Significance
Although immune checkpoint blockade therapies have achieved long-term responses in several malignancies, in colorectal cancer (CRC) patients clinical benefit is observed only in heavily mutated tumors that are mismatch-repair–deficient or have high microsatellite instability. This limitation urges the identification of novel immune escape mechanisms and the design of additional immunotherapeutic modalities. We show that Galectin-1 (Gal-1) confers immune privilege to CRC by increasing the frequency of CD8+CD122+PD-1+ regulatory T cells (Tregs) and accentuating their immunosuppressive activity in experimental models. Accordingly, analysis of CRC patient datasets revealed a “poor prognosis signature” characterized by high Gal-1 expression and elevated CD8+ Treg score. Thus, targeting Gal-1/glycan interactions may represent a potential immunotherapeutic modality for treating CRC by recalibrating the CD8+ Treg compartment.
Keywords: colorectal cancer, immune escape, Galectin-1, CD8+ regulatory T cells
Abstract
Colorectal cancer (CRC) represents the third most common malignancy and the second leading cause of cancer-related deaths worldwide. Although immunotherapy has taken center stage in mainstream oncology, it has shown limited clinical efficacy in CRC, generating an urgent need for discovery of new biomarkers and potential therapeutic targets. Galectin-1 (Gal-1), an endogenous glycan-binding protein, induces tolerogenic programs and contributes to tumor cell evasion of immune responses. Here, we investigated the relevance of Gal-1 in CRC and explored its modulatory activity within the CD8+ regulatory T cell (Treg) compartment. Mice lacking Gal-1 (Lgals1−/−) developed a lower number of tumors and showed a decreased frequency of a particular population of CD8+CD122+PD-1+ Tregs in the azoxymethane-dextran sodium sulfate model of colitis-associated CRC. Moreover, silencing of tumor-derived Gal-1 in the syngeneic CT26 CRC model resulted in reduced number and attenuated immunosuppressive capacity of CD8+CD122+PD-1+ Tregs, leading to slower tumor growth. Moreover, stromal Gal-1 also influenced the fitness of CD8+ Tregs, highlighting the contribution of both tumor and stromal-derived Gal-1 to this immunoregulatory effect. Finally, bioinformatic analysis of a colorectal adenocarcinoma from The Cancer Genome Atlas dataset revealed a particular signature characterized by high CD8+ Treg score and elevated Gal-1 expression, which delineates poor prognosis in human CRC. Our findings identify CD8+CD122+PD-1+ Tregs as a target of the immunoregulatory activity of Gal-1, suggesting a potential immunotherapeutic strategy for the treatment of CRC.
Colorectal cancer (CRC) is the third most commonly diagnosed malignancy and the second leading cause of worldwide cancer-related deaths (1). In developed countries, early detection through screening has improved the 5-y survival of patients with CRC, but ∼25% of patients still present stage 4 disease, and an additional 25 to 50% present with early-stage disease but go on to develop metastatic disease (2). In CRC, immune checkpoint blockade therapies, targeting cytotoxic T-lymphocyte antigen-4 (CTLA-4) and programmed cell death protein-1 (PD-1)/programmed death ligand-1 (PD-L1) pathways received regulatory approval for the treatment of heavily mutated tumors that are mismatch-repair–deficient (dMMR) or have high levels of microsatellite instability (MSI-H) (∼15%). In contrast, immune checkpoint blockade therapies are ineffective in tumors that are MMR-proficient and are microsatellite-stable or have low levels of microsatellite instability (∼85%). In these tumors, low tumor mutational burden and the lack of immune cell infiltration have been proposed as mechanisms of immune resistance (3). Thus, there is an impending need to investigate novel mechanisms of immune evasion in CRC, to identify novel biomarkers of clinical response and to develop novel immunotherapeutic strategies.
Several immune regulatory pathways, commonly used by the immune system to maintain homeostasis and prevent exuberant inflammation, are coopted by cancer cells to evade or re-edit antitumor immune responses, thus fostering local and systemic immunosuppressive microenvironments (4). A well-documented mechanism of immune escape includes the recruitment, expansion, and differentiation of immunosuppressive cell populations, including regulatory T cells (Tregs), which have a detrimental role in cancer by suppressing antitumor effector T cells, natural killer (NK) cells, and antigen-presenting cells (5, 6). While CD4+CD25+Foxp3+ Tregs have been extensively studied, limited information is available on the fate and function of CD8+ Tregs (7). The intrinsic differences between CD4+ and CD8+ T cells regarding antigen-driven activation suggest that CD4+ and CD8+ Tregs may have nonredundant, and probably complementary immunomodulatory functions in the context of tumor-immune escape (7, 8). Nevertheless, and even though CD8+ Tregs were originally described in the early 1970s (9), scarce information is available regarding their role in tumor-immune escape (7, 8). Moreover, definition of CD8+ Tregs remains a major drawback given the paucity of specific markers and functional, transcriptional, or epigenetic programs that could distinguish these cells from conventional cytotoxic and memory CD8+ T cells (10, 11).
Recent studies brought CD8+ Tregs back into the spotlight through the identification of distinct populations expressing a different set of cell surface markers and displaying potent immunosuppressive profiles (7, 8). Among them, a particular population of antigen-elicited CD8+ Tregs expressing CD25, Foxp3, and CTLA-4 has been detected in prostate, breast, and CRC (12). Moreover, a group of inducible CD8+CD28− Tregs that promote CD4+ T cell anergy has been identified in cancer patients (13–15). Additionally, CD8+CD122+ Tregs have been described in mice in different cancer models (16–20). This regulatory population, characterized by high interleukin (IL)-10 production, blunts IL-17 and interferon (IFN)-γ secretion from activated CD4+ T cells (8, 16) and displays cytotoxic activity against effector T cells through Fas ligand (Fas L)-dependent mechanisms (16, 17, 21). Accordingly, administration of anti-CD122 monoclonal antibody (mAb) significantly reduced tumor growth and induced long-term survival in experimental models of CRC and melanoma (22). Further studies revealed that CD8+CD122+ Tregs included subsets of regulatory and memory T cells and suggested the use of PD-1 to discriminate between these subpopulations. Whereas CD8+CD122+PD-1+ cells function as Tregs, CD8+CD122+PD-1− cells exhibit features of memory T cells (11). Interestingly, a CD8+CXCR3+ Treg population secreting high amounts of IL-10, but lacking CD122 expression, has been described in humans as an equivalent of murine CD8+CD122+PD-1+ T cells (23).
Programmed remodeling of cell surface glycans controls tumor progression by influencing different hallmarks of cancer cells, including cell adhesion, invasion, angiogenesis, and immune escape (24). Galectins, a family of endogenous glycan-binding proteins, serve as “glyco-checkpoints” that reprogram the tumor microenvironment (TME) and recalibrate tumor-driven circuits through intracellular and extracellular mechanisms (25). Galectin-1 (Gal-1), a proto-type member of this family specifically recognizes N-acetyllactosamine [LacNAc; Galβ (1-4)-GlcNAc] structures on glycosylated receptors present on the surface of different cells (26). Targeting Gal-1/glycan interactions suppresses tumor progression by thwarting tumor-immune escape (27–29), attenuating aberrant angiogenesis (30–32), preventing metastasis (33), and circumventing resistance to different anticancer therapies, including immunotherapy (34, 35). Mechanistically, Gal-1 limits antitumor immunity by promoting differentiation of tolerogenic dendritic cells (DCs) (36), inducing apoptosis of effector T cells (27, 29), and favoring expansion of CD4+Foxp3+ Tregs (28, 33). In this regard, Gal-1 has emerged as a critical regulator of CD4+ Treg biology by recalibrating the fate and function of these cells (28, 33, 37, 38). Particularly in CRC, higher Gal-1 expression correlates with tumor growth, survival, and progression (39–41). Moreover, recent findings demonstrated that branched N-glycans, which commonly serve as galectin ligands, may be coopted by CRC cells to evade antitumor immunity through inhibition of IFN-γ production (42). However, the relevance of Gal-1 in CRC-driven immunosuppression has not yet been explored.
Here we show that Gal-1 controls tumor growth in CRC by shaping antitumor immunity through modulation of CD8+CD122+PD-1+ Tregs. Using the syngeneic CT26 CRC model and the colitis-associated CRC model chemically induced by azoxymethane (AOM) and dextran sodium sulfate (DSS), we demonstrate that both tumor- and stromal-derived Gal-1 influence tumor growth by controlling the frequency and suppressive activity of CD8+ Tregs. Accordingly, bioinformatic analysis of colorectal adenocarcinoma (COAD) patient datasets (The Cancer Genome Atlas, TCGA) revealed a “poor prognosis signature” characterized by high Gal-1 expression and an elevated CD8+ Treg score, defined by phenotypic and functional markers of these regulatory cells. Thus, targeting Gal-1/glycan interactions may represent a potential immunotherapeutic modality for treating CRC patients by recalibrating the CD8+ Treg compartment.
