Significance
Small GTPases are binary proteins that rapidly switch between active and inactive states through the actions of GEF- and GTPase-activating proteins (GAPs), respectively. GAPs play significant roles in cellular signaling, and their dysregulation is linked to numerous cancers. Here, we show that the BNIP-2 and Cdc42GAP Homology (BCH) domain of p50RhoGAP, known to autoinhibit the adjacent GAP domain, adopts an intertwined, dimeric structure with unique RhoA interactions. The β5-strand of the BCH domain plays a crucial role in the autoinhibition of the GAP domain. A mutation in the β5-strand destabilizes this autoinhibition and leads to RhoGAP activation. Our studies on the dynamics of the RhoGAP BCH domain will clarify its potential role in cancer and other diseases.
Keywords: GTPase-activating protein, BCH domain, Rho, Sec14, signaling
Abstract
Spatiotemporal regulation of signaling cascades is crucial for various biological pathways, under the control of a range of scaffolding proteins. The BNIP-2 and Cdc42GAP Homology (BCH) domain is a highly conserved module that targets small GTPases and their regulators. Proteins bearing BCH domains are key for driving cell elongation, retraction, membrane protrusion, and other aspects of active morphogenesis during cell migration, myoblast differentiation, and neuritogenesis. We previously showed that the BCH domain of p50RhoGAP (ARHGAP1) sequesters RhoA from inactivation by its adjacent GAP domain; however, the underlying molecular mechanism for RhoA inactivation by p50RhoGAP remains unknown. Here, we report the crystal structure of the BCH domain of p50RhoGAP Schizosaccharomyces pombe and model the human p50RhoGAP BCH domain to understand its regulatory function using in vitro and cell line studies. We show that the BCH domain adopts an intertwined dimeric structure with asymmetric monomers and harbors a unique RhoA-binding loop and a lipid-binding pocket that anchors prenylated RhoA. Interestingly, the β5-strand of the BCH domain is involved in an intermolecular β-sheet, which is crucial for inhibition of the adjacent GAP domain. A destabilizing mutation in the β5-strand triggers the release of the GAP domain from autoinhibition. This renders p50RhoGAP active, thereby leading to RhoA inactivation and increased self-association of p50RhoGAP molecules via their BCH domains. Our results offer key insight into the concerted spatiotemporal regulation of Rho activity by BCH domain–containing proteins.
Small GTPases are molecular switches that cycle between an active GTP-bound state and an inactive GDP-bound state and are primarily involved in cytoskeletal reorganization during cell motility, morphogenesis, and cytokinesis (1, 2). These small GTPases are tightly controlled by activators and inactivators, such as guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs), respectively (3, 4), which are multidomain proteins that are themselves regulated through their interactions with other proteins, lipids, secondary messengers, and/or by posttranslational modifications (5–7). Despite our understanding of the mechanisms of action of GTPases, GAPs, and GEFs, little is known about how they are further regulated by other cellular proteins in tightly controlled local environments.
The BNIP-2 and Cdc42GAP Homology (BCH) domain has emerged as a highly conserved and versatile scaffold protein domain that targets small GTPases, their GEFs, and GAPs to carry out various cellular processes in a spatial, temporal, and kinetic manner (8–15). BCH domain–containing proteins are classified into a distinct functional subclass of the CRAL_TRIO/Sec14 superfamily, with ∼175 BCH domain–containing proteins (in which 14 of them are in human) present across a range of eukaryotic species (16). Some well-studied BCH domain–containing proteins include BNIP-2, BNIP-H (CAYTAXIN), BNIP-XL, BNIP-Sα, p50RhoGAP (ARHGAP1), and BPGAP1 (ARHGAP8), with evidence to show their involvement in cell elongation, retraction, membrane protrusion, and other aspects of active morphogenesis during cell migration, growth activation and suppression, myoblast differentiation, and neuritogenesis (17–21). Aside from interacting with small GTPases and their regulators, some of these proteins can also associate with other signaling proteins, such as fibroblast growth factor receptor tyrosine kinases, myogenic Cdo receptor, p38-MAP kinase, Mek2/MP1, and metabolic enzymes, such as glutaminase and ATP-citrate lyase (17–26). Despite the functional diversity and versatility of BCH domain–containing proteins, the structure of the BCH domain and its various modes of interaction remain unknown. The BCH domain resembles the Sec14 domain (from the CRAL-TRIO family) (16, 27, 28), a domain with lipid-binding characteristics, which may suggest that the BCH domain could have a similar binding strategy. However, to date, the binding and the role of lipids in BCH domain function remain inconclusive.
Of the BCH domain–containing proteins, we have focused on the structure and function of p50RhoGAP. p50RhoGAP comprises an N-terminal BCH domain and a C-terminal GAP domain separated by a proline-rich region. We found that p50RhoGAP contains a noncanonical RhoA-binding motif in its BCH domain and is associated with GAP-mediated cell rounding (13). Further, we showed previously that deletion of the BCH domain dramatically enhanced the activity of the adjacent GAP domain (13); however, the full dynamics of this interaction is unclear. Previously, it has been reported that the BCH and other domains regulate GAP activity in an autoinhibited manner (18, 21, 29, 30) involving the interactions of both the BCH and GAP domains, albeit the mechanism remains to be investigated. It has also been shown that a lipid moiety on Rac1 (a Rho GTPase) is necessary for its inactivation by p50RhoGAP (29, 31), which may imply a role in lipid binding. An understanding of how the BCH domain coordinates with the GAP domain to affect the local activity of RhoA and other GTPases would offer a previously unknown insight into the multifaceted regulation of Rho GTPase inactivation.
To understand the BCH domain–mediated regulation of p50RhoGAP and RhoA activities, we have determined the crystal structure of a homologous p50RhoGAP BCH domain from S. pombe for functional interrogation. We show that the BCH domain adopts an intertwined dimeric structure with asymmetric monomers and harbors a unique RhoA-interacting loop and a lipid-binding pocket. Our results show that the lipid-binding region of the BCH domain helps to anchor the prenylation tail of RhoA while the loop interacts directly with RhoA. Moreover, we show that a mutation in the β5-strand releases the autoinhibition of the GAP domain by the BCH domain. This renders the GAP domain active, leading to RhoA inactivation and the associated phenotypic effects in yeast and HeLa cells. The released BCH domain also contributes to enhanced p50RhoGAP–p50RhoGAP interaction. Our findings offer crucial insights into the regulation of Rho signaling by BCH domain–containing proteins.
Results
Structure of the S. Pombe BCH Domain (YBCH).
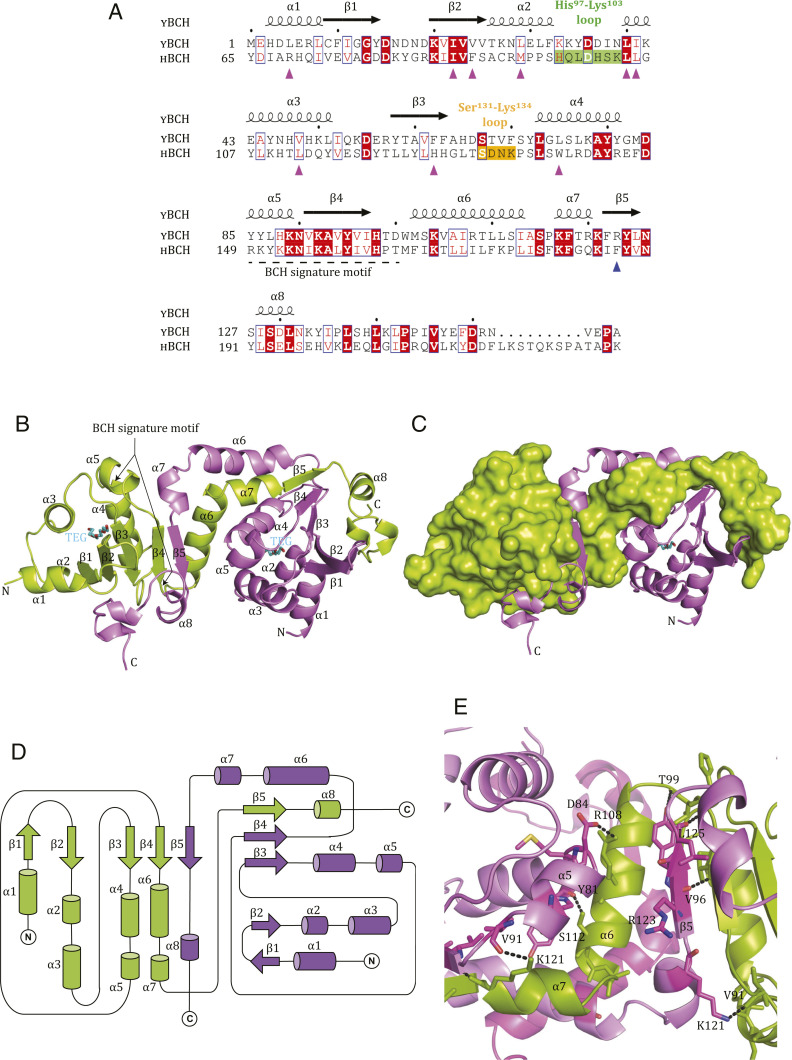
Our initial attempts to crystallize the BCH domain of human p50RhoGAP were unsuccessful. Thus, to understand the architecture of the BCH domain, we crystallized its S. pombe homolog, SPAC1565.02c (1 to 156 aa; YBCH; Y denotes yeast whereas H denotes human). Sequence alignment of the BCH domains of Homo sapiens and S. pombe p50RhoGAP showed 46% sequence similarity and conservation of a BCH domain signature motif: R(R/K)h(R/K)(R/K)NL(R/K)xhhhhHPs (“h” refers to large and hydrophobic residues, and “s” refers to small and weakly polar residues) (Fig. 1A).
