Significance
The cognitive, perceptive, and motor capabilities of the mammalian cerebral cortex depend on assembly of circuit connectivity during development. Subplate neurons, strategically located at the junction of gray and white matter, orchestrate the wiring of cortical circuits. Using a genetic approach to study gene necessity and sufficiency in subplate neurons, we uncover an essential role for chromatin remodeler Arid1a in subplate neuron gene expression and axon guidance functions. Cortical deletion of Arid1a disrupts subplate-dependent formation of corpus callosum, targeting of thalamocortical axons, and development of sensory maps. Together, our study identifies Arid1a as a central regulator of subplate-dependent axon pathfinding, establishes subplate function as essential to callosum development, and highlights noncell-autonomous mechanisms in neural circuit formation and disorders thereof.
Keywords: cerebral cortex, development, chromatin regulation, axon pathfinding, neural circuits
Abstract
Loss-of-function mutations in chromatin remodeler gene ARID1A are a cause of Coffin-Siris syndrome, a developmental disorder characterized by dysgenesis of corpus callosum. Here, we characterize Arid1a function during cortical development and find unexpectedly selective roles for Arid1a in subplate neurons (SPNs). SPNs, strategically positioned at the interface of cortical gray and white matter, orchestrate multiple developmental processes indispensable for neural circuit wiring. We find that pancortical deletion of Arid1a leads to extensive mistargeting of intracortical axons and agenesis of corpus callosum. Sparse Arid1a deletion, however, does not autonomously misroute callosal axons, implicating noncell-autonomous Arid1a functions in axon guidance. Supporting this possibility, the ascending axons of thalamocortical neurons, which are not autonomously affected by cortical Arid1a deletion, are also disrupted in their pathfinding into cortex and innervation of whisker barrels. Coincident with these miswiring phenotypes, which are reminiscent of subplate ablation, we unbiasedly find a selective loss of SPN gene expression following Arid1a deletion. In addition, multiple characteristics of SPNs crucial to their wiring functions, including subplate organization, subplate axon-thalamocortical axon cofasciculation (“handshake”), and extracellular matrix, are severely disrupted. To empirically test Arid1a sufficiency in subplate, we generate a cortical plate deletion of Arid1a that spares SPNs. In this model, subplate Arid1a expression is sufficient for subplate organization, subplate axon-thalamocortical axon cofasciculation, and subplate extracellular matrix. Consistent with these wiring functions, subplate Arid1a sufficiently enables normal callosum formation, thalamocortical axon targeting, and whisker barrel development. Thus, Arid1a is a multifunctional regulator of subplate-dependent guidance mechanisms essential to cortical circuit wiring.
The subplate (SP) is a transient layer of the fetal cerebral cortex essential to the developmental wiring of cortical circuits (1–11). During neurogenesis, cortical neural progenitor cells (NPCs) generate excitatory neurons following an orderly temporal progression, successively giving rise to SP neurons (SPNs), then deep-layer neurons, then upper-layer neurons (12). As the first neurons generated from embryonic cortex, SPNs establish emerging axon tracts and form the earliest synapses (1–4, 13–16). Importantly, SPNs, which are strategically positioned at the interface between postmigratory neurons and developing white matter (WM), serve noncell-autonomous wiring functions in cortical circuit formation. Experimental SP ablation during fetal development causes thalamocortical axon (TCA) misrouting (17–19) and disrupts sensory map formation (20, 21), and perturbed SP function has been hypothesized to contribute to circuit defects in disorders of brain development (22–24). Mechanistically, SPNs noncell-autonomously mediate circuit wiring in part by extending the earliest descending axons, which interact with ascending TCAs during pathfinding (as posited by the “handshake hypothesis”) (25, 26) and contribute to their crossing of the pallial-subpallial boundary (PSB) (27, 28). SPNs also secrete extracellular matrix components that support axon guidance (4, 29) and are required for early oscillatory activity (8, 13). In postnatal ages, some SPNs undergo programmed cell death (30), thereby serving a transient role in cortical development.
Despite the central position of SPNs in orchestrating cortical connectivities, the molecular determinants of SP wiring functions remain largely elusive. Previous studies have focused on genes selectively expressed in SPNs (23, 31) and illuminated the genetic bases of SPN specification, migration, and axon development (32–38). The axon misrouting phenotypes of SP ablation (17–19), however, are not broadly recapitulated in these genetic mutants. The mechanisms underpinning SP guidance functions have remained largely mysterious. Here, by cell type-specific dissection of gene function, we identify Arid1a as a key regulator of multiple SP-dependent axon guidance mechanisms indispensable for cortical circuit wiring.
Arid1a (Baf250a) encodes a subunit of the Brg/Brahma-associated factors (BAF, or mammalian SWI/SNF) ATP-dependent chromatin remodeling complex that mobilizes nucleosomes along DNA, thereby mediating processes such as transcription and DNA repair. Human genetic studies have identified ARID1A loss-of-function mutations in intellectual disability, autism spectrum disorder, and Coffin-Siris syndrome, a developmental disorder characterized by callosal dysgenesis (39). BAF is broadly present in cells and organs (40), and cell type-dependent subunit compositions of the complex play diverse roles in embryonic stem cells, NPCs, and neurons (41). Arid1a, however, is not known to incorporate into BAF cell type specifically, and its potential context-dependent roles remain underexplored.
Here, we leverage a conditional allele to cell type specifically manipulate Arid1a function. We find that pancortical Arid1a deletion leads to extensive intracortical axon mistargeting and corpus callosum (CC) agenesis. Surprisingly, unlike pancortical deletion, sparse Arid1a deletion does not cell-autonomously misroute callosal axons, suggesting axon mistargeting to be a noncell-autonomous consequence of pancortical Arid1a deletion. Supporting this possibility, the axons of thalamocortical neurons, which are not autonomously affected by cortical Arid1a deletion, are also strikingly disrupted in their pathfinding into cortex and innervation of whisker barrels. Arid1a thus plays essential, noncell-autonomous roles in development of cortical axon tracts. By transcriptome analysis, we unbiasedly find a selective loss of SPN gene expression following Arid1a deletion, thus identifying SP as a potential substrate of Arid1a phenotypes. Consistent with this, Arid1a axon misrouting defects are reminiscent of SP ablation (17–19). Furthermore, SPN characteristics crucial to their circuit wiring functions, including SP organization and extracellular matrix, are disrupted following Arid1a deletion. Importantly, descending SP axons are severely attenuated, abrogating their cofasciculation with ascending TCAs. This “handshake” interaction is essential to TCA pathfinding and whisker barrel formation (25, 26), both of which are disrupted by Arid1a deletion. Thus, we find a necessity for Arid1a in orchestrating distinct aspects of SP wiring functions. To empirically test Arid1a sufficiency in SPNs, we use a genetic approach for cortical plate (CP) deletion of Arid1a that spares SPNs. In this model, Arid1a expression in SPNs is sufficient to support SP organization, SP axon “handshake” with TCAs, and SP extracellular matrix. Consistent with these wiring functions, SP Arid1a expression sufficiently enables normal TCA targeting, whisker barrel development, and callosum formation. Together, our study identifies Arid1a as a central regulator of SP-dependent axon pathfinding, unequivocally establishes SP function as essential to callosal development, and highlights noncell-autonomous mechanisms in circuit development and disorders thereof.
Results
Mistargeting of Intracortical, but Not Corticofugal, Axons following Arid1a Deletion from NPCs.
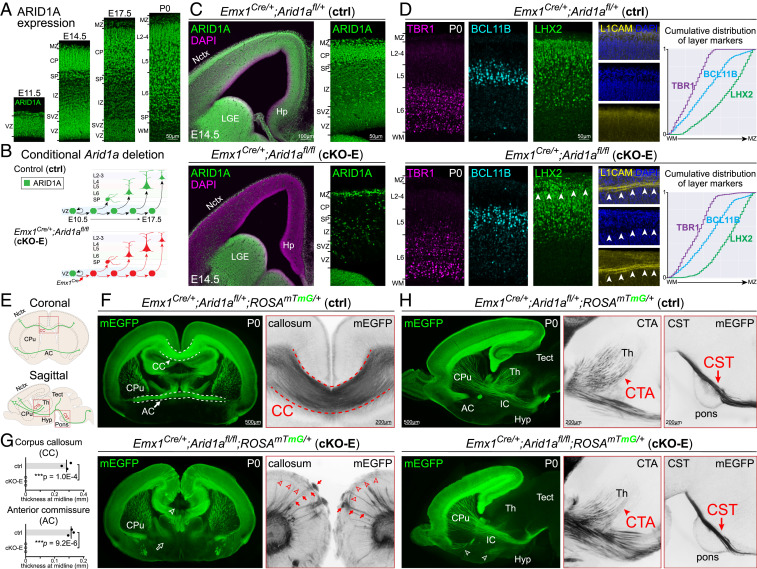
We analyzed ARID1A protein in developing mouse cortex by immunostaining and found broad ARID1A expression in ventricular zone (VZ) and subventricular zone NPCs and CP neurons from embryonic day (E)11.5 to postnatal day (P)0 (Fig. 1A). ARID1A completely colocalized with DAPI DNA staining at E14.5 (SI Appendix, Fig. S1A), which is consistent with ubiquitous ARID1A expression during cortical development. Constitutive Arid1a deletion leads to lethality on E6.5 in mice (42). We therefore leveraged a conditional allele (42) to investigate Arid1a function during brain development.
