Significance
The catalytic cycles of metalloenzymes comprise fleeting intermediates, which are challenging to trap and characterize. Their detection, however, can provide invaluable insights into reaction mechanisms. Herein, we focus on SuiB, a radical S-adenosylmethionine (rSAM) enzyme that introduces a carbon–carbon bond at unactivated positions onto its peptide substrate. We present evidence for an unprecedented intermediate, a transient lysine-cross-linked tryptophan radical and its oxidation by a SuiB-bound Fe-S cluster. Our studies provide evidence for a radical electrophilic aromatic substitution pathway, a mechanistic paradigm that is likely common in metalloenzyme-catalyzed carbon–carbon bond formations. They also carry broad implications for the 165,000 rSAM enzymes that contain auxiliary Fe-S clusters.
Keywords: biochemistry, enzymology, EPR spectroscopy, bioinorganic chemistry, chemical biology
Abstract
The radical S-adenosylmethionine (rSAM) enzyme SuiB catalyzes the formation of an unusual carbon–carbon bond between the sidechains of lysine (Lys) and tryptophan (Trp) in the biosynthesis of a ribosomal peptide natural product. Prior work on SuiB has suggested that the Lys–Trp cross-link is formed via radical electrophilic aromatic substitution (rEAS), in which an auxiliary [4Fe-4S] cluster (AuxI), bound in the SPASM domain of SuiB, carries out an essential oxidation reaction during turnover. Despite the prevalence of auxiliary clusters in over 165,000 rSAM enzymes, direct evidence for their catalytic role has not been reported. Here, we have used electron paramagnetic resonance (EPR) spectroscopy to dissect the SuiB mechanism. Our studies reveal substrate-dependent redox potential tuning of the AuxI cluster, constraining it to the oxidized [4Fe-4S]2+ state, which is active in catalysis. We further report the trapping and characterization of an unprecedented cross-linked Lys–Trp radical (Lys–Trp•) in addition to the organometallic Ω intermediate, providing compelling support for the proposed rEAS mechanism. Finally, we observe oxidation of the Lys–Trp• intermediate by the redox-tuned [4Fe-4S]2+ AuxI cluster by EPR spectroscopy. Our findings provide direct evidence for a role of a SPASM domain auxiliary cluster and consolidate rEAS as a mechanistic paradigm for rSAM enzyme-catalyzed carbon–carbon bond-forming reactions.
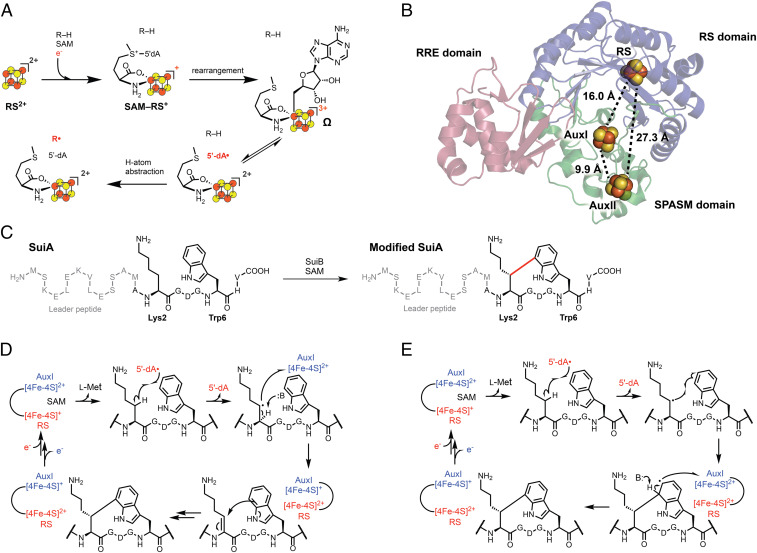
The radical S-adenosylmethionine (rSAM) enzyme superfamily is the largest known in nature, with over 570,000 annotated and predominantly uncharacterized members spanning all domains of life (1–4). The uniting feature of rSAM enzymes is a [4Fe-4S] cluster, usually bound by a CX3CX2C motif that catalyzes reductive cleavage of SAM to form L-Met and a strongly oxidizing 5′-deoxyadenosyl radical (5′-dA•) (5–7). Recent studies on a suite of rSAM enzymes have revealed the presence of a previously unknown organometallic intermediate in this process, termed Ω, in which the 5′-C of 5′-dA• is bound to the unique iron of the [4Fe-4S] cluster (Fig. 1A) (8, 9). Homolysis of the Fe–C bond ultimately liberates 5′-dA•, which abstracts a hydrogen atom from substrate to initiate a profoundly diverse set of chemical reactions in both primary and secondary metabolism, including DNA, cofactor, vitamin, and antibiotic biosynthesis (5, 10–13).
Fig. 1.
(A) Accepted scheme for radical initiation in rSAM enzymes. (B) X-ray crystal structure of SuiB (PDB ID: 5V1T). The RS domain, SPASM domain, and RiPP recognition element are rendered blue, green, and pink, respectively. [4Fe-4S] clusters are shown as spheres with the distances separating them indicated. (C) Lys–Trp cross-link formation (20) catalyzed by SuiB. The carbon–carbon bond installed by SuiB is shown in red. (D and E) Previously proposed EAS (D) and rEAS (E) mechanisms for SuiB-catalyzed Lys–Trp cross-link formation.
Of the 570,000 rSAM enzyme superfamily members, over a quarter (∼165,000 genes from the Enzyme Function Initiative-Enzyme Similarity Tool) possess C-terminal extensions, called SPASM and twitch domains, which bind auxiliary Fe-S clusters (4, 14–19). The SPASM domain typically binds two auxiliary Fe-S clusters and is named after the rSAM enzymes involved in the synthesis of subtilosin, pyrroloquinoline quinone, anaerobic sulfatase, and mycofactocin. The twitch domain is a truncated SPASM domain and only binds one auxiliary cluster (15). Despite the wide prevalence of these domains and the characterization of several different SPASM/twitch rSAM enzymes by spectroscopic and structural studies, direct evidence for their catalytic function(s) has remained elusive.
We previously performed functional and structural characterization on the SPASM rSAM enzyme SuiB (Fig. 1B), which is involved in the biosynthesis of a ribosomal peptide natural product in human and mammalian microbiome streptococci (14, 20–22). SuiB introduces an unusual carbon–carbon bond onto its substrate peptide, SuiA, between the sidechains of Lys2 and Trp6 (Fig. 1C). The mechanism for this transformation is of broader relevance, as a number of enzymes, such as RrrB, PqqE, and MqnC (2, 23, 24), are known to join unactivated aliphatic and aromatic carbons to generate sp3-sp2 cross-links. A general mechanistic paradigm for this class of transformations is not yet available. For SuiB, two pathways have been proposed (20), one through a typical electrophilic aromatic substitution (EAS) mechanism, which is involved in other enzyme-catalyzed indole modifications, such as indole prenylation or flavin adenine dinucleotide (FAD)-enzyme-dependent indole chlorination (25–27). In this pathway, the 5′-dA• generates an alkyl radical, which upon a second one-electron oxidation, creates an α,β-unsaturated amide electrophile with which the indole sidechain reacts via Michael addition (Fig. 1D). Lanthionine cross-links observed in diverse lanthipeptides are built by this general scheme, though via heterolytic chemistry, with Cys acting as the nucleophile (28, 29). Alternatively, a radical electrophilic aromatic substitution (rEAS) reaction has been proposed, wherein the alkyl radical, formed by 5′-dA•, would react with the indole sidechain to generate a radical σ complex, a cross-linked Lys–Trp radical (Lys–Trp•), which upon oxidation and rearomatization would yield product (Fig. 1E). In both mechanisms, AuxI is proposed as an oxidant. Although this role for an rSAM auxiliary cluster has been previously suggested (30, 31), it has yet to be directly demonstrated experimentally. Mechanistic studies have favored the rEAS pathway (20); however, intermediates in the reaction of SuiB and enzymes that catalyze similar reactions have not yet been detected (15).
