Abstract
Domain of unknown function 4228 (DUF4228) proteins are a class of proteins widely found in plants, playing an important role in response to abiotic stresses. However, studies on the DUF4228 family in soybean (Glycine max L.) are sparse. In this study, we identified a total of 81 DUF4228 genes in soybean genome, named systematically based on their chromosome distributions. Results showed that these genes were unevenly distributed on the 20 chromosomes of soybean. The predicted soybean DUF4228 proteins were identified in three groups (Groups I–III) based on a maximum likelihood phylogenetic tree. Genetic structure analysis showed that most of the GmDUF4228 genes contained no introns. Expression profiling showed that GmDUF4228 genes were widely expressed in different organs and tissues in soybean. RNA-seq data were used to characterize the expression profiles of GmDUF4228 genes under the treatments of drought and salt stresses, with nine genes showing significant up-regulation under both drought and salt stress further functionally verified by promoter (cis-acting elements) analysis and quantitative real-time PCR (qRT-PCR). Due to its upregulation under drought and salt stresses based on both RNA-seq and qRT-PCR analyses, GmDUF4228-70 was selected for further functional analysis in transgenic plants. Under drought stress, the degree of leaf curling and wilting of the GmDUF4228-70-overexpressing (GmDUF4228-70-OE) line was lower than that of the empty vector (EV) line. GmDUF4228-70-OE lines also showed increased proline content, relative water content (RWC), and chlorophyll content, and decreased contents of malondialdehyde (MDA), H2O2, and O2–. Under salt stress, the changes in phenotypic and physiological indicators of transgenic plants were the same as those under drought stress. In addition, overexpression of the GmDUF4228-70 gene promoted the expression of marker genes under both drought and salt stresses. Taken together, the results indicated that GmDUF4228 genes play important roles in response to abiotic stresses in soybean.
Keywords: soybean, genome-wide analysis, GmDUF4228 family, stress response mechanism, transgenic hairy root
Introduction
Abiotic stresses are the main limiting factors affecting plant growth and yield, causing significant agricultural and economic losses for farmers worldwide (Ahmad et al., 2014; Sade et al., 2018). For example, 70% of the total annual loss of yield potential is estimated to be due to the imbalance of the physical and chemical environments (Boyer, 1982). Drought and salt stresses are the main abiotic factors affecting the geographical distribution of plants, limiting the crop yield, and threatening food security (Mahalingam and Fedoroff, 2010). In general, plants lack the structures directly functioning in environmental perception, but could respond similarly to environmental changes (Hamant and Haswell, 2017). Plants show extensive defensive responses at the molecular and cellular levels to resist damage to cells caused by stresses (Kim et al., 2014).
Soybean (Glycine max L.) is a major agricultural crop, widely used as food source for humans and livestock due to its rich contents of oil, proteins, and minerals (Papiernik et al., 2005). It is also processed to produce traditional healthy foods in Chinese culture because of its nutritional composition and pharmacological value (Wang and Komatsu, 2018). Legume crops are suitable for growing in a wide range of climatic conditions, though they are extremely sensitive to flooding, drought, and salt (Hossain et al., 2013), especially during its seedling development and early reproductive stages (Gupta et al., 2020; Zhao et al., 2020). Under stress conditions, transcription factors such as bZIP and NAC form a complex regulatory network by binding to specific cis-acting elements and activating stress responsive genes, ultimately enabling plants to resist external stresses (Zong et al., 2016; Ma et al., 2018). The identification of genes that increase drought and salt tolerance of crops is essential for the effective agricultural use of land (Guan et al., 2014) and is of far-reaching significance of broadening the genetic basis of soybeans, enhancing their resistance to stress, and ensuring their sustainable production.
Domain of unknown function (DUF) proteins contain at least one highly conserved DUF domain and are widely found in plants (Bateman et al., 2010). Currently, there are more than 370,0 DUFs listed in the Pfam database, accounting for ∼25% of known domains (Finn et al., 2008). DUF1 and DUF2 were added to the Pfam database by Chris Ponting, who named the DUF domain and proposed naming these domains using a combination of “DUF” and a number (Schultz et al., 1998). Systematic structure analysis clarified that many DUFs may have arisen from the extreme diversification and neofunctionalization of known protein domains (Jaroszewski et al., 2009).
DUF4228 proteins are members of the DUF superfamily. Studies have shown that DUF4228 family members play an important role in response to abiotic stress in plants (Qi et al., 2019). Three members of DUF4228 family in Arabidopsis thaliana, i.e., AT1G10530, AT1G21010, and AT1G28190, are involved in response to drought stress (Yang Q. et al., 2020). Overexpression of the Medicago sativa DUF (MsDUF) gene in tobacco resulted in significantly lowered chlorophyll and soluble sugar contents and increased malondialdehyde (MDA) content. Expression of the MsDUF gene was significantly decreased under the treatments of NaCl, PEG6000, abscisic acid (ABA), and gibberellic acid (GA), indicating that MsDUF played a negative regulatory role in stress resistance of Medicago sativa (Wang et al., 2017). Expression of the Caragana intermedia DUF4228-3 (CiDUF4228-3) gene was significantly up-regulated under dehydration, low temperature, and drought, indicating its involvement in stress response (Na et al., 2016). Recent studies revealed that DUF4228 genes were involved in abiotic stress response and cadmium tolerance. Analysis of AtDUF4228 gene expression under a variety of stress treatments and co-expression network analysis of DUF4228 genes in different species of plants showed that several DUF4228 genes may play a synergistic role in plant defense (Didelon et al., 2020). These results suggest that members of the DUF4228 family may respond to abiotic stresses in soybean.
Although the whole genome of soybean was sequenced 10 years ago, studies investigating the GmDUF4228 gene family in soybean are still lacking. In this study, we identified a total of 81 GmDUF4228 genes and further analyzed their phylogenetic relationships, protein characteristics, gene structures, motifs, promoters, and expression patterns in various tissues and organs under different stress conditions. The expression levels of GmDUF4228-13, –43, –45, –48, –51, –58, –61, –70, and –80 under drought and salt stresses were analyzed by quantitative real-time PCR (qRT-PCR), and the function of the GmDUF4228-70 gene under drought and salt stresses was investigated. The findings provide a scientific foundation for further studies of the functions of GmDUF4228 genes.