Fig. 1.
Tract-dependent misrouting of cortical axons following conditional Arid1a deletion. (A) ARID1A (green) immunostaining on coronal E11.5, E14.5, E17.5, and P0 brain sections revealed widespread ARID1A expression during cortical development. (B) Schematic illustration of conditional Arid1a deletion using Emx1Cre, which mediates recombination in cortical NPCs at E10.5, near the onset of neurogenesis. (C) ARID1A (green) and DAPI (magenta) staining of coronal E14.5 Emx1Cre/+;Arid1afl/fl (cKO-E) brain sections revealed loss of ARID1A from VZ and subventricular zone NPCs and CP neurons. ARID1A expression was unaffected in the ventral forebrain. (D) Layer marker immunostaining on coronal P0 brain sections. TBR1+ (L6, magenta), BCL11B+ (L5, cyan), and LHX2+ (L2 to 5, green) neurons were correctly ordered in cKO-E. Cumulative distribution of layer marker-expressing neurons from WM to marginal zone (MZ) revealed no disruption in cortical lamination in cKO-E (n = 3 animals). In all analyzed cKO-E brains (3/3 animals), but no littermate ctrl brains (0/3 animals), a stereotyped gap (arrowheads) in LHX2 and DAPI (blue) staining was present in upper cortical layers. This gap contained misrouted L1CAM+ (yellow) axons. (E) Schematic illustration of cortical axon tracts on coronal and sagittal brain sections. (F) Coronal sections of P0 brains. mEGFP (green) was expressed Cre dependently from ROSAmTmG, enabling visualization of cortical axons. Callosal agenesis (open arrowhead) was observed in cKO-E (3/3 animals). AC also failed to form (open arrow). cKO-E cortex was characterized by widespread axon misrouting (3/3 animals), including radially directed axons extending to the pia (red arrows) and tangentially directed axons traveling across the upper layers (red arrowheads). (G) Quantification of callosum and AC thickness at midline (data are mean, two-tailed unpaired t test, n = 3 animals). (H) Sagittal sections of P0 brains. In cKO-E, corticofugal axons innervated internal capsule. Corticothalamic axons (CTA, red arrowheads), corticotectal axons, and corticospinal tract (CST) axons (red arrows) were qualitatively reduced but followed normal trajectories without misrouting defects in cKO-E. AC axons were misrouted to the hypothalamus (Hyp, open arrowheads). VZ, ventricular zone; SVZ, subventricular zone; PP, preplate; IZ, intermediate zone; SP, subplate; CP, cortical plate; MZ, marginal zone; Ln, layer n; WM, white matter; Nctx, neocortex; Hp, hippocampus; LGE, lateral ganglionic eminence; CC, corpus callosum; AC, anterior commissure; CPu, caudate putamen; CTA, corticothalamic axons; CST, corticospinal tract; Th, thalamus; Tect, tectum; Hyp, hypothalamus.
To delete Arid1a from developing cortex, we used Emx1Cre, which expresses Cre recombinase in cortical NPCs starting at ∼E10.5, near cortical neurogenesis onset (Fig. 1B) (43). Arid1a deletion was validated in Emx1Cre/+;Arid1afl/fl (cKO-E) by ARID1A immunostaining, which revealed ARID1A loss from cortical NPCs and neurons of the Emx1 lineage (Fig. 1C). cKO-E mice were born at Mendelian ratio, had typical lifespans, and were fertile. At P0, cKO-E cortical area was not significantly different from control (ctrl) littermates (SI Appendix, Fig. S1 B and C).
To assess neocortical layers, we analyzed laminar markers at P0 (Fig. 1D). We found largely normal cortical lamination in cKO-E; TBR1+ layer (L)6, BCL11B+ (CTIP2+) L5, and LHX2+ L2 to 5 neurons were properly ordered and not robustly different from ctrl based on marker quantification (SI Appendix, Fig. S1D) and cumulative distribution of layer markers (Fig. 1D). However, a stereotyped gap in marker and DAPI staining was present in upper layers (L2 to 4) of all analyzed cKO-E brains (arrowheads, Fig. 1D, 3/3 animals) but absent from ctrls (0/3 animals). This gap was characterized by aberrant L1CAM-immunostained axons (arrowheads, Fig. 1D), suggesting the presence of misrouted axons in cKO-E.
We examined potential changes in axonal projections using Cre-dependent reporter ROSAmTmG. Following Emx1Cre recombination, membrane EGFP (mEGFP) was expressed from ROSAmTmG in all cortical excitatory neurons, enabling visualization of their axon projections (schematized in Fig. 1E). In P0 ctrl, mEGFP expression revealed intracortical axon tracts (CC and anterior commissure [AC]) (Fig. 1F). In cKO-E, we found callosal agenesis and widespread axon misrouting, including radially directed axons toward the pia (arrows) and tangentially directed axons through upper layers (arrowheads, Fig. 1F). These misrouting phenotypes were confirmed by L1CAM immunostaining (SI Appendix, Fig. S1E). AC axons were also mistargeted and unable to cross the midline (Fig. 1F and SI Appendix, Fig. S1F). Analysis of callosal and commissural thickness revealed loss of these tracts at the midline in cKO-E (Fig. 1G). Axon misrouting was also present in the hippocampus and accompanied hippocampal hypoplasia and disorganization (SI Appendix, Fig. S1G).
In contrast to intracortical axons, corticofugal tracts (corticothalamic, corticotectal, and corticospinal) were not characterized by gross misrouting deficits in cKO-E (Fig. 1H). These tracts were qualitatively reduced in strength but followed normal trajectories out of cortex and through internal capsule, and innervated their targets in dorsal thalamus, tectum, and medulla. Together, our axonal analyses revealed widespread mistargeting of intracortical but not corticofugal axon tracts following Arid1a deletion, implicating a tract-dependent role for Arid1a.
Noncell-Autonomous Misrouting of TCAs following Arid1a Deletion.
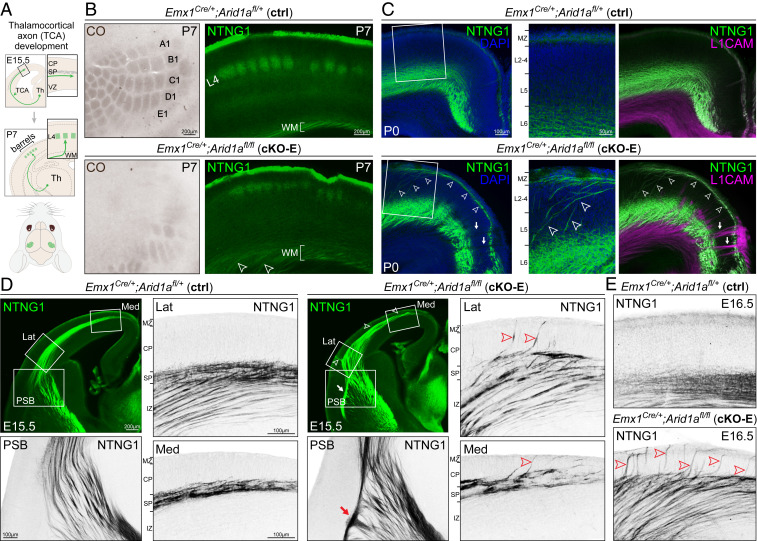
We next analyzed TCAs (schematized in Fig. 2A). In cKO-E, thalamocortical neurons were not autonomously affected by cortical Arid1a deletion; however, their axonal target, the cortex, was broadly affected by Emx1Cre. We used cytochrome oxidase (CO) histochemistry on flattened P7 cortex to visualize whisker barrels, a major target of TCAs. In P7 ctrl, CO staining revealed discrete, stereotyped, and organized whisker barrels in primary somatosensory cortex (Fig. 2B and SI Appendix, Fig. S2A). In cKO-E, barrel formation was severely disrupted; many barrels were missing and remaining barrels were distorted and disorganized. These TCA targeting defects were confirmed by immunostaining for TCA marker NTNG1 (Netrin G1) (Fig. 2B). In addition, analysis of P0 cKO-E revealed that NTNG1+ axons deviated from their normal trajectory in WM (arrowheads, Fig. 2C) and became markedly misrouted, including into tangential bundles through upper cortical layers and toward midline. Interestingly, TCAs largely did not contribute to radially directed aberrant axons in cKO-E, which were revealed by panaxonal marker L1CAM (arrows, Fig. 2C) and ROSAmTmG expression (arrows, SI Appendix, Fig. S2B).