In the current work, we sought to differentiate between the proposed mechanisms by trapping intermediates in the catalytic cycle of SuiB and characterizing them using electron paramagnetic resonance (EPR) spectroscopy. We report observation of three transient reaction intermediates, most importantly the sought-after Lys–Trp•, which is fundamentally different from previously characterized Trp radicals, as it is cross-linked and carries an indole tetrahedral center. We also provide evidence for AuxI as the oxidant of the Lys–Trp• intermediate as well as insights into redox potential changes of Fe-S clusters in SuiB that accompany SuiA binding. Together, our findings support the rEAS pathway for formation of the sp3-sp2 cross-link and carry important implications for other enzymes that catalyze related transformations.
Results
Detection of SuiB Reaction Intermediates.
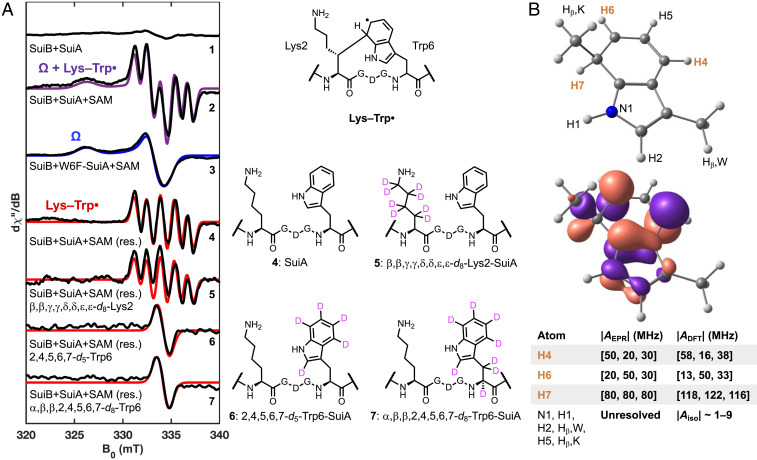
Our previous preparations of SuiB used a hexahistidine purification tag and resulted in incomplete Fe-S cluster incorporation. We undertook rigorous refinement of the SuiB expression and purification protocols and, using an alternative Strep purification tag, obtained higher Fe-S cluster content and improved enzyme quality with which we examined the reaction of SuiB (SI Appendix, Fig. S1). SuiB was reacted with excess sodium dithionite (DT), SuiA, and SAM; the reaction was freeze-quenched at various timepoints and subsequently examined by EPR spectroscopy. In the absence of SAM, no major signals were detected when the spectra were recorded at 50 K (Fig. 2A; spectrum 1). When SAM was included and the reaction quenched after 20 s, we observed accumulation of two paramagnetic intermediates, an organic and an organometallic species (spectrum 2). Two experiments were carried out to aid in identifying these intermediates: Reaction of SuiB with DT, SAM, and only the leader portion of SuiA, which contains residues −14 to −1 and not Lys2 and Trp6, resulted in an increase in the organometallic species and no formation of the organic radical (SI Appendix, Fig. S2). Similarly, reaction with W6F-SuiA, which is not cross-linked by SuiB (21), gave an enhanced organometallic signal and no organic radical intermediate (Fig. 2A, spectrum 3). These data are consistent with the organometallic intermediate preceding the organic radical. We assign the organometallic species, with a g-tensor of [2.054, 2.011, 2.002] as the apparently ubiquitous SAM-based Ω intermediate (9). This species has features similar to Ω previously reported for the pyruvate formate lyase activating enzyme (g||=2.035, g⊥=2.004) (8), and it persists into the steady state, a phenomenon that has also been observed in the rSAM enzyme viperin (32).
Fig. 2.
(A) Characterization of SuiB radical intermediates using X-band EPR at 50 K, 0.10 mT modulation amplitude, and 2.0 mW of nonsaturating microwave power. 1: A sample containing 250 µM SuiB, 10 mM DT, and 2 mM SuiA yields no major signals. 2: Upon addition of 4 mM SAM, the sample was quenched in liquid nitrogen after 20 s on ice, which gave rise to two major signals (sum of components simulated in purple). These were deconvoluted using several substrate variants. 3: A 20 s reaction using the W6F-SuiA suppressed the formation of one of the two paramagnetic species observed with wt SuiA. We classify the remaining signal as the SAM-dependent organometallic species Ω. The blue trace is the simulated spectrum with g = [2.054, 2.011, 2.002]. 4: The organic radical component proposed to be Lys–Trp• is the difference between the previous two spectra. The spectrum can be modeled (red trace) with strong hyperfine coupling to three protons (g = [2.0042, 2.0042, 2.0042], AH = [80, 80, 80; 20, 50, 30; 50, 20, 30] MHz). 5: Use of the isotopologue d8-Lys2-SuiA had no significant effect on the splitting pattern. 6, 7: However, with d5-Trp6-SuiA or d8-Trp6-SuiA, the splitting collapses entirely into a single line of equal line shape (g = [2.0042, 2.0042, 2.0042]). See SI Appendix, Figs. S6–S10 for 1H-NMRs of amino acid isotopologues. (B) Structure and numbering scheme (Top) and the singly occupied molecular orbital (SOMO) generated through a DFT calculation of a doubly methyl-capped analog of the Lys–Trp• (Bottom). The three strongest EPR-based and DFT-predicted hyperfine coupling constants are highlighted and compared in the table. The hyperfine coupling constants of other protons and N1 are too small to resolve by EPR splitting patterns, as is consistent with DFT. See SI Appendix, Table S1 for a list of the smaller DFT-predicted hyperfine coupling constants and SI Appendix, Table S3 for atomic coordinates.
Identification of the Organic Radical.