Materials and Methods
Identification of DUF4228 Genes in Soybean
The Hidden Markov Model (HMM) corresponding to the DUF4228 family (PF14009) was used to identify DUF4228 proteins in the soybean genome using HMMER 3.0. Proteins with an E-value greater than 1e-20 were excluded for further analysis. All qualified GmDUF4228 protein sequences were extracted and aligned to build a soybean-specific HMM profile using the hmmbuild tool in HMMER 3.0. The new HMM profile was used to search a local protein database. Proteins with an E-value less than 0.001 were retained. After removing redundant sequences, the remaining protein sequences were submitted to the SMART1 and NCBI batch CD-search databasesT2 to confirm the presence of the DUF4228 domain (Shan et al., 2020).
Phylogenetic Analysis, Protein Characteristics, Gene Structure, and Motif Analysis of GmDUF4228
The amino acid sequences of DUF4228s derived from Arabidopsis and Medicago truncatula combined with newly identified GmDUF4228s in this study were used to construct phylogenetic trees. Multiple sequence alignment was conducted using the E-INS-I option of MAFFT 7.0 (Katoh and Standley, 2013). Gblocks 0.91b was used to remove ambiguous regions in the alignment with conserved regions used to construct the phylogenetic trees (Castresana, 2000). The maximum likelihood (ML) phylogenetic trees of a total of 135 DUF4228 protein sequences of Arabidopsis, Medicago truncatula, and soybean were reconstructed using MEGA 7.0 based on the JTT + G model with a 4-categories GAMMA distribution. The JTT + G model was identified based on ProTest (Posada, 2011). The bootstrap analysis was conducted with 1,000 replicates. To cluster the genetic structure, motifs, and expression profiles of GmDUF4228 genes in various tissues and organs of soybean, the maximum likelihood (ML) phylogenetic trees of a total of 81 DUF4228 protein sequences of only soybean were reconstructed using MEGA 7.0 based on the JTT + I + G model with a 4-categories GAMMA distribution. The JTT + I + G model was identified based on ProTest. The bootstrap analysis was conducted with 1,000 replicates. ExPASYT3 was used to calculate the molecular weight (MW) and isoelectric point (pI) of each GmDUF4228 protein sequence. The coding sequence (CDS) and cDNA sequences corresponding to each GmDUF4228 gene were submitted to the GSDS databaseT4 to analyze the exon-intron structure. The conserved motifs were analyzed using MEMET5.
Chromosome Localization and Analysis of Potential Cis-Elements
The information of the chromosome locations of GmDUF4228 family was obtained from the Phytozome databaseT6. All GmDUF4228 genes were mapped to the soybean chromosomes based on the genomic annotations. The 2,000 bp regions upstream of the start codon of 9 GmDUF4228 genes showing significant up-regulation in both drought and salt stresses were submitted to PlantCARET7 to identify the cis-acting elements and calculate the number of each element.
Expression Patterns of GmDUF4228 Genes in Different Tissues and Organs of Soybean
The fragments per kilobase of transcript per million mapped reads (FPKM) values for 81 GmDUF4228 family members in nine different tissues and organs (apical meristems, flowers, leaves, nodules, stems, pods, roots, root hairs, and seeds) under normal conditions were retrieved from the Phytozome database (see text footnote 6). TBtools software was used to draw a heatmap of expression data (Chen et al., 2020).
Plant Materials and Growth Conditions
The seeds of soybean Williams 82 were planted in a 10 × 10 cm pot. The leaves of soybean seedlings grown in the greenhouse for 15 days (with a photoperiod cycle of 16 h light/8 h dark, 25°C and/20°C at day and night, respectively, and 60% relative humidity) were used for total RNA extraction and qRT-PCR analysis. For drought stress, seedlings were placed on filter paper. Samples were collected at 0, 0.5, 1, 2, 4, 8, 12, and 24 h after treatment. For salt stress, the roots of seedlings were placed in a 200 mM NaCl solution. Samples were also collected at 0, 0.5, 1, 2, 4, 8, 12, and 24 h after treatment (Su et al., 2020). The collected samples were quickly moved into liquid nitrogen and then stored at −80°C for later use.
RNA Extraction and qRT-PCR
Total RNA was extracted from young plant samples using an RNA extraction and purification kit according to the manufacturer’s protocol (TIANGEN, Beijing, China) and reverse transcribed into cDNA using a PrimeScriptTM RT Reagent Kit (TaKaRa, Japan). The ABI prism 7500 sequence detection system (Applied Biosystems) was used for expression analysis based on qRT-PCR with three biological replicates for each sample. The 2–ΔΔ CT method was used to calculate the relative changes in gene expression (Cui et al., 2018). The qRT-PCR primers used to amplify GmDUF4228 genes were designed using Primer Premier 5.0 software. Soybean actin gene was used as the internal control for qRT-PCR (Zhao et al., 2017; Supplementary Table 1).
Construction of the GmDUF4228-70 Vector
The CDS of GmDUF4228-70 was amplified from Williams 82 cDNA with gene-specific primers containing restriction site sequences for NcoI and BsTEII. The PCR products and the pCAMBIA3301 vector were digested with NcoI and BsTEII (Thermo Fisher Scientific, United States), and the products were ligated to obtain pCAMBIA3301-GmDUF4228-70 (GmDUF4228-70-OE) lines (Kereszt et al., 2007).
Agrobacterium rhizogenes-Mediated Transformation of Soybean Hairy Roots
GmDUF4228-70-OE was studied using soybean hair roots, generated from soybean seedlings using Agrobacterium rhizogenes K599, which contained either the pCAMBIA3301 empty vector (EV) or pCAMBIA3301-GmDUF4228-70-OE and was injected 2 mm below different cotyledonary nodes. When the new roots grew to 5 cm in length, the old roots were excised. After 5 days of cultivation, transgenic plants were prepared for stress treatment as described by Kereszt et al. (2007).
Drought and Salt Stress Assays
After verification, the positive hairy roots of soybean were used in abiotic stress assays. Five transgenic soybean seedlings were cultivated in each pot with each stress treatment repeated for five times. For drought stress, the EV and GmDUF4228-70-OE plants were first dehydrated for 15 days, and then rewatered for 3 days. For salt stress, the EV and GmDUF4228-70-OE plants were treated with 200 mM NaCl for 7 days as described by Yang Y. et al. (2020).