Fig. 2.
Noncell-autonomous disruption of TCA pathfinding following Arid1a deletion. (A) Schematic illustration of TCA development. (B) Whisker barrels in P7 primary somatosensory cortex were visualized by CO staining (brown) on flattened cortices and NTNG1 immunostaining (green) on coronal sections. In cKO-E, barrel formation was severely disrupted (4/4 animals). Many barrels were missing, and the remaining barrels were distorted or disorganized. In cKO-E, NTNG1+ TCAs were defasciculated in cortical WM (open arrowheads). (C) NTNG1 immunostaining (green) on coronal P0 sections. In ctrl, NTNG1+ TCAs were present in WM, L6, and marginal zone (MZ). In cKO-E, NTNG1+ TCAs were markedly misrouted, extending dorsally from WM through the cortical layers (arrowheads). These aberrant axons then traveled tangentially across the upper layers and toward the midline. NTNG1+ TCAs did not contribute to radially directed axon bundles labeled by L1CAM (magenta, arrows) in cKO-E. (D) Analysis of TCA development at E15.5. In ctrl, NTNG1+ TCAs ascended across the PSB and paused within SP during the embryonic “waiting period.” In cKO-E, NTNG1+ TCAs did not cross the PSB along the normal trajectory (3/3 animals). They formed an aberrant bundle parallel to the PSB (red arrow) and entered the cortex via a narrow medial path. Notably, NTNG1+ TCAs prematurely invaded CP in the lateral (Lat) and medial (Med) cortex (red arrowheads) (3/3 animals). (E) Analysis of E16.5 cortex revealed an abundance of NTNG1+ TCAs prematurely invading CP in cKO-E (red arrowheads).
TCAs normally follow a precise developmental path. In ctrl E15.5 cortex, NTNG1+ TCAs had crossed the PSB into pallium and reached SP. Consistent with the “waiting period” (4, 44), TCAs at this age paused their ingrowth within SP and had not entered CP (Fig. 2D). Remarkably, in cKO-E, NTNG1+ axons prematurely invaded CP as aberrant bundles (arrowheads, Fig. 2D). By E16.5, we found an abundance of TCAs prematurely invading CP (Fig. 2E). This bypassing of the waiting period by TCAs is reminiscent of SP ablation (17, 18). In addition, cKO-E NTNG1+ axons did not cross the PSB along the normal path. Instead, they formed an abnormal bundle running parallel to the PSB (arrow, Fig. 2D and SI Appendix, Fig. S2C) and ultimately entered the pallium via a narrow medial trajectory. Impaired PSB crossing of TCAs is also a consequence of disrupted SP function (27, 28). Together, TCA phenotypes following cortex-specific Arid1a deletion in cKO-E revealed noncell-autonomous Arid1a functions in axon guidance.
Correct Callosal Axon Targeting following Sparse Deletion of Arid1a.
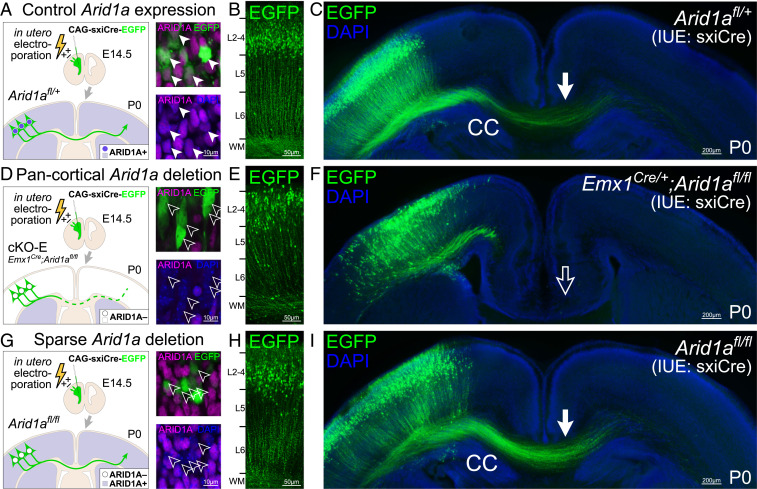
To determine whether intracortical axon misrouting in cKO-E also resulted from disrupted noncell-autonomous Arid1a function, we sought to sparsely delete Arid1a by in utero electroporation (IUE). We generated a self-excising, self-reporting Cre construct (CAG-sxiCre-EGFP) that expresses Cre, and upon Cre recombination, simultaneously excises Cre and turns on EGFP expression (SI Appendix, Fig. S3A). The sxiCre construct was transfected into embryonic cortex by IUE (37) at E14.5 to target NPCs during the genesis of upper-layer neurons, which form the majority of callosal axons. Transfected brains were analyzed at P0. To directly compare the effects of sparse versus widespread loss of Arid1a, we used ctrl, cKO-E (pancortical), and Arid1afl/fl (sparse by Cre transfection) littermates for IUE. In Arid1afl/fl, ARID1A immunostaining revealed 97.67% deletion efficacy by sxiCre in EGFP+ cells (SI Appendix, Fig. S3B). In P0 ctrl, electroporated neurons migrated to upper cortical layers and projected EGFP+ axons across the CC (Fig. 3 A–C). Following broad, genetic deletion of Arid1a in cKO-E, electroporated neurons migrated to upper layers, although their positioning was qualitatively less organized and potentially affected by misrouted axons (Fig. 1D). In cKO-E, axons originating from transfected neurons failed to project into CC (Fig. 3 D–F) and contributed to aberrant radially directed bundles (SI Appendix, Fig. S3C). Following sparse Arid1a deletion by sxiCre IUE in Arid1afl/fl, electroporated neurons migrated to upper layers without deficit (Fig. 3 G and H). Remarkably, despite ARID1A loss, these neurons abundantly extended EGFP+ axons into CC and correctly targeted contralateral cortex in a manner indistinguishable from ctrl (Fig. 3I). Together, our results provided strong support that the callosal and TCA pathfinding defects in cKO-E were noncell-autonomous consequences of broad cortical Arid1a deletion.
Fig. 3.
Correct callosal axon targeting following sparse deletion of Arid1a. A self-excising Cre expression EGFP reporter construct (CAG-sxiCre-EGFP or sxiCre) was transfected into dorsal cortical NPCs of Arid1afl/+ (ctrl, A–C), Emx1Cre/+;Arid1afl/fl (cKO-E, D–F), and Arid1afl/fl (without genetic Cre, G–I) using IUE at E14.5. At P0, ARID1A expression (magenta) was analyzed in EGFP+ transfected cells by immunostaining. ARID1A was present in transfected ctrl EGFP+ cells (solid arrowheads, A) but lost following pancortical genetic Arid1a deletion (cKO-E, open arrowheads, D) or sparse Arid1a deletion (Arid1afl/fl, open arrowheads, G). EGFP+ cells migrated to upper cortical layers in each condition (B, E, and H). EGFP+ axons innervated CC in ctrl (solid arrow, C) but failed to do so following broad Arid1a deletion in cKO-E (open arrow, F). Remarkably, sparse deletion of Arid1a from Arid1afl/fl EGFP+ cells did not disrupt their innervation of CC (solid arrow, I). Loss of ARID1A expression following sparse Arid1a deletion (open arrowheads, G) did not cell-autonomously cause axon misrouting defects.
Selective Disruption of SPN Gene Expression following Arid1a Deletion.
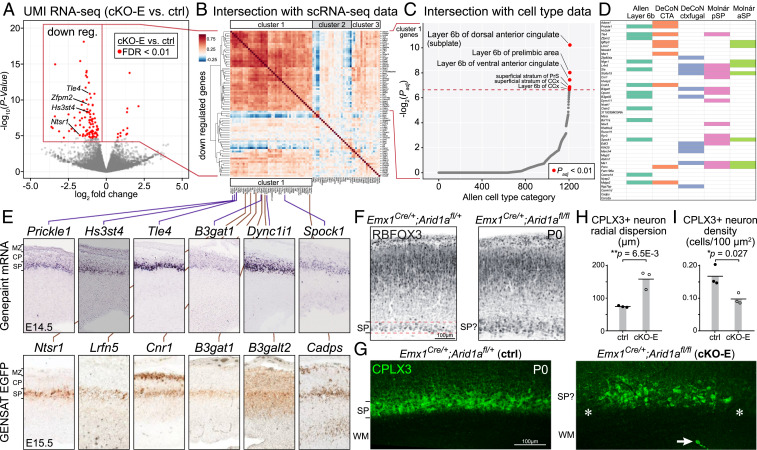
To gain mechanistic insights into the axon guidance roles of Arid1a, we explored its molecular functions. As a chromatin remodeler, ARID1A mediates transcriptional regulation (45, 46). We thus analyzed the transcriptomes of cKO-E and littermate ctrl cortices at E15.5, a developmental stage when cortical axons and TCAs undergo pathfinding (47). We generated libraries for unique molecular identifier (UMI) RNA sequencing (RNA-seq) (48–50) from E15.5 cKO-E and littermate ctrl cortices (ctrl: n = 6, cKO-E: n = 6 animals, SI Appendix, Fig. S4A). Differential gene expression was analyzed using edgeR (51), which revealed significant differential expression of 103 genes in cKO-E compared to ctrl with a stringent false discovery rate of <0.01 (Fig. 4A). Gene expression changes were validated by droplet digital (dd)RT-PCR for two down-regulated genes (Tle4 and Zfpm2) and two genes without significant changes (Lhx2 and Tbr1) and by TLE4 immunostaining (SI Appendix, Fig. S4B).