The organic radical (Fig. 2A, spectrum 4) can be discerned upon subtraction of the Ω signal (spectrum 3) from the composite spectrum (spectrum 2). Reactions with only the leader peptide and with W6F-SuiA above suggest that the organic radical resides in the core sequence of SuiA. We suspected that it may be the lysyl radical (Lys•) or possibly the Lys–cross-linked Trp radical (Lys–Trp•). To test these possibilities, we prepared SuiA containing a sidechain-deuterated d8-Lys2 via solid-phase peptide synthesis. Reaction with SuiB, DT, and SAM under the same conditions did not affect the organic radical feature, ruling out Lys• (spectrum 5). We also synthesized SuiA containing at residue Trp6 either an indole-deuterated Trp isotopologue (d5-Trp6) or uniformly deuterated Trp (d8-Trp6). Upon reacting these site-specifically deuterated SuiA peptides with SuiB, DT, and SAM, we observed collapse of the hyperfine-splitting patterns, which appeared with wt SuiA (spectrum 4), into a single line of similar line width (Fig. 2A, spectra 6 and 7). These results confirm that the radical resides on the indole sidechain. EPR spectral simulations, in conjunction with density functional theory (DFT) calculations using the ZORA-def2-TZVP basis set (33) and the BP86 density functional (34–36), clearly identified three strongly coupled protons in this intermediate, one isotropic out-of-plane proton (H7, A = [80, 80, 80] MHz) and two anisotropic in-plane protons (H4, H6, with A = [20, 50, 30] MHz and [50, 20, 30] MHz, respectively), validating this intermediate as the hypothesized Lys–Trp• (Fig. 2B), and excluding the possibility of the C7-deprotonated radical anion. The discrepancy in the hyperfine coupling constants of H7 between EPR-observed (Aiso ∼80 MHz) and DFT-predicted values (Aiso ∼120 MHz) may be due to an active site hydrogen bonding interaction involving H7. In contrast to Trp radicals characterized before, the Lys–Trp• does not couple strongly to the out-of-plane β-protons, as experiments with d5-Trp6-SuiA and d8-Trp6-SuiA demonstrate. Instead, strong hyperfine coupling to the aromatic protons, especially to the out-of-plane proton at the sp3-hybridized carbon is observed. Identification of the Lys–Trp• intermediate has important implications for the mechanism for SuiB (see below).
Two additionally interesting features of the trapped Lys–Trp• emerged from the studies with the amino acid isotopologues above (Fig. 2A). With d8-Lys2-SuiA, we observed ∼20% diminished Lys–Trp• signal, relative to natural abundance SuiA after 20 s (SI Appendix, Fig. S3). With d5-Trp6-SuiA and d8-Trp6-SuiA, however, we observed ∼60% enhanced Lys–Trp• signal intensity (SI Appendix, Fig. S3). These likely point to the importance of H-atom transfer reactions that precede and follow formation of the Lys–Trp• intermediate (Fig. 2A). With d8-Lys2-SuiA, slower abstraction of deuterium from Lys2, relative to a hydrogen atom, diminishes the rate of Lys–Trp• formation. This step is not rate-determining in multiple turnover, as we have previously shown (22), but it appears to be slow among steps up to formation of the Lys–Trp• intermediate. Moreover, conversion of the Lys–Trp• intermediate to product requires deprotonation of the indole sidechain and one-electron oxidation. Incorporation of deuterium at the indole slows down this reaction, leading to further build-up of the Lys–Trp• intermediate.
The Lys–Trp• intermediate displays an unusual electronic structure that has not been observed before in any indole-based radicals. Trp radicals observed thus far are formed by single electron oxidation. The Lys–Trp•, however, is formed by radical electrophilic addition to Lys• (37–43). Trp radicals observed so far display strong hyperfine coupling constants to one or both β-protons (depending on the dihedral angle) as well as the indole-N1 and H1; coupling constants to other aromatic protons are much weaker (SI Appendix, Table S1). These radicals may compensate for the missing π-system electron density by deprotonation at N1, resulting in the loss of the H1 hyperfine coupling, but an increase in the coupling constant to N1. The hyperfine coupling patterns of typical Trp radicals follow the predicted highest occupied molecular orbital (HOMO) of Trp (44, 45). By contrast, the Lys–Trp• exhibits the opposite effect; it displays strong hyperfine coupling primarily to some aromatic protons, whereas N1 and Hβ splittings are too small to resolve. This pattern is consistent with the predicted lowest unoccupied molecular orbital (LUMO) of Trp. While the C7-deprotonated Lys–Trp• anion is a highly reducing intermediate and has thus far not been observed, the Lys–Trp• is observable likely due to a greater stability of the delocalized Lys–Trp• relative to the initial Lys• and a comparatively slow C7-deprotonation.
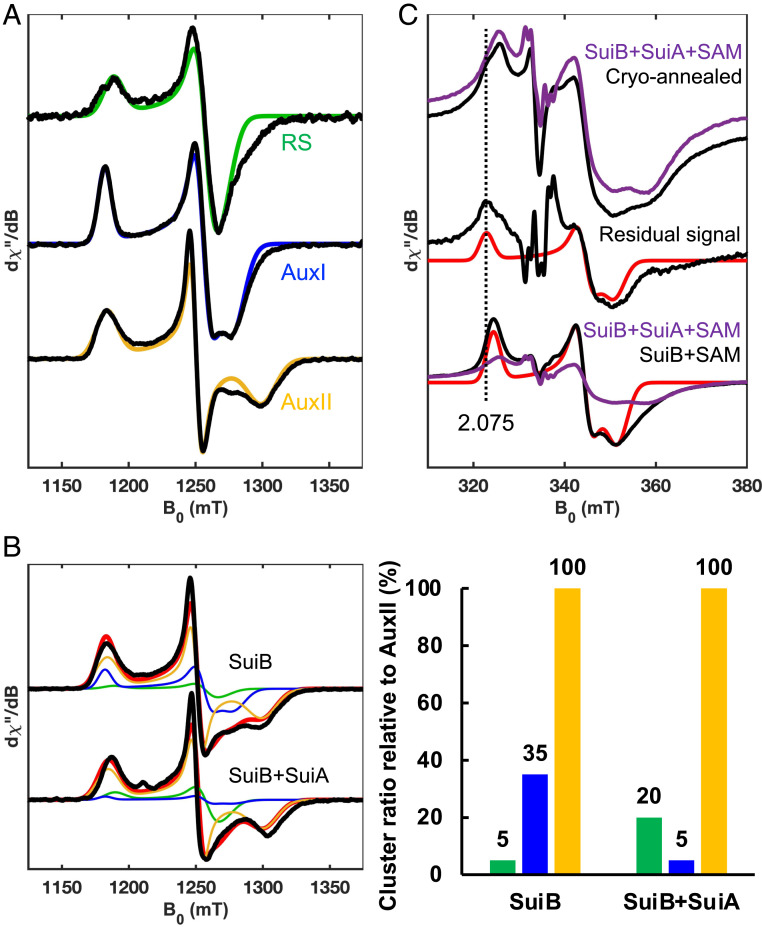
Identification of the Oxidant of the Lys–Trp•.