Relative Water Content and Chlorophyll Content
To determine the relative water content (RWC), fresh leaves of the EV and GmDUF4228-70-OE plants were weighed. The leaves were completely immersed in sterile water for 12 h and then removed. Absorbent paper was used to absorb the excess water on the surface of the leaves and leaves were weighed to obtain the saturated weight. Finally, the leaves were wrapped in tin foil and incubated at 65°C for more than 72 h. After the leaves completely lost moisture, they were weighed to obtain the dry weight. A Sartorius BSA224S-CW 1/10,000 analytical balance (Sartorius, Beijing, China) was used to weigh the samples (Lu et al., 2009). For determination of chlorophyll content, fresh leaves (0.1 g) from EV and GmDUF4228-70-OE plants were cut into strips and immediately placed in a mixture of 50% anhydrous ethanol and 50% acetone and incubated in dark for 12 h. Then, 200 μl extract was extracted from the centrifuge tubes containing different samples and added to the enzyme plates. The absorbance values at 645 nm and 663 nm were measured using the Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific, Waltham, MA, United States). Each measurement was made from three biological replicates.
Measurement of Proline, Malondialdehyde, H2O2, and O2– Contents
The contents of proline, MDA, H2O2, and O2– were measured with the corresponding assay kits (Cominbio, Suzhou, China) based on the manufacturer’s protocols. All measurements were made from three biological replicates.
Trypan Blue, DAB, and NBT Staining
The transgenic soybean seedlings were cultivated without water for 7 days or treated with 200 mM NaCl solution for 3 days. The leaves of the EV and the GmDUF4228-70-OE seedlings were removed and were then submerged in 0.4% trypan blue, DAB (3,3′-diaminobenzidine) (Solarbio, Beijing, China), or NBT (nitroblue tetrazolium) (Solarbio, Beijing, China) staining solution for 12 h, respectively. Finally, the stained leaves were immersed in 75% ethanol until the leaves turned white.
Results
Genome-Wide Identification and Phylogenetic Analysis of GmDUF4228 Family Members
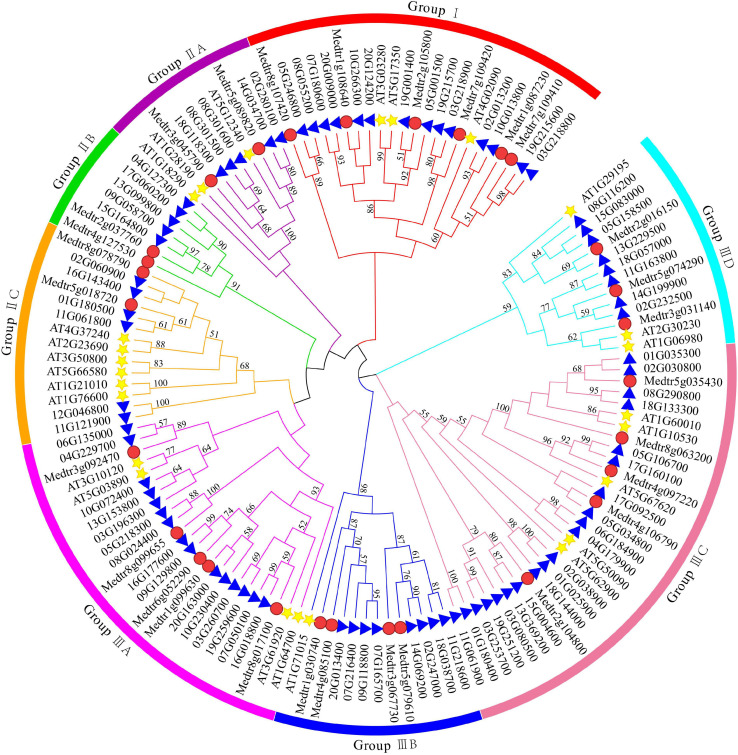
In this study, a total of 81 DUF4228 family members were identified from the soybean genome database and were named GmDUF4228-1 to GmDUF4228-81 according to their chromosomal distributions (Su et al., 2019). A total of 135 amino acid sequences (i.e., 25 AtDUF4228 proteins, 29 MtDUF4228 proteins, and 81 GmDUF4228 proteins) were compared to evaluate their evolutionary relationships, and a phylogenetic tree was built in MEGA 7.0 using the ML method (Figure 1). The topology of the ML tree showed that the 135 GmDUF4228s were identified in three groups (Groups I to III). The predicted GmDUF4228 polypeptide sequences ranged from 141 to 659 amino acids in length, with MW ranging from 15.24 to 75.27 kDa, and pI ranging from 5.59 to 10.13 (Table 1).
FIGURE 1.
Phylogenetic tree of DUF4228 proteins from Arabidopsis, Medicago truncatula, and soybean using the ML method. The colored arcs indicate different groups. The stars, circles, and triangles represent proteins of Arabidopsis Medicago truncatula, and soybean, respectively.
TABLE 1.
Molecular characteristics of the GmDUF4228 genes identified in soybean.