Fig. 4.
Selective disruption of SPN gene expression following Arid1a deletion. (A) Volcano plot of UMI RNA-seq data comparing E15.5 cortex of cKO-E to ctrl littermates (n = 6 animals). For each gene, P value was calculated with likelihood ratio tests, and false discovery rate (FDR) was calculated using the Benjamini-Hochberg procedure. Of 103 differentially expressed genes (FDR < 0.01, red dots), 91 were down-regulated and 12 were up-regulated in cKO-E. (B) Intersectional analysis of significantly down-regulated genes with scRNA-seq data from wild-type embryonic forebrain (52). Unsupervised hierarchical clustering revealed a cluster of 46 down-regulated genes (cluster 1) highly coexpressed at the level of single cells, suggesting that they may be expressed from one cell type. (C) Intersectional analysis of the 46 genes in cluster 1 with a spatiotemporal gene expression dataset covering over 1,200 brain subregions (54). Cluster 1 showed significant overrepresentation of genes selectively expressed in layer 6b (alternative nomenclature for SP). (D) Intersectional analyses with orthogonal datasets (31, 54, 56). (E) SP expression of cluster 1 genes was confirmed by E14.5 in situ hybridization data from the Genepaint database and E15.5 EGFP transgene expression data from the GENSAT consortium (57, 58). (F) RBFOX3 immunostaining (black) on coronal sections of P0 ctrl revealed a distinct and organized SP band positioned beneath the cortical layers. In cKO-E, the SP band was unclearly defined and not distinct from CP. (G) CPLX3 immunostaining (green) on coronal P0 sections. In ctrl, CPLX3+ SPNs were organized into a discrete, continuous band. In cKO-E, CPLX3+ SPNs were more dispersed, characterized by gaps (asterisks), and sometimes aberrantly positioned in WM (arrow). (H and I) Quantification of CPLX3+ SPN radial dispersion and density at P0 (data are mean, two-tailed unpaired t test, n = 3 animals). pSP, posterior subplate; aSP, anterior subplate.
Arid1a has been shown to increase or maintain transcriptional activity (46), which is consistent with our finding that a majority of differentially expressed genes were down-regulated in cKO-E (91/103). To determine whether axon defects in cKO-E may be associated with reduced expression of axon extension or guidance genes, we intersected the 91 down-regulated genes with 253 axon guidance genes (Gene Ontology [GO]: 0007411) and 138 axon extension genes (GO: 0048675). This revealed an overlap of one axon guidance gene (Ablim1) and one axon extension gene (Myo5b), which were validated by ddRT-PCR (SI Appendix, Fig. S4C). Neither group was significantly overrepresented (Phyper = 0.34 and 0.36, respectively).
To determine whether gene expression from particular cell types was preferentially affected in cKO-E, we performed intersectional analysis with single-cell RNA-seq (scRNA-seq) data from wild-type embryonic cortex (52). Unsupervised hierarchical clustering of cKO-E down-regulated genes based on scRNA-seq revealed a cluster with 46/91 genes (cluster 1) that are highly coexpressed at the level of single cells (Fig. 4B), suggesting that these down-regulated genes are normally expressed from one cell type. To validate this important finding, we performed intersectional analysis using an orthogonal dataset from wild-type E14.5 cortex (53), which revealed single-cell coexpression of 58/91 down-regulated genes (cluster A, SI Appendix, Fig. S4D). Remarkably, all 46 genes in cluster 1 were represented in cluster A. This complete overlap provided high confidence that the down-regulated genes in cKO-E reflected selective disruption of a single cell type.
To unbiasedly determine the identity of this cell type, we intersected the 46 cluster 1 genes with a rich spatiotemporal gene expression dataset covering over 1,200 brain subregions (54) using Enrichr (55). This revealed significant enrichment of genes selectively expressed in cortical layer 6b, an alternative nomenclature for SP (Fig. 4C). We next intersected cluster 1 genes with additional datasets orthogonal to the discovery data. We found overrepresentation of cluster 1 genes in genes preferentially expressed in the SP based on microdissection (31) (Fig. 4D). Intersection with a cell type–specific RNA-seq dataset (56) revealed overrepresentation of cluster 1 genes in “corticothalamic group 6” and “corticofugal group 11” (Fig. 4D and SI Appendix, Fig. S4E). Although SPNs were not specifically annotated in that dataset, marker membership suggests that these two groups comprise SPNs. Notably, cluster 1 genes showed no significant overlap with other major cell types (e.g., callosal neurons and corticospinal neurons), suggesting selective dysregulation of SPN genes in cKO-E. Available E14.5 in situ hybridization data (Genepaint) (57) and E15.5 EGFP transgene expression data (Gene Expression Nervous System Atlas [GENSAT]) (58) further supported that cluster 1 genes are expressed in the SP (Fig. 4E). Together, our analyses revealed selective disruption of SPN molecular identity following Arid1a deletion.
Consistent with disruption of SPN gene expression in cKO-E, anatomical development of SPNs was also altered. In P0 ctrl, RBFOX3-labeled SPNs were organized in a distinct, tight band positioned beneath CP (Fig. 4F). In cKO-E, the SP was disorganized and intermingled with CP. In ctrl, SPN marker CPLX3 (59) labeled a continuous band of SPNs (Fig. 4G). In cKO-E, gaps were present in the SP band (asterisks) and some CPLX3+ neurons were mispositioned in WM. In addition, CPLX3+ SPNs were significantly more dispersed (Fig. 4 H and I). Together, these data suggested that SPNs were present in cKO-E but anatomically disorganized and disrupted in cell type-dependent gene expression.
Altered Organization and Morphogenesis of Embryonic SPNs following Arid1a Deletion.
The loss of SPN expression and noncell-autonomous axon phenotypes in cKO-E convergently suggested that Arid1a guidance functions may be centered on SP. SP ablation misroutes cortical axons and causes TCAs to prematurely invade CP in a manner reminiscent of cKO-E (Fig. 2) (17–19). Disrupted SP function also leads to defects in TCA crossing of the PSB and formation of cortical sensory maps (20, 21, 27, 28) similar to Arid1a deletion. We therefore characterized SPN characteristics that contribute to their circuit wiring functions.
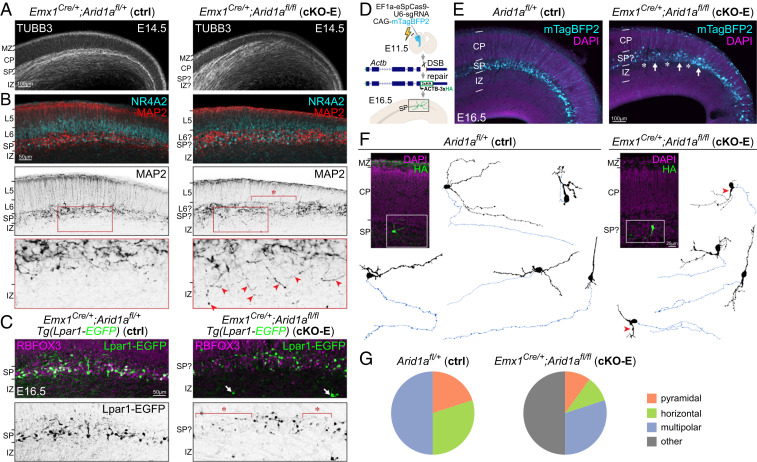
SP mediates axon pathfinding during embryonic ages. We assessed embryonic SP organization by immunostaining of TUBB3 (TUJ1), a neuronal cytoskeleton marker that reveals embryonic cortical layers. At E14.5, TUBB3+ processes were horizontally organized in intermediate zone (IZ) in ctrl but invaded CP diagonally in cKO-E (Fig. 5A). In ctrl, analysis of MAP2, a somatodendritic marker, and NR4A2 (NURR1), an SPN and L6 neuron marker, revealed a distinct, continuous SP layer below CP (Fig. 5B). In cKO-E, SPNs showed abnormal clustering and cell-sparse gaps. Notably, MAP2-labeled dendrites aberrantly projected ventrally into IZ (red arrowheads, Fig. 5B). We further used the Tg(Lpar1-EGFP) transgene, which expresses EGFP in a subset of SPNs (58). In E16.5 and P0 cKO-E cortex, Lpar1-EGFP–labeled SPNs were characterized by cell-sparse gaps (asterisks) and some were aberrantly positioned in IZ or WM (arrows, Fig. 5C and SI Appendix, Fig. S5A).