Results
Mice Lacking Gal-1 Develop Fewer Tumors and Display Lower Frequency of CD8+ Tregs in a Colitis-Associated CRC Model.
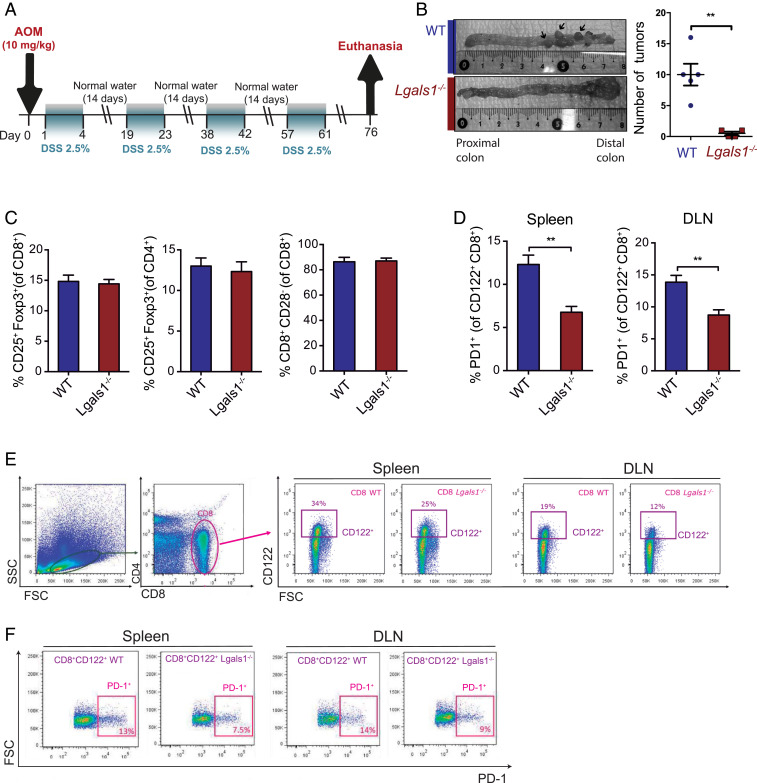
To investigate the immunoregulatory role of Gal-1 in CRC, we first used the AOM-DSS colitis-associated CRC (CACRC) model, which recapitulates the initiation and progression phases of human CACRC (43). Both WT and Gal-1–deficient (Lgals1−/−) mice were subjected to inflammation-driven colon carcinogenesis by treatment with AOM-DSS (Fig. 1A). We found a substantial reduction in the number of tumors developed in the distal colon of Lgals1−/− compared to WT mice (**P < 0.01) (Fig. 1B). Given the involvement of Gal-1 in regulatory immune programs, particularly CD4+ Tregs, in different pathologic settings (33, 37, 44), we analyzed the frequency of different Treg cell populations in tumor-draining lymph nodes (DLNs) and spleen of AOM-DSS–treated animals (Fig. 1 C and D). While we found no substantial differences in the proportions of CD4+CD25+Foxp3+, CD8+CD25+Foxp3+, or CD8+CD28− Tregs between Gal-1–deficient and WT genotypes (Fig. 1C), the percentage of PD-1+ cells on CD8+CD122+ Treg cells was substantially lower in tumor-bearing Lgals1−/− vs. WT mice, both in DLN and spleen (Fig. 1 D and E). Of note, modulation of NKT cells, which also express CD122, was ruled out based on the concomitant analysis of the NKT cell marker CD49b (SI Appendix, Fig. S1). Moreover, study of activated and memory CD4+ and CD8+ T cell populations revealed no differences in the frequency of these cells between WT and Lgals1−/− animals (SI Appendix, Fig. S2). Thus, endogenous Gal-1 favors CACRC tumorigenesis through selective modulation of CD8+CD122+PD-1+ Tregs.
Fig. 1.
Endogenous Gal-1 promotes tumorigenesis in the AOM-DSS CACRC model and fosters an immunosuppressive tumor microenvironment via induction of CD8+ Tregs. (A) Scheme of the AOM-DSS CRC model. (B) Number of tumors in the distal colon of Lgals1−/− compared to WT mice. (C) Analysis of CD4+CD25+Foxp3+, CD8+CD25+Foxp3+, and CD8+CD28− T cells in tumor-DLN of Lgals1−/− versus WT mice. (D) Analysis of PD-1 expression in CD8+CD122+ T cells in DLN and spleen of Lgals1−/− and WT mice. (E and F) Flow cytometry dot plot analysis showing gates for the selected immune populations. Data presented are mean ± SEM from three independent experiments. n = 5 to 6 mice per group. Unpaired Student’s t test, *P < 0.05, **P < 0.01.
Gal-1 Increases the Frequency and Accentuates the Immunosuppressive Capacity of CD8+ Tregs.
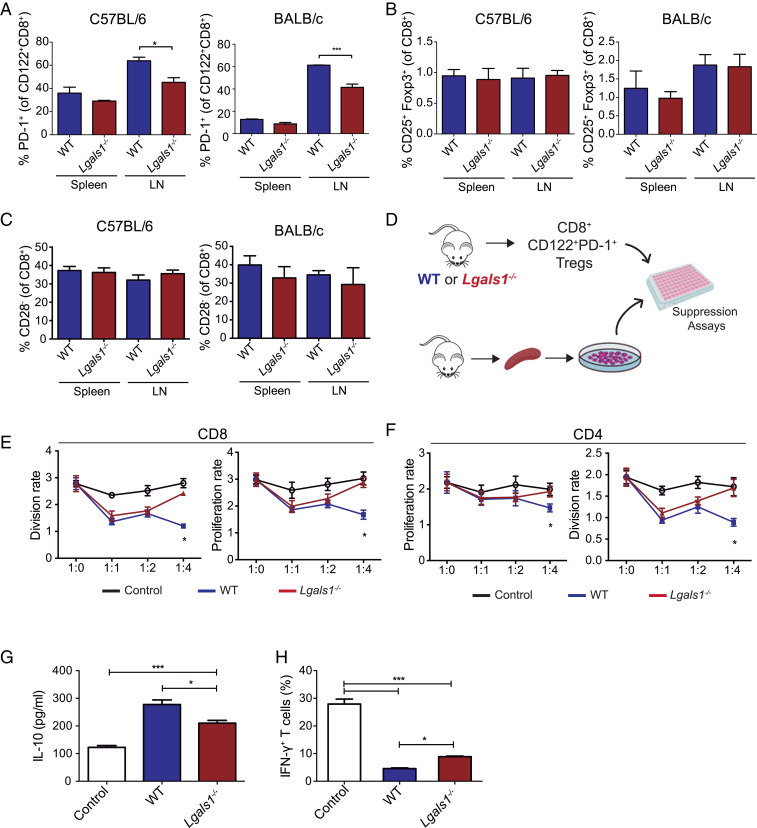
To evaluate whether Gal-1 has an intrinsic ability to control the CD8+ Treg compartment in a nonpathological setting, we studied the frequency and function of CD8+ Tregs in healthy WT and Lgals1−/− mice. Analysis of the frequency of different Treg cell populations showed only a significant reduction in the percentage of CD8+CD122+PD-1+ Tregs in axillary and inguinal lymph nodes of Lgals1−/− compared to WT mice (Fig. 2 A–C), consistent with the findings in the AOM-DSS CACRC model. These results were validated both in the C57BL/6 and the BALB/c strains, suggesting that modulation of this Treg population is strain-independent and intrinsically associated with Gal-1 deficiency. Notably, differences in CD8+ Treg frequency were not verified in the spleen neither in BALB/c nor in C57BL/6 mice (Fig. 2 A–C).
Fig. 2.
Gal-1 augments the frequency and immunosuppressive capacity of CD8+CD122+PD-1+ Tregs. (A–C) Spleen and axillary and inguinal lymph nodes from WT or Lgals1−/− C57BL/6 (Left) or BALB/c mice (Right) were processed and CD8+CD122+PD1+ (A), CD8+CD25+Foxp3+ (B), and CD8+CD28− (C) T cells were analyzed. (D) Schematic representation of immunosuppression assays. CD8+CD122+PD-1+ Tregs were isolated from the spleens of WT or Lgals1−/− C57BL/6 mice and were further cocultured with total labeled splenocytes at different ratios (1:1, 1:2, 1:4). (E and F) Proliferation and division rates of CD4+ and CD8+ responders T cells. (G) Determination of IL-10 in supernatants of coculture assays at 96 h. (H) Frequency of IFN-γ+ T cells in splenocytes cocultured with CD8+CD122+PD1+ Tregs from WT or Lgals1−/− mice detected by flow cytometry. Data presented are the mean ± SEM from three independent experiments. n = 5 to 6 mice per group. ANOVA, Bonferroni multiple comparison test, *P < 0.05 and ***P < 0.001.