| Gene name | Gene ID | CDS (bp) | ORF (aa) | MW (kDa) | pI | Chromosome |
| GmDUF4228-1 | 01G025900 | 729 | 242 | 27.4 | 9.77 | 1 |
| GmDUF4228-2 | 01G035300 | 519 | 172 | 19.14 | 10.11 | 1 |
| GmDUF4228-3 | 01G180400 | 825 | 275 | 30 | 6.88 | 1 |
| GmDUF4228-4 | 01G180500 | 480 | 159 | 16.96 | 8.37 | 1 |
| GmDUF4228-5 | 02G013200 | 543 | 180 | 20.78 | 8.87 | 2 |
| GmDUF4228-6 | 02G030800 | 519 | 172 | 19.19 | 10.11 | 2 |
| GmDUF4228-7 | 02G038900 | 750 | 249 | 27.98 | 9.51 | 2 |
| GmDUF4228-8 | 02G060900 | 498 | 165 | 17.96 | 8.27 | 2 |
| GmDUF4228-9 | 02G232500 | 516 | 171 | 18.68 | 9.86 | 2 |
| GmDUF4228-10 | 02G247000 | 645 | 214 | 23.62 | 9.74 | 2 |
| GmDUF4228-11 | 02G280100 | 687 | 228 | 25.54 | 8.53 | 2 |
| GmDUF4228-12 | 03G080500 | 804 | 267 | 30.93 | 9.8 | 3 |
| GmDUF4228-13 | 03G196300 | 471 | 156 | 17.71 | 8.56 | 3 |
| GmDUF4228-14 | 03G218800 | 600 | 199 | 22.72 | 8.98 | 3 |
| GmDUF4228-15 | 03G218900 | 498 | 165 | 18.12 | 9.46 | 3 |
| GmDUF4228-16 | 03G253700 | 729 | 242 | 27.63 | 10.08 | 3 |
| GmDUF4228-17 | 03G260700 | 615 | 204 | 22.44 | 5.59 | 3 |
| GmDUF4228-18 | 04G127300 | 426 | 141 | 15.24 | 7.73 | 4 |
| GmDUF4228-19 | 04G179900 | 522 | 173 | 19.24 | 9.83 | 4 |
| GmDUF4228-20 | 04G229700 | 447 | 148 | 16.24 | 9.32 | 4 |
| GmDUF4228-21 | 05G001500 | 486 | 161 | 17.91 | 5.79 | 5 |
| GmDUF4228-22 | 05G034800 | 543 | 180 | 20.06 | 9.51 | 5 |
| GmDUF4228-23 | 05G106700 | 543 | 180 | 19.57 | 9.52 | 5 |
| GmDUF4228-24 | 05G158500 | 528 | 175 | 19.51 | 9.94 | 5 |
| GmDUF4228-25 | 05G218300 | 645 | 214 | 23.37 | 8.04 | 5 |
| GmDUF4228-26 | 05G246800 | 504 | 167 | 19.14 | 6.91 | 5 |
| GmDUF4228-27 | 06G135000 | 447 | 148 | 16.29 | 9.56 | 6 |
| GmDUF4228-28 | 06G184900 | 546 | 181 | 20.21 | 9.94 | 6 |
| GmDUF4228-29 | 07G050100 | 594 | 197 | 21.81 | 6.84 | 7 |
| GmDUF4228-30 | 07G165700 | 621 | 206 | 22.57 | 9.87 | 7 |
| GmDUF4228-31 | 07G180600 | 513 | 170 | 19.29 | 7.68 | 7 |
| GmDUF4228-32 | 07G216400 | 633 | 210 | 23.55 | 9.73 | 7 |
| GmDUF4228-33 | 08G024400 | 630 | 209 | 22.91 | 8.54 | 8 |
| GmDUF4228-34 | 08G055200 | 507 | 168 | 19.29 | 6.9 | 8 |
| GmDUF4228-35 | 08G116200 | 528 | 175 | 19.7 | 9.9 | 8 |
| GmDUF4228-36 | 08G290800 | 531 | 176 | 19.65 | 9.81 | 8 |
| GmDUF4228-37 | 08G301500 | 645 | 214 | 24.23 | 8.97 | 8 |
| GmDUF4228-38 | 08G301600 | 651 | 216 | 24.12 | 9.14 | 8 |
| GmDUF4228-39 | 09G058700 | 546 | 181 | 19.79 | 9.54 | 9 |
| GmDUF4228-40 | 09G118800 | 600 | 199 | 21.6 | 9.71 | 9 |
| GmDUF4228-41 | 09G129800 | 741 | 246 | 26.46 | 8.56 | 9 |
| GmDUF4228-42 | 10G013800 | 603 | 200 | 22.7 | 7.74 | 10 |
| GmDUF4228-43 | 10G072400 | 489 | 162 | 18.13 | 8.82 | 10 |
| GmDUF4228-44 | 10G230400 | 544 | 247 | 26.74 | 6.29 | 10 |
| GmDUF4228-45 | 10G266300 | 543 | 180 | 20.27 | 8.79 | 10 |
| GmDUF4228-46 | 11G061800 | 492 | 163 | 17.76 | 8.73 | 11 |
| GmDUF4228-47 | 11G061900 | 444 | 147 | 15.69 | 7.68 | 11 |
| GmDUF4228-48 | 11G121900 | 435 | 144 | 15.65 | 9.1 | 11 |
| GmDUF4228-49 | 11G163800 | 504 | 167 | 18.32 | 9.64 | 11 |
| GmDUF4228-50 | 11G218600 | 645 | 214 | 23.69 | 9.79 | 11 |
