Significance
HER2+ breast cancers (BrCs) are heterogeneous, but they are treated as a single type. Using a mouse model with Erbb2 (the rodent homolog of HER2)-induced BrC and cell lineage-tracing capacity, we found that ERα+Erbb2+ cancer cells proliferate slowly and are nonmetastatic but then progressively lose ERα expression to become fast proliferating and highly metastatic ERα−Erbb2+ cancer cells. ERα−Erbb2+ cancer cells with an ERα− origin proliferate fast, but they metastasize weakly. These findings suggest: 1) ERα expression should be preserved in ERα+HER2+ BrCs to restrict growth and metastasis; 2) ERα−HER2+ BrCs contain a highly metastatic subtype with an ERα+ origin and a weakly metastatic subtype with an ERα− origin, indicating a future need to identify and differentially treat these two subtypes.
Keywords: cell lineage tracing, estrogen receptor, HER2+ breast cancer, cancer cell origin, metastasis
Abstract
HER2-positive (HER2+) breast cancers (BrCs) contain approximately equal numbers of ERα+HER2+ and ERα−HER2+ cases. An enduring obstacle is the unclear cell lineage-related characteristics of these BrCs. Although ERα+HER2+ BrCs could lose ERα to become ERα−HER2+ BrCs, direct evidence is missing. To investigate ERα dependencies and their implications during BrC growth and metastasis, we generated ERαCreRFP-T mice that produce an RFP-marked ERα+ mammary gland epithelial cell (MGEC) lineage. RCAS virus-mediated expression of Erbb2, a rodent Her2 homolog, first produced comparable numbers of ERα+RFP+Erbb2+ and ERα−RFP−Erbb2+ MGECs. Early hyperplasia developed mostly from ERα+RFP+Erbb2+ cells and ERα−RFP−Erbb2+ cells in these lesions were rare. The subsequently developed ductal carcinomas in situ had 64% slow-proliferating ERα+RFP+Erbb2+ cells, 15% fast-proliferating ERα−RFP+Erbb2+ cells derived from ERα+RFP+Erbb2+ cells, and 20% fast-proliferating ERα−RFP−Erbb2+ cells. The advanced tumors had mostly ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ cells and only a very small population of ERα+RFP+Erbb2+ cells. In ERα−RFP+Erbb2+ cells, GATA3 and FoxA1 decreased expression and ERα promoter regions became methylated, consistent with the loss of ERα expression. Lung metastases consisted of mostly ERα−RFP+Erbb2+ cells, a few ERα−RFP−Erbb2+ cells, and no ERα+RFP+Erbb2+ cells. The high metastatic capacity of ERα−RFP+Erbb2+ cells was associated with ERK1/2 activation. These results show that the slow-proliferating, nonmetastatic ERα+RFP+Erbb2+ cells progressively lose ERα during tumorigenesis to become fast-proliferating, highly metastatic ERα−RFP+Erbb2+ cells. The ERα−Erbb2+ BrCs with an ERα+ origin are more aggressive than those ERα−Erbb2+ BrCs with an ERα− origin, and thus, they should be distinguished and treated differently in the future.
The mammary gland (MG) epithelium contains both ERα+ and ERα− luminal epithelial cells (1). Breast cancers (BrCs) may arise from either ERα+ or ERα− MG epithelial cells (MGECs). BrCs are heterogeneous and can be roughly grouped into ERα+, HER2+, and triple negative BrCs. About 70% of BrCs belong to the ERα+ group, which is associated with a relatively good prognosis, and about 20% fall into the HER2+ group with a much worse prognosis. About 50% of HER2+ BrCs express ERα (2), so HER2+ BrCs are accordingly designated as ERα+HER2+ and ERα−HER2+ BrCs. In ERα+HER2+ cancers, the cross-talk between ERα and HER2 signaling pathways and loss of ERα partially account for resistance to endocrine therapy (3, 4). ERα+HER2+ cancers exhibit a wide range of disease relapse time, metastatic potential, and responsiveness to anti-HER2 treatment, while ERα−HER2+ cancers generally are more malignant with earlier relapse, stronger metastatic potential, and much worse prognosis (5–13). ERα and HER2 expression vary during tumor progression, and patients can have both ERα+ primary tumors as well as ERα− metastases (3, 14, 15). In some patients with recurrent BrCs, the ratios of ERα+ to ERα− cancer cells may be reduced (16). These observations suggest that some ERα+HER2+ BrCs may progress to ERα−HER2+ BrCs; yet, direct evidence is missing since cell lineage tracing in people is unethical.
To elucidate the relationships among the HER2+ BrC subtypes, we set out to answer three unresolved biomedical questions: First, do ERα+HER2+ cancers lose ERα to become the more aggressive ERα−HER2+ cancers? Second, do ERα−HER2+ BrCs originate directly from ERα− MGECs or indirectly from ERα+HER2+ cancer cells? Third, are ERα−HER2+ cancers derived from preexisting ERα+HER2+ cancers and do ERα−HER2+ cancers that stem from ERα− MGECs have different cell proliferation rates and metastatic capabilities? To answer these important questions, we developed a trigenic mouse model that allows for in vivo tracing of the ERα+ and ERα− MGEC lineages during tumorigenesis and metastasis. In this model, breast carcinogenesis was induced by Erbb2 (the rodent homolog of HER2) expression in both ERα+ and ERα− MGECs in adulthood, and all tumor cells arising from the ERα+ MGEC lineage were traced with red fluorescent protein (RFP) expression during the entire process of cancer initiation, progression, and metastasis. ERα expression history, cell proliferation rate, and metastatic capability were compared and characterized among different subtypes of BrC cells.