Next, we evaluated the impact of endogenous Gal-1 on the immunosuppressive capacity of CD8+CD122+PD-1+ Tregs using an in vitro lymphocyte proliferation assay. Mouse splenocytes were fluorescently labeled and cocultured with increasing ratios of sorted CD8+CD122+PD-1+ Tregs obtained from WT or Lgals1−/− C57BL/6 mice and analyzed by flow cytometry (Fig. 2 D–F). Notably, CD8+CD122+PD-1+ Tregs from Lgals1−/− mice showed a significantly lower suppressive capacity on both CD4+ and CD8+ effector T cells as compared to WT CD8+ Tregs (Fig. 2 E and F). Similar results were obtained using Lgals1−/− and WT splenocytes from BALB/c mice (SI Appendix, Fig. S3). Thus, although no significant changes were observed in the proportion of CD8+CD122+PD-1+ Tregs in the spleen, substantial differences were found in their immunosuppressive capacity.
Given the central role of IL-10 as a mediator of the immunosuppressive capacity of CD8+CD122+PD1+Tregs, we analyzed IL-10 secretion in supernatants from coculture experiments. Remarkably, CD8+CD122+PD1+ Tregs obtained from Lgals1−/− mice secreted lower amounts of IL-10 compared to CD8+CD122+PD1+ WT Tregs (Fig. 2G). A marked decline in IFN-γ–producing T cells was observed in cocultures of splenocytes with CD8+CD122+PD1+ Tregs from both WT and Lgals1−/− mice, compared to splenocytes cultured in the absence of Tregs. However, the percentage of IFN-γ–producing T cells was significantly higher when splenocytes were exposed to Lgals1−/− CD8+CD122+PD1+ Tregs compared to their WT counterpart (Fig. 2H). Thus, Gal-1 deficiency lowers the frequency of CD8+CD122+PD-1+ Tregs and attenuates their immunosuppressive capacity.
Tumor-Derived Gal-1 Fosters Immunosuppression in a Syngeneic CRC Model through Induction of CD8+ Tregs.
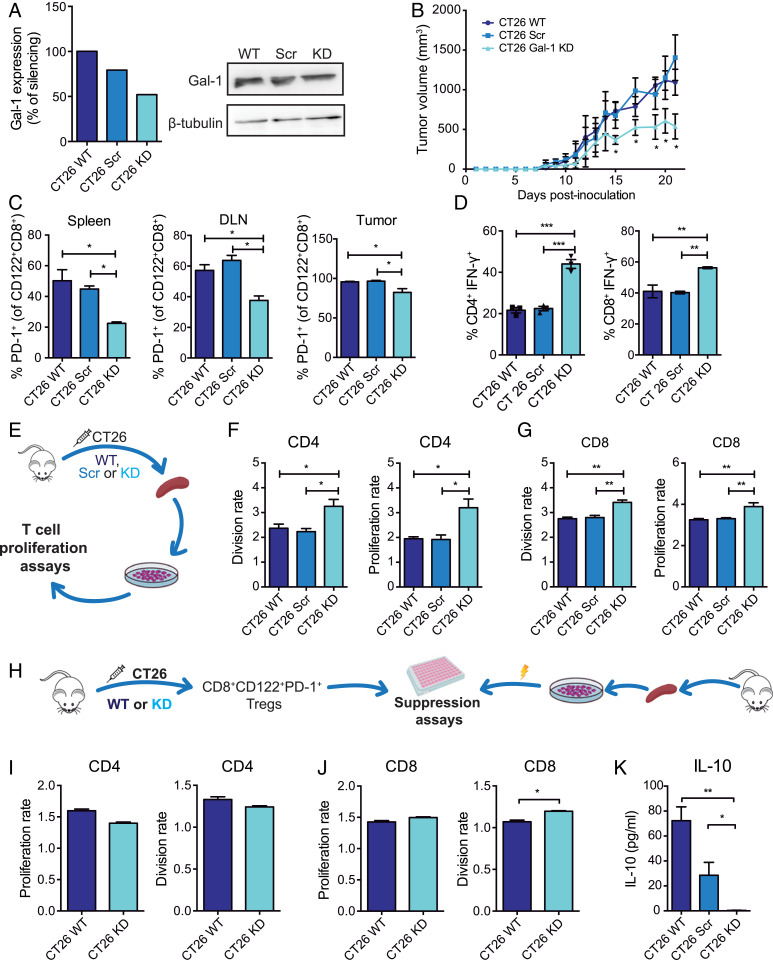
Since Gal-1 deletion controls the frequency and suppressive activity of CD8+ Tregs and reduces tumor growth in the AOM-DSS model, we then investigated the role of tumor-derived Gal-1 in modulating the CD8+ Treg compartment using the syngeneic CT26 CRC model. Gal-1–deficient and control CT26 cell lines were generated by stable transfection with a Gal-1–specific small-hairpin RNA (shRNA) (CT26 Gal-1 knockdown, KD) or a scramble unspecific sequence (CT26 Scr). Gal-1 KD resulted in ∼50% reduction of protein expression, as determined by Western blot (Fig. 3A). Gal-1 KD, Scr, and WT CT26 cells were subcutaneously injected into WT mice. Tumor growth was significantly reduced in mice inoculated with Gal-1 KD cells compared to mice receiving WT or Scr cells (P < 0.05) (Fig. 3B). Moreover, mice injected with Gal-1 KD cells showed a significant reduction in the percentage of PD-1+ cells within the CD8+CD122+ T cell population in tumor-infiltrating lymphocytes, spleen, and DLN, as compared to mice receiving WT or Scr CT26 cells (Fig. 3C). In contrast, no significant differences were detected in the frequencies of CD8+CD28−, CD8+CD25+Foxp3+, or CD4+CD25+Foxp3+ Tregs obtained from animals inoculated with Gal-1 KD or WT CT26 cells (SI Appendix, Fig. S4). Finally, WT mice inoculated with Gal-1 KD cells showed higher percentages of CD4+IFN-γ+ and CD8+IFN-γ+ T cells (Fig. 3D). Of note, Gal-1 silencing did not affect intrinsic proliferation or survival of CT26 cells (SI Appendix, Fig. S5), as previously described in other tumor cells, including melanoma (29), lung cancer (30), Kaposi’s sarcoma (31), and breast adenocarcinoma cells (33). These results highlight the immune inhibitory activity of tumor-derived Gal-1 in CRC by favoring expansion of CD8+CD122+PD-1+ Tregs.
Fig. 3.
Tumor-derived Gal-1 increases the frequency of CD8+CD122+PD-1+ Tregs and accentuates their immunosuppressive capacity in the syngeneic CRC model. (A) Gal-1 expression detected by Western blot in CT26 WT cells stably transfected with shRNA (CT26 Scr) or with Gal-1–specific shRNA (CT26 Gal-1 KD). (B) Tumor growth kinetics in BALB/c mice subcutaneously inoculated with WT, Scr or Gal-1 KD CT26 cells. (C) Analysis of PD-1+ cells within the CD8+CD122+ T cell population isolated from the spleen, DLN, and tumors from mice inoculated with CT26 WT, Scr, or Gal-1 KD cells. (D) Flow cytometry analysis of intracytoplasmic IFN-γ in CD8+- and CD4+-activated T cells. (E) Schematic representation of proliferation assays performed with CFSE-labeled splenocytes. (F and G) Analysis of CD4+ and CD8+T cell proliferation and division index. (H) Schematic representation of suppression assays involving CD8+CD122+PD-1+ Tregs and CD4+ and CD8+ T cells. (I and J) Proliferation and division index of CD4+ and CD8+ responder T cells. (K) Determination of IL-10 by ELISA in coculture supernatants at 96 h. Data presented are mean ± SEM from a representative experiment. n = 5 mice per group. One- or two-way ANOVA, Bonferroni multiple comparison test (A–G), or unpaired Student’s t test (H–I), *P < 0.05; **P < 0.01; ***P < 0.001.
To evaluate the role of tumor-derived Gal-1 on T cell fitness, we isolated splenocytes from mice inoculated with Gal-1 KD, Scr, or WT CT26 cells and analyzed their proliferative response (Fig. 3 E–G). Interestingly, CD4+ T cells obtained from mice bearing Gal-1 KD tumors showed enhanced proliferative response compared to those isolated from mice bearing WT or Scr tumors (Fig. 3F). Moreover, a significant increase in proliferation and division rates was also observed in CD8+ T cells from Gal-1 KD tumor-bearing mice (Fig. 3G).