With the intermediates identified, we next sought to examine the fate of the Lys–Trp• species. To do so, we revisited the characterization of the [4Fe-4S] clusters in SuiB by EPR spectroscopy. Three reduced [4Fe-4S]+ clusters are observed in the resting state, corresponding to the radical SAM (RS) cluster, AuxI and AuxII (Figs. 1B and 3A), when SuiB is reduced and monitored by EPR spectroscopy at 10 K. The deconvolution and assignment of these was facilitated by recording EPR spectra at Q-band microwave frequency (∼33 GHz), which gives greater g value resolution (Fig. 3A), and by using site-directed SuiB mutants in which binding of the RS cluster or the AuxII cluster was abolished (SI Appendix, Fig. S4). The observed RS cluster signal in SuiB exhibits a near-axial g-tensor of [2.048, 1.936, 1.916], which is typical for RS clusters in rSAM enzymes (5, 46). In contrast, the two tetracysteine-ligated [4Fe-4S] clusters in SuiB, AuxI and AuxII, exhibit more rhombic g-tensors of [2.060, 1.939, 1.904] and [2.057, 1.946, 1.872], respectively. EPR signals from the three reduced [4Fe-4S]+ clusters are readily distinguishable by their g3 values, and this feature was used to deconvolute the contribution of each Fe-S cluster to the spectra below (SI Appendix, Fig. S4).
Fig. 3.
(A) Analysis of [4Fe-4S] clusters in SuiB by Q-band echo-detected field sweeps at 10 K. Individual cluster components determined from the spectra of wild-type (wt) SuiB and the RS and AuxII knockout mutants reduced by DT (SI Appendix, Fig. S4). Predominantly single-component RS (g = [2.048, 1.936, 1.916], green), AuxI (g = [2.060, 1.939, 1.904], blue) and AuxII (g = [2.057, 1.946, 1.872], yellow) clusters could be separated and simulated. See SI Appendix, Fig. S4 for details. (B) The wt SuiB spectrum at 10 K was simulated using individual cluster components with and without addition of substrate SuiA. Upon binding of SuiA, the AuxI cluster signal diminishes, and the RS cluster signal increases. To the right of the spectra is a bar graph quantifying each cluster component used to model the spectra in panel B compared to the relatively static AuxII signal intensity. The effect of the AuxI cluster is also apparent using the RS knockout mutant (SI Appendix, Fig. S11). (C) Cryo-annealing of the Lys–Trp• measured at X-band frequency, 35 K, 0.80 mT modulation amplitude, and 1.0 mW of nonsaturating microwave power. Anaerobic cryo-annealing of a reaction sample (purple trace to black trace) at 200 K for 10 min removes the Lys–Trp• signal completely and gives rise to a residual [4Fe-4S] feature. No significant effect was observed when using the W6F-SuiA mutant (SI Appendix, Fig. S12). The residual [4Fe-4S] feature (g = [2.075 1.944 1.908]) compared to a control reaction sample lacking SuiA yields an AuxI signal (g = [2.066, 1.946, 1.905]). The slight g-tensor discrepancy can be attributed to presence of SuiA. See SI Appendix, Fig. S5 for structural comparison and SI Appendix, Table S4 for summary of SuiB [4Fe-4S] cluster g-tensors.
We established a relative ratio of the three [4Fe-4S]+ clusters in the resting state of SuiB after reduction with DT. While the AuxII cluster contributed the major signal, and its intensity was nearly static upon substrate binding, to our surprise, the RS cluster, which is active in the reduced state, was barely observable (∼5% relative to the AuxII signal), whereas a notable fraction of total signal corresponded to AuxI (∼35% relative to the AuxII signal) (Fig. 3B). Upon binding of SuiA, however, we observed a striking decrease in the AuxI signal (to ∼5% relative to the AuxII signal), suggesting it had been converted to the [4Fe-4S]2+ form. This shift occurred concomitantly with an increase in the signal for the RS cluster (to ∼20% relative to the AuxII signal), indicating it was now in the reduced [4Fe-4S]+ form upon SuiA binding. These results suggest that substrate binding sets the redox states of the RS and AuxI clusters. The RS cluster, now reduced, is poised to react with SAM. The AuxI cluster, now oxidized, could serve as the low-potential oxidant of the radical intermediate.
Under our conditions, the trapped Lys–Trp• was stable at 77 K or below. The maximum intensity was achieved when freeze-trapping at ∼20 s (SI Appendix, Fig. S2). Samples frozen at reaction times longer than 20 s had reduced amounts of the trapped Lys–Trp• signal. To allow the intermediate to react further, freeze-quenched SuiB samples, prepared with either wt SuiA or W6F-SuiA, were subjected to anaerobic cryo-annealing, a method that has previously been used to allow for characterization of difficult-to-trap intermediates in metalloenzyme reactions (47–49). Comparison of the resulting spectra revealed, only in the reaction with wt SuiA, disappearance of the Lys–Trp• signal concomitant with formation of a [4Fe-4S]+ feature very closely resembling AuxI, suggesting that the Lys–Trp• intermediate had been oxidized by AuxI (Fig. 3C). An interesting feature of the trapped AuxI cluster in the reduced form is its remarkably long relaxation time resulting in sharp signals even at elevated temperatures. Hence, at 35 K the AuxI signal retains a sharp line width, while the RS and AuxII cluster signals broaden substantially. The g-tensor obtained for the SuiA-bound reduced Aux I (g = [2.075, 1.944, 1.908]) diverged slightly from the resting state AuxI in the absence of substrate (g = [2.066, 1.946, 1.905]). The most distinct feature is the higher g1 = 2.075 compared to the resting state AuxI with g1 = 2.066. We attribute the slightly altered AuxI g-tensor to the presence of the substrate peptide, which in a cocrystal structure has been shown to bind adjacent to AuxI (SI Appendix, Fig. S5) (14). DT-treated, SuiA-bound AuxI could previously not be detected by EPR spectroscopy, as the AuxI cluster oxidized upon addition of SuiA (concomitant with reduction of the RS cluster). Importantly, the AuxI cluster could never be reduced when SuiB and SuiA were treated with DT, suggesting its redox potential is lower than that of DT (E°′ = −0.66 V versus normal hydrogen electrode; NHE). Thus, the strongly reducing Lys–Trp•, presumably upon deprotonation, is required for reduction of the SuiA-bound AuxI cluster during catalysis. Likewise, it appears the AuxI potential, upon peptide binding, is matched to react with the reducing, deprotonated Lys–Trp• intermediate.
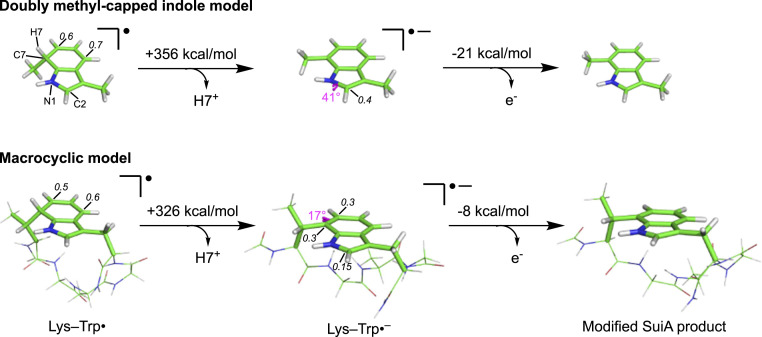
Modeling the Oxidation Reaction of Lys–Trp•.