Fig. 1.
Crystal structure of BCH domain from SPAC1565.02c (1 to 156 aa), S. pombe (YBCH denotes yeast BCH). (A) Sequence alignment of residues 1 to 156 aa of YBCH protein (SPAC1565.02c) and its homologous region in p50RhoGAP (HBCH, residues 65 to 229). The regions such as His97-Lys103 loop (green) and Ser131-Lys134 loop (orange) are highlighted. The R123 in YBCH (and F187 in p50RhoGAP) in the β5 are indicated (blue up arrow). The BCH signature motif covers the α5 to β4 (dashed underline). The residues that form the lipid-binding pocket are indicated (pink up arrow). The secondary structure of the YBCH (SPAC1565.02c) is shown on top of the sequence. (B) Ribbon representation of WT YBCH dimer; the monomers are shown in green and violet colors. The BCH signature motif corresponds to the region from α5 to β4. TEG molecules present in the lipid-binding site of both monomers are shown as stick representation in cyan. (C) The surface (green) and ribbon (violet) representation of the monomers is shown. (D) Topology diagram of dimeric structure of YBCH. For all these figures, the N and C termini of YBCH are labeled. (E) The dimer interface hydrogen bonding contacts are shown. For clarity, only a few key hydrogen bonds were shown from the key dimer interface secondary structures such β5, α5, α6, and α7 (for the full contact details, please refer to SI Appendix, Table S1).
The structure of the YBCH domain was determined at 2.8 Å resolution (Fig. 1 B and C and Table 1). The asymmetric unit has four molecules, consisting of a dimer of dimers. The molecules are well defined in the electron density map, except for seven to nine residues at the C terminus. Each monomer consists of eight α-helices and five β-strands that form two regions: 1) a globular structure (1 to 102 aa), comprising α1 to α5 and β1 to β4 and 2) an extended structure (103 to 146 aa), consisting of α6 to α8, β5, and the C-terminal loop. The structure of each monomer is intertwined with the other monomer. Specifically, each monomer has a core β-sheet, comprising β1↓- β2↑- β3↑- β4↑ from molecule A and β5↓ from molecule B; these are surrounded by α1 to α5 helices of molecule A on one side (Fig. 1 B and D) and by α8 and the C-terminal loop from molecule B on the other. The electron density map shows the presence of one tetra ethylene glycol (TEG) molecule bound per monomer at the core N-terminal region (1 to 102 aa) of the YBCH domain.
Table 1.
Crystallographic data collection and refinement for YBCH
| YBCH | ||
| Data collection | Peak | Native |
| Cell parameters (Å, °) | a = b = 109.26, | a = b = 108.39, |
| c = 244.31 | c = 250.41 | |
| α = β = 90, γ = 120 | α = β = 90, γ = 120 | |
| Space group | P61 | P61 |
| Resolution range (Å)* | 50.0 to 3.0 (3.11 to 3.00) | 50.0 to 2.80 (2.90 to 2.80) |
| Wavelength (Å) | 0.979 | 1.5418 |
| Observed reflections >1σ | 163,368 | 147,115 |
| Unique reflections | 29,647 | 36,910 |
| Completeness (%) | 90.1 | 87.7 |
| Overall (I/σ (I)) | 14.5 | 12.6 |
| RSym† (%) | 6.3 | 8.9 |
| Refinement and quality‡ | ||
| Resolution range (Å) | 49.7 to 2.80 (2.87 to 2.80) | |
| Rwork§ (no. of reflections) | 0.228 (36099) | |
| Rfree¶ (no. of reflections) | 0.260 (2143) | |
| rmsd bond lengths (Å) | 0.005 | |
| rmsd bond angles (°) | 0.83 | |
| No. atoms | ||
| Protein | 4,910 | |
| Ligand/ions | 52 | |
| B-factors (Å2) | ||
| Protein | 98.2 | |
| Ligand/ion | 117.7 | |
| Ramachandran plot | ||
| Ramachandran favored (%) | 92.06 | |
| Ramachandran allowed (%) | 7.94 | |
| Ramachandran outliers (%) | 0 | |
The high-resolution bin details are in the parentheses.
RSym = ∑|Ii – <I>|/|Ii| in which Ii is the intensity of the ith measurement, and <I> is the mean intensity for that reflection.
Reflections with I > σ were used in the refinement.
Rwork = |Fobs – Fcalc|/|Fobs| in which Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rwork, but for 6% of the total reflections chosen at random and omitted from refinement.
Asymmetric Dimer of the YBCH Domain Has a Unique and Intertwined Architecture.
The YBCH dimer consists of two nonidentical—and, therefore, asymmetric—monomers (Fig. 1B). The N-terminal residues 1 to 102 of YBCH form α1 to α5 and β1 through β4, which adopt an identical structure in both monomers. However, after β4, the orientation of the molecule changes significantly, due to a change in the direction of the loop between α6 and α7 in the C-terminal region (SI Appendix, Fig. S1 A and B). This conformational change allows for dimerization of two asymmetric monomers (SI Appendix, Fig. S1B). The different orientations of the C-terminal regions of monomer A and monomer B are highlighted following superposition of the structurally nonidentical monomers of the dimer (monomer A and monomer B; 1.3 Å rmsd for 101 Cα atoms, from the N-terminal YBCH), with the change occurring at Pro116. Yet, notably, the C-terminal regions (117 to 146 aa) of the two monomers superimpose well, with rmsd of 0.55 Å (SI Appendix, Fig. S2A). The electron density map for the region 111 to 119 aa of the monomers shows that the structure is well defined (SI Appendix, Fig. S2 B and C). The average B-factors of the C-terminal regions (113 to 146 aa) of monomers A and B are 101.7 Å2 and 104.9 Å2, respectively, suggesting similar flexibility of the C-terminal regions of the asymmetric monomers (SI Appendix, Fig. S3A). Further, the dimeric structure of YBCH is held together by several hydrogen bonding contacts between the monomers. In particular, an intermolecular β-sheet is formed between β4 of monomer A and β5 of monomer B (SI Appendix, Table S1 and Fig. 1E). Of the 34 hydrogen-bonding contacts made, 14 are asymmetrical because of the nonidentical monomers. The total buried area of the dimer interface is 2,895 Å2, in which β5 contributes 335 Å2. The two dimers of the asymmetric unit of the YBCH structure are identical (rmsd 0.47 Å for all Cα atoms) (SI Appendix, Fig. S3B). Moreover, when evaluated by analytical ultracentrifugation, YBCH exists as a mixed population of monomers and dimers at 2.4 mg/mL concentration (a similar concentration was used for the crystallization of YBCH) (SI Appendix, Fig. S4A). Size-exclusion chromatography shows an increase in the dimer population for YBCH in a concentration-dependent manner (SI Appendix, Fig. S4B). Finally, YBCH was ectopically expressed in mammalian cells and coimmunoprecipitated, confirming that YBCH dimerization occurs in cells; indeed, YBCH forms a self-associated complex in cells (SI Appendix, Fig. S5).
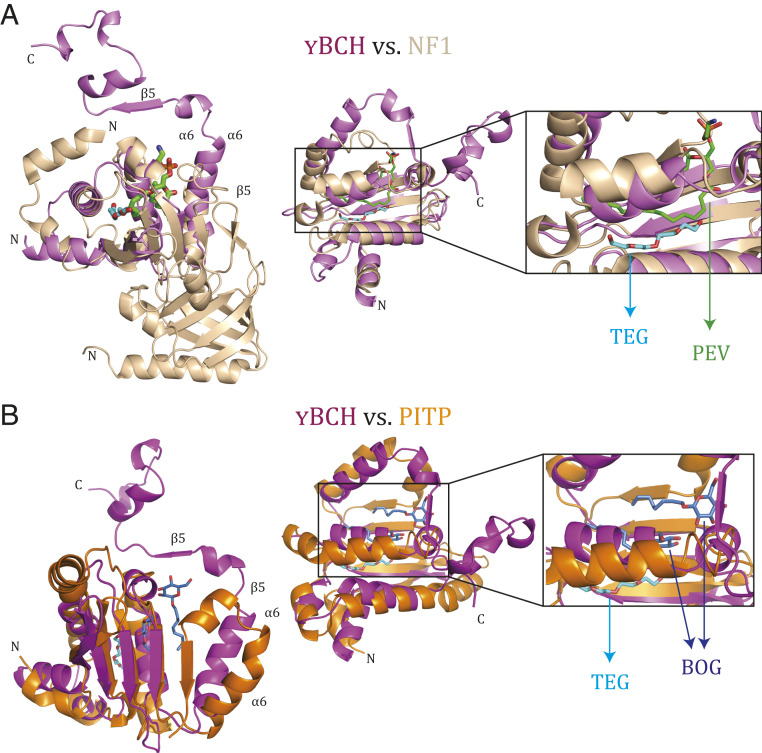
A search for structural homologs in the Protein Data Bank (PDB) database using the DALI (Distance-Matrix Alignment Method) server (32) was performed independently for the N-terminal (1 to 102) region, the C-terminal (103 to 146) region, and the monomer and dimer of YBCH. The search for the N-terminal region showed that part of this domain is similar to the Sec14 domain of neurofibromin (1,560 to 1,670 aa) (PDB: 3P7Z) (Fig. 2A and SI Appendix, Fig. S6A). The search for structural homologs of the monomeric and dimeric YBCH yielded no matches. These findings suggest that YBCH adopts an intertwined fold with a unique dimer formation. Structure- and sequence-based analyses of YBCH with various BCH domains suggests that the structure of BCH domains could be well conserved (SI Appendix, Fig. S6B).