To study the role of tumor-derived Gal-1 in the immunosuppressive capacity of CD8+CD122+PD-1+ Tregs in the CT26 CRC model, we performed ex vivo immunosuppressive assays using CD8+CD122+PD-1+ Tregs obtained from the spleens of mice inoculated with Gal-1 KD or WT CT26 cells cocultured with splenocytes obtained from healthy WT mice (Fig. 3H). Remarkably, CD8+CD122+PD-1+ Tregs obtained from mice bearing Gal-1 KD CT26 tumors showed impaired T cell inhibitory activity, evidenced by a significant increase in the division rate of CD8+ T cells (Fig. 3 I and J), as compared to CD8+CD122+PD-1+ Tregs isolated from mice bearing WT tumors. This effect was also reflected by a significant drop in IL-10 secretion in supernatants of splenocytes cocultured with CD8+CD122+PD-1+ Tregs isolated from mice bearing Gal-1 KD tumors (Fig. 3K). Thus, tumor-derived Gal-1 increases the frequency of CD8+CD122+PD-1+ Tregs and accentuates their immunosuppressive capacity in the syngeneic CRC model.
Stromal-Derived Gal-1 Impacts the Immunosuppressive Capacity of CD8+ Tregs In Vivo.
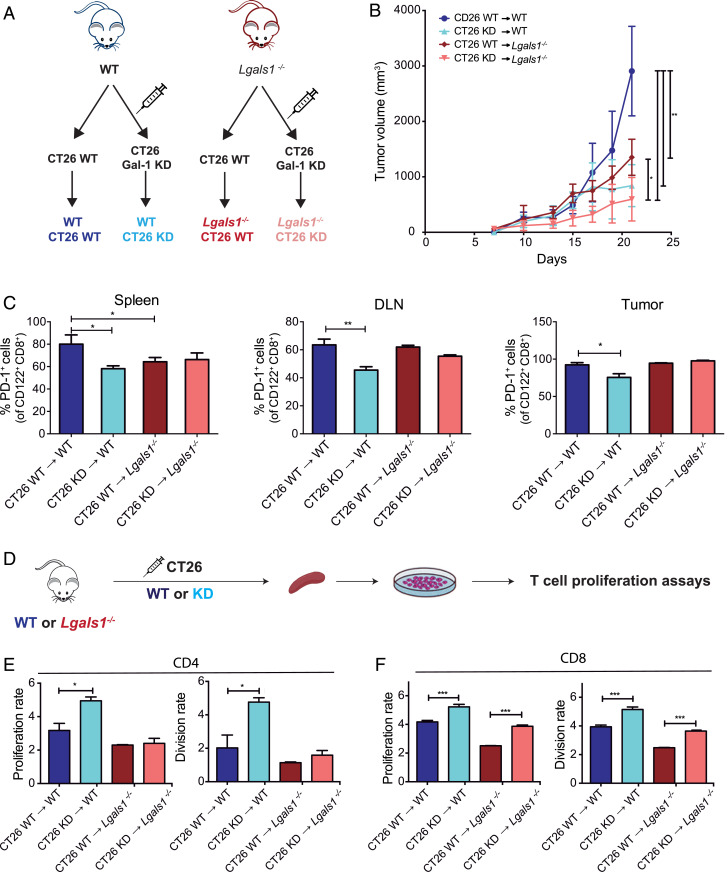
Since Gal-1 is produced not only by tumor cells but also by stromal cells, including fibroblasts, endothelial cells, and immune cells (26, 28, 41), we next sought to evaluate the impact of stromal-derived Gal-1 by analyzing the growth of Gal-1 KD or WT CT26 tumor cells in Lgals1−/− or WT mice (Fig. 4A). Notably, the slowest tumor growth rate was observed in Lgals1−/− animals inoculated with CT26 KD cells (Fig. 4B), highlighting the contribution of both tumor and stromal-derived Gal-1 to this effect.
Fig. 4.
Tumor- and stromal-derived Gal-1 contributes to the immunosuppressive microenvironment in CRC. (A) Experimental design. (B) Kinetics of tumor growth in WT or Lgals1−/− BALB/c mice subcutaneously inoculated with WT or Gal-1 KD CT26 cells. (C) Analysis of PD-1 expression in the spleen, DLN, and tumor-infiltrating CD8+CD122+ T cells detected by flow cytometry. (D) Schematic representation of the proliferation assays of CFSE-labeled splenocytes purified from WT or Lgals1−/− mice bearing CT26 WT or Gal-1 KD tumors. (E and F) Flow cytometry analysis of CD4+ and CD8+ proliferation and division index. Data presented are mean ± SEM from three independent experiments. n = 5 to 6 mice per group. ANOVA, Bonferroni multiple comparison test, *P < 0.05; **P < 0.01; and ***P < 0.001.
When we analyzed the impact of tumor-derived Gal-1 in modulating CD8+ Tregs, we verified a reduction in the proportion of CD8+CD122+PD-1+ T cells in the spleens, DLN, and tumors of animals inoculated with CT26 Gal-1 KD cells, compared to mice receiving CT26 WT cells (Fig. 4C), as previously observed in Fig. 3C. Moreover, when comparing WT and Lgals1−/− mice inoculated with CT26 WT cells, Gal-1–deficient mice showed a lower proportion of CD8+CD122+PD-1+ T cells in the spleen (Fig. 4C), highlighting the impact of endogenous stromal-derived Gal-1 in the modulation of this CD8+ Treg population. Remarkably, CD8+ T cells obtained from the spleen of mice bearing Gal-1 KD CT26 tumors showed increased proliferative capacity, as reflected by higher proliferation and division index, irrespective of the host genotype. These results underscore the relevance of tumor-derived Gal-1 in distant T cell-mediated immunoregulation (Fig. 4 D and E). Notably, stromal Gal-1 did not show any role in proliferation and division rates of CD4+ T cells obtained from mice inoculated with either CT26 Gal-1 KD cells or WT tumors (Fig. 4F). Taken together, these results suggest that both tumor- and stromal-derived Gal-1 affect the rate and proliferative capacity of CD8+ T cells and influence tumor growth.
Prognosis of Human CRC Is Dictated by Gal-1 Expression and a CD8+ Treg Score.
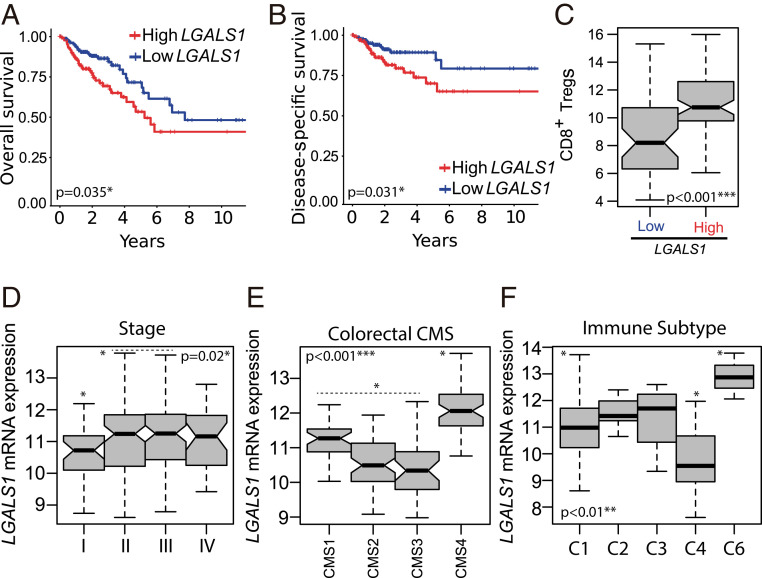
Given the role of Gal-1 in modulating the proliferation and immunosuppressive capacity of CD8+ Tregs in experimental CRC models, we next examined whether high Gal-1 expression could correlate with heightened infiltration of CD8+ Tregs in a COAD patients TCGA dataset. Moreover, we sought to investigate whether these parameters could be associated with poor prognosis and survival. For this, we analyzed Gal-1 gene (LGALS1) expression in these tumors and defined a CD8+ Treg score as a gene signature involving key markers of human CD8+ Tregs (11). This allowed categorization of human tumors according to the StepMiner one-step algorithm (SI Appendix, Fig. S6A). High LGALS1 expression was associated with poor prognosis in human COAD, evidenced by lower overall survival and disease-specific survival (Fig. 5 A and B). Moreover, we found a significant increase of the CD8+ Treg score in COAD samples displaying higher LGALS1 expression (P < 0.001) (Fig. 5C). Next, we evaluated LGALS1 expression during the course of CRC (Fig. 5D). Interestingly, early-stage primary invasive adenocarcinomas showed significantly lower LGALS1 expression than tumor stages II/III (P < 0.02) (Fig. 5D). In addition, we found higher LGALS1 expression on the consensus molecular subtype (CMS)-4 (Fig. 5E) and on the poorly immunogenic immune cancer subtype C6 (Fig. 5F), characterized by a proangiogenic phenotype, high stromal infiltration, and TGF-β activation (45, 46). Conversely, we found no significant association between LGALS1 expression and MSI status or hypermethylation in human COAD samples (SI Appendix, Fig. S6 B and C). However, there was a slight increase in LGALS1 expression in MSI tumors compared to microsatellite stable tumors (SI Appendix, Fig. S6D). Thus, a dynamic signature characterized by high LGALS1 expression and elevated CD8+ Treg score delineates poor prognosis in human CRC.