To gain further insights into the oxidation reaction of Lys–Trp• by the AuxI cluster, DFT calculations (M06-2X/6–31+G(d,p)) (50) were performed using a doubly methyl-capped indole model (Fig. 4, Top) as well as a macrocyclic Lys–Trp• model (Fig. 4, Bottom). One viable mechanism for product formation involves initial deprotonation of H7 of Lys–Trp• to form a radical anion, Lys–Trp•–, followed by oxidation by the AuxI cluster to give the cross-linked SuiA product. The results of our DFT calculations indicate that the macrocyclic Lys–Trp• model is substantially more acidic than the doubly methyl-capped indole model. The computed gas phase acidities differ by ∼30 kcal/mol, suggesting that the macrocyclic ring promotes deprotonation, likely as a result of a change in macrocyclic ring strain upon deprotonation (51). On the other hand, the macrocyclic Lys–Trp•– radical anion is predicted to be less likely to lose an electron (by ∼13 kcal/mol; based on computed electron affinities) than the doubly methyl-capped indole Lys–Trp•–, but both are predicted to be much more likely to lose an electron than an indole radical anion (by ∼8 and ∼21 kcal/mol, respectively). The electron affinity of indole is predicted here (SI Appendix, Table S2) and observed experimentally to be ∼0 kcal/mol (52), implying that electron loss from Lys–Trp•– is inherently thermodynamically feasible. The geometries and electronic structures of the doubly methyl-capped indole Lys–Trp• model and macrocyclic Lys–Trp• model also differ from each other (Fig. 4). For the former, spin density is largely concentrated on the five-membered ring (note the pyramidalization of N1 and adjacent C2, as well as spin densities, in Fig. 4). For the latter, spin density is instead largely concentrated on the six-membered ring (note the pyramidalization of C7 that connects to Lys2, as well as spin densities, in Fig. 4). Thus, the macrocycle may itself be a necessary component of the catalytic pathway. While we cannot rule out proton-coupled electron transfer, the simulation of which is beyond our current capabilities, our results indicate that stepwise deprotonation and electron transfer is energetically viable.
Fig. 4.
Results from DFT calculations on the oxidation of the Lys–Trp• intermediate as a two-step process. When using either a doubly methyl-capped indole model (Top) or macrocyclic model (Bottom), a compensatory pyramidalization effect is seen in the anionic radical intermediate, Lys–Trp•–. While the site of pyramidalization in the doubly methyl-capped model is at N1 and C2 in the five-membered ring, it changes to C7 in the six-membered ring in the macrocycle. Hence, the macrocycle may be involved in stabilizing the pyramidal geometry at C7 in Lys–Trp•–, thereby leading to a more acidic H7. In each case, the subsequent oxidation step appears to be thermodynamically downhill relative to similar compounds (SI Appendix, Table S2). The largest Mulliken spin densities are labeled on atoms. For the macrocyclic anion, the spin density at C2 is shown for comparison. Since the substrate portion of SuiA is not resolvable in the SuiA-bound SuiB crystal structure (PDB ID: 5V1T, SI Appendix, Fig. S5), it is unclear specifically how the enzyme catalyzes these steps.
Detailed Mechanism for SuiB.
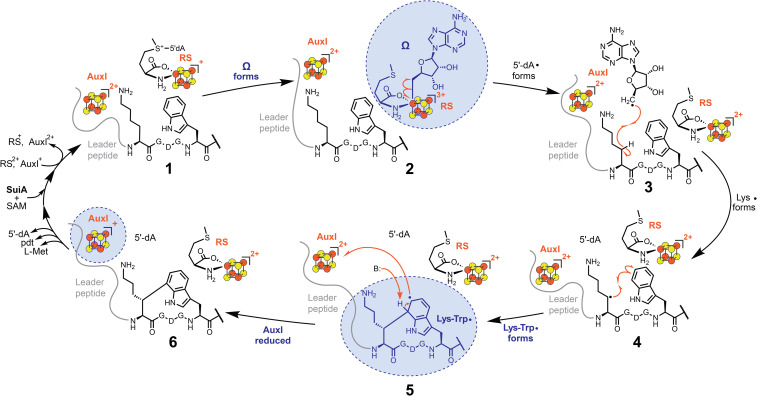
Our findings shed light onto the catalytic cycle of SuiB. Together, they visualize three paramagnetic intermediates and provide direct evidence in favor of the previously proposed rEAS mechanism (Fig. 5) (20, 22). In our current model, the catalytic cycle commences with binding of SuiA to SuiB, which results in a decrease of reduced AuxI and an increase of reduced RS, thus readying AuxI as an oxidant during the reaction and RS as a reductant to initiate SAM cleavage (Fig. 5, state 1). This feature, redox tuning by a substrate, has been reported in other metalloenzymes as well, notably cytochrome P450 enzymes (53). The organometallic Ω is an observable intermediate (Fig. 5, state 2; Fig. 2A, spectrum 3) followed by the Lys–Trp• via two transient radical intermediates: 5′-dA• (Fig. 5, state 3) and Lys• (Fig. 5, state 4). The formation of Lys• has been verified based on formation of 5′-2H-5′-dA upon reaction of SuiB with sidechain-deuterated d8-Lys2-SuiA (20, 22). The Lys• then reacts with Trp6 to generate the Lys–Trp•, which under our conditions is stable in the protonated form (Fig. 5, state 5; Fig. 2A, spectrum 4). H7 is likely the gatekeeper of the ensuing reduction reaction, as the Lys-Trp•– is highly reducing and can reduce the low-potential AuxI cluster to generate rearomatized product and reduced AuxI (Fig. 5, state 6; Fig. 3C). Release of product and binding of substrate would reset the redox states of the RS and AuxI clusters, thereby permitting multiple turnovers.
Fig. 5.
Proposed rEAS mechanism for the SuiB-catalyzed carbon–carbon bond formation between the Lys2 and Trp6 sidechains. The spectroscopically observed paramagnetic intermediates, Ω (Fig. 2A, spectrum 3), Lys–Trp• (Fig. 2A, spectrum 4) and AuxI (Fig. 3C), are highlighted in blue. See the Detailed Mechanism for SuiB section for details.
While the easily reducible AuxII cluster could conceivably be the final destination for the electron in vivo, this cluster is kept reduced by DT throughout our work in vitro. Consistent with reports that SuiB can recycle electrons for several catalytic cycles (21), the observed binding effects of SuiA may be involved in redirecting electrons back to the RS cluster prior to another round of turnover. In this model, AuxII is not required for the reaction, but it could conceivably provide a “safety net,” a repository of a reducing equivalent in case of off-target reactions at AuxI or the RS cluster. It may also form a conduit for introduction of reducing equivalents from external protein redox partners in the cytosol. Given that AuxI is the direct oxidant of the Lys–Trp• intermediate, AuxI and AuxII likely fulfill different roles during catalysis.