Fig. 2.
Structural comparison between YBCH domain and Sec14 domains. (A) Superposition of crystal structure of Sec14 domain of neurofibromin NF1 (1,560 to 1,670 aa) (3P7Z) (light brown) and YBCH (pink) (rmsd of 2.9 Å for 101 Cα atoms), ligands PEV (green) and TEG (cyan) are shown in stick representation. The region around C13 to C16 of PEV of the Sec14 domain of the neurofibromin occupies a similar position as TEG in the YBCH domain. (Left) Orientation one, (Middle) orientation two, and (Right) zoomed view of the ligand binding region. (B) Superposition of crystal structure of Sec14 domain of PITP (98 to 245 aa) (1AUA) (dark brown) and YBCH (pink) (rmsd 4.4 Å for 101 Cα atoms). (Left) Orientation one, (Middle) orientation two, and (Right) zoomed view of the ligand binding region. Structural difference appeared beyond α6 of YBCH with respect to Sec14 domain, due to which the orientation of β5 of YBCH (marked with arrow) has changed. This led to the dimer formation of YBCH, which is not observed in Sec14 domain and making the structure of YBCH unique. The two BOG in PITP is shown in blue and TEG bound to YBCH is in cyan. One of the two BOG is in the TEG binding site while the other one is in a nearby area. All ligands are shown in stick representation.
Potential Lipid-Binding Region in YBCH.
Previously it was shown that BNIP-2 binds to phosphatidylserine (a phospholipid) via its BCH domain, which implies that the BCH domain has lipid-binding properties (33). Moreover, the Sec14 domain of neurofibromin was highlighted in our N-terminal domain structural search as being homologous with the BCH domain. Sec14 domains are known to mediate lipid-based interactions. Thus, we suspected that YBCH may similarly be involved in lipid binding. However, sequence identity was low between the N-terminal regions of the YBCH domain and two Sec14 domain–containing proteins (neurofibromin, 15% homology [rmsd 2.9 Å for 101 Cα atoms]; Phosphatidylinositol transfer protein [PITP], 6% homology [rmsd 4.4 Å for 101 Cα atoms]) (Fig. 2). Furthermore, whereas the β5-strand of the YBCH domain forms part of an intermolecular β-sheet, the β5 equivalents of the Sec14 domains of neurofibromin and PITP instead form a β-sheet within the same molecule. Of interest, the lipid-binding regions of the Sec14 domains of both proteins aligned with the N-terminal core of the YBCH domain (1 to 102 aa) at the site of TEG binding in the crystal structure (Fig. 2 and SI Appendix, Fig. S7A). TEG was bound in a hydrophobic pocket of ∼20 × 10 × 15 Å involving α1, β2, α3, α4, β3, and α5. Specifically, this pocket is formed by Leu5, Ile22, Val24, Leu29, Leu40, Ile41, Val48, Phe62, and Leu75 of the core YBCH domain (key residues are underlined; refer to SI Appendix, Fig. S7B). These amino acids are conserved among BCH domains from different proteins (SI Appendix, Fig. S6B).
Additionally, the β-strand secondary structures and some of the hydrophobic residues of the lipid-binding region are conserved in the YBCH and Sec14 domains (Fig. 2 and SI Appendix, Fig. S6A). Notably, PEV (phosphatidylethanolamine; 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine), which binds the core region of the Sec14 domain of neurofibromin (PDB: 3P7Z), is stabilized by hydrophobic residues from β-strands and an α-helix equivalent to β3, α5, and β4 of the YBCH domain. Similarly, the crystal structure of the Sec14 of PITP (28, 34) (PDB: 1AUA) shows two β-octyl glucoside (BOG) molecules bound in the core domain of Sec14, where it is stabilized by β-strands and an α-helix equivalent to β4, β5, and α5 of the YBCH domain. One of these BOG molecules is present in the TEG pocket while the other is bound in a nearby region. Overall, these findings suggest that TEG in YBCH occupies a similar hydrophobic pocket as that of PEV in neurofibromin and β-octyl glucoside in PITP, respectively (Fig. 2). Therefore, the core region of the BCH domain (N-terminal region 1 to 100 aa) may have lipid-binding properties.
The Lipid-Binding Pocket of HBCH Domain Is Likely Responsible for Anchoring the Prenyl Tail of RhoA.
A model of the human BCH domain (HBCH) of p50RhoGAP was built in i-TASSER (Iterative Threading Assembly Refinement) (35) using the YBCH crystal structure to predict the equivalent key regions in human p50RhoGAP (Fig. 1A and SI Appendix, Figs. S6B and S8). The key residues of the lipid-binding pocket in YBCH domain are Ile22, Val24, and Val29; the corresponding residues in HBCH are Ile86, Phe88, and Met93. We mutated these residues to alanine in a triple-alanine mutant (3A-NBCH; NBCH refers to N-terminal of p50RhoGAP from aa 1 to 217). Also of note, because RhoA GTPases are prenylated as a posttranslational modification at the C terminus, specifically at the “CAAX” motif (36), we deleted this conserved region (RhoA ΔCAAX) to verify its role in the interaction between RhoA and the BCH domain.
We show that the HBCH triple-alanine mutant (3A-NBCH) was unable to capture the wild-type (WT) RhoA (SI Appendix, Fig. S9). Consistently, there was drastically reduced binding between WT NBCH (with an intact lipid-binding region) and RhoA ΔCAAX (SI Appendix, Fig. S9). These results are tempting to suggest that prenylation of RhoA and the lipid-binding region in the HBCH domain are required for anchoring the prenylated RhoA.
His97-Lys103 Loop Forms a Unique Rho-Binding Region in BCH Domain.
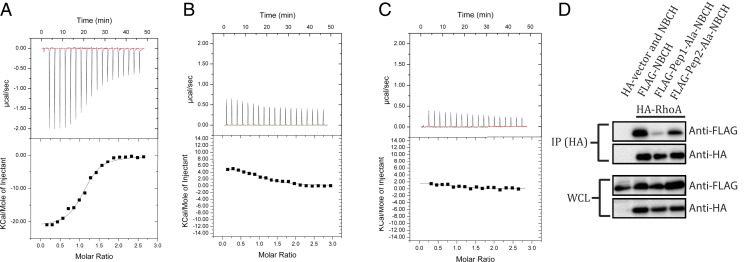
We previously showed that a putative Rho-binding motif (aa 85 to 120; 85RBM120) in HBCH is crucial for the interaction between RhoA and p50RhoGAP (13). Moreover, structural comparison of YBCH and HBCH (model) showed differences in two loop regions: His97-Lys103 and Ser131-Lys134 (HBCH nomenclature) (SI Appendix, Fig. S8B). Using isothermal titration calorimetry (ITC), we thus sought to examine the role of these regions in the interactions between RhoA and the BCH domain. As the HBCH protein was unstable and could not be purified, HBCH short peptides were instead created, designated as Pep1 (85IIVFSACRMPPSHQ-LDHSKLLGYLKHTLDQYVESDY120, HQLDHSK is the His97-Lys103 loop) and Pep2 (126HHGLTSDNKPS136, SDNK is the Ser131-Lys134 loop), where key loop regions are underlined. When titrated against RhoA, Pep1 had a Kd of 1.4 ± 0.5 µM, whereas Pep2 showed no significant interaction (Fig. 3 A and B). We observed that Pep1 of HBCH is involved in the binding with RhoA. This was confirmed with an alanine mutant of Pep1 peptide (Pep1-Ala) (85IIVFSACRMPPSAALAAAALLGYLKHTLDQYVESDY120), which failed to interact with RhoA (Fig. 3C). Thus, the surface-exposed His97-Lys103 loop (SI Appendix, Fig. S10) has a critical role in the interaction between HBCH and RhoA. Of note, RhoA for ITC was produced in Escherichia coli and thus was not posttranslationally modified; therefore, this binding was independent of RhoA prenylation.
Fig. 3.
Interaction of RhoA with HBCH. The upper panels show the raw ITC data for injection of BCH peptide into the sample cell containing RhoA. The peaks were normalized to the peptide: protein molar ratio and were integrated as shown in the bottom panels. Solid dots indicate the experimental data, and their best fit was obtained from a nonlinear least squares method, using a single site binding model depicted by a continuous line. (A) The titration of the Pep1 peptide (85IIVFSACRMPPSHQLDHSKLLGYLKHTLDQYVESDY120, His97-Lys103 loop shown in underline) against RhoA had a Kd of 1.4 ± 0.5 μM, (B) Pep2 peptide (126HHGLTSDNKPS136, Ser131-Lys134 loop shown in underline) and (C) Pep1-Ala mutant (85IIVFSACRMPPSAALAAAALLGYLKHTLDQYVESDY120) show no binding with RhoA. (D) Coimmunoprecipitation of FLAG-NBCH (WT and mutants) by HA-RhoA using anti-HA magnetic beads. 293T cells were transfected with the expression vectors HA-RhoA or HA-vector and the FLAG-NBCH or its mutants as indicated. Bound protein complexes were resolved on SDS-PAGE and detected by the antibodies indicated. Equal loading of the lysates were demonstrated on the whole-cell lysate section.
Next, we verified these results in the presence of prenylated RhoA using the WT and alanine-mutated Pep1 and Pep2 from the BCH domain. We found an apparent reduction in the coimmunoprecipitation levels of HRhoA with the FLAG-tagged Pep1-Ala-NBCH mutant as compared to that with WT-NBCH or the Pep2-Ala-NBCH mutant. These results confirm the importance of the His97-Lys103 loop in this HBCH/RhoA interaction (Fig. 3D) and suggest that this loop is equally crucial for binding as the lipid-binding pocket, with both regions contributing to the stability of the RhoA–BCH interaction.