Fig. 5.
In silico analysis of LGALS1 mRNA expression according to clinical, prognostic, and tumor-immune features of primary invasive human colorectal adenocarcinomas (TCGA-COAD dataset). (A) Kaplan–Meier plot depicting overall survival of two colorectal tumor categories based on LGALS1 expression. (B) Kaplan–Meier plot depicting disease-specific survival of two colorectal tumor categories based on LGALS1 expression. (C) CD8+ Treg score in low and high LGALS1 expression tumor categories. (D–F) LGALS1 expression across colon adenocarcinoma samples categorized according to tumor stages I to IV (D), four consensus molecular subtypes described by Guinney et al. (45) (E), and six immune phenotypes described by Thorsson et al. (46) (F). ANOVA, Bonferroni multiple comparison test, *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Although immunotherapy has achieved long-term durable responses in several malignancies, including melanoma, lung adenocarcinoma, head and neck squamous cell carcinoma (HNSCC), and metastatic renal cancer, in CRC this treatment modality is restricted to patients with dMMR or MSI-H tumors (3). This limitation urges the investigation of additional immune escape mechanisms and the development of novel immunotherapeutic approaches.
Aberrant glycosylation has emerged as a critical hallmark of cellular oncogenesis and tumor progression, since altered glycan structures play relevant roles in cancer signaling, migration, immunomodulation, and metastasis (24, 47). These tumor-associated carbohydrates involve not only the under- or overexpression of naturally occurring glycans, but also the neo-expression of glycan structures (24, 48). In CRC, changes in the tumor N-glycome may include increased sialylation, decreased overall fucosylation, increased β(1−6) branching of N-linked glycans and higher abundance of poly-LacNAc chains, while O-glycans are altered by reduced expression of core 3 and core 4 structures and higher levels of (sialyl) T-antigen (core 1) and (sialyl) Tn-antigen (49, 50). Recent advances in structural and functional characterization of glycans (51) uphold the potential role of altered glycosylation as a promising therapeutic strategy in cancer (52, 53). In this scenario, galectins have emerged as regulatory glyco-checkpoints that control antitumor immunity by inducing T cell exhaustion, limiting T cell survival and enhancing expansion of CD4+ Tregs (38). These extracellular activities are mostly mediated by recognition of permissive glycan structures on the surface of immune cells. Interestingly, previous studies proposed the assembly of a “galectin-glycan lattice” a dynamic spatial array that may control galectin binding, cross-linking, and signaling. These multivalent structures can regulate tumor progression by shaping antitumor immunity, modulating tumor cell invasion and promoting aberrant angiogenesis (25, 38, 54).
In this work, using the colitis-associated CRC model chemically induced by AOM-DSS and the syngeneic CT26 CRC model, we identified a role of Gal-1 in CRC progression and immune escape by modulating the frequency and immunosuppressive capacity of CD8+CD122+PD-1+ Tregs. Through glycosylation-dependent mechanisms, Gal-1 can reprogram the immune landscape of different tumor types, including melanoma (29, 55), lung adenocarcinoma (27, 30), pancreatic adenocarcinoma (28), glioblastoma (56), breast adenocarcinoma (33), and HNSCC (34) by dismantling the effector function of CD4+ and CD8+ T cells, inducing tolerogenic DCs, and favoring the expansion of CD4+CD25+Foxp3+ Tregs (57). Similarly, other members of the galectin family—including Gal-3, Gal-8, and Gal-9—play key roles in the control of CD4+ Tregs (58–60). However, the relevance of Gal-1 in modulating the fate and function of CD8+ Tregs has not yet been explored. Moreover, although Gal-1 is highly up-regulated in the circulation and TME in early- and late-stage CRC patients (40, 41, 61), the contribution of this lectin to CRC progression and underlying immunosuppression is uncertain. We found that both tumor- and stromal-derived Gal-1 accelerate tumor growth and promote immune escape in two different experimental CRC models. Moreover, bioinformatic analysis revealed a strong association between LGALS1 expression, CD8+ Treg score, and poor prognosis in a COAD database, as reflected by lower overall survival and disease-specific survival of human CRC patients. In this regard, recent findings showed that branched N-glycans, which serve as Gal-1–specific ligands, are coopted by CRC tumors to evade immune recognition through inhibition of IFN-γ (42). Further studies are warranted to elucidate whether a Gal-1–N-glycan lattice plays a central role in this immune inhibitory circuitry.
To explore the molecular mechanism underlying the protumoral activity of Gal-1 in CRC, we studied the immunoregulatory function of this lectin in AOM-DSS colitis-associated CRC. The molecular, immunological and mutational landscape of this model recapitulates that associated with the CMS4 of CRC, which, interestingly, exhibited the highest Gal-1 expression among the four CMS analyzed in TCGA-COAD samples (Fig. 5). CMS4 represents poorly immunogenic tumors associated with bad prognosis and characterized by high stromal infiltration, TGF-β activation, and angiogenesis (45). Accordingly, high Gal-1 expression also correlated with the poorly immunogenic immune cancer subtype C6, which is portrayed by increased TGF-β signaling, negative prognosis, and tumor-promoting immune infiltrate (46).
Notably, Lgals1−/− mice developed significantly fewer tumors than WT mice in the AOM-DSS CRC model, indicating a critical role for Gal-1 in CACRC progression. Since Gal-1 modulates the fate and function of CD4+ Tregs in different physiologic and pathologic settings, including pregnancy (62), autoimmune inflammation (44, 63), and breast cancer metastasis (33), we speculated that this lectin could impact the Treg cell compartment in CRC. Whereas Gal-1 targeting did not induce any change in the proportion of CD4+ Tregs in the tumor, DLN, and spleen of CRC-bearing mice and no alterations were detected in inducible CD8+CD25+Foxp3+ or CD8+CD28− Tregs, we found a selective modulation of CD8+CD122+PD-1+ Tregs in Lgals1−/− compared to WT mice.
To further analyze whether induction of CD8+CD122+PD-1+ Tregs is tumor-dependent, we further explored the impact of Gal-1 deficiency in the proportion of CD4+ and CD8+ Treg cell populations in healthy animals. Notably, analysis of lymph nodes of healthy Lgals1−/− mice showed a statistically significant reduction of CD8+CD122+PD-1+ T cells compared to WT mice, highlighting the relevance of endogenous Gal-1 in shaping the CD8+ Treg compartment even in the absence of pathologic stimuli. Accordingly, CD8+CD122+PD-1+ Tregs obtained from Lgals1−/− mice showed lower immunosuppressive capacity compared to CD8+CD122+PD-1+ Tregs obtained from WT mice. These results suggest that, in addition to other immunoregulatory effects, Gal-1 can also reprogram the function of CD8+CD122+PD-1+ Tregs. Notably, an elevated CD8+ Treg score was associated with high LGALS1 expression, independently of the MSI status in a TCGA-COAD database (Fig. 5), highlighting the connection between Gal-1 and CD8 Tregs and their correlation with poor prognosis in CRC patients.