Fig. 5.
Disrupted SP organization and SPN morphology following Arid1a deletion. (A) TUBB3 (TUJ1) immunostaining (white) on E14.5 sections. In ctrl, TUBB3+ processes were horizontally organized in IZ and SP and radially organized in CP. In cKO-E, TUBB3+ axons became defasciculated in IZ and invaded CP diagonally. (B) MAP2 (red, black) and NR4A2 (cyan) immunostaining on E14.5 sections. In ctrl, MAP2+/NR4A2+ SPNs were organized within a clearly delineated layer below CP. In cKO-E, SPNs were characterized by abnormal clustering and cell-sparse gaps (asterisk), and misoriented MAP2+ dendrites aberrantly projected ventrally into IZ (red arrowheads, Inset). (C) RBFOX3 (NEUN) immunostaining (magenta) on E16.5 brains carrying the Lpar1-EGFP transgene. In cKO-E, Lpar1-EGFP+ SPNs (green, black) were characterized by cell-sparse gaps (asterisks), and some Lpar1-EGFP+ neurons were aberrantly positioned in IZ (arrows). (D) Schematic illustration of sparse SPN labeling by in utero genome editing. A DNA break was induced by CRISPR-Cas9 within the coding region of Actb near the C terminus. A reporter repair template was designed such that correct repair would lead to ACTB-3xHA expression. CRISPR-Cas9, reporter repair, and mTagBFP2 expression constructs were cotransfected into cortical NPCs at E11.5 by IUE. Electroporated brains were analyzed at E16.5. (E) mTagBFP2 (cyan) successfully targeted SPNs in electroporated brains. In cKO-E, labeled SPNs showed disorganization with abnormal cell clusters (arrows) and cell-sparse gaps (asterisks). (F) HA immunostaining (green) revealed complete morphology of ACTB-3xHA-labeled SPNs. Neurons were reconstructed based on confocal Z-stacks. Dendrites are indicated in black. Axons are indicated in blue. In cKO-E, some SPNs were characterized by a dendrite ventrally directed into IZ (red arrowheads). (G) Quantification of SPN morphological subclasses.
To analyze SPN morphology, we leveraged an in vivo genome editing method that targets the actin gene Actb for sparse, whole-cell labeling. We used CRISPR-Cas9 to generate a DNA break at the C terminus of Actb and a homology-independent repair template containing a 3xHA epitope tag (50). Nonhomologous end joining that incorporates the template in forward orientation would lead to in-frame expression of ACTB-3xHA. To perform this assay in vivo, we used IUE to cotransfect CRISPR-Cas9, repair template, and CAG-mTagBFP2 into cortical NPCs at E11.5, the peak of SP neuronogenesis (schematized in Fig. 5D). Analyzed at E16.5, mTagBFP2 expression revealed successful targeting of SPNs (Fig. 5E). mTagBFP2-labeled SPNs were organized in a continuous narrow band positioned beneath CP in ctrl but showed aberrant clustering and gaps in cKO-E.
Reconstruction of confocal Z-stacks revealed diverse morphologies in ACTB-3xHA-labeled SPNs in ctrl, including pyramidal, horizontal, and multipolar (Fig. 5 F and G). Diverse subclasses in SPN morphology have been documented (7, 60, 61), although embryonic SPN morphologies have not been extensively characterized. In cKO-E, SPN morphologies were also diverse; however, many deviated from the morphological subclasses observed in normal SP. Some SPNs were characterized by a ventrally directed dendrite extending into IZ (red arrowheads, Fig. 5F). Unlike SPNs, cKO-E pyramidal neurons were morphologically indistinguishable from ctrl (SI Appendix, Fig. S5B). Thus, our data revealed a cell type-dependent Arid1a function in SPN morphogenesis.
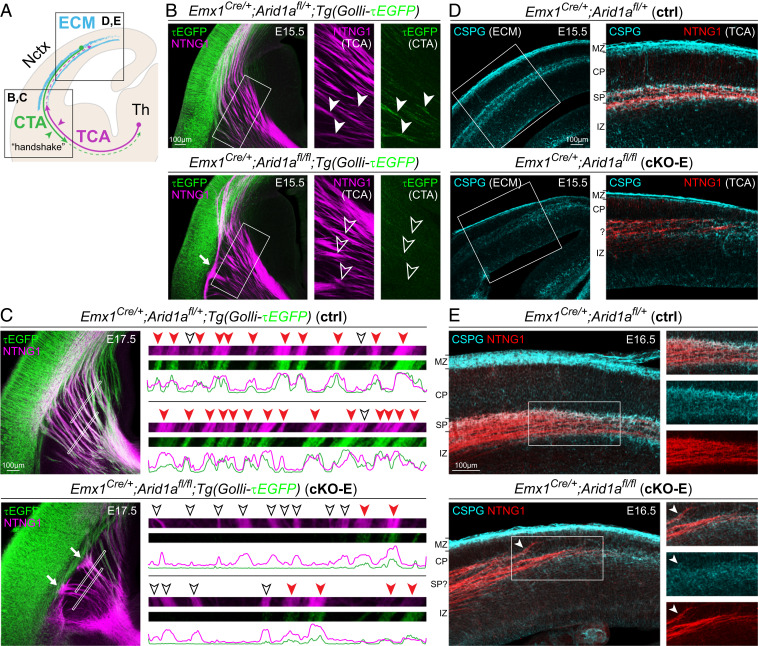
Disrupted Extracellular Matrix and SP Axon-TCA Cofasciculation following Arid1a Deletion.
SPNs are the first neurons to extend corticofugal axons. These descending axons contribute to guidance of thalamocortical ascending axons via cofasciculation in a model known as the “handshake hypothesis” (25, 62) (schematized in Fig. 6A). To visualize SPN corticofugal axons, we used the Tg(Golli-tau-EGFP) transgene, which expresses TAU (τ)EGFP in SPNs and L6 neurons (63). In E15.5 ctrl, an abundance of τEGFP+ axons had extended across the PSB. In subpallium, these descending axons were closely apposed with NTNG1+ TCAs, consistent with cofasciculation (solid arrowheads, Fig. 6B). In contrast, in cKO-E, subpallial innervation by τEGFP+ axons was attenuated and no cofasciculation with NTNG1+ axons was found (open arrowheads, Fig. 6B). By E17.5, extensive cofasciculation of τEGFP+ and NTNG1+ axons was present in ctrl but was largely absent from cKO-E (Fig. 6C). In P0 cKO-E, some NTNG1+ axons followed the aberrant trajectories of misrouted τEGFP+ axons, whereas other NTNG1+ axons, without cofasciculation, were misrouted (SI Appendix, Fig. S6A). Concomitant with loss of cofasciculation in cKO-E, NTNG1+ TCAs were consistently unable to correctly traverse the PSB (arrows, Fig. 6 B and C and SI Appendix, Fig. S6B, 5/5 animals), entered cortex via a narrow medial path, became defasciculated in IZ, and prematurely invaded CP. Notably, earlier in development in cKO-E, TCAs did not show misrouting defects at E13.5, prior to PSB crossing (SI Appendix, Fig. S6C), suggesting that their misrouting followed the loss of subpallial cofasciculation with SP axons. Our data are thus consistent with the model posited by the “handshake hypothesis” (7).
Fig. 6.
Aberrant SPN axon projections and extracellular matrix following Arid1a deletion. (A) Schematic illustration of SPN functions. (B and C) NTNG1 immunostaining (magenta) on E15.5 (B) and E17.5 (C) brains carrying the Golli-τEGFP transgene. In E15.5 ctrl, τEGFP+ (green) descending axons from SPNs closely cofasciculated (solid arrowheads) with ascending NTNG1+ TCAs (magenta). In E15.5 cKO-E, τEGFP+ axons largely have not descended across the PSB. Ascending NTNG1+ TCAs, without cofasciculation with descending τEGFP+ axons (open arrowheads), did not cross the PSB along the normal trajectory and formed an aberrant bundle parallel to the boundary (solid arrow). In E17.5 ctrl, we found frequent cofasciculation (red arrowheads, Inset) of τEGFP+ and NTNG1+ axons, consistent with the “handshake hypothesis.” In E17.5 cKO-E, most NTNG1+ TCAs did not cofasciculate with τEGFP+ axons (open arrowheads) and were unable to cross the PSB (solid arrows). (D and E) CSPG (cyan) and NTNG1 (red) immunostaining on E15.5 (D) and E16.5 (E) brain sections. In ctrl, NTNG1+ TCAs tangentially traversed the embryonic cortex within a SP/IZ corridor neatly delineated by extracellular matrix (ECM) component CSPG. In cKO-E, CSPG expression was reduced and the CSPG corridor had collapsed. NTNG1+ TCAs were not confined within SP/IZ, deviated from their normal trajectory, and prematurely invaded CP (arrowhead). CTA, corticothalamic axon.