Discussion
A number of rSAM enzymes catalyze cross-coupling reactions to link unactivated sp2 and sp3 carbons. Generally speaking, these can follow an EAS pathway, in which the enzyme first generates an electrophile that is captured by the aromatic nucleophile, or alternatively an rEAS pathway in which an electrophilic radical reacts with an electron-rich aromatic system. Our motivation to differentiate between these mechanisms led us to conduct studies with SuiB with the aim of freeze-trapping and characterizing reaction intermediates. Perhaps the most compelling result is the identification of a Lys–Trp•, which we characterize with extensive EPR spectroscopy and DFT-based calculations. The necessity to work in a glovebox with an enzyme that contains three Fe-S clusters have made rigorous demonstration of chemical and kinetic competence of the Lys–Trp• intermediate challenging. Nonetheless, the absence of this species with W6F-SuiA and other SuiA variants and its ability to react with and reduce AuxI strongly suggest that the intermediate is on-pathway. Its formation in 20 s suggests kinetic competence (kcat ∼0.18 min−1 for SuiB). A rigorous kinetic analysis remains to be carried out in the future via anaerobic rapid freeze-quenching techniques.
The Lys–Trp• is fundamentally different from Trp radicals that have been observed before. Trp radicals, best characterized in the cationic form (Trp•+), are generated by one-electron oxidation (41). The Lys–Trp•, however, is formed by conversion of a π-bond into a σ-bond and a p-orbital occupying single electron. Unlike Trp•+, the Lys–Trp• carries a tetrahedral center, and the unpaired electron couples strongly to H7 at this center but not to the β-protons. The identification of the Lys–Trp• together with previous mechanistic studies rule out the EAS pathway for SuiB. Aside from SuiB, the rEAS pathway likely underpins catalysis by other rSAM enzymes that carry out similar transformations, and it will be interesting to search for cross-linked aromatic radicals in PqqE, RrrB, and similar enzymes (51, 54).
A second key insight is the role of AuxI in the catalytic cycle of SuiB as an oxidant of the Lys–Trp• intermediate, consistent with previous hypotheses (22, 30, 31). Particularly of interest and broader significance is the redistribution of electrons between AuxI and the RS cluster upon binding of SuiA. This result suggests that substrate binding modulates the redox potential of AuxI and RS. The molecular basis underlying this process remains to be determined, but the binding of SuiA in close vicinity to AuxI is in line with this notion. The role of AuxII remains to be determined. The obvious hypothesis that it shuttles electrons in and out of SuiB needs to be investigated with the appropriate protein redox partners. Given that SuiB can recycle electrons, AuxII may act as a reserve of reducing equivalents for the infrequent scenario of an off-pathway reaction at the active site but otherwise may be dispensable for Lys-Trp cross-link formation.
Our results will guide mechanistic studies in other rSAM enzymes that catalyze similar transformations. Likewise, they provide a starting point for investigating roles of Aux clusters in diverse SPASM/twitch-bearing rSAM enzymes. Redox reactions will certainly form one important function of Aux clusters in the broader SPASM/twitch enzymes. Given the breadth of this family, however, additional roles will likely be borne out as well. Especially noteworthy here are the thioether-building rSAM enzymes, which likely activate the Cys-thiol via a Lewis acid role while also serving a redox function (55). Indeed, with thousands of rSAM enzymes yet to be characterized, it is safe to assume that new reactions and mechanistic paradigms will emerge in the future.
Materials and Methods
Synthesis of SuiA and SuiA Variants.
The precursor peptide SuiA along with its variants were synthesized as previously described (21, 56). The protocol used for the synthesis of protected isotopologue amino acids is also described. See SI Appendix, Figs. S6–S10 for 1H-NMR spectra of protected, deuterated amino acids.
Site-Directed Mutagenesis.
SuiB cluster knockout mutants as well as the substitution of the His6 tag for a Strep tag were generated using the QuikChange Lightning (Agilent) site-directed mutagenesis kit as previously described (57, 58).
Expression, Purification, and Fe-S Cluster Reconstitution of His6-SuiB.
SuiB bearing a His6 tag (on a pET-28a vector) was expressed, purified, and reconstituted as previously described (21, 56).
Expression of Strep-SuiB.
Experiments were performed using the SuiB construct in a pET-28a vector as in previous studies (14, 20, 21, 56), but with the N-terminal residues HHHHHH substituted for the residues WSHPQFEK. The modified pET-28a plasmid bearing the Strep-SuiB gene was cotransformed with the pDB1282 plasmid (59) bearing the isc operon into Escherichia coli BL21(DE3), allowed to grow in SOC Outgrowth Medium (New England BioLabs) for 1 h at 37 °C before transfer onto an LB agar plate containing 100 µg/mL of ampicillin and 50 µg/mL of kanamycin and incubation overnight at 37 °C. On the following day, a single colony was used to inoculate 100 mL of LB containing the same concentrations of antibiotics, which was incubated in a shaker at 37 °C and shaken at 180 rpm overnight. On the following day, 15 mL of the preculture was used to inoculate 1.5 L of LB containing the same concentrations of antibiotics as well as 1 mM (NH4)2Fe(SO4)2. The culture was grown at 37 °C and 200 rpm until the optical density at 600 nm (OD600) ∼0.3, at which point the culture was supplemented with 0.2% L-arabinose and 5 mM L-cysteine. The culture was further grown to OD600 ∼0.6, at which point the temperature was lowered to 18 °C. After 1 h, 250 µM of IPTG was added, the shaking speed was lowered to 90 rpm, and the cell pellets were harvested on the following morning by centrifugation at 6,000 rpm for 15 min. Harvested cell pellets were flash frozen in liquid nitrogen and stored in a freezer at −80 °C.
Purification of Strep-SuiB.
To ensure high and consistent integrity of iron–sulfur clusters in the enzyme, the following protocol was executed on a single day in an anaerobic chamber with predeoxygenated solutions. Leftover enzyme was discarded. An ∼4 g of frozen cell pellet (from 1.5 L of cell culture) was brought into an anaerobic chamber and mixed with 40 mL of buffer A (300 mL KCl, 50 mM Hepes [pH 7.5]). The mixture was supplemented with 15 µL of Benzonase Nuclease (Millipore), 1 µL of rLysozyme (Millipore) and 1 Complete Protein Inhibitor Mixture tablet (Millipore). To induce lysis, 5 mL of BugBuster 10× Protein Extraction Reagent (Millipore) was added to the mixture, and the mixture was stirred for at room temperature. After 1 h, the solution was transferred into ultracentrifuge tubes and sealed prior to bringing them out of the anaerobic chamber. The solutions were ultracentrifuged at 35,000 g for 1 h at 4 °C. After this, the solution was transferred back into the anaerobic chamber and the supernatant was loaded onto 4 mL of Strep-Tactin XT resin (IBA Lifesciences). The resin was washed with 10 column volumes of buffer A. The protein was eluted by gentle addition of 3 column volumes of buffer A + 5 mM D-biotin. In our experience, the highest integrity of iron–sulfur clusters was obtained when subsequent concentration via centrifugation was avoided. Therefore, the darkest fractions of the eluant were collected separately and used to prepare EPR samples immediately, typically yielding ∼1 mL of ∼300 µM purified SuiB.
EPR Sample Preparation.