Results and Discussion
In Vivo Cell Lineage Tracing Revealed a Progressive Loss of ERα Expression in Erbb2+ Tumor Cells during MG Tumor Growth and Progression.
Heterozygous trigenic ERCreRFP-T mice were generated by cross-breeding ERα-F2A-Cre mice with Cre expression in ERα-positive cells, Rosa26-LoxP-STOP-LoxP-tdRFP mice with a Cre-activated RFP expression cassette, and mouse mammary tumor virus long terminal repeat-tumor virus A (MMTV-TVA) mice (17–19) (SI Appendix, Fig. S1A). Replication-competent avian leukosis virus long terminal repeat with splice acceptor-Erbb2HA (RCAS-Erbb2HA) virus was introduced into the MG ductal lumens of 9-wk-old ERCreRFP-T mice as described previously (19, 20). In ERCreRFP-T mice, Cre expressed in ERα+ MGECs activates RFP expression driven by the Rosa26 locus and is used as a lineage-tracing marker. No ERα−RFP+ MGECs were detected, indicating that normal ERα+ MGECs always maintain ERα expression (SI Appendix, Fig. S1B). TVA, a receptor for RCAS avian virus, was expressed in both ERα+RFP+ and ERα−RFP− MGECs, allowing the RCAS-Erbb2HA virus to infect both types of MGECs. The RCAS-Erbb2HA virus mediates stable expression of the hemagglutinin antibody epitope (HA)-tagged rodent Erbb2 active protein (21). Palpable MG tumors were detected 14 wk after RCAS-Erbb2HA virus injection into the MG lumens of ERCreRFP-T mice, and these tumors grew rapidly (SI Appendix, Fig. S2A). Immunohistochemistry (IHC) staining for HA-tagged Erbb2 protein identified Erbb2+ tumor cells in ductal carcinoma in situ (DCIS) lesions at week 13, and Erbb2+ poorly differentiated tumor cells in advanced large MG tumors at week 27. Luminal epithelial markers such as K8 and E-cadherin were expressed in these Erbb2+ tumor cells at both stages. Progesterone receptor (PR), which usually is coexpressed with ERα in MGECs, was detected in DCIS cells at week 13, but not in most of the Erbb2+ tumor cells at week 27. K14, a myoepithelial marker, was observed only in Erbb2− myoepithelial cells. Vimentin, a mesenchymal cell marker, was not detected in any of the Erbb2+ tumor cells (SI Appendix, Fig. S2B). These results suggest that these MG tumors emerged as highly differentiated DCIS lesions and then progressed into poorly differentiated luminal-type BrCs.
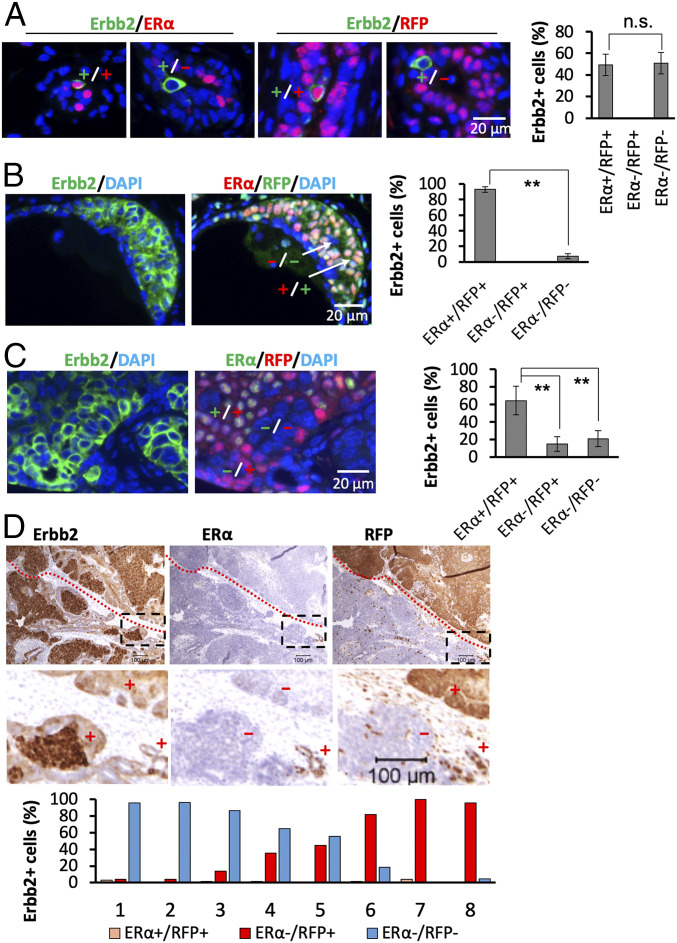
One week after viral infection, the HA-tagged Erbb2 protein was detected in comparable numbers of ERα+RFP+ and ERα−RFP− MGECs, indicating that RCAS-Erbb2HA virus infected both types of MGECs with similar efficiency. There were no detectable ERα−RFP+Erbb2+ cells at this stage (Fig. 1A). Four weeks after infection, we observed atypical hyperplastic lesions that consisted of 93% ERα+RFP+Erbb2+ cells and only 7% ERα−RFP−Erbb2+ cells. ERα−RFP+Erbb2+ cells still were not found at this stage (Fig. 1B). These observations indicate that ERα+RFP+Erbb2+ cells detected at week 1 survived better or proliferated faster than ERα−RFP−Erbb2+ cells, and that ERα expression is well maintained at this stage. At 13 wk, we detected DCIS lesions that contained 64.2% ERα+RFP+Erbb2+ and 20.4% ERα−RFP−Erbb2+ tumor cells, as well as 15.4% ERα−RFP+Erbb2+ tumor cells that often were colocalized with ERα+RFP+Erbb2+ tumor cells (Fig. 1C and SI Appendix, Fig. S3). These results indicate that a subset of ERα+RFP+Erbb2+ cells detected at weeks 1 and 4 have lost ERα expression to form a new ERα−RFP+Erbb2+ tumor cell population. At 27 wk, most tumor volumes reached 13,500 mm3 at an experimental endpoint (SI Appendix, Fig. S2A). In these advanced large tumors, the number of ERα+RFP+Erbb2+ tumor cells became quite few, averaging only 1.8% of the total Erbb2+ tumor cells, and clusters of ERα−RFP+Erbb2+ or ERα−RFP−Erbb2+ tumor cells were frequently observed (Fig. 1D). However, the ratio of ERα−RFP+Erbb2+ to ERα−RFP−Erbb2+ tumor cells varied significantly in the eight examined tumors: Tumor Nos. 1 through 3 and Nos. 6 through 8 consisted of mostly ERα−RFP−Erbb2+ and ERα−RFP+Erbb2+ tumor cells, respectively; No. 4 had 62% ERα−RFP−Erbb2+ and 38% ERα−RFP+Erbb2+ tumor cells; and No. 5 had similar numbers of ERα−RFP−Erbb2+ and ERα−RFP+Erbb2+ tumor cells (Fig. 1D). Although ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells originated from different luminal cell lineages, they exhibited indistinguishable morphologies. These results demonstrate that the predominant ERα+RFP+Erbb2+ cell population at week 13 has become a minor cell population at week 27, while ERα−RFP+Erbb2+ or ERα−RFP−Erbb2+ cells have become dominant cell populations at week 27. These cell lineage-tracing data prove that ERα+RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells originated from ERα+ and ERα− MGECs, respectively, and that a significant proportion of the ERα+RFP+Erbb2+ cells progressively abrogated ERα expression to become ERα−RFP+Erbb2+ cells that proliferate and become a substantial cell population in many advanced tumors. Although the initial ERα−RFP−Erbb2+ cells do not multiply well at an early stage, their numbers also progressively increased at later stages to become one of the two major tumor cell populations.