| GmDUF4228-51 | 12G046800 | 432 | 143 | 15.63 | 8.91 | 12 |
| GmDUF4228-52 | 13G099800 | 516 | 171 | 18.69 | 8.62 | 13 |
| GmDUF4228-53 | 13G153800 | 486 | 161 | 18.07 | 9.28 | 13 |
| GmDUF4228-54 | 13G229500 | 588 | 195 | 21.9 | 9.11 | 13 |
| GmDUF4228-55 | 13G369200 | 753 | 250 | 27.93 | 9.47 | 13 |
| GmDUF4228-56 | 14G034700 | 660 | 219 | 24.3 | 8.63 | 14 |
| GmDUF4228-57 | 14G069200 | 645 | 214 | 23.54 | 9.6 | 14 |
| GmDUF4228-58 | 14G199900 | 504 | 167 | 18.69 | 9.37 | 14 |
| GmDUF4228-59 | 15G004600 | 768 | 255 | 28.49 | 8.98 | 15 |
| GmDUF4228-60 | 15G083000 | 531 | 176 | 19.71 | 9.83 | 15 |
| GmDUF4228-61 | 15G164800 | 537 | 178 | 19.52 | 9.57 | 15 |
| GmDUF4228-62 | 16G018800 | 600 | 199 | 22.04 | 6.17 | 16 |
| GmDUF4228-63 | 16G143400 | 516 | 171 | 18.33 | 8.71 | 16 |
| GmDUF4228-64 | 16G177600 | 735 | 244 | 26.58 | 9.11 | 16 |
| GmDUF4228-65 | 17G060200 | 513 | 170 | 18.48 | 8.93 | 17 |
| GmDUF4228-66 | 17G092500 | 549 | 182 | 20.15 | 9.75 | 17 |
| GmDUF4228-67 | 17G160100 | 537 | 178 | 19.51 | 9.73 | 17 |
| GmDUF4228-68 | 18G038700 | 657 | 218 | 24.26 | 9.83 | 18 |
| GmDUF4228-69 | 18G057000 | 498 | 165 | 18.09 | 9.74 | 18 |
| GmDUF4228-70 | 18G118300 | 594 | 197 | 22.09 | 8.71 | 18 |
| GmDUF4228-71 | 18G133300 | 525 | 174 | 19.2 | 9.91 | 18 |
| GmDUF4228-72 | 18G144000 | 1980 | 659 | 75.27 | 8.79 | 18 |
| GmDUF4228-73 | 19G001400 | 489 | 162 | 18.16 | 6.84 | 19 |
| GmDUF4228-74 | 19G215600 | 600 | 199 | 22.79 | 8.96 | 19 |
| GmDUF4228-75 | 19G215700 | 441 | 146 | 16.05 | 9 | 19 |
| GmDUF4228-76 | 19G251200 | 678 | 225 | 25.88 | 10.13 | 19 |
| GmDUF4228-77 | 19G259600 | 645 | 214 | 23.71 | 6.45 | 19 |
| GmDUF4228-78 | 20G009000 | 513 | 170 | 19.1 | 5.95 | 20 |
| GmDUF4228-79 | 20G013400 | 639 | 212 | 23.47 | 9.59 | 20 |
| GmDUF4228-80 | 20G124200 | 534 | 177 | 19.67 | 8.96 | 20 |
| GmDUF4228-81 | 20G163000 | 708 | 235 | 25.41 | 6.75 | 20 |
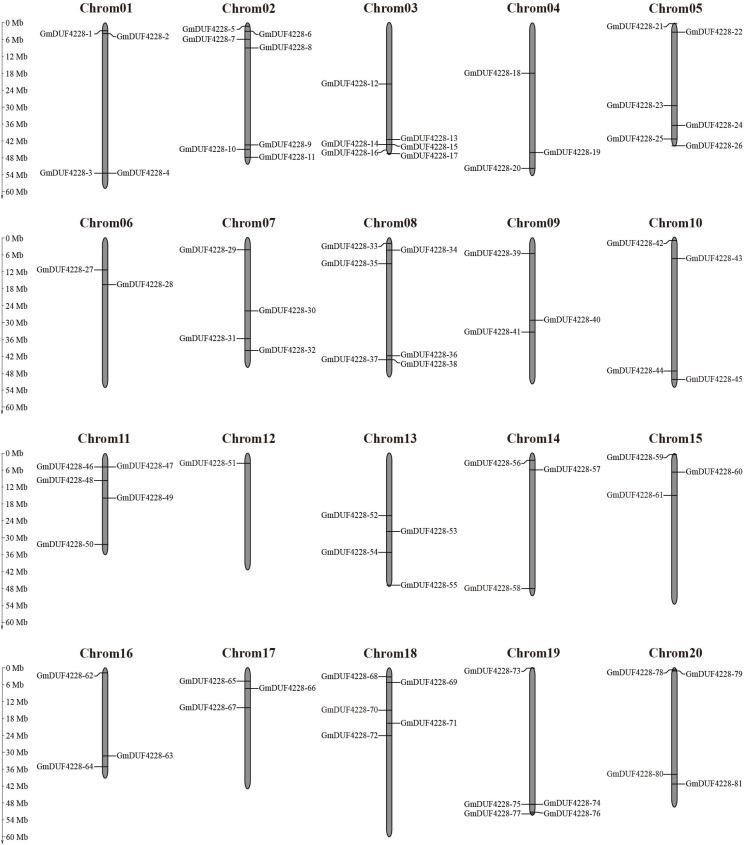
The 81 GmDUF4228 genes were unevenly distributed on the twenty chromosomes of soybean with chromosome 2 containing the largest number of genes (7) (Figure 2). Six genes were located on chromosomes 3, 5, and 8, respectively, while only one gene (GmDUF4228-51) was found on chromosome 12. Each of other chromosomes contained 2–5 genes.
FIGURE 2.
Chromosomal distribution of GmDUF4228 genes. The scale bar indicates the length (Mb) of soybean chromosomes.
Structure and Motif Analyses of GmDUF4228 Genes
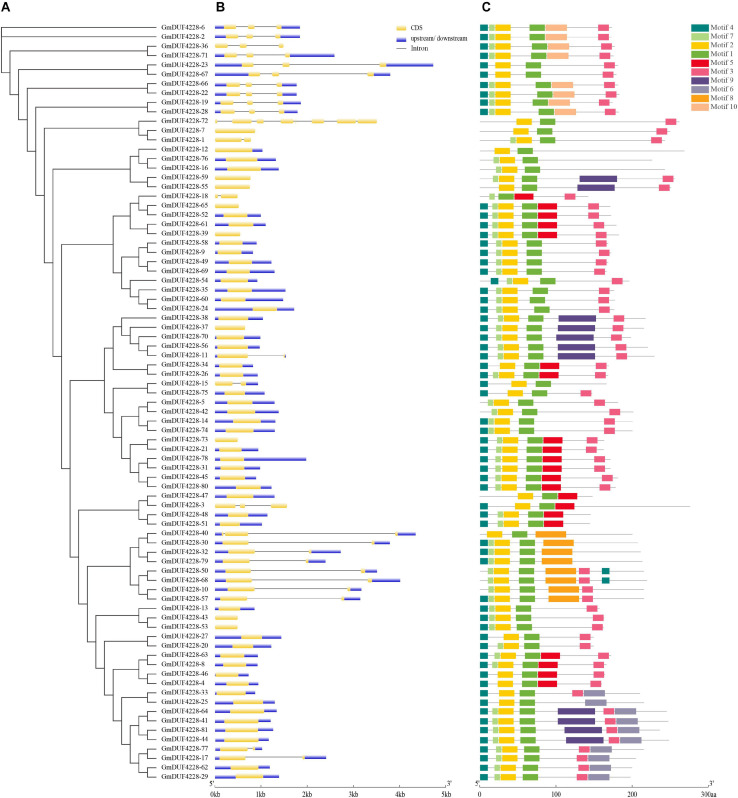
The phylogenetic tree based on the 81 amino acid sequences of GmDUF4228 proteins was constructed using ML method. The gene structure and motifs were clustered according to the ML phylogenetic tree (Figure 3A). Diversification of gene structure plays an important role in gene family evolution (Schmutz et al., 2010). Our results showed that most GmDUF4228 genes contained no or 1–2 introns with GmDUF4228-72 containing 7 introns (Figure 3B). Specifically, no intron was detected in 55 GmDUF4228 genes (68%), while 1 and 2 introns were identified in 13 (16%) and 12 GmDUF4228 genes (15%), respectively.
