Two conserved processes express the genetic information of all organisms. First, DNA is transcribed into a messenger RNA (mRNA) by the multisubunit enzyme RNA polymerase (RNAP). Second, the mRNA directs protein synthesis, when the ribosome translates its nucleotide sequence to amino acids using the genetic code. Because these two processes are so fundamental, a multitude of regulatory processes have evolved to regulate them. Most examples involve regulation of either transcription or translation. In PNAS, Chatterjee et al. (1) instead describe a complex and intricate regulatory process in which transcription and translation are concurrently regulated by each other.
Transcription and translation are commonly viewed as separate. In eukaryotes, their respective confinement to the nucleus and cytoplasm enforces this. Yet, prokaryotes have no such barrier, and newly synthesized mRNAs are translated while they are still being transcribed. RNAP and the trailing ribosome are therefore in close spatial proximity, allowing each to influence the activity of the other. The possibility of a physical connection that could support functional coupling was proposed in 1964 by Marshall Nirenberg’s laboratory based on biochemical experiments (2). They highlighted the potential importance of regulatory processes that simultaneously affect both transcription and translation. Electron micrographs of ruptured Escherichia coli cells, commonly termed “Miller spreads,” confirmed the close proximity between RNAP and the trailing ribosome (3).
The role and mechanism of coupling have received renewed interest over the past 10 y. Biochemical and structural approaches alongside new measurements of gene expression rates in vivo have clarified several important aspects. Early studies had demonstrated that translation can release RNAP from regulatory pauses (4). This mechanism, part of a process known as attenuation, had been described in the context of the leader sequences of specific operons. Yet more recent evidence points to additional genome-wide mechanisms of translation promoting transcription: the trailing ribosome pushing RNAP forward along the gene (5, 6). RNAP pauses regularly when it encounters specific DNA sequences and can slide backward. A forward translocating ribosome could thus minimize the formation and aid in the release of transcriptional pauses. This could explain the synchronization of transcription and translation rates observed in E. coli (5). It is also essential to fitness, as translation maintains genome stability by releasing arrested transcription complexes that would otherwise interfere with DNA replication (7).
The molecular architecture that occurs during pause release likely resembles recent structures of ribosome–RNAP complexes determined with short intervening mRNAs (8–10). This supramolecular assembly has been termed the “expressome.” The expressome is dynamic and adopts a distinct arrangement when the transcription factor NusG is present (9, 10). By simultaneously binding the ribosome and RNAP, NusG acts as a physical bridge. This minimizes the formation of mRNA secondary structures that could inhibit transcription and translation and also sequesters a NusG domain that promotes transcription termination. Impairment of the NusG–ribosome interaction impacts coupling in vivo (11). RfaH, another member of the NusG family, also has the ability to bind both RNAP and the ribosome, but the consequences are less-well-understood (12).
In PNAS, Chatterjee et al. (1) shed further light with an analysis of factors that drive the establishment of coupled translation at an early stage of transcription. Using biochemical and single-molecule fluorescence analyses, the interplay between ribosome recruitment and rates of transcription and translation are examined. Further, how each of these is modulated by a regulatory riboswitch is tested. Previous work has focused on the regulation of RNAP by translation elongation, and here a link is demonstrated between RNAP and translation initiation: an interesting aspect of transcription–translation coupling.