SPNs are rich in extracellular matrix (64, 65), and secreted molecules derived from SPNs are thought to guide axons traversing the SP (66). During normal pathfinding, TCAs innervate cortex via a WM corridor delineated by extracellular matrix component chondroitin sulfate proteoglycan (CSPG) (29), which we observed in E15.5 and E16.5 ctrl (Fig. 6 D and E). In cKO-E, CSPG expression was reduced, and the CSPG corridor had collapsed (Fig. 6 D and E). Concomitant with CSPG corridor disruption, NTNG1+ TCAs were not confined within WM, deviated from their normal trajectory, and prematurely invaded CP. Together, our analyses revealed deficits in SPN morphogenesis, SP axon cofasciculation with TCAs, and SP extracellular matrix following Arid1a deletion.
SP-Spared CP Deletion of Arid1a Extensively Abrogated Axon Misrouting.
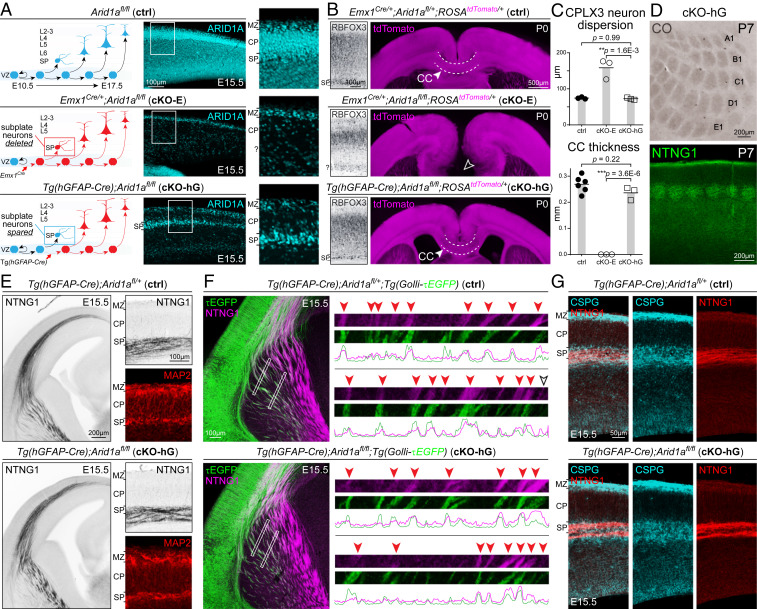
To empirically test the hypothesis that SPNs mediate the axon guidance functions of Arid1a, we sought to determine whether Arid1a expression in SPNs was sufficient to support axon pathfinding from cortical neurons that lacked Arid1a. We generated an Arid1a cKO using Tg(hGFAP-Cre) (67). Tg(hGFAP-Cre) mediates Cre recombination in cortical NPCs starting at E12.5, after the majority of SPNs have been generated (Fig. 7A and SI Appendix, Fig. S7A); therefore, Arid1a would be deleted from CP neurons, whereas SPNs would be spared. SP-spared cortical deletion was confirmed by ARID1A immunostaining in Tg(hGFAP-Cre);Arid1afl/fl (cKO-hG) (Fig. 7A). Importantly, Tg(hGFAP-Cre) mediated Arid1a deletion from L6 neurons by E13.5 (Fig. 7A and SI Appendix, Fig. S7 B and C), thus uncoupling the effects of SPNs from molecularly similar L6 neurons. In P0 cKO-hG, sparing SPNs from Arid1a deletion led to correct anatomical formation of the SP band (Fig. 7 B and C). Next, we analyzed cortical axon tracts using Cre-dependent reporter ROSAtdTomato (Fig. 7B). Remarkably, in cKO-hG, axons arising from tdTomato-labeled, Arid1a-deleted, CP neurons correctly formed CC and projected into contralateral cortex (Fig. 7 B and C). In addition to intracortical axons, TCAs were also normal in pathfinding in cKO-hG. At P7, CO staining on flattened cKO-hG cortices revealed typical organization of barrel cortex (Fig. 7D and SI Appendix, Fig. S7D, 3/3 animals), which was confirmed by NTNG1 (Fig. 7D). Thus, Arid1a expression in SPNs was sufficient for normal SP organization, callosum formation, and TCA targeting.
Fig. 7.
SP-spared CP deletion of Arid1a. (A) Schematic illustration of SP-spared CP deletion of Arid1a. Emx1Cre mediates Cre recombination in cortical NPCs starting at E10.5, prior to SPN genesis. ARID1A immunostaining (cyan) in E15.5 cKO-E revealed loss of ARID1A from SPNs and CP neurons. Tg(hGFAP-Cre) mediates Cre recombination in cortical NPCs starting at E12.5, after the majority of SPNs have been generated. In E15.5 Tg(hGFAP-Cre);Arid1afl/fl (cKO-hG), ARID1A was lost from CP neurons but present in SPNs. (B) SP and axon tract analyses on P0 brain sections. RBFOX3 immunostaining revealed in cKO-hG an organized, distinct SP band positioned just beneath CP. tdTomato (magenta) was expressed Cre dependently from ROSAtdTomato, enabling visualization of cortical axons. Callosal agenesis (open arrowhead) was observed in cKO-E. However, the CC formed without gross defect in cKO-hG (solid arrowhead, 3/3 animals). (C) Quantitative analyses revealed no significant changes in CPLX3+ SPN radial dispersion or callosal thickness at midline in cKO-hG compared to ctrl (data are mean, ANOVA with Tukey’s post hoc test, n ≥ 3 animals). (D) Whisker barrels in P7 primary somatosensory cortex were visualized by CO staining (brown) on flattened cortices and NTNG1 immunostaining (green) on coronal sections. Whisker barrels formed without defect in cKO-hG (3/3 animals). (E) Analysis of TCAs and SPNs in E15.5 cortex. In cKO-hG, NTNG1+ TCAs extended along a normal trajectory across the PSB, without forming an aberrant bundle parallel to the boundary. Upon reaching the cortex, NTNG1+ axons correctly paused within SP without prematurely invading CP in cKO-hG. MAP2 immunostaining (red) revealed that SPNs were organized within a continuous and clearly delineated layer below CP. (F) NTNG1 immunostaining (magenta) on E15.5 brains carrying the Golli-τEGFP transgene. In cKO-hG, τEGFP+ (green) descending axons from SPNs closely cofasciculated (red arrowheads) with ascending NTNG1+ TCAs (magenta). (G) CSPG (cyan) and NTNG1 (red) immunostaining on E15.5 brain sections. In cKO-hG, NTNG1+ TCAs traveled within an SP/IZ corridor neatly delineated by extracellular matrix component CSPG in a manner indistinguishable from ctrl.
Next, we assessed whether correct formation of axon tracts in cKO-hG was coincident with normal SP wiring functions. At E15.5, MAP2 immunostaining in cKO-hG revealed typical organization of SPNs indistinguishable from ctrl (Fig. 7E). NTNG1+ TCAs entered the cortex along a normal trajectory without premature invasion of CP (Fig. 7E). Golli-τEGFP-labeled corticofugal axons descended without defect (SI Appendix, Fig. S7E) and extensively cofasciculated with NTNG1-labeled TCAs in E15.5 cKO-hG (Fig. 7F). Furthermore, a CSPG corridor delineating the path of TCAs was present (Fig. 7G). Therefore, despite absence of ARID1A from CP neurons, SP ARID1A expression was sufficient for normal SP organization, SP axon-TCA cofasciculation, and extracellular matrix. Consistent with these wiring functions, SP Arid1a sufficiently enabled normal callosum formation, TCA targeting, and whisker barrel development.
Together, Arid1a cKO-E and cKO-hG supported examination of cell- and noncell-autonomous Arid1a functions. In SPNs, Arid1a was required for transcription of SP genes and gave rise to SPN organization, cofasciculation with TCAs, and extracellular matrix. By regulating the identity and functions of SPNs, Arid1a noncell-autonomously controlled the wiring of callosal and thalamocortical connectivities via the axon guidance roles of SP (schematized in SI Appendix, Fig. S8). Arid1a is thus a central regulator of multiple SP-dependent axon guidance mechanisms essential to cortical circuit assembly.
Discussion
Despite the central role of SPNs in cortical circuit assembly and their potential contribution to neurodevelopmental disorders (5, 22, 24), they are relatively understudied compared to their CP counterparts. Previous studies have focused on SP-enriched genes (23, 31) and characterized important molecular determinants of SPN specification, migration, and axon projections (32–38). The severe axon misrouting phenotypes of SP ablation (17–19) are, however, not broadly recapitulated in these genetic mutants. Here, we leverage cortical Arid1a deletion, which causes axon misrouting defects strikingly reminiscent of SP ablation, to gain mechanistic insights into the noncell-autonomous wiring functions of SPNs in assembling cortical connectivities.