FIGURE 3.
Phylogenetic relationships, gene structures, and conserved motifs of GmDUF4228. (A) Phylogenetic tree of DUF4228 proteins from soybean constructed using the ML method. (B) Exon/intron organization of GmDUF4228 genes. Yellow boxes represent exons and black lines represent introns. The upstream/downstream regions of GmDUF4228 genes are indicated by blue boxes. The scale at the bottom is used to infer the lengthes of exons. (C) Distribution of conserved motifs in GmDUF4228 genes. Ten putative motifs are indicated by colored boxes (Table 2).
Amino acid sequences were analyzed using MEME in order to further characterize the structure of the GmDUF4228 proteins (Figure 3C). A total of 10 conserved motifs ranging in length from 8 to 50 amino acids were identified (Table 2).
TABLE 2.
List of the putative motifs in GmDUF4228 proteins.
| Motif number | Length (aa) | Best possible match |
| 1 | 21 | RIKPLPPDEELQPGQVYFLLP |
| 2 | 21 | VEELKTPITASEVMKENPGHV |
| 3 | 15 | KSRVWKPSLETISEE |
| 4 | 11 | MGNCLSCDLAT |
| 5 | 26 | MSRLNSPVSPVDMAALAVKATSAAKK |
| 6 | 29 | LLEILSQEARTEALIESVRTVAKCGNGAP |
| 7 | 8 | KVVHPBGK |
| 8 | 41 | RVRGGRGMNAKDRLESLVLSKRSVSDLTJMK QGPNPENDGP |
| 9 | 50 | SVPNKSTIKFGRCSFEYLKGSNGRVLIK VSPEFITRLISRSKRPCCNSSN |
| 10 | 29 | TQEVMKGLKAKKHAKMRKPLAESAZKPNL |
Expression Profiles of the GmDUF4228 Genes in Various Tissues and Organs of Soybean
In order to explore the expression pattern of GmDUF4228 genes in different tissues and organs during soybean development, we extracted the transcriptome data of GmDUF4228 genes from nine soybean tissues and organs, including apical meristems, flowers, leaves, nodules, stems, pods, roots, root hairs, and seeds, from the publically available RNA-seq data in Phytozome database. We also used the amino acid sequences of 81 GmDUF4228 proteins to construct a phylogenetic tree using ML method to cluster the expression profiles of soybean GmDUF4228 genes in different tissues and organs (Figure 4).
FIGURE 4.
Expression heatmap of GmDUF4228 genes in different tissues and organs of soybean constructed with TBtools. Fragments per kilobase of transcript per million mapped reads (FPKM) values of GmDUF4228 genes are transformed by log10. Red and green represent high and low expressions, respectively. Genes are ordered according to the phylogenetic tree of soybean DUF4228 proteins shown in Figure 3.
Results showed that GmDUF4228-10 and –46 showed high expression levels in all nine tissues and organs of soybean, while some genes were highly expressed in only one tissue. For example, GmDUF4228-5, –14, –26, and –75 showed high expression levels in pods, nodules, root hairs, and leaves, respectively, while GmDUF4228-56, –70, and –73 were expressed mainly in roots. These transcription profiles suggested that GmDUF4228 genes may be involved in the development of soybean plants.
Responses of GmDUF4228 Genes to Drought and Salt Treatments
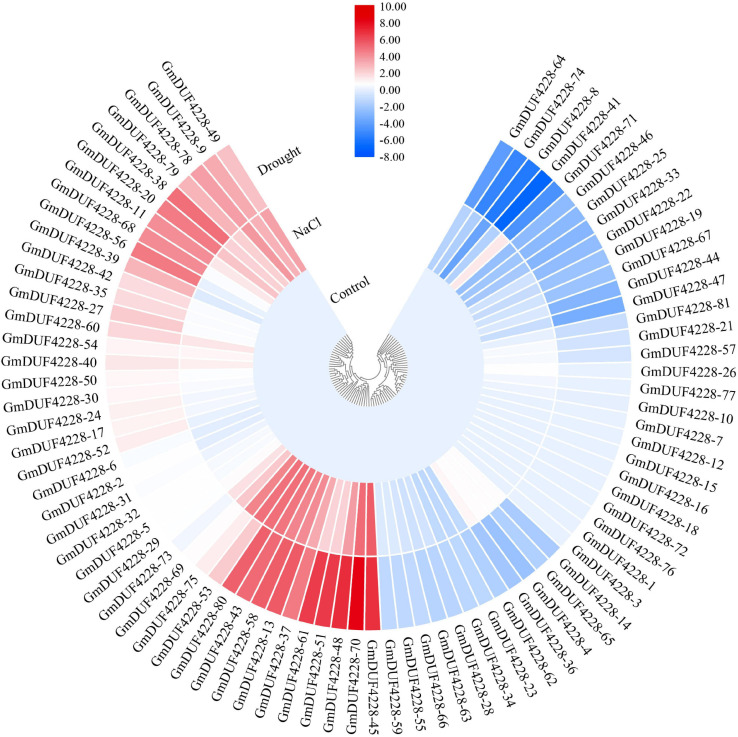
To investigate the potential functions of GmDUF4228 genes under abiotic stress, we examined the transcription levels of GmDUF4228 genes under drought and salt stresses based on previously published RNA-seq data (Shi et al., 2018). The results showed that most of the GmDUF4228 genes were induced by drought and salt (Figure 5). A total of 22 genes were up-regulated under drought stress (based on the criterion of “fold change ≥ 2”), such as GmDUF4228-45,–48, –51, –61, and –70, while 15 genes were down-regulated (based on the criterion of “fold change ≤ 2”), such as GmDUF4228-8, –41, –64, –71, and –74. A total of 18 genes were up-regulated under salt stress, such as GmDUF4228-43, –45, –53, –70, and –80, and 4 genes were down-regulated, including GmDUF4228-8, −25, −33, and –46. It was worth mentioning that four genes (i.e., GmDUF4228-4, –36, –39, and –71) showed opposite expression patterns between the treatments of drought and salt stresses. To further confirm the expression of GmDUF4228 genes under abiotic stresses, we selected nine genes (GmDUF4228-13, –43, 8-45, –48, –51, –58, –61, –70, and –80) significantly up-regulated under both drought and salt stresses for further functional verification based on promoter (cis-acting element) analysis and gene expression analysis using qRT-PCR.