Stock solutions of Na2S2O4 (1 M in water), SAM (105 mM, dissolved and diluted in a 0.5 M KOH solution until pH ∼7) and SuiA (20 mM in buffer A) were freshly prepared in an anaerobic chamber. EPR samples were prepared by addition to 50 µL for Q-band samples (100 µL for X-band samples) of ∼250 µM SuiB in the sequence: i) 10 mM Na2S2O4, ii) 2 mM SuiA, iii) 4 mM SAM. Samples were loaded into clear fused quartz capillaries (VitroCom; 2.4 mm outer diameter, 2.0 mm inner diameter for Q-band samples; Wilmad-LabGlass; 4.0 mm outer diameter, 3.0 mm inner diameter for X-band samples). Mixtures were incubated for 1 min at room temperature before addition of SAM, and incubation for 20 s on ice (unless stated otherwise) after addition of SAM, followed by flash freezing in liquid nitrogen.
Anaerobic Cryo-Annealing.
Anaerobic cryo-annealing was performed on frozen reaction samples in the EPR cavity by increasing the temperature to 200 K while maintaining a steady flow of helium and thus anaerobic conditions. It took ∼1 min to increase the temperature from 35 K to 200 K. The sample was incubated at 200 K for 10 min, after which the temperature was lowered to 35 K.
Continuous Wave EPR Experiments.
Continuous wave EPR experiments were performed at X-band (9.37 GHz) using a Bruker (Billerica, MA) EleXsys E500 spectrometer equipped with a superhigh Q resonator (ER4122SHQE) with a conversion time of 60 ms and modulation frequency of 100 kHz. Other settings are given in the corresponding figure captions. Cryogenic temperatures were achieved and controlled using an ESR900 liquid helium cryostat in conjunction with a temperature controller (Oxford Instruments ITC503) and a gas flow controller.
Pulse EPR Experiments.
To minimize signal overlap experienced at X-band for cluster characterization, echo-detected field sweeps were collected at Q-band (33.9 GHz) using a Bruker EleXsys E580 spectrometer equipped with a 10 W amplifier and an R. A. Isaacson-built cylindrical TE011 resonator mounted in an Oxford CF935 cryostat. The standard Hahn echo sequence (π/2–τ–π–τ–echo) was applied to each sample via the XEPR software at 10 K and with π = 24 ns and τ = 300 ns at various magnetic field values. The resulting field sweeps were pseudomodulated using a modulation amplitude of 50 G.
EPR Spectral Analysis.
Simulations of spectra were performed using EasySpin 5.2.27 toolbox (60, 61) within the Matlab 2018b software suite (Mathworks Inc.).
DFT Calculations.
The geometry optimization and EPR parameter calculation of the doubly methyl-capped Lys–Trp• radical shown in Fig. 2B were carried out using the ORCA 4.0.1 quantum chemistry program (62) using the ZORA-def2-TZVP basis set (33) and the BP86 density functional (34, 35) as previously described (36). See SI Appendix, Table S3 for optimized atomic coordinates. Geometries of the Lys–Trp• models shown in Fig. 4 were optimized and free energies were calculated at the M06-2X/6–31+G(d,p) level using Gaussian09 (50, 63).
Supplementary Material
Acknowledgments
We thank Troy A. Stich and Daniel L. M. Suess for helpful discussions about spectroscopic assignments. This work was funded through an NSF CAREER Award (1847932, to M.R.S.), an NIH R35 Grant (1R35GM126961-01, to R.D.B.), an NSF XSEDE Grant (CHE-030089, to D.J.T.), and an NSERC Postdoctoral Fellowship (to A.R.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101571118/-/DCSupplemental.
Data Availability
The coordinates of the computed structures in Fig. 4 are available at https://www.iochem-bd.org/handle/10/222141. All other data are available in the article text or SI Appendix.
References
- 1.Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., Miller N. E., Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caruso A., Martinie R. J., Bushin L. B., Seyedsayamdost M. R., Macrocyclization via an arginine-tyrosine crosslink broadens the reaction scope of radical S-adenosylmethionine enzymes. J. Am. Chem. Soc. 141, 16610–16614 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Landgraf B. J., McCarthy E. L., Booker S. J., Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 85, 485–514 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Seyedsayamdost M. R., Caruso A., Davis K. M., “The chemistry and structural enzymology of RiPP-modifying radical SAM metalloenzymes” in Comprehensive Natural Products III, Begley T., Liu B., Eds. (Elsevier, 2020), pp. 49–64. [Google Scholar]
- 5.Broderick J. B., Duffus B. R., Duschene K. S., Shepard E. M., Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayler R. I., et al., Trapping and electron paramagnetic resonance characterization of the 5'dAdo• radical in a radical S-adenosyl methionine enzyme reaction with a non-native substrate. ACS Cent. Sci. 5, 1777–1785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H., et al., The elusive 5′-deoxyadenosyl radical: Captured and characterized by electron paramagnetic resonance and electron nuclear double resonance spectroscopies. J. Am. Chem. Soc. 141, 12139–12146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horitani M., et al., Radical SAM catalysis via an organometallic intermediate with an Fe-[5′-C]-deoxyadenosyl bond. Science 352, 822–825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byer A. S., et al., Paradigm shift for radical S-Adenosyl-l-methionine reactions: The organometallic intermediate Ω is central to catalysis. J. Am. Chem. Soc. 140, 8634–8638 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkovitch F., Nicolet Y., Wan J. T., Jarrett J. T., Drennan C. L., Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grove T. L., et al., A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science 332, 604–607 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Klinman J. P., Bonnot F., Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chem. Rev. 114, 4343–4365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flühe L., et al., The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 8, 350–357 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Davis K. M., et al., Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition. Proc. Natl. Acad. Sci. U.S.A. 114, 10420–10425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grell T. A., Goldman P. J., Drennan C. L., SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. J. Biol. Chem. 290, 3964–3971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barr I., et al., X-ray and EPR characterization of the auxiliary Fe-S clusters in the radical SAM enzyme PqqE. Biochemistry 57, 1306–1315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grell T. A. J., et al., Structural and spectroscopic analyses of the sporulation killing factor biosynthetic enzyme SkfB, a bacterial AdoMet radical sactisynthase. J. Biol. Chem. 293, 17349–17361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grove T. L., et al., Structural insights into thioether bond formation in the biosynthesis of sactipeptides. J. Am. Chem. Soc. 139, 11734–11744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haft D. H., Basu M. K., Biological systems discovery in silico: Radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J. Bacteriol. 