Cortical Arid1a deletion and previous experimental SP ablation (17–20) phenotypically converge on misrouted TCAs, which prematurely invade CP and ultimately fail to innervate their L4 targets with correct topology. Several aspects of SP function may contribute to correct TCA pathfinding. First, the “handshake hypothesis” posits that close cofasciculation between descending SP axons and ascending TCAs is important for guidance of both tracts and formation of reciprocal connectivity (25, 62, 68). Following Arid1a deletion in cKO-E, SP corticofugal axons are markedly reduced, and their cofasciculation with TCAs is lost. Consistent with the “handshake hypothesis,” TCAs, in the absence of cofasciculation, are impaired in PSB crossing, enter cortex via a narrow medial path, and become defasciculated and misrouted after entering the cortex. These phenotypes are reminiscent of misrouting defects that follow disrupted SP function (27, 28) or abnormal shifting of SP due to piriform cortex expansion (69). Second, SP is characterized by a rich extracellular matrix (4, 44), which contributes to axon guidance by interacting with growth cones and supporting signaling (5). During circuit formation, cortical afferent and efferent axons extend along a WM corridor delineated by matrix component CSPG (29). Following cortical Arid1a deletion, CSPG expression is reduced and the corridor collapses. Concomitantly, TCAs become defasciculated and prematurely invade CP, a phenotype reminiscent of SP ablation (17). Notably, sparing SPNs from Arid1a deletion in cKO-hG is sufficient to support both SP axon-TCA cofasciculation and the CSPG corridor and enables correct TCA pathfinding. The roles of Arid1a in thalamocortical tract formation are therefore centered on SPNs. Interestingly, corticothalamic axons from CP neurons are largely intact following Arid1a deletion in cKO-E, despite reduced SP-thalamic axons. Thus, we do not find an Arid1a-dependent pioneering role for SP axons in guiding corticothalamic axons from CP neurons.
Unlike the better-known roles of SPNs in TCA guidance, SP contribution to intracortical tract development is less established. Early studies suggest that SPNs pioneer CC formation by extending the first callosal axons (60, 70–72). Some subsequent studies, however, find this possibility unlikely (73, 74). We find that pancortical Arid1a deletion in cKO-E leads to CC agenesis and mistargeting of intracortical axons. Sparse Arid1a deletion, however, does not autonomously misroute callosal axons, indicating that callosal agenesis is a noncell-autonomous consequence of pancortical Arid1a deletion. Remarkably, SP expression of Arid1a in cKO-hG is sufficient for callosum formation. Thus, we unequivocally establish that SP function is essential to callosum development. We note that Tg(hGFAP-Cre) is active in indusium griseum and glial wedge (75, 76). It is therefore unlikely that Arid1a expression in these structures could contribute to callosum formation in cKO-hG. Diverse developmental disorders are characterized by agenesis or dysgenesis of CC (77). Our study highlights a potential contribution of SP dysfunction to callosal defects in disease.
One barrier to molecular study of SP function is the lack of specific genetic access to embryonic SPNs during critical stages of circuit wiring. Although several published Cre lines show SPN specificity, Cre expression occurs too late for study of circuit development (78). Here, we describe a genetic strategy to target SPNs. We find that Emx1Cre mediates gene deletion from all cortical NPCs, including those that give rise to SPNs, whereas Tg(hGFAP-Cre) mediates deletion from NPCs after SPNs have been generated. Importantly, Tg(hGFAP-Cre) mediates recombination in a majority of L6 neurons, thereby enabling potential effects of SPNs to be uncoupled from closely related L6 neurons. By comparing “pancortical deletion” (Emx1Cre) versus “subplate-spared deletion” (Tg[hGFAP-Cre]), this approach enables interrogation of gene necessity and sufficiency in SP-mediated circuit wiring.
In cortical NPCs, we find that Arid1a deletion selectively disrupts SPN gene expression. Despite ubiquitous ARID1A expression during cortical development, the effects of Arid1a deletion are surprisingly cell type-dependent. A recent study showed that Arid1b expression following Arid1a depletion partially supported BAF function (45). In our cKO-E, expression of Arid1b may have attenuated the effects of Arid1a loss from CP neurons, thus contributing to SP-selective deficits.
Recent human genetic findings have convergently implicated altered chromatin function in disorders of brain development (79, 80). These studies identified loss-of-function mutations in ARID1A in intellectual disability, autism spectrum disorder, and Coffin-Siris syndrome, a developmental disorder characterized by callosal dysgenesis (39). The mechanisms by which chromatin dysregulation contributes to brain disorders are an active field of study. An important implication of our work is that deficits in SPNs may be an underappreciated contributor to neural circuit miswiring in neurodevelopmental disorders, including those associated with chromatin dysregulation.
Materials and Methods
Further experimental details can be found in SI Appendix, Supplementary Materials and Methods.
Animals.
All experiments were carried out in compliance with a protocol approved by the University of Michigan Institutional Animal Care & Use Committee. Strain details are provided in SI Appendix.
IUE.
Approximately 2 μL DNA was transfected into VZ NPCs by IUE. Experimental details are provided in SI Appendix.
UMI RNA-Seq.
Data were deposited to National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO): GSE163273. Details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the Kwan laboratory for discussions and comments on the study, critical reading of the manuscript, and scientific discussions and colleagues in the Michigan Neuroscience Institute (MNI) and Department of Human Genetics for insightful suggestions. This work was supported by the NIH (R01 NS097525 to K.Y.K., F31 NS110206 to D.Z.D., and T32 GM007544 to O.H.F.), the Brain Research Foundation (BRFSG-2016-04 to K.Y.K.), March of Dimes Foundation (No. 5-FY15-33 to K.Y.K.), and Simons Foundation Autism Research Initiative (402213 and 324586 to K.Y.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100686118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in NCBI GEO (GSE163273).
References
- 1.Allendoerfer K. L., Shatz C. J., The subplate, a transient neocortical structure: Its role in the development of connections between thalamus and cortex. Annu. Rev. Neurosci. 17, 185–218 (1994). [DOI] [PubMed] [Google Scholar]
- 2.McConnell S. K., Ghosh A., Shatz C. J., Subplate neurons pioneer the first axon pathway from the cerebral cortex. Science 245, 978–982 (1989). [DOI] [PubMed] [Google Scholar]
- 3.McConnell S. K., Ghosh A., Shatz C. J., Subplate pioneers and the formation of descending connections from cerebral cortex. J. Neurosci. 14, 1892–1907 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostović I., Rakic P., Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J. Comp. Neurol. 297, 441–470 (1990). [DOI] [PubMed] [Google Scholar]
- 5.Hoerder-Suabedissen A., Molnár Z., Development, evolution and pathology of neocortical subplate neurons. Nat. Rev. Neurosci. 16, 133–146 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Wang W. Z., et al., Subplate in the developing cortex of mouse and human. J. Anat. 217, 368–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molnár Z., Adams R., Goffinet A. M., Blakemore C., The role of the first postmitotic cortical cells in the development of thalamocortical innervation in the reeler mouse. J. Neurosci. 18, 5746–5765 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molnár Z., Luhmann H. J., Kanold P. O., Transient cortical circuits match spontaneous and sensory-driven activity during development. Science 370, eabb2153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostović I., Molliver M. E., A new interpretation of the laminar development of cerebral cortex: Synaptogenesis in different layers of neopallium in the human fetus. Anat. Rec. 178, 395 (1974). [Google Scholar]
- 10.Kostović I., The enigmatic fetal subplate compartment forms an early tangential cortical nexus and provides the framework for construction of cortical connectivity. Prog. Neurobiol. 194, 101883 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Molnár Z., et al., New insights into the development of the human cerebral cortex. J. Anat. 235, 432–451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwan K. Y., Sestan N., Anton E. S., Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development 139, 1535–1546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanold P. O., Luhmann H. J., The subplate and early cortical circuits. Annu. Rev. Neurosci. 33, 23–48 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Friauf E., McConnell S. K., Shatz C. J., Functional synaptic circuits in the subplate during fetal and early postnatal development of cat visual cortex. J. Neurosci. 10, 2601–2613 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wess J. M., Isaiah A., Watkins P. V., Kanold P. O., Subplate neurons are the first cortical neurons to respond to sensory stimuli. Proc. Natl. Acad. Sci. U.S.A. 114, 12602–12607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtaka-Maruyama C., et al., Synaptic transmission from subplate neurons controls radial migration of neocortical neurons. Science 360, 313–317 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Ghosh A., Shatz C. J., A role for subplate neurons in the patterning of connections from thalamus to neocortex. Development 117, 1031–1047 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Ghosh A., Antonini A., McConnell S. K., Shatz C. J., Requirement for subplate neurons in the formation of thalamocortical connections. Nature 347, 179–181 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Ghosh A., Shatz C. J., Involvement of subplate neurons in the formation of ocular dominance columns. Science 255, 1441–1443 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Kanold P. O., Shatz C. J., Subplate neurons regulate maturation of cortical inhibition and outcome of ocular dominance plasticity. Neuron 51, 627–638 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Kanold P. O., Kara P., Reid R. C., Shatz C. J., Role of subplate neurons in functional maturation of visual cortical columns. Science 301, 521–525 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Serati M., et al., The role of the subplate in schizophrenia and autism: A systematic review. Neuroscience 408, 58–67 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Hoerder-Suabedissen A., et al., Expression profiling of mouse subplate reveals a dynamic gene network and disease association with autism and schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 110, 3555–3560 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostović I., Judaš M., Sedmak G., Developmental history of the subplate zone, subplate neurons and interstitial white matter neurons: Relevance for schizophrenia. Int. J. Dev. Neurosci. 29, 193–205 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Blakemore C., Molnár Z., Factors involved in the establishment of specific interconnections between thalamus and cerebral cortex. Cold Spring Harb. Symp. Quant. Biol. 55, 491–504 (1990). [DOI] [PubMed] [Google Scholar]