FIGURE 5.
Heatmap based on hierarchical cluster analysis of 81 GmDUF4228 gene expression profiles under drought and salt stresses and control conditions. Red and blue represent high and low expressions, respectively.
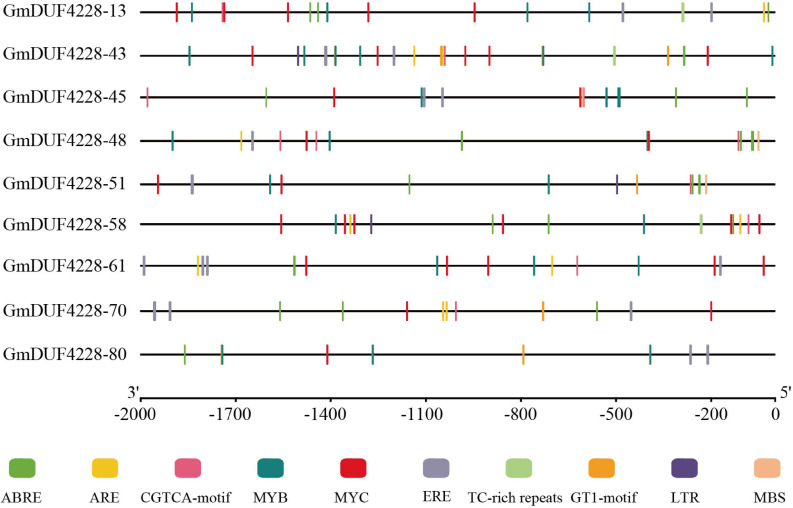
Analysis of Cis-Acting Elements in GmDUF4228 Promoters
The 2,000 bp regions upstream of the start codon in the nine GmDUF4228 genes were analyzed for cis-acting elements (Figure 6). Ten elements related to abiotic stress were identified to further investigate the role of these nine genes in response to abiotic stresses. These elements included the ABA-responsive element (ABRE), anaerobic induction element (ARE), methyl jasmonate-responsive element (CGTCA-motif), ethylene-responsive element (ERE), low temperature response element (LTR), three drought and salt-responsive elements (MYB, MYC, and MBS), defense and stress responsive elements (TC-rich repeats), and light-responsive element (GT1-motif). Results showed that each gene promoter contained at least five or more cis-acting elements related to abiotic stress, with five cis-acting elements (i.e., the ABRE, CGTCA-motif, MYB, MYC, and ERE) revealed in most of the GmDUF4228 gene promoter regions. These results indicated that these nine candidate GmDUF4228 genes might be involved in response to abiotic stresses.
FIGURE 6.
Predicted cis-acting elements in GmDUF4228 promoters. Promoter sequences (2,000 bp) of nine GmDUF4228 genes were analyzed by PlantCARE. The scale at the bottom is used to infer the length of upstream sequences starting from the starting site of translation.
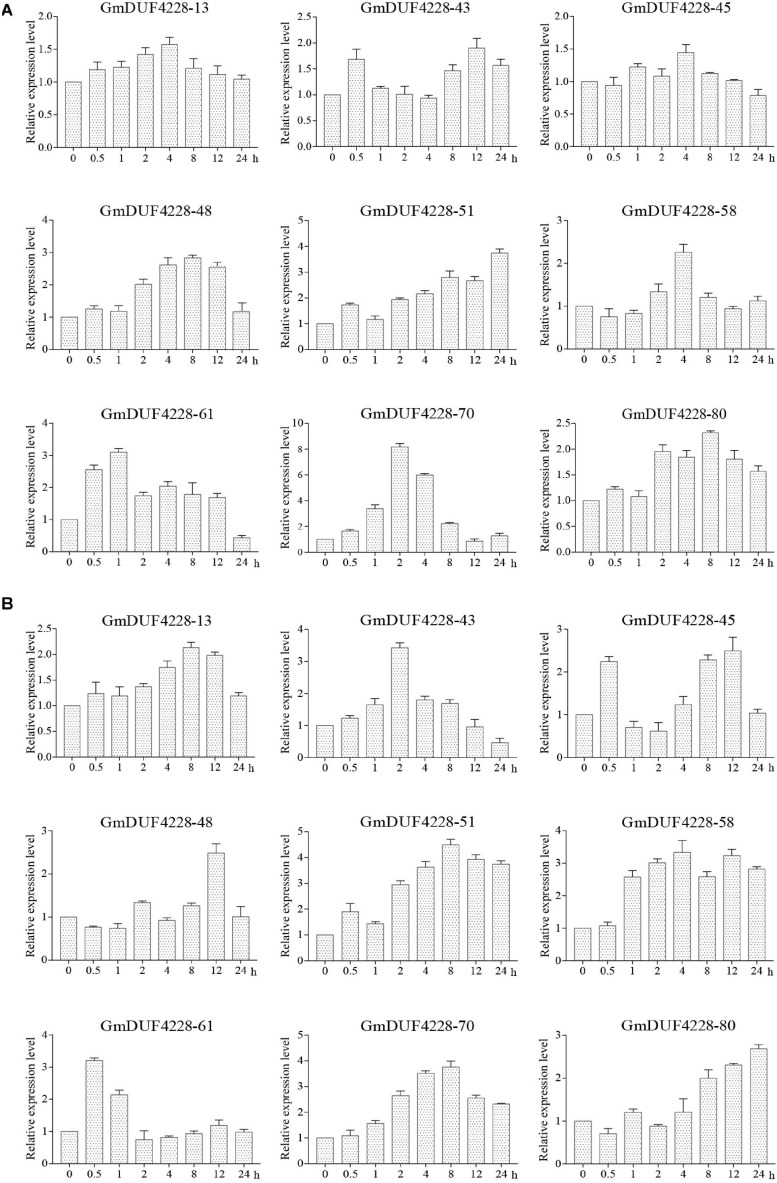
qRT-PCR Analysis of GmDUF4228 Genes
To investigate the potential functions of the nine GmDUF4228 genes in response to different stimuli, qRT-PCR was used to analyze the expression patterns of these genes in plants treated under drought and salt stresses (Figure 7). Under drought stress (Figure 7A), the expression levels of GmDUF4228-48 and –80 reached their peaks at 8 h and those of GmDUF4228-51, –58, –61, and –70 peaked at 24, 4, 1, and 2 h (> 2-fold), respectively, while GmDUF4228-70 showed expression level more than seven times higher than that of the control. Under salt stress, the expression levels of genes GmDUF4228-13, -45, –48, –51, and –70 reached their peaks at 8–12 h, while those of GmDUF4228-43, –58, –61, and –80 peaked at 2, 4, 0.5, and 24 h, respectively (Figure 7B). These results indicated that the transcription levels of most of these nine GmDUF4228 genes were affected by drought and salt stresses. GmDUF4228-70 was selected for further functional studies due to its relatively evident transcription changes under both drought and salt stresses.