Fig. 1.
ERα+RFP+Erbb2+ tumor cells progressively cease ERα expression while they transform into ERα−RFP+Erbb2+ tumor cells during MG tumor progression. (A) Analysis of Erbb2+ cells by double IF at week 1 after viral infection. Data collected from 12 analyzed sections prepared from six MGs with two sections/MG showed 17 ± 3 Erbb2+ERα+, 15 ± 5 Erbb2+RFP+, 20 ± 7 Erbb2+ERα−, and 15 ± 6 Erbb2+RFP− cells per MG. The ERα+RFP+Erbb2+ cell number represents both ERα+Erbb2+ and RFP+Erbb2+ cells since these cells overlap as confirmed by double IF for ERα and RFP at this stage. ERα−Erbb2+ and RFP−Erbb2+ cells also overlap. The percentages of each cell type numbers compared to total ERα+RFP+Erbb2+ and ERα−RFP−Erbb2+ cell number are presented. ERα−RFP+Erbb2+ cells were not observed. (B) At week 4, IF staining for Erbb2 and double IF staining for ERα and RFP were performed on adjacent sections. Six sections from each MG and a total of six MGs were examined. For each section, the Erbb2+ hyperplasia regions were identified, and the numbers of ERα+RFP+ and ERα−RFP− cells in these regions were counted. The data are presented as average percentages of the indicated cell type numbers compared to total Erbb2+ cell number. ERα−RFP+ cells were not observed in Erbb2+ hyperplasia regions. (C) At week 13, adjacent sections were prepared, IF staining was performed, and data were collected and presented as described in B. (D) At week 27, five pieces of tissues (1 cm × 1 cm × 0.3 cm) were sampled from different regions of each large tumor (n = 8), and sections were prepared from each tissue piece. IHC was performed to detect Erbb2, ERα, and RFP on each set of three serial sections. All stained sections were imaged for analysis. ImageJ software was used to quantify ERα+RFP+Erbb2+, ERα−RFP+Erbb2+, and ERα−RFP+Erbb2+ cells in the tumor sections. Shown are average numbers relative to the respective areas each cell type occupied in the tumor sections. The red-dotted line demarcates the regions with ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ cells. The boxed areas in the Upper panels with ER+RFP+Erbb2+ cells are amplified in the Lower panels. “+” and “−” indicate regions with positive and negative immunoreactivities of the indicated proteins. n.s. in A, not significant (P > 0.05), and ** in B and C, P < 0.01, by Student’s t test (A and B) or one-way ANOVA test (C).
FoxA1 and GATA3 are required for normal ERα expression (22, 23). FoxA1 and GATA3 protein levels were high in ERα+RFP+Erbb2+ cells, but much lower in ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ cells (SI Appendix, Fig. S4). DNA methylation has been implicated as a mechanism to silence ERα expression in ERα− BrC (24). Three CpG islands are predicted in the ERα promoter regions (SI Appendix, Fig. S5A). Our DNA methylation assays revealed moderately methylated CpG sites in island 1 in normal ERα− MGECs and ERα−RFP−Erbb2+ tumor cells, and no methylation in this island in normal ERα+ MGECs and ERα−RFP+Erbb2+ tumor cells (SI Appendix, Fig. S5B).). In island 2, no methylation was detected in normal ERα+ MGECs, but normal ERα− MGECs showed moderate levels of CpG methylation. Moderate to high levels of methylation were observed in ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ cells, with relatively high levels at CpG sites −113 and −108 (SI Appendix, Fig. S5B). No CpG methylation was detected in island 3 in all examined cells. In agreement with the methylated CpG islands 1 and/or 2, ERα mRNA levels were high in normal ERα+ MGECs, but extremely low in normal ERα− MGECs, ERα−RFP+Erbb2+ cells, and ERα−RFP−Erbb2+ cells (SI Appendix, Fig. S5C). Together, these results suggest that decreased FoxA1 and GATA3 expression and ERα promoter methylation indicatively contribute to the loss of ERα expression during the progression of ERα+RFP+Erbb2+ cells to ERα−RFP+Erbb2+ cells.