β5-Strand Is Crucial for Regulating p50RhoGAP Activity in S. pombe.
The binding between RhoA and the HBCH domain is crucial for presenting the bound RhoA for GAP-mediated inactivation. The GAP domain must be active and available to inactivate RhoA. Previous work shows that the HBCH domain maintains the GAP domain in an autoinhibited state, which impedes the activity of p50RhoGAP such that it cannot inactivate RhoA (29). We employed S. pombe cell growth and mutational assays to investigate this further.
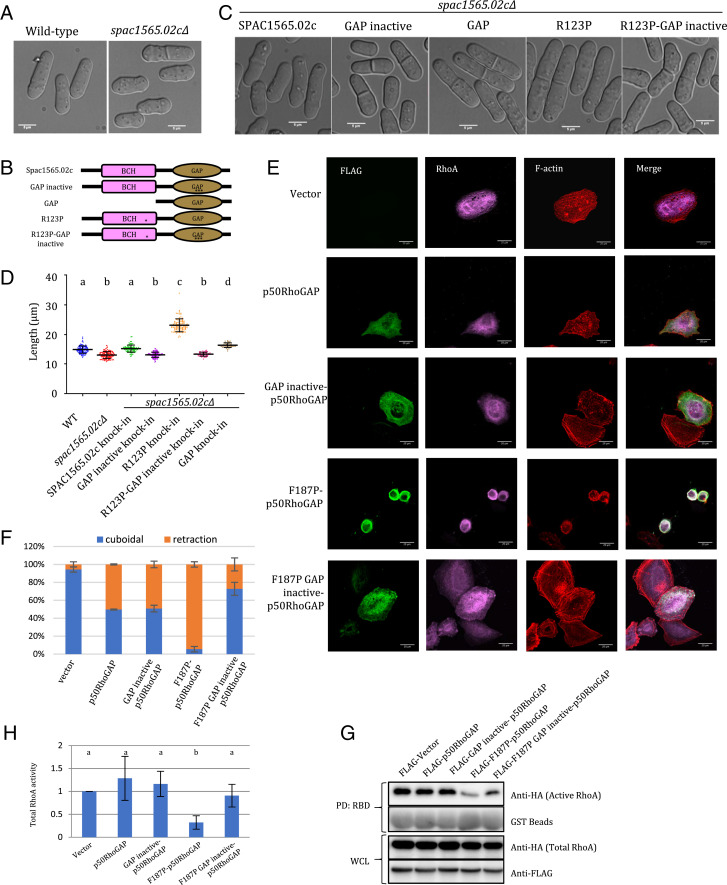
Spac1565.02c, a homolog of p50RhoGAP in S. pombe, carries an N-terminal BCH domain (amino acids 1 to 156; YBCH) and a C-terminal GAP domain (aa 170 to 352). spac1565.02cΔ cells, which lack p50RhoGAP, display mild morphological defects and are ∼12% shorter than WT cells (13.0 ± 1.1 μm versus 14.8 ± 1.2 μm at the time of division; Fig. 4 A and D and Table 2). Knock-in of SPAC1565.02c into spac1565.02cΔ cells can restore the original cell size (Fig. 4 C and D and Table 2), indicating that yeast p50RhoGAP is required to maintain cell shape and size. In contrast, when spac1565.02cΔ cells were transformed with inactive yeast GAP (R200A/K201A/N308A) [equivalent to the mutations of the catalytic triad of the GAP domain of p50RhoGAP (13)], the cells were ∼8% shorter than the WT (13.3 ± 0.8 μm versus 14.4 ± 1.0 μm) (Fig. 4 B–D). Comparatively, knock-in of the GAP domain alone generated a longer phenotype (16.3 ± 0.8 μm versus 14.4 ± 1.0 μm at the time of cell division), implying that the absence of the BCH domain results in an uninhibited, active GAP. These results further support the notion that the BCH and GAP domains of yeast p50RhoGAP work in concert for the proper maintenance of cell size in S. pombe.
Fig. 4.
Disruption of the β5 via R123P (YBCH) or F187P (HBCH) mutation activates GAP activity of Spac1565.02c (yeast p50RhoGAP) and p50RhoGAP, respectively. (A) Differential interference contrast images of WT (n = 107) and spac1565.02cΔ (n = 124) S. pombe cells (measurements from three independent experiments). spac1565.02cΔ cells were 12% shorter than WT cells. (B) Pictorial representation of various SPAC1565.02c constructs as indicated in B. * represents R123P, and *** represents Arg finger mutation (R200A/K201A/N308A). (C) spac1565.02cΔ cells with knocked-in constructs containing SPAC1565.02c (n = 93), GAP inactive (n = 116), GAP alone (n = 47), R123P (n =100), or R123P-GAP inactive (n = 57). (D) Dot plot of the length of the different yeast cell populations as described in A and C. Measurements from three independent experiments. Different letter denotes statistical significance at P < 0.05. Data was plotted and analyzed by Prism, Graphpad. (E) HeLa cells were cotransfected with HA-RhoA and FLAG-p50RhoGAP or its mutants as labeled. Immunostaining was performed. Representative images are shown. (Scale bar, 20 μm.) (F) The ratios of cuboidal and round cells were scored with at least 100 transfected cells counted per sample per experiment. The scores for “shrinkage” and “rounded” cells as “retraction” group. Data are means ± SD (n = 3). Scoring of the transfected cells were based on the criteria set by the study of Zhou et al. (13). (G) 293T cells were transfected with the expression vectors HA-RhoA and the FLAG-p50RhoGAP or its mutants as indicated. Cell lysates were incubated with immobilized GST-Rho–binding domain of rhotekin. Bound active RhoA were resolved by SDS-PAGE and detected by anti-HA antibody. Whole-cell lysate indicates equal loading. (H) Densitometry analysis of the active RhoA (RhoA-GTP bound by RBD assay) with the vector set as reference. Error bars represent SDs. Different letters denote statistical significance at P < 0.05. n = 5. Student’s t test was performed.
Table 2.
Cell dimensions of different yeast constructs
| Strain | Length (μm) | Width (μm) |
| WT (n = 107 cells) | 14.8 ± 1.2 | 4.1 ± 0.4 |
| spac1565.02cΔ (n = 124 cells) | 13.0 ± 1.1 | 4.1 ± 0.4 |
| SPAC1565.02c knock-in (n = 93 cells) | 14.4 ± 1.0 | 4.0 ± 0.3 |
| GAP inactive knock-in (n = 116 cells) | 13.3 ± 0.8 | 4.0 ± 0.5 |
| R123P knock-in (n = 100 cells) | 23.1 ± 1.7 | 4.6 ± 0.4 |
| R123P-GAP inactive knock-in (n = 57 cells) | 13.3 ± 0.7 | 3.6 ± 0.4 |
| GAP alone knock-in (n = 47 cells) | 16.3 ± 0.8 | 3.7 ± 0.4 |
Taking clues from the previous studies by us as well as others (13, 29) and from the crystal structure presented here, we hypothesized that the β5-strand of the BCH domain might play a crucial role in mediating this autoinhibition and that disrupting it may affect the autoinhibition of the GAP domain and thus RhoA activity in yeast and human cells. To investigate this, we used a proline substitution in the β-strand at Arg123 in YBCH (Phe187 in HBCH) to destabilize the secondary structure (37) of the β5-strand in yeast p50RhoGAP into spac1565.02cΔ. This mutation led to a drastic increase in cell size (60% longer and 15% broader) (23.1 ± 1.7 μm versus 14.4 ± 1.0 μm [length]; 4.6 ± 0.4 μm versus 4.0 ± 0.3 μm [width]). Most importantly, knock-in with a R123P-GAP inactive mutant reverted the phenotype to that of spac1565.02cΔ and that of the knock-in with the inactive GAP (13.3 ± 0.7 μm) (Fig. 4 B–D). These results clearly show that the R123P mutation in YBCH releases the GAP domain from autoinhibition thereby producing the phenotype associated with an active GAP.
β5-Strand Is Crucial for Regulating p50RhoGAP Activity in HeLa Cells.
Next, we sought to confirm whether the equivalent residue in human p50RhoGAP, F187, could similarly enhance GAP activity in human cell assays. First, an F187R mutant of human p50RhoGAP was designed (R from YBCH sequence, SI Appendix, Fig. S6B) to note the importance of the unconserved residues in the β5-strand. This F187R substitution did not affect the interaction between p50RhoGAP and RhoA or the oligomerization of p50RhoGAP (SI Appendix, Fig. S11 A and B), as the β-sheet is maintained by backbone hydrogen bonding contacts. This suggested little impact of the side chains on the BCH domain–mediated autoinhibition of GAP. However, only the Pro mutation in this position is associated with increased GAP activity in cells.
Next, HeLa cells were cotransfected with HA-RhoA and human FLAG-p50RhoGAP or various mutants (in the BCH and GAP regions, SI Appendix, Fig. S12), and we quantified the extent of cell retraction (cell shrinkage and rounding) and the associated Rho and RhoGAP activities, as previously described (13). HeLa cells transfected with p50RhoGAP bearing the F187P mutation exhibited extensive cell retraction, which is reminiscent of hyper-GAP activity in the cell (13) (Fig. 4E). It is noteworthy to mention that the cell retraction phenotype is not the result of decreased cell viability (SI Appendix, Fig. S13). This retraction could be rescued by introducing a GAP inactive (R282A/R283A/N391A) mutation into the F187P-p50RhoGAP mutant (F187P/R282A/R283A/N391A; “FLAG-187P GAP inactive p50RhoGAP mutant”; Fig. 4 E and F). Consistently, the level of active RhoA was greatly reduced by the introduction of the F187P mutation, and this could be abated by the GAP inactive/F187P mutant (Fig. 4 G and H). Taken together, our results demonstrate that the β5-strand regulates the autoinhibition of the full-length p50RhoGAP. The destabilization of the β5-strand due to a F187P mutation could release the GAP domain from this autoinhibition by the BCH domain. This released GAP domain therefore is capable of effectively inactivating RhoA as it is now “exposed” and functional. The release of autoinhibition might also expose the BCH domain in a similar manner for RhoA sequestration and self-association.