193, 2745–2755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schramma K. R., Forneris C. C., Caruso A., Seyedsayamdost M. R., Mechanistic investigations of lysine-tryptophan cross-link formation catalyzed by streptococcal radical S-adenosylmethionine enzymes. Biochemistry 57, 461–468 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Schramma K. R., Seyedsayamdost M. R., Lysine-tryptophan-crosslinked peptides produced by radical SAM enzymes in pathogenic streptococci. ACS Chem. Biol. 12, 922–927 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Schramma K. R., Bushin L. B., Seyedsayamdost M. R., Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat. Chem. 7, 431–437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barr I., et al., Demonstration that the radical S-adenosylmethionine (SAM) enzyme PqqE catalyzes de Novo carbon-carbon cross-linking within a peptide substrate PqqA in the presence of the peptide chaperone PqqD. J. Biol. Chem. 291, 8877–8884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahanta N., Fedoseyenko D., Dairi T., Begley T. P., Menaquinone biosynthesis: formation of aminofutalosine requires a unique radical SAM enzyme. J. Am. Chem. Soc. 135, 15318–15321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh E., Garneau S., Walsh C. T., Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 102, 3960–3965 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh E., Blasiak L. C., Koglin A., Drennan C. L., Walsh C. T., Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases. Biochemistry 46, 1284–1292 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Miyamoto K., et al., A 7-dimethylallyl tryptophan synthase from a fungal neosartorya sp.: Biochemical characterization and structural insight into the regioselective prenylation. Bioorg. Med. Chem. 22, 2517–2528 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Repka L. M., Chekan J. R., Nair S. K., van der Donk W. A., Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 117, 5457–5520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortega M. A., van der Donk W. A., New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem. Biol. 23, 31–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grove T. L., Ahlum J. H., Sharma P., Krebs C., Booker S. J., A consensus mechanism for radical SAM-dependent dehydrogenation? BtrN contains two [4Fe-4S] clusters. Biochemistry 49, 3783–3785 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanz N. D., Booker S. J., Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochim. Biophys. Acta 1853, 1316–1334 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Honarmand Ebrahimi K., et al., The radical-SAM enzyme Viperin catalyzes reductive addition of a 5′-deoxyadenosyl radical to UDP-glucose in vitro. FEBS Lett. 591, 2394–2405 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Van Lenthe E., Ehlers A., Baerends E.-J., Geometry optimizations in the zero order regular approximation for relativistic effects. J. Chem. Phys. 110, 8943–8953 (1999). [Google Scholar]
- 34.Perdew J. P., Density-functional approximation for the correlation energy of the inhomogeneous electron gas. Phys. Rev. B Condens. Matter 33, 8822–8824 (1986). [DOI] [PubMed] [Google Scholar]
- 35.Becke A. D., Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A Gen. Phys. 38, 3098–3100 (1988). [DOI] [PubMed] [Google Scholar]
- 36.Tao L., Lai T. Y., Power P. P., Britt R. D., Germanium hydride radical trapped during the photolysis/thermolysis of diarylgermylene. Inorg. Chem. 58, 15034–15038 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Bleifuss G., et al., Tryptophan and tyrosine radicals in ribonucleotide reductase: A comparative high-field EPR study at 94 GHz. Biochemistry 40, 15362–15368 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Pogni R., et al., Tryptophan-based radical in the catalytic mechanism of versatile peroxidase from Bjerkandera adusta. Biochemistry 44, 4267–4274 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Pogni R., et al., A tryptophan neutral radical in the oxidized state of versatile peroxidase from Pleurotus eryngii: A combined multifrequency EPR and density functional theory study. J. Biol. Chem. 281, 9517–9526 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Stoll S., et al., Hydrogen bonding of tryptophan radicals revealed by EPR at 700 GHz. J. Am. Chem. Soc. 133, 18098–18101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sivaraja M., Goodin D. B., Smith M., Hoffman B. M., Identification by ENDOR of Trp191 as the free-radical site in cytochrome c peroxidase compound ES. Science 245, 738–740 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Li Y. F., Heelis P. F., Sancar A., Active site of DNA photolyase: tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochemistry 30, 6322–6329 (1991). [DOI] [PubMed] [Google Scholar]
- 43.Han W. G., Liu T., Lovell T., Noodleman L., Active site structure of class I ribonucleotide reductase intermediate X: A density functional theory analysis of structure, energetics, and spectroscopy. J. Am. Chem. Soc. 127, 15778–15790 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Mariadasse R., Choubey S. K., Jeyakanthan J., Insights into exogenous tryptophan-mediated allosteric communication and helical transition of TRP protein for transcription regulation. J. Chem. Inf. Model. 60, 175–191 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Compagnon I., Allouche A. R., Bertorelle F., Antoine R., Dugourd P., Photodetachment of tryptophan anion: An optical probe of remote electron. Phys. Chem. Chem. Phys. 12, 3399–3403 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Tao L., Zhu W., Klinman J. P., Britt R. D., Electron paramagnetic resonance spectroscopic identification of the Fe-S clusters in the SPASM domain-containing radical SAM enzyme PqqE. Biochemistry 58, 5173–5187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davydov R., Hoffman B. M., Active intermediates in heme monooxygenase reactions as revealed by cryoreduction/annealing, EPR/ENDOR studies. Arch. Biochem. Biophys. 507, 36–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davydov R. M., et al., Probing the ternary complexes of indoleamine and tryptophan 2,3-dioxygenases by cryoreduction EPR and ENDOR spectroscopy. J. Am. Chem. Soc. 132, 5494–5500 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukoyanov D., et al., A confirmation of the quench-cryoannealing relaxation protocol for identifying reduction states of freeze-trapped nitrogenase intermediates. Inorg. Chem. 53, 3688–3693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y., Truhlar D. G., The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 120, 215–241 (2008). [Google Scholar]
- 51.Nguyen T. Q. N., et al., Post-translational formation of strained cyclophanes in bacteria. Nat. Chem. 12, 1042–1053 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Linstrom P. J., NIST chemistry webbook. https://webbook.nist.gov/ (2005). Accessed 23 November 2020.
- 53.Hosseinzadeh P., Lu Y., Design and fine-tuning redox potentials of metalloproteins involved in electron transfer in bioenergetics. Biochim. Biophys. Acta 1857, 557–581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imai Y., et al., A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caruso A., Bushin L. B., Clark K. A., Martinie R. J., Seyedsayamdost M. R., Radical approach to enzymatic β-thioether bond formation. J. Am. Chem. Soc. 141, 990–997 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Bushin L. B., Seyedsayamdost M. R., Guidelines for determining the structures of radical SAM enzyme-catalyzed modifications in the biosynthesis of RiPP natural products. Methods Enzymol. 606, 439–460 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Balo A. R., Feyrer H., Ernst O. P., Toward precise interpretation of DEER-based distance distributions: Insights from structural characterization of V1 spin-labeled side chains. Biochemistry 55, 5256–5263 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Balo A. R., Lee J., Ernst O. P., Stationary phase EPR spectroscopy for monitoring membrane protein refolding by conformational response. Anal. Chem. 91, 1071–1079 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Lanz N. D., et al., RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods Enzymol. 516, 125–152 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Stoll S., Schweiger A., EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 178, 42–55 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Stoll S., Britt R. D., General and efficient simulation of pulse EPR spectra. Phys. Chem. Chem. Phys. 11, 6614–6625 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Neese F., Wennmohs F., Becker U., Riplinger C., The ORCA quantum chemistry program package. J. Chem. Phys. 152, 224108 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Frisch M. J.et al., Gaussian 09, Revision D.01 (Gaussian, Inc., Wallingford, CT, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the computed structures in Fig. 4 are available at https://www.iochem-bd.org/handle/10/222141. All other data are available in the article text or SI Appendix.