The Loss of ERα Expression in ERα+RFP+Erbb2+ Tumor Cells Is Associated with Robustly Increased Cell Proliferation.
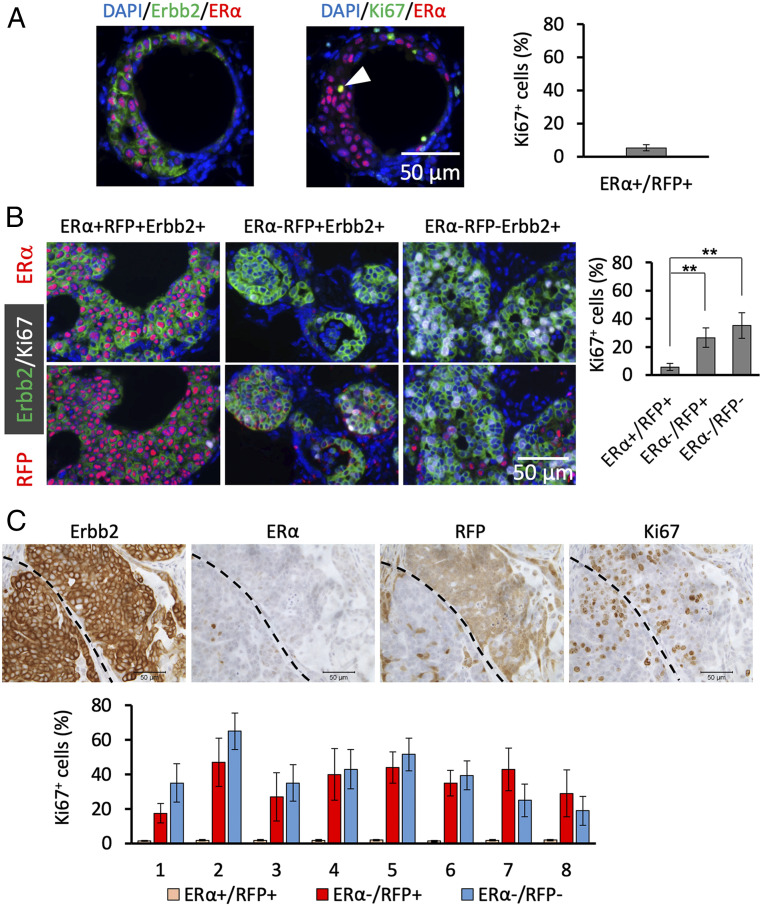
One week after viral infection, both ERα+RFP+Erbb2+ and ERα−RFP−Erbb2+ MGECs were not proliferating and appeared as individual cells among normal MGECs (Fig. 1A). At week 4, Ki67 IHC revealed proliferating cells in about 5% of ERα+RFP+Erbb2+ cells in atypical hyperplastic lesions (Fig. 2A). The very low number of ERα−RFP−Erbb2+ cells at this stage foiled reliable quantification of their proliferating cells. In DCIS lesions analyzed at week 13, Ki67 was expressed in 5% of ERα+RFP+Erbb2+ cells and in 25% of ERα−RFP+Erbb2+ cells, indicating that loss of ERα expression is associated with dramatically increased cell proliferation. The proliferation rate of ERα−RFP−Erbb2+ cells was higher than the rates of ERα+RFP+Erbb2+ and ERα−RFP+Erbb2+ cells (Fig. 2B). This high proliferation rate of ERα−RFP−Erbb2+ cells explained the increase in this cell population at week 13 compared to what we found at week 4. In advanced tumors at week 27, Ki67 immunostaining and BrdU incorporation assays revealed that very few ERα+RFP+Erbb2+ tumor cells were proliferating, while ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells were highly proliferative (Fig. 2C and SI Appendix, Fig. S6). These results indicate that the initial ERα+RFP+Erbb2+ tumor cells originating from ERα+ MGECs multiply slowly, but while they progressively lose ERα expression they become fast-proliferating ERα−RFP+Erbb2+ tumor cells and, consequently, largely replace the population of slow-proliferating ERα+RFP+Erbb2+ tumor cells of early hyperplastic lesions. On the other hand, ERα−RFP−Erbb2+ tumor cells that originated from ERα− MGECs are fast-proliferating tumor cells throughout the cancer progression process.
Fig. 2.