β5-Strand Mutation Enhances p50RhoGAP–RhoA Interaction.
In coimmunoprecipitation assays, we observed more F187P-p50RhoGAP binding with RhoA as compared with the WT-p50RhoGAP (Fig. 5A). We surmise that this is because the proline mutation releases the autoinhibition and allows the BCH domain to freely interact with RhoA. Comparatively, weak interactions were observed for the WT p50RhoGAP in its binding with RhoA; this is consistent with the role of the BCH domain in maintaining the GAP domain in the autoinhibited state.
Fig. 5.
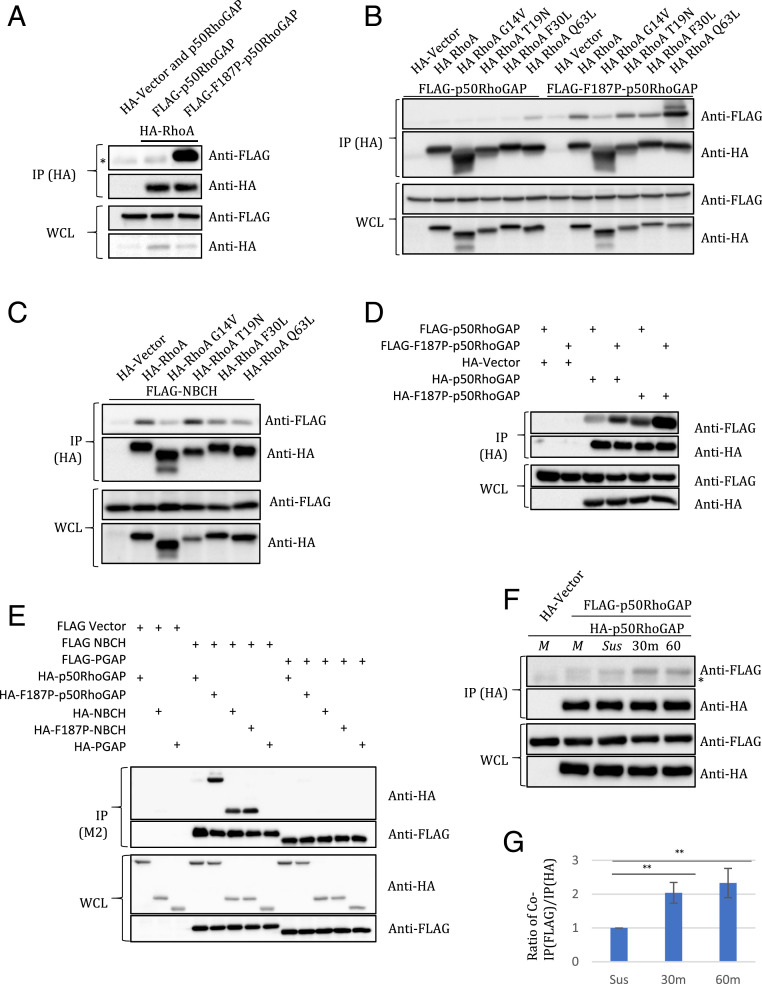
Intermolecular β5 sheet of BCH domain regulates p50RhoGAP interaction with RhoA and its homodimerization for cell spreading. (A) Coimmunoprecipitation of FLAG-p50RhoGAP or F187P-p50RhoGAP by HA-RhoA using anti-HA magnetic beads. 293T cells were transfected with the expression vectors HA-RhoA and the FLAG-p50RhoGAP or its mutants as indicated. Cells were lysed and immunoprecipitated with anti-HA magnetic beads. (B) Coimmunoprecipitation of FLAG-p50RhoGAP or FLAG-F187P-p50RhoGAP by HA-RhoA or its mutants using anti-HA magnetic beads. 293T cells were transfected with the expression vectors FLAG-p50RhoGAP or FLAG-F187P-p50RhoGAP and the HA-RhoA or its mutants as indicated. Cells were lysed and immunoprecipitated with anti-HA magnetic beads. (C) Coimmunoprecipitation of FLAG-NBCH by HA-RhoA or its mutants using anti-HA magnetic beads. 293T cells were transfected with the expression vectors FLAG-NBCH and the HA-RhoA or its mutants as indicated. Cells were lysed and immunoprecipitated with anti-HA magnetic beads. (D) Coimmunoprecipitation of FLAG-p50RhoGAP or FLAG-F187P-p50RhoGAP by HA-p50RhoGAP or HA-F187P-p50RhoGAP using anti-HA magnetic beads. 293T cells were transfected with the expression vectors HA-p50RhoGAP or HA-F187P-RhoGAP and the FLAG-p50RhoGAP or FLAG-F187P-RhoGAP as indicated. The loss of inhibition leads to apparent increase in self-association. Bound protein complexes were resolved on SDS-PAGE and detected by the antibodies indicated. (E) Coimmunoprecipitation of HA-p50RhoGAP, HA-F187P-p50RhoGAP, HA-NBCH, HA-F187P-NBCH, or HA-PGAP by FLAG-NBCH or FLAG-PGAP using anti-FLAG M2 beads. 293T cells were transfected with the expression vectors FLAG-Vector, FLAG-NBCH, or FLAG-PGAP and the HA-p50RhoGAP, HA-F187P-p50RhoGAP, HA-NBCH, HA-F187P-NBCH, or HA-PGAP as indicated. For A–E, bound protein complexes were resolved on SDS-PAGE and detected by the antibodies indicated. Equal loading of the lysates was demonstrated on the whole-cell lysate (WCL) section. (F) Coimmunoprecipitation of FLAG-p50RhoGAP by HA-p50RhoGAP using anti-HA magnetic beads. HeLa cells were cotransfected with the expression vectors HA-Vector or HA-p50RhoGAP and the FLAG-p50RhoGAP as indicated. HeLa cells were trypsinized and allowed to recover prior to seeding on collagen-coated surfaces. Unattached cells were washed off, and attached cells were lysed for Co-IP at the indicated time-points. “Sus” denotes cell in suspension. “M” denotes monolayer. Bound protein complexes were resolved on SDS-PAGE and detected by the antibodies indicated. Equal loading of the lysates was demonstrated on the WCL section. (G) Quantification of the band intensities as measured by Chemidoc (Bio-Rad) of F. The graph shows the ratio of the band intensity of coimmunoprecipitated FLAG-p50RhoGAP by the band intensity of the immunoprecipitated HA-p50RhoGAP. Error bars denote SD; n = 4; and ** denotes P value less than 0.01. All blots (A–F) are representative of at least three independent experiments. * denotes Ig heavy chain.
To examine the nucleotide specificity of RhoA binding, we performed coimmunoprecipitation of the BCH domain and p50RhoGAP with various RhoA functional mutants: constitutively active (CA)-RhoA-G14V, RhoA-Q63L, dominant negative (DN)-RhoA-T19N, and fast-cycling RhoA-F30L (Fig. 5 B and C). The coimmunoprecipitation results showed that the BCH domain preferentially bound the DN-RhoA-T19N mutant followed by WT RhoA over the CA and fast-cycling mutants (Fig. 5C). The full-length WT p50RhoGAP interacts weakly with the RhoA mutants, whereas the F187P-p50RhoGAP interacts strongly with RhoA-Q63L but not with the RhoA-G14V (Fig. 5B). Because there was no enhanced interaction between the BCH domain alone and RhoA-Q63L, the increased association between the full-length F187P and RhoA-Q63L may be due to sustained interaction of RhoA-Q63L with the GAP domain, consistent with the notion that failed GTP hydrolysis of RhoA Q63L means that it would be a preferred substrate for the GAP domain.
Next, we studied the interaction between RhoA-GDP and RhoA-GTP mutants with BCH peptides in vitro using biolayer interferometry experiments. We found that the biotinylated Pep1 peptide showed no preference toward the different nucleotide (GTP or GDP)-bound forms of HRhoA (SI Appendix, Fig. S14), suggesting that the preference identified in the coimmunoprecipitation assays was not conferred by the 85RBM120 region and that any preference is likely attributed to the whole BCH domain or even the full-length p50RhoGAP.
Finally, we show that the GAP domain alone interacts too transiently for detection with RhoA (SI Appendix, Fig. S15). Thus, it is plausible that different nucleotide-bound forms of RhoA can have varied preferential interactions with the GAP domain and that these interactions may be stabilized by the BCH domain. Taken together, these results are consistent with the results showing higher GAP activity for the Proline mutant (active form) compared to the WT protein (autoinhibited form) (Fig. 4). The results suggest that the BCH domain is the main interacting and stabilizing domain for RhoA. It confirms that the BCH domain is available for RhoA binding upon release of autoinhibition. The preference for a specific nucleotide form of RhoA may be attributed by regions beyond the BCH domain.
β5-Strand Mutation Enhances Intermolecular p50RhoGAP–p50RhoGAP Interactions.
BCH domain–containing proteins can undergo oligomerization with other BCH domain–containing proteins (23). To investigate how the release of the autoinhibition (by the F187P mutant; F187 is in the β5-strand) will affect the self-dimerization of p50RhoGAP, we performed coimmunoprecipitation experiments (Fig. 5D). We show that the loss of autoinhibition results in an apparent increase in p50RhoGAP self-association.