To discern the effect of stromal- and tumor-derived Gal-1 in CRC biology and immunosuppressive potential, we explored the role of this lectin in the syngeneic CT26 CRC model. Silencing of tumoral Gal-1 suppressed tumor growth and reduced expansion of CD8+CD122+PD-1+ Tregs. In this regard, Gao et al. (64) recently reported CD8+ T cell-dependent inhibition of CRC metastasis using PD-1 blocking agents by reducing Treg cell differentiation and increasing IFN-γ production. In addition, Gal-1 silencing increased the proliferative capacity of CD4+ and CD8+ splenic T cells, thus confirming the role of tumor-derived Gal-1 in CRC-driven immune escape. Furthermore, splenocytes cocultured with CD8+CD122+PD-1+ Tregs obtained from mice bearing CT26 Gal-1 KD tumors secreted lower amounts of IL-10, as previously observed in other tumor types (27, 30), thus substantiating the immunosuppressive capacity of this lectin. Thus, Gal-1 secreted by CRC contributes to immune escape through distinct mechanisms, including selective expansion of the CD8+CD122+PD-1+ Tregs, enhancement of their immunosuppressive capacity via IL-10, and inhibition of CD4+ and CD8+ T cell proliferation. Since Gal-1 interacts with the T cell receptor (TCR) complex and CD45 and influences downstream signaling pathways on effector T cells (65, 66), further studies should examine whether tumor Gal-1 may differentially control TCR signaling and CD45 phosphatase activity in CD8+ regulatory versus effector T cells. In this context, Gal-3 has been shown to bind to immune checkpoint molecules, including CTLA-4 and lymphocyte antigen-3 (LAG-3) on CD8+ T cells, thus regulating their signaling threshold, activation and differentiation (67, 68). In this regard, the precise mechanisms implicated in Gal-1–driven induction of CD8+ Tregs and the control of their functionality should be explored in detail, including a full characterization of the glycosylation signature of these cells as well as the transcriptional and epigenetic programs underlying these immunoregulatory effects.
Finally, in addition to the contribution of tumoral Gal-1, our findings also demonstrate the importance of stromal-derived Gal-1 by comparing injection of CT26 WT and Gal-1 KD cells into WT or Lgals1−/− mice. While tumoral Gal-1 showed a more pronounced effect in regulating the proportion of CD8+ Tregs, both tumor- and stromal-derived Gal-1 influenced the proliferation of CD8+ T cells and the immunosuppressive capacity of CD8+CD122+PD-1+ Tregs. Lower Gal-1 expression, whether of stromal or tumor origin, resulted in reduced tumor growth. Accordingly, Lgals1−/− mice bearing Gal-1–silenced CT26 tumors showed the lowest tumor growth rate among the different experimental conditions explored.
In the past years, Gal-1 has emerged as a multifunctional mediator of cancer-driven immunosuppression, a reliable biomarker of tumor prognosis, and a potential therapeutic target in different cancer types (35). In this study we identified a role for Gal-1 in CRC by shaping antitumor immunity through modulation of the frequency and immunosuppressive function of CD8+CD122+PD-1+ Tregs. Moreover, bioinformatic analysis of TCGA-COAD datasets revealed strong association between Gal-1 expression, CD8+ Treg score and the prognosis of CRC patients. Thus, therapeutic targeting of Gal-1 might contribute to overcome CRC-driven immunosuppression and improve the clinical outcome of CRC patients. Further exploration of combinatorial strategies involving Gal-1 blockade together with immunotherapy, chemotherapy, or antiangiogenic therapy might help increase the clinical effectiveness of these treatments. In this regard, several Gal-1 inhibitors have already been developed, including small-molecule glycan inhibitors (52, 69, 70), antagonistic peptides (32, 71), and specific neutralizing mAb (31, 72).
Materials and Methods
Cell Lines and Cell Culture.
The mouse colon carcinoma CT26 cell line was purchased from the American Type Culture Collection (ATCC CRL-2638) and maintained in RPMI 1640 (Sigma Aldrich) supplemented with 10% fetal bovine serum (FBS, Biochrom AG) and 1% penicillin-streptomycin (PenStrep, Gibco). Gal-1–specific shRNA was designed as previously described (31), cloned into the pSIREN-RetroQ vector, and delivered using lipofectamine (Invitrogen). After infection, cells were subjected to puromycin selection (1 µg/mL) and further cultured in the presence of 0.1 µg/mL of this antibiotic.
Annexin V Binding Assay.
CT26 cells (1 × 106 cells/mL) were incubated in RPMI medium supplemented with 10% FBS and antibiotics. Apoptotic cells were identified by staining with APC-conjugated annexin V (BD Biosciences) and propidium iodide (PI). Cell death was determined as percent of annexin V+PI+ cells.
Tumor Cell Proliferation Assay.
CT26 cells (50,000 cells/mL) were cultured in a 24-well plate in RPMI medium supplemented with 10% FBS and antibiotics. Cells were sequentially harvested with Triple Enzyme express (Gibco), washed, centrifuged, and resuspended. Live cells were counted for 7 d using a Countess automated cell counter (Thermo Fisher).
Mouse Strains.
Mice were bred at the animal facility of the Instituto de Biología y Medicina Experimental (IBYME) according to NIH guidelines. Gal-1–deficient (Lgals1−/−) C57BL/6 mice were kindly provided by Francoise Poirier, Jacques Monod Institute, Paris, France. BALB/c WT mice were obtained from the Animal House of the School of Exact and Natural Sciences, University of Buenos Aires (Argentina). Lgals1−/− BALB/c mice were previously generated by crossing C57BL/6 Lgals1−/− mice into BALB/c background for N9 generations. Purity of the BALB/c Lgals1−/− strain was confirmed by analyzing a panel of specific microsatellites as short tandem repeats or simple sequence-length polymorphism at Instituto Nacional de Tecnología Agropecuaria, Buenos Aires, Argentina. All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee of IBYME.
Cell Isolation and Flow Cytometry.
After killing, tumors, spleen, and lymph nodes were extracted and transferred to cold RPMI 1640 culture medium (Gibco). Next, organs were homogenized in RPMI 1640 culture medium supplemented with 10% FBS and antibiotics (2 U/mL penicillin, 2 μg/mL streptomycin, and 5 ng/mL amphotericin B). Cells were then harvested by centrifugation at 1,500 rpm for 5 min. To enrich tumor-infiltrating lymphocytes, a Percoll (Sigma-Aldrich) density gradient was performed. Cells were stained with the corresponding mAbs against CD3, CD4, CD8, CD25, CD28, CD44, CD62L, CD122, Foxp3, and PD-1 (BD Biosciences). Isotype-matched irrelevant mAbs were used as negative controls. Intracytoplasmic IFN-γ and Foxp3 staining was performed using fixation and permeabilization buffers according to manufacturer’s instructions (eBioscience). Acquisition of cell events was performed on a FACSCanto II flow cytometer (BD Biosciences) and data were analyzed using FlowJo software.
Syngeneic CT26 CRC Model.
WT, Gal-1 KD, or scramble CT26 cells were injected into 6- to 8-wk-old BALB/c mice (5 × 105 cells in 100 µL PBS, subcutaneously, right flank). Tumor volume was calculated as follows: length × (width)2 × 0.5 (73). After 3 wk, or when primary tumors reached an average volume of 1,000 mm3, animals were killed, tumors were surgically removed and weighted.
CACRC Model.
Eight- to 10-wk-old C57BL/6 mice received an intraperitoneal injection (10 mg/kg body weight) of AOM (Sigma- Aldrich) followed by 5 d of DSS treatment (2.5%) and 14 d of recovery, as described previously (74). This initial 19-d cycle was repeated three times. Body weight, presence of rectal bleeding, and stool consistency were assessed three times per week, recorded, and averaged to generate a clinical score (75). After the fourth DSS cycle, mice were killed, colons were extracted, and the number of intestinal tumors was assessed.
Immunoblotting.
Immunoblotting was performed essentially as described previously (33). In brief, equal amounts of protein were resolved by SDS/PAGE and blotted onto nitrocellulose membranes (GE Healthcare). After blocking, membranes were incubated with rabbit antiactin mAb (I-19; Santa Cruz Biotechnology) and rabbit anti–Gal-1 polyclonal IgG, obtained as previously described (31). Membranes were then incubated with horseradish peroxidase (HRP)-labeled anti-rabbit secondary antibody (Bio-Rad) and developed using ECL Prime Western Blotting Detection Reagent (Amersham Biosciences). Protein bands were analyzed with ImageJ software.
Cell Sorting of CD8+ Tregs.
After killing, spleens were removed and cells were isolated after by mechanical disaggregation and filtered, generating unicellular suspensions. Erythrocytes were lysed by incubation in ACK lysis buffer (NH4Cl 0.15 M, KHCO3 10 mM, Na2EDTA 0.1 mM, pH 7.2 to 7.4) for 3 min. After centrifugation, CD8+ T cells were purified using Negative Selection DynaBeads Untouched Mouse CD8 cells kit (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s instructions. CD8+ T cells were resuspended in PBS buffer supplemented with 1 mM EDTA, 25 mM Hepes pH 7.0, and 1% FBS, at a concentration of 100 × 106 cells/mL. Cells were incubated during 30 min on ice with anti–CD8-APC (BD Biosciences), anti–CD3-PE-Cy5 (BD Biosciences), anti–CD122-FITC (eBiosciences), and anti–PD-1-PE (BioLegend) labeled mAb. After 30 min, cells were washed, centrifuged, and resuspended in the same buffer at a concentration of 10 to 25 × 106 cells/mL. Finally, CD8+CD122+PD-1+ T cells were sorted on a FACS Aria II (BD) flow cytometer.