Loss of ERα expression is associated with a robust increase in cell proliferation. (A) At week 4 after viral infection, adjacent sections were prepared from six MGs and double IF was performed for Erbb2/ERα and for Ki67/ERα. At this stage, all ERα+Erbb2+ cells were RFP positive (Fig. 1B). The numbers of Erbb2+ERα+ cells and Ki67+ERα+ cells in hyperplastic regions were identified and counted, and 972 ± 162 ERα+Erbb2+ and 51 ± 9 Ki67+ERα+ cells per section were identified. ERα−Erbb2+ and Ki67+/ERα− cells were rare and their proliferation rate could not be determined at this stage. The arrowhead indicates a Ki67+/ERα+ cell. (B) At week 13, adjacent sections were prepared from six MGs for Erbb2/ERα/Ki67 and Erbb2/RFP/Ki67 triple IF staining. The numbers of Ki67+ cells were counted in ERα+RFP+ (n = 4,469), ERα−/RFP+ (n = 7,290), and ERα−/RFP− (n = 6,960) cell populations. **P < 0.01 by one-way ANOVA test. (C) At week 27, five pieces of tumor tissues with a volume of 1 × 1 × 0.3 (cm) per piece were sampled from different regions of each tumor. Serial tumor sections were prepared from eight tumors as described in Fig. 1D. IHC was performed with sets of four serial sections for Erbb2, ERα, RFP, and Ki67. Images were obtained by scanning the stained sections. Quantitative image analysis was performed by identifying the tumor regions containing Erbb2+ERα+RFP+, Erbb2+ERα−RFP+, and Erbb2+ERα−RFP− cells and then determining the relative number of Ki67+ cells in each region.
In normal MGs of adult mice, ERα+ MGECs barely proliferate, while ERα− MGECs are highly proliferative in response to hormonal stimulation (25). The slow- and fast-proliferating features of ERα+RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells may be inherited from their parental ERα+ and ERα− MGECs. Together, these findings suggest that ERα plays an important role in restricting the proliferation of both normal ERα+ MGEC and ERα+RFP+Erbb2+ tumor cells.
The Loss of ERα Expression in ERα+RFP+Erbb2+ Cells Is Associated with Distant BrC Metastasis.
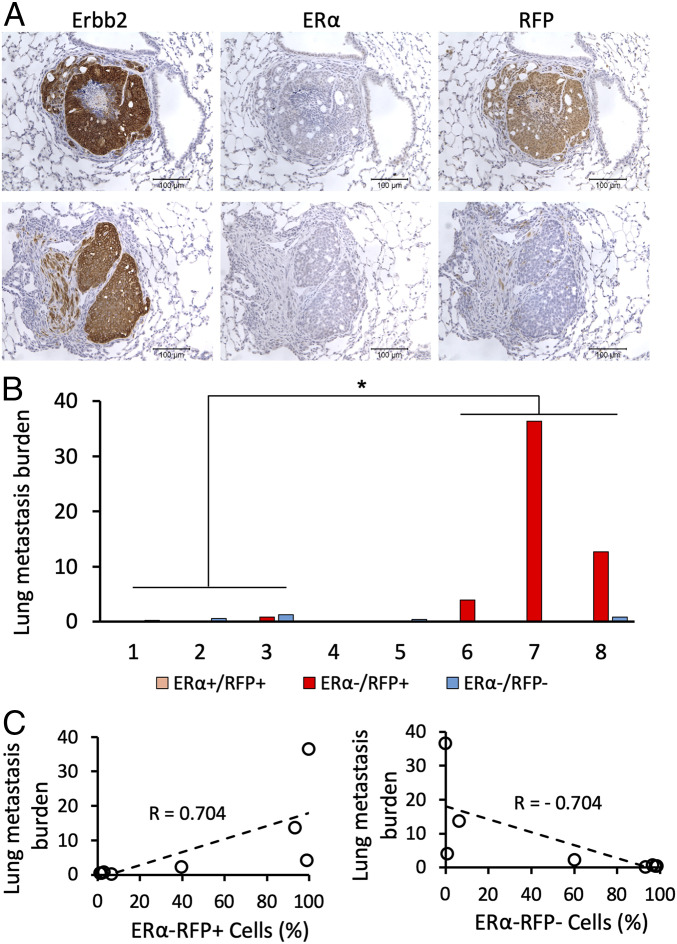
To determine the metastatic potentials of ERα+RFP+Erbb2+, ERα−RFP+Erbb2+, and ERα−RFP−Erbb2+ cells, we sectioned through the lungs of eight ERCreRFP-T mice with large MG tumors at week 27 after viral infection. We examined Erbb2, ERα, and RFP expressions in tumor cells on adjacent sections by IHC and calculated the percentage of areas occupied by ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells relative to total lung areas examined. We found that most lung metastases were made of ERα−RFP+Erbb2+ tumor cells, and only a few metastatic nodules contained ERα−RFP−Erbb2+ cells. Three mice (Nos. 6 through 8) bearing MG tumors with mostly ERα−RFP+Erbb2+ cells developed extensive lung metastases containing the same type of tumor cells, while other mice (Nos. 1 through 5) carrying similar size MG tumors with predominantly ERα−RFP−Erbb2+ tumor cells only developed low grade lung metastases (Figs. 3 A and B and 1D and SI Appendix, Fig. S2A). We did not find any ERα+RFP+Erbb2+ cells in all examined lungs, suggesting that these tumor cells do not metastasize (Fig. 3 A and B). Interestingly, the ratios of ERα−RFP+Erbb2+ to ERα−RFP−Erbb2+ cells showed a positive correlation, while ERα−RFP−Erbb2+ to ERα−RFP+Erbb2+ cell ratios showed a negative correlation with lung metastasis burdens (Fig. 3C).
Fig. 3.