Next, we sought to determine which domain (BCH domain or GAP domain) is responsible for this elevated p50RhoGAP–p50RhoGAP self-association. We cotransfected HEK293T cells with FLAG-tagged NBCH and either HA-tagged p50RhoGAP, HA-tagged F187P-p50RhoGAP, HA-tagged NBCH, or HA-tagged F187P-NBCH (SI Appendix, Fig. S12). A significantly enhanced interaction was observed between NBCH and F187P-p50RhoGAP as compared with NBCH and WT p50RhoGAP (Fig. 5E). A similar result was observed between WT p50RhoGAP and F187P-p50RhoGAP as compared with the WT p50RhoGAP and WT p50RhoGAP (Fig. 5D). These results confirm that the F187P mutation indeed releases the GAP domain from autoinhibition, and this allows the released p50RhoGAP to self-associate via the BCH domain (Fig. 5D). Similar observation was made for BCH domain alone (SI Appendix, Fig. S16). The binding between F187P-NBCH and NBCH was similar to that for NBCH with NBCH, suggesting that the F187P mutation in the BCH domain does not confer a binding preference as compared with the WT BCH domain (Fig. 5E). Despite perturbation to the β5-strand, our data suggest that the WT and mutant protein structures are the same, and thus, the BCH–BCH interaction occurs without preference toward the WT or F187P proteins (Fig. 5 D and E). As expected, the GAP domain (present in the PGAP construct; residues 218 to 439 of HBCH; refer to SI Appendix, Fig. S12) does not interact with p50RhoGAP, F187P-p50RhoGAP, NBCH, F187P-NBCH, or itself (Fig. 5E), confirming that the GAP domain does not participate in p50RhoGAP oligomerization. Taken together, these results suggest that the F187P mutation in p50RhoGAP disrupts the intramolecular interaction with the GAP domain possibly through the formation of a “kink,” thus releasing the autoinhibition on the p50RhoGAP molecule and leading to enhanced RhoA binding and self-association.
Finally, to confirm that p50RhoGAP–p50RhoGAP interactions takes place under physiological conditions, we expressed different epitope-tagged p50RhoGAP molecules in HeLa cells. After sufficient cell spreading, we examined complex formation using coimmunoprecipitation assays. Cell spreading is regularly used to study cell dynamics governed by cytoskeleton rearrangement, which is subjected to regulation by Rho GTPases (13). We noted that the p50RhoGAP–p50RhoGAP association increased significantly over the 30 min of cell spreading (Fig. 5 F and G) in which Rho inactivation is evident (38). Consistently, it shows that the BCH domain becomes free and capable of self-associating upon release. This suggests that GAP activity positively correlates with BCH homo-oligomerization. Thus, similar to the case of GEF domain in p115RhoGEF (39), oligomerization (possibly the higher order of dimers) in the BCH domain may be crucial for the regulation of GAP activity.
Discussion
BCH domain–containing proteins comprise a family of highly conserved scaffolding proteins that regulate GTPases and their signaling partners and regulators, with critical roles in numerous aspects of active morphogenesis. Here, we reveal how the BCH domain can interact with RhoA via two distinct sites and how the release of the autoinhibition of the adjacent GAP domain can regulate GAP domain activity and thereafter lead to Rho inactivation. Despite a limited structural similarity, it appears that the BCH domain has evolved from the Sec14 domain (16) to adopt a unique architecture with additional features to regulate Rho activity. The lipid-binding pocket that is required, but not sufficient, for RhoA binding highlights the multiple levels of regulation that exist for RhoA binding to its target protein, in addition to the well-known function of the lipidated tails of GTPases for membrane insertion. Notably, the BCH domain’s preference for different nucleotide-bound or mutant RhoA is similar, but not identical, to that of the Rho GDP-dissociation inhibitor (RhoGDI), which acts to extract lipidated GTPases from the membrane (40, 41). Indeed, while it remains unclear whether p50RhoGAP itself can extract RhoA from the membrane in a manner similar to that of other RhoGDIs, it is plausible that p50RhoGAP accomplishes the capture, retention, and inactivation mechanism proposed in the present study: RhoA binding to both the lipid-binding pocket and the other Rho-binding motif maintains a necessary local concentration of RhoA for its inactivation by the adjacent GAP domain following an appropriate signal.
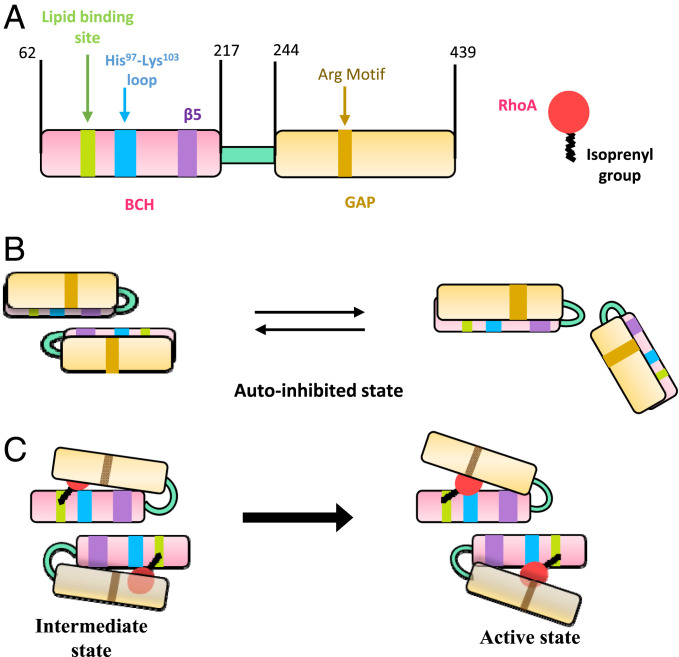
As previously proposed by Moskwa et al. and as also observed in our parallel studies with another homolog of p50RhoGAP, BPGAP1 (18, 21, 29), the BCH domain forms the intramolecular interaction with the GAP domain to control RhoGAP activity. While we do not yet have an atomic-level structure for the interplay between these two adjacent domains in the full-length p50RhoGAP, our extensive mutagenesis, functional, and morphological studies in both yeast and HeLa cells suggest how autoinhibition is maintained and how its “release” is governed by the integrity of the β5-strand located at the dimerization region of BCH domain. This suggest that the GAP activation essentially relies on the relative disposition between the BCH domain and GAP domain (Fig. 6).
Fig. 6.
A mechanistic model of p50RhoGAP regulation via its BCH domain. (A) Schematic representation of FL p50RhoGAP and RhoA. The BCH domain is shown in pink with the lipid-binding site, His97-Lys103 loop, and β5-strand in green, blue, and purple, respectively. The GAP domain is shown in yellow with the catalytic arginine motif in brown. The RhoA is shown as a red balloon/circle, while the C-terminal isoprenylation is shown as a black line. (B) The WT p50RhoGAP could be existing in the monomer–dimer equilibrium in the inactive state. In this state, BCH domain interacts with the GAP domain and keeps it autoinhibited, and therefore, no RhoA inactivation occurs. (C) RhoA is isoprenylated at its C terminus (CAAX motif). This isoprenyl group binds to the lipid-binding pocket present in the BCH domain of p50RhoGAP. The BCH domain dimer is a bystander in the regulation of GAP. As the isoprenyl group anchors on the BCH domain in this intermediate state, RhoA could interact with His97-Lys103 loop in BCH domain. In the autoinhibited state (even in the intermediate state), we believe that the GAP catalytic residue (Arg motif) is being occluded and incapable of RhoA inactivation, and it only becomes accessible upon the release of autoinhibition of GAP from the BCH domain. The lipid binding to BCH domain might initiate the transition for the release of GAP domain from the inactive state to the active state of p50RhoGAP. In other words, these binding events trigger the release of the GAP domain (here, the GAP domain is more transparent [schematically] for better RhoA visibility) from autoinhibition (at β5 shown in BCH domain). This leads to the activation of the GAP domain of p50RhoGAP. Upon activation of GAP domain, the Arginine motif would then aid in the GAP-mediated GTPase inactivation of RhoA. Thus, the p50RhoGAP becomes capable of regulating active RhoA population and thereby controls the cell morphology.
Zhu et al. (42) reported the need for onco-Dbl homo-oligomerization through its Dbl homology domain to generate a signaling complex that augments its Rho GTPase-activating potential. Similarly, the association of the BCH domains could serve to activate p50RhoGAP and further facilitate the presentation of RhoA to the GAP domain for its inactivation (Fig. 6). Notably, dimers are concentration dependent and of low affinity. In addition, they are most likely to be stabilized when present at high local concentrations, for example when localized on a membrane. We observed that oligomers (or possibly higher-order dimers) were accompanied by good GAP activity, as in the case of the mutant p50RhoGAP protein. Further, a variety of posttranslational modifications, such as phosphorylation and lipidation, will possibly regulate this oligomerization and GAP activity. For example, p122RhoGAP phosphorylation by AKT kinase leads to low-activity monomers while PKA phosphorylation facilitates GAP active dimers (43, 44). These reports show that posttranslational modification-dimerization relationships are not simplistic mechanisms but are governed by multiple factors. Other Rho regulatory proteins, such as GAPs and GEFs, target GTPases at their switch I and II regions and the P-loop (where the GTP/GDP molecule is bound) to enhance either GTPase activity or GDP/GTP exchange (45, 46). The p50RhoGAP prefers the RhoA Q63L mutant, which suggests that, in the full-length protein, the BCH domain locks onto the GTP-bound form of RhoA. This further facilitates the GAP-activated GTPase to carry out the hydrolysis of GTP to GDP (therefore, inactivating RhoA) before the complex falls apart. Further, we observed that the RhoA Q63L mutant binds tightly to p50RhoGAP but not to the BCH domain alone, indicating that regions beyond the BCH domain might contribute to this interaction. Sequence alignment of the BCH domain with GAP and GEF domains shows no significant sequence similarity among these RhoA-interacting motifs (SI Appendix, Fig. S17 A and B), further highlighting the significance of a unique motif that has evolved within part of the BCH domain.