Lymphocyte Proliferation Assay.
After killing, spleen, and lymph nodes were extracted and immediately transferred to cold RPMI medium 1640 (Gibco). Tissues were homogenized, filtered, and cells were collected by centrifugation at 1,500 rpm for 8 min. Splenocytes were resuspended in PBS and labeled with 1 μM carboxyfluorescein N-succinimidyl ester (CFSE) for 7 min at 37 °C. Reaction was stopped and cells were collected by centrifugation, washed, and resuspended in RPMI medium 1640 supplemented with 10% FBS. Cells were transferred to U-bottom P-96 culture plates. To stimulate lymphocyte proliferation, plates were previously coated with 100 μL of 1 μg/mL anti-CD3 mAb (BioXCell, 2 h at 37 °C). Cells were incubated at 37 °C and 5% CO2 during 96 h. Then, cells were collected, washed, and stained in PBS pH = 7.4 supplemented with 0.5% BSA with anti–CD4-PE-Cy5 (BD Biosciences) and anti–CD8-PE (30 min at 4 °C) antibodies (eBiosciences). Proliferation of responder cells was determined by CFSE dilution and analyzed by flow cytometry. Accordingly, division rate was calculated as the number of total divisions per number of total original cells and the proliferation rate was determined as the number of total divisions per number of divided original cells.
Suppression Assay.
CD3+CD8+CD122+PD-1+ T cells were sorted as previously described from the spleens of WT or Lgals1−/− mice. Responder T cells were obtained from spleen homogenates from WT mice, resuspended in PBS, labeled with 1 μM CFSE during 7 min at 37 °C, and cocultured with WT or Lgals1−/− CD8+ Tregs in 10% FBS RPMI (Treg:responder T cells ratio 1:1, 1:2, and 1:4). Controls included cultures performed in the absence of Tregs. After 96 h at 37 °C, cells were harvested, washed, and stained in PBS buffer pH = 7.4 supplemented with 0.5% BSA with anti–CD4-PE-Cy-5 and (BD Biosciences) anti–CD8-PE antibodies (eBiosciences) for 30 min at 4 °C. Proliferation of responder T cells was analyzed in a FACS Aria II (BD) flow cytometer.
ELISA.
Mouse IL-10 was determined in supernatants using a specific ELISA kit (BD Biosciences) according to the manufacturer’s instructions and relativized to total protein.
Bioinformatics Analysis.
In silico analysis of LGALS1 mRNA in association to a defined CD8+ Treg score and clinical data were performed in tumor and immune cell populations using data obtained from a TCGA-COAD cohort. Clinical data and preprocessed RNA-sequencing normalized mRNA data of 259 primary invasive colon adenocarcinomas were obtained from the University of California, Santa Cruz (UCSC) Xena browser (https://xenabrowser.net/). Univariate and bivariate expression analysis among cases were performed with the R software. To further explore the prognostic value of the gene-expression signature driven by LGALS1 (high or low), we evaluated the colon adenocarcinomas dataset. Briefly, cancer patients with gene-expression signature and follow-up data were divided into two subgroups for the survival analyses of LGALS1 expression (low Gal-1 or high Gal-1). Discretization of the LGALS1 gene expression data into low- or high-expression levels was performed according the StepMiner one-step algorithm of their respective profiles (http://genedesk.ucsd.edu/home/public/StepMiner/). These two groups were then compared based on the overall survival and disease-specific survival (Kaplan–Meier curves and log-rank test) using the Survival and Survminer R packages. Cancer-associated immune subtypes (C1 to C6) were directly retrieved from the UCSC Xena resource and CD8+ Treg score was computed as a weighted sum of the CD8+ Treg signature of the following algebraic expression: CD8A + CXCR3 + IL2RA + FOXP3 − CD28 − CD4. Curated molecular subtypes and associated categories (consensus molecular subtypes, molecular subtypes, hypermethylation categories, and MSI status) were retrieved using the TCGA biolinks R/Bioconductor package (76).
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism 5.0 Software (GraphPad). Two-way ANOVA and Bonferroni posttests were used for multiple comparisons. Two groups were compared with Student’s t test for unpaired data. P values of 0.05 or less were considered significant.
Supplementary Material
Acknowledgments
We thank Pablo Hockl for support and Florencia Moses for assistance in development of Lgals1−/− BALB/c mice. A.J.C., A.G.B., M.C.A., M.S., D.O.C., K.V.M. and G.A.R. are members of the Research Career of The Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). M.L.G. and A.M.C. received doctoral fellowships from CONICET. This work was supported by Argentinean Agency for Promotion of Science and Technology Grants PICT 2017-0494 (to G.A.R.) and PICT 2015-0564 (to K.V.M.). We also thank the Sales, Bunge & Born, Baron and Richard Lounsbery foundations for support; and the Ferioli, Ostry, and Caraballo families for generous donations.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102950118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Bray F., et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Siegel R., Ma J., Zou Z., Jemal A., Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Ganesh K., et al., Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 16, 361–375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian S. L., Drake C. G., Pardoll D. M., Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plitas G., Rudensky A. Y., Regulatory T cells: Differentiation and function. Cancer Immunol. Res. 4, 721–725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucca L. E., Dominguez-Villar M., Modulation of regulatory T cell function and stability by co-inhibitory receptors. Nat. Rev. Immunol. 20, 680–693 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Flippe L., Bézie S., Anegon I., Guillonneau C., Future prospects for CD8+ regulatory T cells in immune tolerance. Immunol. Rev. 292, 209–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y., et al., Recent advances in CD8+ regulatory T cell research. Oncol. Lett. 15, 8187–8194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gershon R. K., Kondo K., Cell interactions in the induction of tolerance: The role of thymic lymphocytes. Immunology 18, 723–737 (1970). [PMC free article] [PubMed] [Google Scholar]
- 10.Mathews D. V., et al., CD122 signaling in CD8+ memory T cells drives costimulation-independent rejection. J. Clin. Invest. 128, 4557–4572 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang X., Kumar V., Advances in the study of CD8+ regulatory T cells. Crit. Rev. Immunol. 39, 409–421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosmi L., et al., Human CD8+CD25+ thymocytes share phenotypic and functional features with CD4+CD25+ regulatory thymocytes. Blood 102, 4107–4114 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Filaci G., et al., CD8+ CD28- T regulatory lymphocytes inhibiting T cell proliferative and cytotoxic functions infiltrate human cancers. J. Immunol. 179, 4323–4334 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Vuddamalay Y., et al., Mouse and human CD8(+) CD28(low) regulatory T lymphocytes differentiate in the thymus. Immunology 148, 187–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colovai A. I., et al., Induction of xenoreactive CD4+ T-cell anergy by suppressor CD8+CD28- T cells. Transplantation 69, 1304–1310 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Endharti A. T., et al., Cutting edge: CD8+CD122+ regulatory T cells produce IL-10 to suppress IFN-γ production and proliferation of CD8+ T cells. J. Immunol. 175, 7093–7097 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Akane K., Kojima S., Mak T. W., Shiku H., Suzuki H., CD8+CD122+CD49dlow regulatory T cells maintain T-cell homeostasis by killing activated T cells via Fas/FasL-mediated cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 113, 2460–2465 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai H., et al., Cutting edge: Programmed death-1 defines CD8+CD122+ T cells as regulatory versus memory T cells. J. Immunol. 185, 803–807 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Wang L.-X., et al., CD122+CD8+ Treg suppress vaccine-induced antitumor immune responses in lymphodepleted mice. Eur. J. Immunol. 40, 1375–1385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S., et al., A naturally occurring CD8(+)CD122(+) T-cell subset as a memory-like Treg family. Cell. Mol. Immunol. 11, 326–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rifa’i M., et al., CD8+CD122+ regulatory T cells recognize activated T cells via conventional MHC class I-alphabetaTCR interaction and become IL-10-producing active regulatory cells. Int. Immunol. 20, 937–947 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Villarreal D. O., et al., Targeting of CD122 enhances antitumor immunity by altering the tumor immune environment. Oncotarget 8, 109151–109160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Z., et al., Human CD8+CXCR3+ T cells have the same function as murine CD8+CD122+ Treg. Eur. J. Immunol. 