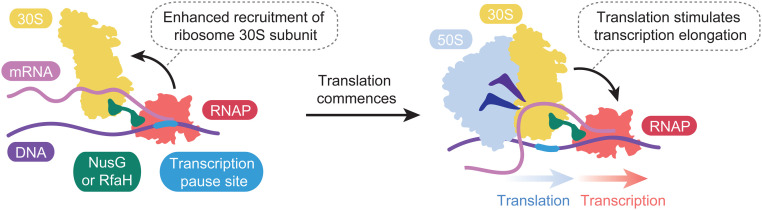
Transcription of the mRNA studied by Chatterjee et al. (1) undergoes a programmed pause after ∼100 nucleotides have been synthesized, a common feature of E. coli transcripts (13). RNAP waits for translation initiation to occur and coupling to be established. This is analogous to the pause sequences upstream of attenuation sites, where coupling controls transcription termination (14). Chatterjee et al. reveal that recruitment of the small ribosomal subunit to the mRNA is stimulated by the paused RNAP and further promoted by NusG. This extends the regulatory role of NusG beyond transcription to translation initiation. Chatterjee et al. also present evidence that the translating ribosome then stimulates RNAP release from the pause and increases transcription rates. This is consistent with the notion that the transcriptional pause allows time for translation to commence (Fig. 1) and supports observations made in vivo (5, 15).
Fig. 1.
Model of the establishment of transcription–translation coupling. Recruitment of the ribosome 30S subunit is enhanced by paused RNAP and further promoted by coupling factors NusG or RfaH that bind both complexes. Translation then prompts RNAP to resume transcription with an increased rate.
The contribution of RNAP and NusG or RfaH relative to the mRNA ribosome binding site is examined by Chatterjee et al. (1) using a riboswitch. Riboswitches are structured regulatory RNA elements involved in sensing pH, temperature, and the presence of metabolites. Riboswitches can affect transcription and translation as a function of their ligands (16–18). The Bacillus subtilis riboswitch used by Chatterjee et al. senses the concentration of a nucleotide precursor preQ1. Upon preQ1-induced folding, the riboswitch masks the ribosome binding site. This reduces translation and, in part as a consequence of the role of translation in stimulating transcription, reduces transcription. Comparisons of the unfolded and folded riboswitch also revealed that the role of NusG in recruiting the small ribosomal subunit depends on the ribosome binding site of the mRNA’s being accessible. Interestingly, recruitment of the small subunit by the NusG paralog RfaH did not depend on this. This is suggestive of an unexpected mechanistic difference between the factors.
The results presented by Chatterjee et al. (1) add another layer to the intricate relationship between RNAP and the ribosome. The relationship evidently extends beyond a simple cooperative forward push of the two machineries along the gene. Many exciting questions arise. How does RNAP promote ribosome recruitment? An interaction between RNAP and the small ribosomal subunit was previously structurally characterized (19) and could be involved. The importance of the transcription factors NusG, which was investigated here, and others such as NusA, which was not, remains to be elucidated. How the ribosome releases RNAP from paused states remains an important outstanding question. Transcription pausing occurs in at least three ways: inhibitory mRNA hairpins such as those located upstream of attenuators, RNAP back-tracking, and thermodynamically driven “consensus” pauses. The ribosome’s ability to release RNAP from hairpin-induced and back-tracked pauses has previously been demonstrated (4, 6). Chatterjee et al. now present evidence of release from a pause sequence resembling the consensus. It remains to be tested whether the ribosome releases RNAP from each pause type by shared or distinct mechanisms.
Physical coupling of RNAP to the ribosome is not the only mechanism of functional coordination between them. For example, reduced global translation rates direct reduced transcription via the alarmone (p)ppGpp (20, 21). The contribution of different types of coupling in different contexts is an important question. The ubiquity of transcription–translation coupling across species also remains unresolved. Concurrent transcription–translation was observed in archaea (22), suggesting similarities across prokaryotic domains, yet not all bacteria display physical coupling. In B. subtilis, transcription outpaces the trailing ribosome (23). Expressome formation in Mycoplasma pneumonia appears to predominantly involve the transcription factor NusA and not NusG, as it does in E. coli (24). Addressing these and further questions will require complementary approaches. Classical biochemistry and single-molecule studies will need to be combined with high-resolution structural biology and genome-wide approaches to study the effects of coupling and the effect of its disruption.