The number of ERα−RFP+Erbb2+ tumor cells in the primary MG tumors was positively associated with lung metastasis. (A) IHC analysis of Erbb2, ERα, and RFP expression in adjacent sections of lung metastases at week 27 after viral infection. (B) Quantitative analysis of lung metastasis developed from the indicated types of tumor cells in eight ERCreRFP-T mice at week 27 after viral infection. Each whole lung was sectioned, and sets of three serial sections with a 50-μm interval between each set were used for IHC to assay Erbb2, ERα, and RFP expression. Digital images were taken from each stained section for quantifying areas with ERα+RFP+, ERα−RFP+Erbb2+, and ERα−RFP−Erbb2+ metastatic tumor cells using ImageJ software. The sum of metastatic areas of each indicated tumor cell type on all examined sections was used to represent the metastasis burden of each mouse lung. The primary MG tumors in mice Nos. 1 through 3 and 6 through 8 mainly consisted of ERα−RFP−Erbb2+ and ERα−RFP+Erbb2+ tumor cells, respectively. The lung metastasis burdens developed in mice Nos. 6 through 8 were significantly more severe than that in mice Nos. 1 through 3 (*P < 0.05 by one-way ANOVA test). (C) The percentages of ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells in the advanced MG tumors at week 27 were positively and negatively correlated with the lung metastasis burdens, respectively.
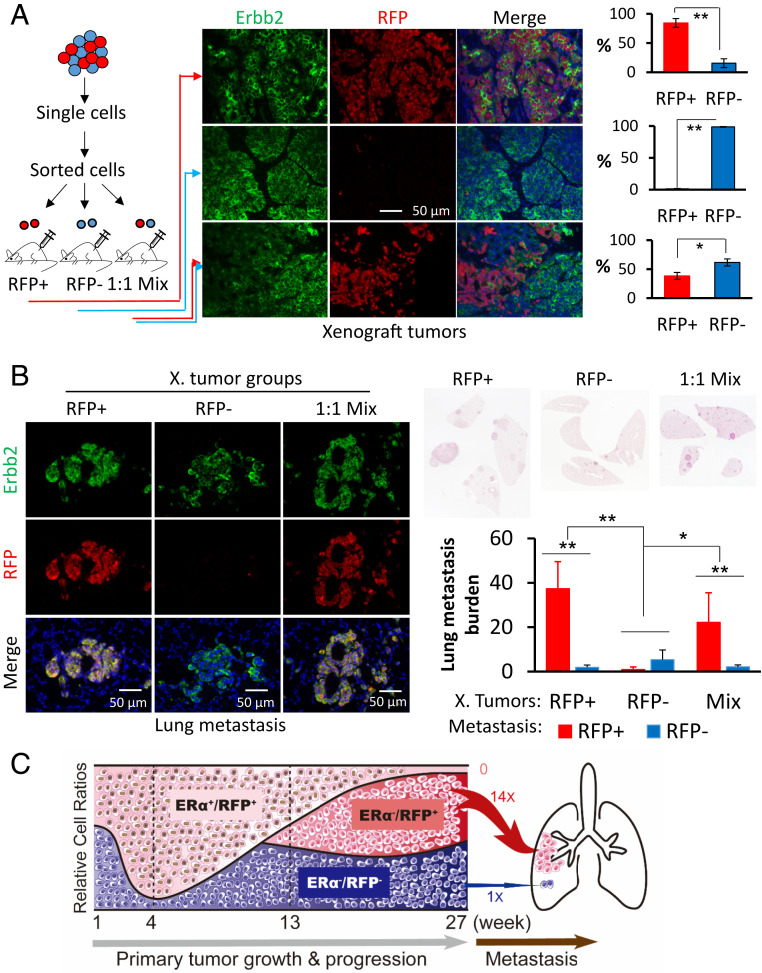
To confirm the significantly different metastatic competence of ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ cells, we then grew cell line-specific tumors in mouse xenograft models. Since the advanced MG tumors at week 27 mainly consisted of ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells with only a few ERα+RFP+ tumor cells (Fig. 1D), we used flow cytometry to isolate RFP+ and RFP− tumor cells from these large tumors. We orthotopically inoculated a half million cells of each RFP type as well as a 1:1 mixture of both into the MG fat pads of severe combined immunodeficiency (SCID) mice (Fig. 4A). The growth rates of xenograft tumors developed from these three groups were similar (SI Appendix, Fig. S7A). As expected, we did not find any ERα+Erbb2+ tumor cells in these tumors, and the tumors derived from RFP+ and RFP− cell groups mainly had ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells, respectively. The tumors derived from the cell mixture averaged about 40% ERα−RFP+Erbb2+ and 60% ERα−RFP−Erbb2+ cells (Fig. 4A and SI Appendix, Fig. S7B). The xenograft tumors with predominantly ERα−RFP+Erbb2+ cells produced the most lung metastases within ERα−RFP+Erbb2+ nodules, while the ones with mostly ERα−RFP−Erbb2+ cells developed the fewest lung metastases. The xenografts with mixed ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ cells produced the second most lung metastases with nodules that consisted of mainly ERα−RFP+Erbb2+ cells (Fig. 4B). We did not find any ERα+RFP+Erbb2+ tumor cells in the lung metastases of all examined mice (SI Appendix, Fig. S8). These results demonstrate that: 1) ERα+RFP+Erbb2+ tumor cells originating from ERα+ MGECs are nonmetastatic; 2) ERα−RFP−Erbb2+ tumor cells originating from ERα− MGECs are weakly metastatic; and 3) ERα−RFP+Erbb2+ tumor cells that derived from ERα+RFP+Erbb2+ tumor cells after losing ERα are extremely metastatic. Accordingly, these findings defined an interesting hierarchy for metastatic capacity that is determined by both ERα expression status and origin of the BrC cell lineage.
Fig. 4.