- 26.Molnár Z., Blakemore C., How do thalamic axons find their way to the cortex? Trends Neurosci. 18, 389–397 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Magnani D., Hasenpusch-Theil K., Theil T., Gli3 controls subplate formation and growth of cortical axons. Cereb. Cortex 23, 2542–2551 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Magnani D., Theil T., Pratt T., Price D. J., Evidence that descending cortical axons are essential for thalamocortical axons to cross the pallial-subpallial boundary in the embryonic forebrain. PLoS One 7, e33105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bicknese A. R., Sheppard A. M., O’Leary D. D., Pearlman A. L., Thalamocortical axons extend along a chondroitin sulfate proteoglycan-enriched pathway coincident with the neocortical subplate and distinct from the efferent path. J. Neurosci. 14, 3500–3510 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price D. J., Aslam S., Tasker L., Gillies K., Fates of the earliest generated cells in the developing murine neocortex. J. Comp. Neurol. 377, 414–422 (1997). [PubMed] [Google Scholar]
- 31.Oeschger F. M., et al., Gene expression analysis of the embryonic subplate. Cereb. Cortex 22, 1343–1359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han W., et al., TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc. Natl. Acad. Sci. U.S.A. 108, 3041–3046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hevner R. F., et al., Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Tiong S. Y. X., et al., Kcnab1 is expressed in subplate neurons with unilateral long-range inter-areal projections. Front. Neuroanat. 13, 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arai Y., et al., Evolutionary gain of Dbx1 expression drives subplate identity in the cerebral cortex. Cell Rep. 29, 645–658.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Ratie L., et al., Loss of Dmrt5 affects the formation of the subplate and early corticogenesis. Cereb. Cortex 30, 3296–3312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwan K. Y., et al., SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. U.S.A. 105, 16021–16026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna W. L., et al., Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J. Neurosci. 31, 549–564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosho T., Okamoto N.; Coffin-Siris Syndrome International Collaborators , Genotype-phenotype correlation of Coffin-Siris syndrome caused by mutations in SMARCB1, SMARCA4, SMARCE1, and ARID1A. Am. J. Med. Genet. C. Semin. Med. Genet. 166C, 262–275 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Olave I., Wang W., Xue Y., Kuo A., Crabtree G. R., Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 16, 2509–2517 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son E. Y., Crabtree G. R., The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am. J. Med. Genet. C. Semin. Med. Genet. 166C, 333–349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao X., et al., ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. U.S.A. 105, 6656–6661 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorski J. A., et al., Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kostović I., Judas M., The development of the subplate and thalamocortical connections in the human foetal brain. Acta Paediatr. 99, 1119–1127 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Trizzino M., et al., The tumor suppressor ARID1A controls global transcription via pausing of RNA polymerase II. Cell Rep. 23, 3933–3945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathur R., et al., ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 49, 296–302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antón-Bolaños N., Espinosa A., López-Bendito G., Developmental interactions between thalamus and cortex: A true love reciprocal story. Curr. Opin. Neurobiol. 52, 33–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Routh A., Head S. R., Ordoukhanian P., Johnson J. E., ClickSeq: Fragmentation-free next-generation sequencing via click ligation of adaptors to stochastically terminated 3′-azido cDNAs. J. Mol. Biol. 427, 2610–2616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi L., Qalieh A., Lam M. M., Keil J. M., Kwan K. Y., Robust elimination of genome-damaged cells safeguards against brain somatic aneuploidy following Knl1 deletion. Nat. Commun. 10, 2588 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keil J. M., et al., Symmetric neural progenitor divisions require chromatin-mediated homologous recombination DNA repair by Ino80. Nat. Commun. 11, 3839 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuzwa S. A., et al., Developmental emergence of adult neural stem cells as revealed by single-cell transcriptional profiling. Cell Rep. 21, 3970–3986 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Loo L., et al., Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 10, 134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sunkin S. M., et al., Allen brain atlas: An integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 41, D996–D1008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuleshov M. V., et al., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molyneaux B. J., et al., DeCoN: Genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visel A., Thaller C., Eichele G., GenePaint.org: An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–D556 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong S., et al., A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Hoerder-Suabedissen A., et al., Novel markers reveal subpopulations of subplate neurons in the murine cerebral cortex. Cereb. Cortex 19, 1738–1750 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Hoerder-Suabedissen A., Molnár Z., Morphology of mouse subplate cells with identified projection targets changes with age. J. Comp. Neurol. 520, 174–185 (2012). [DOI] [PubMed] [Google Scholar]
- 61.De Carlos J. A., O’Leary D. D., Growth and targeting of subplate axons and establishment of major cortical pathways. J. Neurosci. 12, 1194–1211 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molnár Z., Garel S., López-Bendito G., Maness P., Price D. J., Mechanisms controlling the guidance of thalamocortical axons through the embryonic forebrain. Eur. J. Neurosci. 35, 1573–1585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piñon M. C., Jethwa A., Jacobs E., Campagnoni A., Molnár Z., Dynamic integration of subplate neurons into the cortical barrel field circuitry during postnatal development in the Golli-tau-eGFP (GTE) mouse. J. Physiol. 587, 1903–1915 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kostović I., Išasegi I. Ž., Krsnik Ž., Sublaminar organization of the human subplate: Developmental changes in the distribution of neurons, glia, growing axons and extracellular matrix. J. Anat. 235, 481–506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Judas M., Milosević N. J., Rasin M. R., Heffer-Lauc M., Kostović I., Complex patterns and simple architects: Molecular guidance cues for developing axonal pathways in the telencephalon. Prog. Mol. Subcell. Biol. 32, 1–32 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Kondo S., Al-Hasani H., Hoerder-Suabedissen A., Wang W. Z., Molnár Z., Secretory function in subplate neurons during cortical development. Front. Neurosci. 9, 100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhuo L., et al., hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 31, 85–94 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Hevner R. F., Miyashita-Lin E., Rubenstein J. L., Cortical and thalamic axon pathfinding defects in Tbr1, Gbx2, and Pax6 mutant mice: Evidence that cortical and thalamic axons interact and guide each other. J. Comp. Neurol. 447, 8–17 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Amaniti E.-M., et al., Expansion of the piriform cortex contributes to corticothalamic pathfinding defects in Gli3 conditional mutants. Cereb. Cortex 25, 460–471 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Chun J. J., Nakamura M. J., Shatz C. J., Transient cells of the developing mammalian telencephalon are peptide-immunoreactive neurons. Nature 325, 617–620 (1987). [DOI] [PubMed] [Google Scholar]
- 71.Antonini A., Shatz C. J., Relation between putative transmitter phenotypes and connectivity of subplate neurons during cerebral cortical development. Eur. J. Neurosci. 2, 744–761 (1990). [DOI] [PubMed] [Google Scholar]
- 72.deAzevedo L. C., Hedin-Pereira C., Lent R., Callosal neurons in the cingulate cortical plate and subplate of human fetuses. J. Comp. Neurol. 386, 60–70 (1997). [DOI] [PubMed] [Google Scholar]
- 73.Ozaki H. S., Wahlsten D., Timing and origin of the first cortical axons to project through the corpus callosum and the subsequent emergence of callosal projection cells in mouse. J. Comp. Neurol. 400, 197–206 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Koester S. E., O’Leary D. D., Axons of early generated neurons in cingulate cortex pioneer the corpus callosum. J. Neurosci. 14, 6608–6620 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benadiba C., et al., The ciliogenic transcription factor RFX3 regulates early midline distribution of guidepost neurons required for corpus callosum development. PLoS Genet. 8, e1002606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith K. M., et al., Midline radial glia translocation and corpus callosum formation require FGF signaling. Nat. Neurosci. 9, 787–797 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Paul L. K., et al., Agenesis of the corpus callosum: Genetic, developmental and functional aspects of connectivity. Nat. Rev. Neurosci. 8, 287–299 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Hoerder-Suabedissen A., et al., Subset of cortical layer 6b neurons selectively innervates higher order thalamic nuclei in mice. Cereb. Cortex 28, 1882–1897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanders S. J.et al.; Autism Sequencing Consortium , Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Rubeis S.et al.; DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in NCBI GEO (GSE163273).