A reductionist approach has proved powerful to the study of gene expression mechanisms. Transcription and translation have been studied in great detail for several decades in isolation. However, many key enzymes in gene expression and beyond are organized in multicomponent, supramolecular assemblies. Here new functions emerge, which are not necessarily predictable from the sum of their parts. It is exciting to see that we no longer neglect this aspect and begin to address the next level of structural organization to learn about the biological roles and functional implications of these higher-order assemblies.
Acknowledgments
M.W.W. and A.W.’s research is supported by an EMBO (European Molecular Biology Organisation) long-term fellowship to M.W.W., by the European Research Council starting grant TRANSREG (679734) to A.W., and by the Agence Nationale de la Recherche grant PrTxConf (ANR-17-CE11-0042). We are also supported by the French Infrastructure for Integrated Structural Biology (FRISBI ANR-10-INBS-05, Instruct-ERIC, and grant ANR-10-LABX-0030- INRT, a French State fund managed by the Agence Nationale de la Recherche under the program Investissements d’Avenir ANR-10- IDEX-0002-02).
Footnotes
The authors declare no competing interest.
See companion article, “A translational riboswitch coordinates nascent transcription–translation coupling,” 10.1073/pnas.2023426118.
References
- 1.Chatterjee S., Chauvier A., Dandpat S. S., Artsimovitch I., Walter N. G., A translational riboswitch coordinates nascent transcription–translation coupling. Proc. Natl. Acad. Sci. U.S.A. 118, e2023426118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne R., Levin J. G., Bladen H. A., Nirenberg M. W., The in vitro formation of a DNA–ribosome complex. Proc. Natl. Acad. Sci. U.S.A. 52, 140–148 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O. L. Miller, Jr, Hamkalo B. A., C. A. Thomas, Jr, Visualization of bacterial genes in action. Science 169, 392–395 (1970). [DOI] [PubMed] [Google Scholar]
- 4.Landick R., Carey J., Yanofsky C., Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc. Natl. Acad. Sci. U.S.A. 82, 4663–4667 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proshkin S., Rahmouni A. R., Mironov A., Nudler E., Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson-Jones F., Woodgate J., Castro-Roa D., Zenkin N., Ribosome reactivates transcription by physically pushing RNA polymerase out of transcription arrest. Proc. Natl. Acad. Sci. U.S.A. 117, 8462–8467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta D., Shatalin K., Epshtein V., Gottesman M. E., Nudler E., Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146, 533–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler R., Mooney R. A., Mills D. J., Landick R., Cramer P., Architecture of a transcribing-translating expressome. Science 356, 194–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C., et al., Structural basis of transcription-translation coupling. Science 369, 1359–1365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webster M. W., et al., Structural basis of transcription-translation coupling and collision in bacteria. Science 369, 1355–1359 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Saxena S., et al., Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol. Microbiol. 108, 495–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burmann B. M., et al., An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 150, 291–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson M. H., et al., A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1042–1047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbough C. L., Regulation of bacterial gene expression by transcription attenuation. Microbiol. Mol. Biol. Rev. 83, e00019-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel U., Jensen K. F., The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 176, 2807–2813 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winkler W., Nahvi A., Breaker R. R., Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Mironov A. S., et al., Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111, 747–756 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Sherwood A. V., Henkin T. M., Riboswitch-mediated gene regulation: Novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Demo G., et al., Structure of RNA polymerase bound to ribosomal 30S subunit. eLife 6, 94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer S., Le D., Park B. R., Kim M., Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli. Nat. Microbiol. 3, 741–748 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Zhu M., Mori M., Hwa T., Dai X., Disruption of transcription-translation coordination in Escherichia coli leads to premature transcriptional termination. Nat. Microbiol. 4, 2347–2356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.French S. L., Santangelo T. J., Beyer A. L., Reeve J. N., Transcription and translation are coupled in Archaea. Mol. Biol. Evol. 24, 893–895 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Johnson G. E., Lalanne J. B., Peters M. L., Li G. W., Functionally uncoupled transcription-translation in Bacillus subtilis. Nature 585, 124–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Reilly F. J., et al., In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]