ERα−RFP+Erbb2+ cancer cells were much more metastatic than ERα−RFP−Erbb2+ cancer cells. (A) RFP+ and RFP− tumor cells were isolated from MG tumors at week 27 after viral infection and orthotopically inoculated into female SCID mice as sketched. Xenografts were collected 8 wk after inoculation. Five tissue pieces from different regions of each tumor were sampled for preparing sections. Double IF for Erbb2 and RFP was performed on sections of all tissue pieces to determine the percentage of RFP+Erbb2+ tumor cells to RFP−Erbb2+ tumor cells. *P < 0.05, and **P < 0.01 by Student’s t test. n = 8, 6, and 6 assayed tumors for Upper, Middle, and Lower bar graphs, respectively. (B) H&E staining and double IF for Erbb2 and RFP were performed with sets of serial sections prepared from each lung of SCID mice bearing RFP+Erbb2+ (n = 8 mice), RFP−Erbb2+ (n = 6 mice), and 1:1 mixture (n = 6 mice) xenografts. Stained sections were imaged by scanning. The lung tissue and the RFP+Erbb2+ and RFP−Erbb2+ cancer cell areas (pixels) in all section sets were measured using ImageJ software. The percentage of tumor cell area compared to lung area is presented as lung metastasis burden. * and **P < 0.05 and P < 0.01 by one-way ANOVA test. (C) A model depicting dynamic changes of the relative ratios of ERα+RFP+Erbb2+, ERα−RFP−Erbb2+, and ERα−RFP+Erbb2+ tumor cell populations during primary MG tumor growth, progression, and lung metastasis, and estimating lung metastasis capacity for each cancer cell lineage. Our quantitative data suggest that the relative lung-metastatic potencies of ERα+RFP+Erbb2+, ERα−RFP−Erbb2+, and ERα−RFP+Erbb2+ tumor cells could be assigned as 0, 1, and 14 folds, respectively.
The ERK1/2 MAPKs Are Activated in ERα−RFP+Erbb2+ Tumor Cells but Not in ERα−RFP−Erbb2+ Tumor Cells.
We compared the transcriptomes of ERα−RFP+Erbb2+ and ERα−RFP−Erbb2+ tumor cells and identified 230 up-regulated and 143 down-regulated transcripts in ERα−RFP+Erbb2+ cells (SI Appendix, Fig. S9A and Table S1). Gene set enrichment analysis revealed several cancer-related pathways, including Ras, cell adhesion, and PI3K-AKT signaling pathways (SI Appendix, Fig. S9B). The increased levels of Rasgrf1 and Fgf13 expression in the Ras pathway and Prlr, LamC3, and Itga7 expression in the PI3K-AKT pathway were validated in ERα−RFP+Erbb2+ cells versus ERα−RFP−Erbb2+ cells by RT-qPCR assays (SI Appendix, Fig. S9C). The PI3K-AKT pathway and the Ras pathway that activates MAPK ERK1/2 are known to promote cancer growth and metastasis. We found that the expression levels of AKT and ERK1/2 mRNAs as well as the phosphorylated active AKT showed no changes in these cells. However, levels of phosphorylated ERK1/2 were high in ERα+RFP+Erbb2+ and ERα−RFP+Erbb2+ cells in 13-wk hyperplasia and in ERα−RFP+Erbb2+ cells in 27-wk tumors, but were detected only in a very small proportion of ERα−RFP−Erbb2+ cells in tumors at both stages (SI Appendix, Figs. S10 and S11). ERK1/2 phosphorylation activates these kinases to translocate into the nucleus where they phosphorylate nuclear targets. Our results suggest that the high ERK1/2 activity is associated with the ERα+ tumor cell lineage, which may be responsible in part for the fast-proliferating and strong-metastatic capabilities of the subsequent ERα−RFP+Erbb2+ cancer cells. Further future studies are needed to understand why the slow-proliferating and nonmetastatic ERα+RFP+Erbb2+ tumor cells also have high ERK1/2 activities.
In summary, overexpression of Erbb2 can transform ERα+ and ERα− mouse MGECs into BrCs. ERα+Erbb2+ tumor cells from the ERα+ cell lineage have high ERK1/2 activities, but proliferate slowly and do not metastasize if they maintain ERα expression. However, due to decreased FoxA1 and GATA3 expression and ERα promoter methylation, these cells with high ERK1/2 activities progressively lose ERα to become fast-proliferating and highly metastatic ERα−Erbb2+ cancer cells. ERα−Erbb2+ tumor cells from the ERα− cell lineage proliferate fast, but have much weaker metastatic capability compared to ERα−Erbb2+ cancer cells with an ERα+ origin (Fig. 4C). These findings indicate that cell lineage origin and ER expression status are crucial factors contributing to the heterogenous growth and metastasis features of HER2+ BrCs. A future BrC treatment objective could be to prevent the transformation of ERα+Erbb2+ cancer cells into aggressive ERα−Erbb2+ cancer cells with an ERα+ origin while treating ERα+Erbb2+ cancers. Our results also suggest that the ERα−HER2+ human BrCs that are currently treated as a single type can consist of a more aggressive ERα−HER2+ subtype with an ERα+ origin and a less aggressive ERα−HER2+ subtype with an ERα− origin. A new objective then should involve the discovery of molecular markers specific to each subtype for a more distinguished diagnosis. Indeed, our findings warrant future studies to develop inherent molecular markers for these two BrC subtypes and to identify subtype-specific molecular targets for precision therapy.
Materials and Methods
Mouse Models.
