. Rev. Genet. 9, 868–882 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Heim N. A., et al., Hierarchical complexity and the size limits of life. Proc. Biol. Sci. 284, 20171039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anfinsen C. B., Principles that govern the folding of protein chains. Science 181, 223–230 (1973). [DOI] [PubMed] [Google Scholar]
- 8.Dill K. A., Chan H. S., From Levinthal to pathways to funnels. Nat. Struct. Biol. 4, 10–19 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Englander S. W., Mayne L., The nature of protein folding pathways. Proc. Natl. Acad. Sci. U.S.A. 111, 15873–15880 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han J. H., Batey S., Nickson A. A., Teichmann S. A., Clarke J., The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 8, 319–330 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Finka A., Mattoo R. U., Goloubinoff P., Experimental milestones in the discovery of molecular chaperones as polypeptide unfolding enzymes. Annu. Rev. Biochem. 85, 715–742 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., Hartl F. U., Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Caplan A. J., What is a co-chaperone? Cell Stress Chaperones 8, 105–107 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schopf F. H., Biebl M. M., Buchner J., The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18, 345–360 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Duncan E. J., Cheetham M. E., Chapple J. P., van der Spuy J., The role of HSP70 and its co-chaperones in protein misfolding, aggregation and disease. Subcell. Biochem. 78, 243–273 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Tokuriki N., Tawfik D. S., Protein dynamism and evolvability. Science 324, 203–207 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Bogumil D., Dagan T., Cumulative impact of chaperone-mediated folding on genome evolution. Biochemistry 51, 9941–9953 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Alvarez-Ponce D., Aguilar-Rodríguez J., Fares M. A., Molecular chaperones accelerate the evolution of their protein clients in yeast. Genome Biol. Evol. 11, 2360–2375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedges S. B., Marin J., Suleski M., Paymer M., Kumar S., Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margulis L., Chapman M., Guerrero R., Hall J., The last eukaryotic common ancestor (LECA): Acquisition of cytoskeletal motility from aerotolerant spirochetes in the Proterozoic Eon. Proc. Natl. Acad. Sci. U.S.A. 103, 13080–13085 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Duve C., The origin of eukaryotes: A reappraisal. Nat. Rev. Genet. 8, 395–403 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Martin W., et al., Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Dombrowski N., Lee J. H., Williams T. A., Offre P., Spang A., Genomic diversity, lifestyles and evolutionary origins of DPANN archaea. FEMS Microbiol. Lett. 366, fnz008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battistuzzi F. U., Feijao A., Hedges S. B., A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4, 44 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burki F., Roger A. J., Brown M. W., Simpson A. G. B., The new tree of eukaryotes. Trends Ecol. Evol. 35, 43–55 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Simpson A. G., et al., Evolutionary history of “early-diverging” eukaryotes: The excavate taxon Carpediemonas is a close relative of Giardia. Mol. Biol. Evol. 19, 1782–1791 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Mayer M. P., Gymnastics of molecular chaperones. Mol. Cell 39, 321–331 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Henzler-Wildman K., Kern D., Dynamic personalities of proteins. Nature 450, 964–972 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Lafita A., Tian P., Best R. B., Bateman A., Tandem domain swapping: Determinants of multidomain protein misfolding. Curr. Opin. Struct. Biol. 58, 97–104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu K., Maciuba K., Kaiser C. M., The ribosome cooperates with a chaperone to guide multi-domain protein folding. Mol. Cell 74, 310–319.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong Y., et al., An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: Implications to protein folding pathways in the cell. Mol. Syst. Biol. 5, 275 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn R. D., et al., Pfam: The protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaeffer R. D., Liao Y., Cheng H., Grishin N. V., ECOD: New developments in the evolutionary classification of domains. Nucleic Acids Res. 45, D296–D302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pawar A. P., et al., Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J. Mol. Biol. 350, 379–392 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Halaby D. M., Mornon J. P., The immunoglobulin superfamily: An insight on its tissular, species, and functional diversity. J. Mol. Evol. 46, 389–400 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Andrade M. A., Perez-Iratxeta C., Ponting C. P., Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 134, 117–131 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Sormanni P., Aprile F. A., Vendruscolo M., The CamSol method of rational design of protein mutants with enhanced solubility. J. Mol. Biol. 427, 478–490 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Wang C., Uversky V. N., Kurgan L., Disordered nucleiome: Abundance of intrinsic disorder in the DNA- and RNA-binding proteins in 1121 species from Eukaryota, Bacteria and Archaea. Proteomics 16, 1486–1498 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Xue B., Dunker A. K., Uversky V. N., Orderly order in protein intrinsic disorder distribution: Disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 30, 137–149 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Van de Peer Y., Maere S., Meyer A., The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Berkemer S. J., McGlynn S. E., A new analysis of Archaea-Bacteria domain separation: Variable phylogenetic distance and the tempo of early evolution. Mol. Biol. Evol. 37, 2332–2340 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saibil H., Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 14, 630–642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Techtmann S. M., Robb F. T., Archaeal-like chaperonins in bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 20269–20274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sousa F. L., Nelson-Sathi S., Martin W. F., One step beyond a ribosome: The ancient anaerobic core. Biochim. Biophys. Acta 1857, 1027–1038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss M. C., et al., The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Kriehuber T., et al., Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 24, 3633–3642 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Fernández-Fernández M. R., Valpuesta J. M., Hsp70 chaperone: A master player in protein homeostasis. F1000 Res. 7 1497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenzweig R., Nillegoda N. B., Mayer M. P., Bukau B., The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 20, 665–680 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Battistuzzi F. U., Hedges S. B., A major clade of prokaryotes with ancient adaptations to life on land. Mol. Biol. Evol. 26, 335–343 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Hemmingsen S. M., et al., Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333, 330–334 (1988). [DOI] [PubMed] [Google Scholar]