39, 2106–2119 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Pinho S. S., Reis C. A., Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Girotti M. R., Salatino M., Dalotto-Moreno T., Rabinovich G. A., Sweetening the hallmarks of cancer: Galectins as multifunctional mediators of tumor progression. J. Exp. Med. 217, e20182041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cutine A. M., et al., Tissue-specific control of galectin-1-driven circuits during inflammatory responses. Glycobiology, 10.1093/glycob/cwab007 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Banh A., et al., Tumor galectin-1 mediates tumor growth and metastasis through regulation of T-cell apoptosis. Cancer Res. 71, 4423–4431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orozco C. A., et al., Targeting galectin-1 inhibits pancreatic cancer progression by modulating tumor-stroma crosstalk. Proc. Natl. Acad. Sci. U.S.A. 115, E3769–E3778 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubinstein N., et al., Targeted inhibition of galectin-1 gene expression in tumor cells results in heightened T cell-mediated rejection; A potential mechanism of tumor-immune privilege. Cancer Cell 5, 241–251 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Croci D. O., et al., Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell 156, 744–758 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Croci D. O., et al., Disrupting galectin-1 interactions with N-glycans suppresses hypoxia-driven angiogenesis and tumorigenesis in Kaposi’s sarcoma. J. Exp. Med. 209, 1985–2000 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thijssen V. L. J. L., et al., Galectin-1 is essential in tumor angiogenesis and is a target for antiangiogenesis therapy. Proc. Natl. Acad. Sci. U.S.A. 103, 15975–15980 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalotto-Moreno T., et al., Targeting galectin-1 overcomes breast cancer-associated immunosuppression and prevents metastatic disease. Cancer Res. 73, 1107–1117 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Nambiar D. K., et al., Galectin-1-driven T cell exclusion in the tumor endothelium promotes immunotherapy resistance. J. Clin. Invest. 129, 5553–5567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro P., Martínez-Bosch N., Blidner A. G., Rabinovich G. A., Impact of galectins in resistance to anticancer therapies. Clin. Cancer Res. 26, 6086–6101 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Ilarregui J. M., et al., Tolerogenic signals delivered by dendritic cells to T cells through a galectin-1-driven immunoregulatory circuit involving interleukin 27 and interleukin 10. Nat. Immunol. 10, 981–991 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Garín M. I., et al., Galectin-1: A key effector of regulation mediated by CD4+CD25+ T cells. Blood 109, 2058–2065 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Blidner A. G., Méndez-Huergo S. P., Cagnoni A. J., Rabinovich G. A., Re-wiring regulatory cell networks in immunity by galectin-glycan interactions. FEBS Lett. 589, 3407–3418 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Barrow H., Rhodes J. M., Yu L.-G., The role of galectins in colorectal cancer progression. Int. J. Cancer 129, 1–8 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Gopalan V., et al., The expression profiles of the galectin gene family in colorectal adenocarcinomas. Hum. Pathol. 53, 105–113 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Bacigalupo M. L., Carabias P., Troncoso M. F., Contribution of galectin-1, a glycan-binding protein, to gastrointestinal tumor progression. World J. Gastroenterol. 23, 5266–5281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva M. C., et al., Glycans as immune checkpoints: Removal of branched N-glycans enhances immune recognition preventing cancer progression. Cancer Immunol. Res. 8, 1407–1425 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Beaugerie L., Itzkowitz S. H., Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 372, 1441–1452 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Toscano M. A., et al., Galectin-1 suppresses autoimmune retinal disease by promoting concomitant Th2- and T regulatory-mediated anti-inflammatory responses. J. Immunol. 176, 6323–6332 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Guinney J., et al., The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorsson V.et al.; Cancer Genome Atlas Research Network , The immune landscape of cancer. Immunity 48, 812–830.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magalhães A., Duarte H. O., Reis C. A., Aberrant glycosylation in cancer: A novel molecular mechanism controlling metastasis. Cancer Cell 31, 733–735 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Rodrigues Mantuano N., Natoli M., Zippelius A., Läubli H., Tumor-associated carbohydrates and immunomodulatory lectins as targets for cancer immunotherapy. J. Immunother. Cancer 8, e001222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holst S., Wuhrer M., Rombouts Y., Glycosylation characteristics of colorectal cancer. Adv. Cancer Res. 126, 203–256 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Boyaval F., et al., N-glycomic signature of stage II colorectal cancer and its association with the tumor microenvironment. Mol. Cell. Proteomics 20, 100057 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kailemia M. J., et al., Recent advances in the mass spectrometry methods for glycomics and cancer. Anal. Chem. 90, 208–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cagnoni A. J., Pérez Sáez J. M., Rabinovich G. A., Mariño K. V., Turning-off signaling by siglecs, selectins, and galectins: Chemical inhibition of glycan-dependent interactions in cancer. Front. Oncol. 6, 109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mereiter S., Balmaña M., Campos D., Gomes J., Reis C. A., Glycosylation in the era of cancer-targeted therapy: Where are we heading? Cancer Cell 36, 6–16 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Nabi I. R., Shankar J., Dennis J. W., The galectin lattice at a glance. J. Cell Sci. 128, 2213–2219 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Yazawa E. M., et al., Melanoma cell galectin-1 ligands functionally correlate with malignant potential. J. Invest. Dermatol. 135, 1849–1862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baker G. J., et al., Natural killer cells eradicate galectin-1-deficient glioma in the absence of adaptive immunity. Cancer Res. 74, 5079–5090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabinovich G. A., Conejo-García J. R., Shaping the immune landscape in cancer by galectin-driven regulatory pathways. J. Mol. Biol. 428, 3266–3281 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Fermino M. L., et al., Galectin-3 negatively regulates the frequency and function of CD4(+) CD25(+) Foxp3(+) regulatory T cells and influences the course of Leishmania major infection. Eur. J. Immunol. 43, 1806–1817 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Sampson J. F., Suryawanshi A., Chen W.-S., Rabinovich G. A., Panjwani N., Galectin-8 promotes regulatory T-cell differentiation by modulating IL-2 and TGFβ signaling. Immunol. Cell Biol. 94, 213–219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu C., et al., Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity 41, 270–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hittelet A., et al., Upregulation of galectins-1 and -3 in human colon cancer and their role in regulating cell migration. Int. J. Cancer 103, 370–379 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Blois S. M., et al., A pivotal role for galectin-1 in fetomaternal tolerance. Nat. Med. 13, 1450–1457 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Martínez Allo V. C., et al., Suppression of age-related salivary gland autoimmunity by glycosylation-dependent galectin-1-driven immune inhibitory circuits. Proc. Natl. Acad. Sci. U.S.A. 117, 6630–6639 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao C. E., Zhang M., Song Q., Dong J., PD-1 inhibitors dependent CD8+ T cells inhibit mouse colon cancer cell metastasis. OncoTargets Ther. 12, 6961–6971 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chung C. D., Patel V. P., Moran M., Lewis L. A., Miceli M. C., Galectin-1 induces partial TCR ζ-chain phosphorylation and antagonizes processive TCR signal transduction. J. Immunol. 165, 3722–3729 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Earl L. A., Bi S., Baum L. G., N- and O-glycans modulate galectin-1 binding, CD45 signaling, and T cell death. J. Biol. Chem. 285, 2232–2244 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lau K. S., et al., Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Kouo T., et al., Galectin-3 shapes antitumor immune responses by suppressing CD8+ T cells via LAG-3 and inhibiting expansion of plasmacytoid dendritic cells. Cancer Immunol. Res. 3, 412–423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giguère D., et al., Synthesis of stable and selective inhibitors of human galectins-1 and -3. Bioorg. Med. Chem. 16, 7811–7823 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Pal K. B., Mahanti M., Leffler H., Nilsson U. J., A galactoside-binding protein tricked into binding unnatural pyranose derivatives: 3-deoxy-3-methyl-gulosides selectively inhibit galectin-1. Int. J. Mol. Sci. 20, 3786–3808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Astorgues-Xerri L., et al., OTX008, a selective small-molecule inhibitor of galectin-1, downregulates cancer cell proliferation, invasion and tumour angiogenesis. Eur. J. Cancer 50, 2463–2477 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Pérez Sáez J. M., et al., Characterization of a neutralizing anti-human galectin-1 monoclonal antibody with angioregulatory and immunomodulatory activities. Angiogenesis 24, 1–5 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Chou L.-C., et al., The synthesized 2-(2-fluorophenyl)-6,7-methylenedioxyquinolin-4-one (CHM-1) promoted G2/M arrest through inhibition of CDK1 and induced apoptosis through the mitochondrial-dependent pathway in CT-26 murine colorectal adenocarcinoma cells. J. Gastroenterol. 44, 1055–1063 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Allen I. C., et al., The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207, 1045–1056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siegmund B., et al., Adenosine kinase inhibitor GP515 improves experimental colitis in mice. J. Pharmacol. Exp. Ther. 296, 99–105 (2001). [PubMed] [Google Scholar]
- 76.Mounir M., et al., New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 15, e1006701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.