ERα-F2A-Cre mice were cross-bred with Rosa26-LoxP-STOP-LoxP-tdRFP and MMTV-TVA mouse lines (17–19) to generate heterozygous trigenic ERCreRFP-T mice. Mouse genotypes were assayed by PCR using allele-specific oligonucleotide primers as described previously (17–19). RCAS-Erbb2HA virus was produced in DF-1 chicken fibroblasts, and 1 million viral particles were introduced into the ductal lumina of every fourth MG in 9-wk-old female ERCreRFP-T mice as described previously (19, 20). After viral infection, these female ERCreRFP-T mice were maintained to allow MG tumor development. For xenograft models, flow cytometry-sorted mouse tumor cells were injected into the fourth pair of MG fat pads of 8-wk-old female SCID mice. Each fat pad received 5 × 105 cells in 100 µL Matrigel (354230, Corning). Tumor growth, histopathology, and lung metastasis were examined as described previously (20, 26). The animal protocol was approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Tissue collection, H&E staining, immunohistochemistry, immunofluorescence (IF), DNA methylation assay, flow cytometry, transcriptome analysis, RT-qPCR, and statistical analysis were performed as described in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Dr. David Lonard for discussion and Jarrod Martinez, Mu Yang, Suoling Zhou, and Wen Bu for experimental assistance. We thank the Cytometry and Cell Sorting Core at Baylor College of Medicine for assisting with cell sorting and Zhongming Zhao, Xian Chen, and Chunying Yang in the Cancer Genomics Center at the University of Texas Health Science Center for RNA-Seq. This study is partially supported by NIH grant CA193455 (to J.X.). J.X. is also partially supported by the Gordon Cain Endowed Professorship in Cell Biology at Baylor College of Medicine.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100673118/-/DCSupplemental.
Data Availability
The FASTQ file for RNA-Seq data has been deposited in the BioProject category of the NCBI Sequence Read Archive database, accession no. PRJNA713819 (27). All other study data are included in the article and/or supporting information.
References
- 1.Rodilla V., et al., Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol. 13, e1002069 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quénel N., et al., The prognostic value of c-erbB2 in primary breast carcinomas: A study on 942 cases. Breast Cancer Res. Treat. 35, 283–291 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Gutierrez M. C., et al., Molecular changes in tamoxifen-resistant breast cancer: Relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J. Clin. Oncol. 23, 2469–2476 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Schiff R., et al., Cross-talk between estrogen receptor and growth factor pathways as a molecular target for overcoming endocrine resistance. Clin. Cancer Res. 10, 331S–336S (2004). [DOI] [PubMed] [Google Scholar]
- 5.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) , Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 365, 1687–1717 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Kennecke H., et al., Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 28, 3271–3277 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Lee H. J., et al., Two histopathologically different diseases: Hormone receptor-positive and hormone receptor-negative tumors in HER2-positive breast cancer. Breast Cancer Res. Treat. 145, 615–623 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Dowsett M., et al., Relationship between quantitative estrogen and progesterone receptor expression and human epidermal growth factor receptor 2 (HER-2) status with recurrence in the Arimidex, Tamoxifen, Alone or in Combination trial. J. Clin. Oncol. 26, 1059–1065 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Piccart-Gebhart M. J.et al.; Herceptin Adjuvant (HERA) Trial Study Team , Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 353, 1659–1672 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Baselga J.et al.; NeoALTTO Study Team , Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): A randomised, open-label, multicentre, phase 3 trial. Lancet 379, 633–640 (2012).Corrected in: Lancet379, 616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey L. A., et al., Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J. Clin. Oncol. 34, 542–549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goutsouliak K., et al., Towards personalized treatment for early stage HER2-positive breast cancer. Nat. Rev. Clin. Oncol. 17, 233–250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massarweh S., et al., Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 68, 826–833 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Ignatov A., Eggemann H., Burger E., Ignatov T., Patterns of breast cancer relapse in accordance to biological subtype. J. Cancer Res. Clin. Oncol. 144, 1347–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong Y., Han E. Y., Guo M., Pusztai L., Sneige N., Stability of estrogen receptor status in breast carcinoma: A comparison between primary and metastatic tumors with regard to disease course and intervening systemic therapy. Cancer 117, 705–713 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Lindström L. S., et al., Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J. Clin. Oncol. 30, 2601–2608 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Lee H., et al., Scalable control of mounting and attack by Esr1+ neurons in the ventromedial hypothalamus. Nature 509, 627–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luche H., Weber O., Nageswara Rao T., Blum C., Fehling H. J., Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37, 43–53 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Du Z., et al., Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc. Natl. Acad. Sci. U.S.A. 103, 17396–17401 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y., et al., Breast tumor cell-specific knockout of Twist1 inhibits cancer cell plasticity, dissemination, and lung metastasis in mice. Proc. Natl. Acad. Sci. U.S.A. 114, 11494–11499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bargmann C. I., Weinberg R. A., Oncogenic activation of the neu-encoded receptor protein by point mutation and deletion. EMBO J. 7, 2043–2052 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernardo G. M., et al., FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development 137, 2045–2054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eeckhoute J., et al., Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 67, 6477–6483 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Williams K. E., et al., DNA methylation in breast cancers: Differences based on estrogen receptor status and recurrence. J. Cell. Biochem. 120, 738–755 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Clarke R. B., Howell A., Potten C. S., Anderson E., Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 57, 4987–4991 (1997). [PubMed] [Google Scholar]
- 26.Wang S., et al., Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc. Natl. Acad. Sci. U.S.A. 106, 151–156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Y., Xu J., RNA-Seq profiling of ER-/Erbb2+ mouse breast cancer cells with ER+ and ER- cellular origins. BioProject category of the NCBI Sequence Read Archive database. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA713819 (Deposited 12 March 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The FASTQ file for RNA-Seq data has been deposited in the BioProject category of the NCBI Sequence Read Archive database, accession no. PRJNA713819 (27). All other study data are included in the article and/or supporting information.