- 51.Martin W., et al., Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bogumil D., Alvarez-Ponce D., Landan G., McInerney J. O., Dagan T., Integration of two ancestral chaperone systems into one: The evolution of eukaryotic molecular chaperones in light of eukaryogenesis. Mol. Biol. Evol. 31, 410–418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kampinga H. H., et al., Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones 24, 7–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Day J., Passecker A., Beck H. P., Vakonakis I., The Plasmodium falciparum Hsp70-x chaperone assists the heat stress response of the malaria parasite. FASEB J. 33, 14611–14624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shonhai A., Maier A. G., Przyborski J. M., Blatch G. L., Intracellular protozoan parasites of humans: The role of molecular chaperones in development and pathogenesis. Protein Pept. Lett. 18, 143–157 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Dahl J. U., Gray M. J., Jakob U., Protein quality control under oxidative stress conditions. J. Mol. Biol. 427, 1549–1563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niforou K., Cheimonidou C., Trougakos I. P., Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2, 323–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarkar N. K., Kundnani P., Grover A., Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 18, 427–437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smock R. G., Yadid I., Dym O., Clarke J., Tawfik D. S., De novo evolutionary emergence of a symmetrical protein is shaped by folding constraints. Cell 164, 476–486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lynch M., Evolution of the mutation rate. Trends Genet. 26, 345–352 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lynch M., Bobay L. M., Catania F., Gout J. F., Rho M., The repatterning of eukaryotic genomes by random genetic drift. Annu. Rev. Genomics Hum. Genet. 12, 347–366 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Light S., Sagit R., Sachenkova O., Ekman D., Elofsson A., Protein expansion is primarily due to indels in intrinsically disordered regions. Mol. Biol. Evol. 30, 2645–2653 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Maisnier-Patin S., et al., Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat. Genet. 37, 1376–1379 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Bukau B., Horwich A. L., The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Scheibel T., Weikl T., Buchner J., Two chaperone sites in Hsp90 differing in substrate specificity and ATP dependence. Proc. Natl. Acad. Sci. U.S.A. 95, 1495–1499 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rüdiger S., Germeroth L., Schneider-Mergener J., Bukau B., Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tawfik D. S., Gruic-Sovulj I., How evolution shapes enzyme selectivity—Lessons from aminoacyl-tRNA synthetases and other amino acid utilizing enzymes. FEBS J. 287, 1284–1305 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Kampinga H. H., et al., Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14, 105–111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hipp M. S., Kasturi P., Hartl F. U., The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 20, 421–435 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Jayaraj G. G., Hipp M. S., Hartl F. U., Functional modules of the proteostasis network. Cold Spring Harb. Perspect. Biol. 12, a033951 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taipale M., et al., A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 158, 434–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haslberger T., et al., Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat. Struct. Mol. Biol. 15, 641–650 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Rosenzweig R., Moradi S., Zarrine-Afsar A., Glover J. R., Kay L. E., Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339, 1080–1083 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Glover J. R., Lindquist S., Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Veinger L., Diamant S., Buchner J., Goloubinoff P., The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J. Biol. Chem. 273, 11032–11037 (1998). [DOI] [PubMed] [Google Scholar]
- 76.Morán Luengo T., Kityk R., Mayer M. P., Rüdiger S. G. D., Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell 70, 545–552.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Genest O., Hoskins J. R., Camberg J. L., Doyle S. M., Wickner S., Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc. Natl. Acad. Sci. U.S.A. 108, 8206–8211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wohlgemuth S. L., Crawford B. D., Pilgrim D. B., The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev. Biol. 303, 483–492 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Woodford M. R., et al., Tumor suppressor Tsc1 is a new Hsp90 co-chaperone that facilitates folding of kinase and non-kinase clients. EMBO J. 36, 3650–3665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Echtenkamp F. J., et al., Global functional map of the p23 molecular chaperone reveals an extensive cellular network. Mol. Cell 43, 229–241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melnyk A., Rieger H., Zimmermann R., Co-chaperones of the mammalian endoplasmic reticulum. Subcell. Biochem. 78, 179–200 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Craig E. A., Marszalek J., How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 42, 355–368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benson D. A., et al., GenBank. Nucleic Acids Res. 41, D36–D42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jorda J., Kajava A. V., T-REKS: Identification of tandem repeats in sequences with a K-means based algorithm. Bioinformatics 25, 2632–2638 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Sormanni P., Amery L., Ekizoglou S., Vendruscolo M., Popovic B., Rapid and accurate in silico solubility screening of a monoclonal antibody library. Sci. Rep. 7, 8200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dosztányi Z., Csizmók V., Tompa P., Simon I., The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 347, 827–839 (2005). [DOI] [PubMed] [Google Scholar]
- 87.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 88.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang M., Herrmann C. J., Simonovic M., Szklarczyk D., von Mering C., Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15, 3163–3168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.