Based on our results, we propose the following model for BCH regulation of GAP activity: 1) p50RhoGAP is present in an autoinhibited state (Fig. 6). 2) RhoA binds to two regions within the BCH domain: a) the His97- Lys103 loop and b) the potential lipid-binding region. This potential lipid-binding region possibly captures the prenylation moiety of RhoA (Fig. 2 and SI Appendix, Figs. S6A, S7, S9, and S10); this supposition builds on previous findings by Moskwa et al. (29) that the adjacent N-terminal BCH domain binding to prenylated small GTPases releases the autoinhibition of the GAP domain. 3) Anchoring RhoA to p50RhoGAP releases the GAP domain from its autoinhibited state. Of note, an F187P mutation in the BCH domain will also release p50RhoGAP from autoinhibition. 4) This loss of inhibition greatly enhances GAP activity and promotes p50RhoGAP–p50RhoGAP interactions (dimerization or higher order oligomerization) through the exposed BCH domains. 5) This oligomerization may help to sustain p50RhoGAP in its active form for RhoA binding and for its inactivation by the GAP domain. Clarifying these mechanisms will be instrumental in developing potential therapeutic approaches to disengage or restore BCH function, as implicated in various cancers and disease (47–50).
Materials and Methods
Cloning, Expression, and Purification of YBCH Domain.
YBCH (aa 1 to 156) was cloned into a modified pET32a vector that carries a (His)6-SlyD tag followed by PreScission protease cleavage site. The protein was purified using Ni-NTA affinity chromatography. Prior to crystallization and analytical ultracentrifugation experiments, the tag was cleaved with PreScission protease and the cation exchange chromatography (HiTrap SP HP [GE Healthcare]) was performed. More details are provided in the SI Appendix.
Crystallization and Structure Determination.
Purified YBCH at a concentration of 2.4 mg/mL was crystallized using hanging drop vapor diffusion method at room temperature (22 °C). The optimized crystallization condition consists of 0.1 M Bis-Tris propane pH 7.0 and 2.1 M NaCl. The crystals were dehydrated and cryoprotected prior to data collection. More details are provided in the SI Appendix.
Analytical Ultracentrifugation.
WT (2.4 mg/mL) YBCH was subjected to sedimentation velocity experiments using analytical ultracentrifugation to verify oligomerization. Sedimentation velocity profiles were collected by monitoring the absorbance at 280 nm. The samples were sedimented at 40,000 rpm at 24 °C for 5 h in a Beckman Optima XL-I centrifuge (Beckman Coulter Inc.) fitted with a four-hole AN-60 rotor and double-sector aluminum center pieces and equipped with absorbance optics. A total of 95 scans were collected and analyzed using Sedfit.
RhoA Purification.
Human RhoA (P61586; 1 to 182 amino acids) was purified with a hexa-histidine tag followed by size exclusion chromatography on 16/60 HiLoad Superdex 75 column (GE Healthcare) in gel filtration buffer (20 mM Tris 7.5, 100 mM NaCl, and 2 mM β-mercaptoethanol). More details are provided in the SI Appendix.
ITC.
The binding affinity between purified RhoA and different peptides of BCH domain were characterized using MicroCal iTC200 system. A total of 15 μM of RhoA was used in the sample cell. Titrations were done using 220 μM of peptides. All peptides used in this study were purchased from GL Biochem Ltd. All the samples were thoroughly degassed and centrifuged to remove any precipitates. Volumes of 4 μL per injection were used for all experiments and consecutive injections were separated by 4 min to allow the peak to return to baseline. The data were analyzed on Origin MicroCal iTC200 software.
Site-Directed Mutagenesis.
Site-directed mutagenesis on different genes described in this paper was achieved via inverse PCR technique (51) using the Kapa HiFi DNA polymerase Kit (KAPA Biosystems). Positive plasmids were verified by DNA sequencing.
S. pombe Strains, Media, and Growth Conditions.
Yeast strains used in this study are listed in SI Appendix, Table S2. PCR-based deletion of endogenous genes was employed to generate spac1565.02cΔ mutant as described by Janke et al. (52) Cells were routinely grown and maintained in the yeast extract medium or Edinburgh minimal medium with appropriate supplements, as described by Moreno et al. (53)
S. pombe Transformation.
Yeast transformation was performed using lithium acetate (LiAc)/Dimethyl sulfoxide (DMSO) method (54) with slight modification. In brief, 50 mL overnight culture (OD595 ∼ 0.5) was harvested at 3,000 × g for 1 min and washed with 1 mL LiAc/TE buffer (100 mM lithium acetate pH 7.5, 10 mM Tris⋅HCl, and 1 mM EDTA [ethylenediaminetetraacetic acid] pH 7.5). After suspending cells in 100 μL LiAc/TE buffer, 10 μL (10 mg/mL) salmon sperm carrier DNA (Sigma) and 1 to 2 μg DNA fragments (linearized) or plasmids were added to the suspended cells and incubated at room temperature for 10 min. Next, 260 μL polyethylene glycol (PEG)/LiAc/TE (40% wt/vol PEG 4000 in LiAc/TE buffer) was added to cells followed by gentle mixing. After incubating them at 30 °C for 45 to 60 min, 43 μL of DMSO was added to the cells followed by gentle shaking. Cells were heated at 42 °C for 5 min and washed with sterile water. Cells were plated on the selective agar plates.
Wide-field Fluorescence Microscopy.
Samples harvested from time-point indicated in the relevant section were observed directly without fixation using an IX81 wide-field fluorescence microscope (Olympus) with 60× NA 1.4 oil lens and 1.5× optivar. Fluorescence microscopy filter sets were purchased from Semrock and Omega. Cell images were captured using a complementary metal–oxide–semiconductor camera (Hamamatsu), and image acquisition was controlled by Metamorph (Molecular Devices). ImageJ (NIH) was used for image processing.
Cell Culture and Transfection.
Human 293T cells and HeLa cells were maintained in Roswell Park Memorial Institute-1640 medium and DMEM (Dulbecco’s Modified Eagle Medium) (high glucose), respectively. Both media were supplemented with 10% (vol/vol) Fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin (all from Gibco and Thermo Fisher). Cells were chemically transfected with indicated plasmids expression vector(s) using Lipofectamine 2000 (Invitrogen) or TransIT-LT1 (Mirus Bio), according to manufacturers’ protocol.
Construction of Expression Plasmids.
pXJ40-tagged RhoA and p50RhoGAP expression plasmids were obtained as described in ref. 13. Mutants of p50RhoGAP were constructed as described in the site-directed mutagenesis section.
Bio-Imaging.
HeLa cells were cultured on coverslip and transfected with indicated plasmids. Cells were fixed with 4% PFA and labeled with anti-FLAG (Sigma-Aldrich), anti-HA, and phalloidin (Invitrogen and Thermo Fisher). Cells were imaged with Nikon AIR model inverted confocal microscopy (Nikon).
Coimmunoprecipitation Studies and Western Blot Analyses.
Transfected cells were lysed in modified RIPA buffer (150 mM sodium chloride, 50 mM Tris, pH 7.3, 0.25 mM EDTA, 1% sodium deoxycholate, 1% Triton-X 100, 0.2% sodium fluoride, 5 mM sodium orthovanadate, 25 mM sodium glycerophosphate, and mixture protease inhibitors) (Roche Applied Science). Anti-FLAG M2 beads (Sigma-Aldrich) or Magnetic anti-HA beads (Pierce and Thermo Fisher) were used to immunoprecipitate FLAG-tagged or HA-tagged protein, respectively. Bound protein partners of the precipitated proteins were analyzed by Western blotting. Blots were probed with anti-FLAG (Sigma-Aldrich) and anti-HA (Invitrogen). For studies of p50RhoGAP dimerization during cell spreading, HeLa cells were transfected with expression vectors for 24 h as indicated via the use of Lipofectamine 2000 (according to manufacturer’s protocol—Thermo Fisher). HeLa cells were trypsinized and allowed to recover in 0.5% bovine serum albumin/DMEM for 1 h. Suspended cells were seeded onto collagen-coated surfaces and allowed to spread. Cells were washed with phosphate-buffered saline and harvested for coimmunoprecipitation (co-IP) at the stated time points.
Active RhoA Assay.
Detection for active RhoA populations in transfected cells were performed as described in ref. 12. Briefly, cells were cotransfected with HA-RhoA and FLAG-p50RhoGAP or its mutants for 24 h. Cells were lysed, and active-RhoA was pulled down by glutathione S-transferase (GST)-Rho–binding domain of Rhotekin (kind gift from Simone Schoenwaelder, Monash University, Australia). Bound active RhoA were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and detected by Western blotting.
Bio-Layer Interferometry (BLI).
BLI was used to characterize the affinity between purified RhoA and biotinylated peptides pep1, pep2, and pep1-ala from HBCH domain. More details are provided in the SI Appendix.
Cell Viability Assay.
The viability transfected cells were determined using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega). HeLa cells were seeded in a 96-well plate at a density of 2,500 cells/well. HeLa cells were then transfected with expression vector. After a 24 h transfection, 20 μL of CellTiter 96 AQueous One Solution Reagent was added to each well in 100 μL of culture medium. The plate was incubated at 37 °C for 4 h. The plate was then read at 490 nm using the Tecan Infinite M200 Microplate reader (Tecan Trading AG).
Supplementary Material
Acknowledgments
This work was supported by Ministry of Education (MOE) Singapore Tier 2 Grant R154-000-625-112 and National University of Singapore Academic Research Fund (AcRF) Tier 1 Grants R154-000-683-112, R154-000-C07-114, and R154-000-A72-115 (to J.S.). B.C.L. was supported by the Mechanobiology Institute Singapore, and by Singapore Ministry of Education AcRF Tier 3 Grant MOE2016-T3-1-002. The X-ray diffraction data were collected at the National Synchrotron Radiation Research Center Beamline 13B1 and Northeastern Collaborative Access Team Beamline 24IDC at Advanced Photon Source, Argonne National Laboratory, funded by the National Institute of General Medical Sciences from the NIH (Grant P30 GM124165). This research used the resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. Y.M.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014242118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. Atomic coordinates have been deposited in the Protein Data Bank: 7E0W.
References
- 1.Ridley A. J., Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Heasman S. J., Ridley A. J., Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Symons M., Settleman J., Rho family GTPases: More than simple switches. Trends Cell Biol. 10, 415–419 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Hodge R. G., Ridley A. J., Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 17, 496–510 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Jaffe A. B., Hall A., Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Bos J. L., Rehmann H., Wittinghofer A., GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129, 865–877 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Moon S. Y., Zheng Y., Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13, 13–22 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Pan C. Q., Low B. C., Functional plasticity of the BNIP-2 and Cdc42GAP Homology (BCH) domain in cell signaling and cell dynamics. FEBS Lett. 586, 2674–2691 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Mayer B. J., The discovery of modular binding domains: Building blocks of cell signalling. Nat. Rev. Mol. Cell Biol. 16, 691–698 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Ting Zhou Y., Guy G. R., Low C., BNIP-2 induces cell elongation and membrane protrusions by interacting with Cdc42 via a unique Cdc42-binding motif within its BNIP-2 and Cdc42GAP homology domain. Exp. Cell Res. 303, 263–274 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Soh U. J. K., Low B. C., BNIP2 extra long inhibits RhoA and cellular transformation by Lbc RhoGEF via its BCH domain. J. Cell Sci. 121, 1739–1749 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y. T., Guy G. R., Low B. C., BNIP-Salpha induces cell rounding and apoptosis by displacing p50RhoGAP and facilitating RhoA activation via its unique motifs in the BNIP-2 and Cdc42GAP homology domain. Oncogene 25, 2393–2408 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y. T., Chew L. L., Lin S. C., Low B. C., The BNIP-2 and Cdc42GAP homology (BCH) domain of p50RhoGAP/Cdc42GAP sequesters RhoA from inactivation by the adjacent GTPase-activating protein domain. Mol. Biol. Cell 21, 3232–3246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang X., Zhou Y. T., Low B. C., Concerted regulation of cell dynamics by BNIP-2 and Cdc42GAP homology/Sec14p-like, proline-rich, and GTPase-activating protein domains of a novel Rho GTPase-activating protein, BPGAP1. J. Biol. Chem. 278, 45903–45914 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Pan M., et al., BNIP-2 retards breast cancer cell migration by coupling microtubule-mediated GEF-H1 and RhoA activation. Sci. Adv. 6, eaaz1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta A. B., Wee L. E., Zhou Y. T., Hortsch M., Low B. C., Cross-species analyses identify the BNIP-2 and Cdc42GAP homology (BCH) domain as a distinct functional subclass of the CRAL_TRIO/Sec14 superfamily. PLoS One 7, e33863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buschdorf J. P., et al., Brain-specific BNIP-2-homology protein Caytaxin relocalises glutaminase to neurite terminals and reduces glutamate levels. J. Cell Sci. 119, 3337–3350 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Jiang T., Pan C. Q., Low B. C., BPGAP1 spatially integrates JNK/ERK signaling crosstalk in oncogenesis. Oncogene 36, 3178–3192 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Yi P., et al., KIF5B transports BNIP-2 to regulate p38 mitogen-activated protein kinase activation and myoblast differentiation. Mol. Biol. Cell 26, 29–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J., et al., BNIP-H recruits the cholinergic machinery to neurite terminals to promote acetylcholine signaling and neuritogenesis. Dev. Cell 34, 555–568 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Ravichandran A., Low B. C., SmgGDS antagonizes BPGAP1-induced Ras/ERK activation and neuritogenesis in PC12 cell differentiation. Mol. Biol. Cell 24, 145–156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Low B. C., Seow K. T., Guy G. R., Evidence for a novel Cdc42GAP domain at the carboxyl terminus of BNIP-2. J. Biol. Chem. 275, 14415–14422 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Low B. C., Seow K. T., Guy G. R., The BNIP-2 and Cdc42GAP homology domain of BNIP-2 mediates its homophilic association and heterophilic interaction with Cdc42GAP. J. Biol. Chem. 275, 37742–37751 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Low B. C., Lim Y. P., Lim J., Wong E. S. M., Guy G. R., Tyrosine phosphorylation of the Bcl-2-associated protein BNIP-2 by fibroblast growth factor receptor-1 prevents its binding to Cdc42GAP and Cdc42. J. Biol. Chem. 274, 33123–33130 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Kang J. S., et al., A Cdo-Bnip-2-Cdc42 signaling pathway regulates p38α/β MAPK activity and myogenic differentiation. J. Cell Biol. 182, 497–507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan C. Q., Liou Y. C., Low B. C., Active Mek2 as a regulatory scaffold that promotes Pin1 binding to BPGAP1 to suppress BPGAP1-induced acute Erk activation and cell migration. J. Cell Sci. 123, 903–916 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Panagabko C., et al., Ligand specificity in the CRAL-TRIO protein family. Biochemistry 42, 6467–6474 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Welti S., et al., Structural and biochemical consequences of NF1 associated nontruncating mutations in the Sec14-PH module of neurofibromin. Hum. Mutat. 32, 191–197 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Moskwa P., Lè Ne Paclet M.-H., Dagher M.-C., Bet Ligeti E., Autoinhibition of p50 Rho GTPase-activating protein (GAP) is released by prenylated small GTPases. J. Biol. Chem. 280, 6716–6720 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Eberth A., et al., A BAR domain-mediated autoinhibitory mechanism for RhoGAPs of the GRAF family. Biochem. J. 417, 371–377 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Molnár G., Dagher M. C., Geiszt M., Settleman J., Ligeti E., Role of prenylation in the interaction of Rho-family small GTPases with GTPase activating proteins. Biochemistry 40, 10542–10549 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Holm L., Sander C., Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Akamatsu R., Ishida-Kitagawa N., Aoyama T., Oka C., Kawaichi M., BNIP-2 binds phosphatidylserine, localizes to vesicles, and is transported by kinesin-1. Genes Cells 20, 135–152 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Li X., et al., Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol. Biol. Cell 11, 1989–2005 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts P. J., et al., Rho family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 283, 25150–25163 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph P. R. B., et al., Proline substitution of dimer interface β-strand residues as a strategy for the design of functional monomeric proteins. Biophys. J. 105, 1491–1501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren X.-D., Kiosses W. B., Schwartz M. A., Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18, 578–585 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chikumi H., et al., Homo- and hetero-oligomerization of PDZ-RhoGEF, LARG and p115RhoGEF by their C-terminal region regulates their in vivo Rho GEF activity and transforming potential. Oncogene 23, 233–240 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Mata R., Boulter E., Burridge K., The ‘invisible hand’: Regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 12, 493–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golding A. E., Visco I., Bieling P., Bement W. M., Extraction of active RhoGTPases by RhoGDI regulates spatiotemporal patterning of RhoGTPases. eLife 8, e50471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu K., Debreceni B., Bi F., Zheng Y., Oligomerization of DH domain is essential for Dbl-induced transformation. Mol. Cell. Biol. 21, 425–437 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ko F. C. F., et al., PKA-induced dimerization of the RhoGAP DLC1 promotes its inhibition of tumorigenesis and metastasis. Nat. Commun. 4, 1618 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Tripathi B. K., et al., Receptor tyrosine kinase activation of RhoA is mediated by AKT phosphorylation of DLC1. J. Cell Biol. 216, 4255–4270 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rittinger K., et al., Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature 388, 693–697 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Abdul Azeez K. R., Knapp S., Fernandes J. M. P., Klussmann E., Elkins J. M., The crystal structure of the RhoA-AKAP-Lbc DH-PH domain complex. Biochem. J. 464, 231–239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Münter S., Way M., Frischknecht F., Signaling during pathogen infection. Sci. STKE 2006, re5 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Sahai E., Marshall C. J., RHO-GTPases and cancer. Nat. Rev. Cancer 2, 133–142 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Vega F. M., Ridley A. J., Rho GTPases in cancer cell biology. FEBS Lett. 582, 2093–2101 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Zandvakili I., Lin Y., Morris J. C., Zheng Y., Rho GTPases: Anti- or pro-neoplastic targets? Oncogene 36, 3213–3222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dominy C. N., Andrews D. W., Site-directed mutagenesis by inverse PCR. Methods Mol. Biol. 235, 209–223 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Janke C., et al., A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Moreno S., Klar A., Nurse P., Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 (1991). [DOI] [PubMed] [Google Scholar]
- 54.Murray J. M., Watson A. T., Carr A. M., Transformation of Schizosaccharomyces pombe: Lithium acetate/dimethyl sulfoxide procedure. Cold Spring Harb. Protoc. 2016, prot090969 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Atomic coordinates have been deposited in the Protein Data Bank: 7E0W.