FIGURE 7.
Expression patterns based on qRT-PCR of nine GmDUF4228 genes under drought and salt stresses. (A) Drought treatment under 0, 0.5, 1, 2, 4, 8, 12, and 24 h. (B) Salt treatment (200 mM NaCl) for 0, 0.5, 1, 2, 4, 8, 12, and 24 h. Data are presented as mean ± standard deviation (n = 3).
Overexpression of GmDUF4228-70 Improved Drought and Salt Tolerance
To study the biological function of GmDUF4228-70 in soybean, a GmDUF4228-70 overexpression vector (GmDUF4228-70-OE) and empty vector (EV) were transferred into soybean hairy roots by Agrobacterium-mediated transformation, and the phenotypes of seedlings under drought and salt stresses were evaluated. Results of the qRT-PCR analysis showed that the GmDUF4228-70 transcripts in the hairy roots of GmDUF4228-70-OE soybean seedlings were significantly higher than those in seedlings of EV control under normal conditions (Supplementary Figure 1).
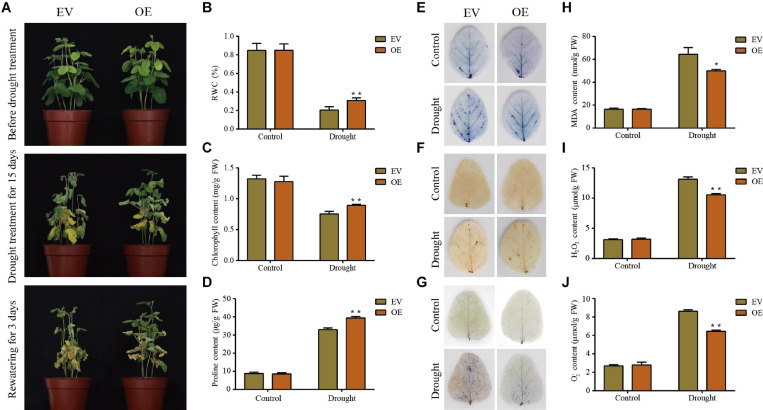
Prior to the treatment with drought or salt, the EV and the GmDUF4228-70-OE soybean seedlings showed similar growth phenotypes. Once treated under drought and salt stresses, the GmDUF4228-70-OE plants grew better than the EV lines, as indicated by significantly less leaf wilting, curling, and chlorosis (Figure 8A). After 3 days of drought or salt treatment, the EV lines started to show curled and wilted leaves, while the wilting in the GmDUF4228-70-OE lines showed trivial change. At the seventh day of salt treatment, all of the leaves in EV lines were wilted, while only some of the leaves in the GmDUF4228-70-OE lines became wilted (Figure 9A).
FIGURE 8.
Phenotypic and physiological analyses of the transgenic empty vector (EV) lines and GmDUF4228-70-OE (OE) transgenic lines under drought stress. (A) Phenotypes of EV and GmDUF4228-70-OE transgenic soybean plants treated under drought stress for 15 days and rewatering for 3 days. Relative water content (RWC) (B), chlorophyll content (C), proline content (D), trypan blue staining (E) DAB staining (F), NBT staining (G), Malondialdehyde (MDA) content (H) H2O2 content (I), and O2– content (J) of the leaves of EV and GmDUF4228-70-OE plants grown under drought or control conditions for 7 days. Data are shown as mean ± standard deviation (n = 3). Significant differences based on ANOVA are set at p < 0.05 (*) and p < 0.01 (**), respectively.
FIGURE 9.
Phenotypic and physiological analyses of the transgenic empty vector (EV) lines and GmDUF4228-70-OE (OE) transgenic lines under salt stress. (A) Phenotypes of EV and GmDUF4228-70-OE transgenic soybean plants treated under salt stress for 3 and 7 days, respectively. Relative water content (RWC) (B), chlorophyll content (C), proline content (D), trypan blue staining (E), DAB staining (F), NBT staining (G), malondialdehyde (MDA) content (H), H2O2 content (I), and O2– content (J) of the leaves of EV and GmDUF4228-70-OE plants grown under salt treatment or normal control conditions for 3 days. Data are shown as mean ± standard deviation (n = 3). Significant differences based on ANOVA are set at p < 0.05 (*) and p < 0.01 (**), respectively.
Two physiological indices, i.e., chlorophyll content as an indicator of photosynthetic capacity (Tanaka and Tanaka, 2006) and RWC as an indicator of plant water status, were measured to quantify the effects of salt and drought on the plant development (Meher et al., 2018). The changes in RWC (Figures 8B, 9B) and chlorophyll content (Figures 8C, 9C) of plant leaves in response to drought and salt were consistent with the observation of leaf phenotypes. Specifically, the leaves of GmDUF4228-70-OE lines showed a higher RWC and chlorophyll content than those of the EV lines, indicating that the GmDUF4228-70-OE lines showed an evident growth advantage compared with the EV lines under drought and salt stresses.
The contents of proline, MDA, H2O2, and O2– are important indicators used to measure the effects of abiotic stress on plant growth (Wang D. et al., 2019). Proline is a protective agent against osmotic stress, MDA reflects the degree of lipid oxidative damage, while both H2O2 and O2– play a role of immunity and signal transduction, though excessive accumulation may cause cell membrane damage (Du et al., 2018; Zhang X. Z. et al., 2019). To investigate the underlying physiological mechanisms of GmDUF4228-70-OE enhancing plant stress tolerance, we measured the contents of proline, MDA, H2O2, and O2– in the EV lines and GmDUF4228-70-OE lines under normal and stress conditions.
The results showed that under drought and salt stresses, the GmDUF4228-70-OE plants showed higher proline contents (Figures 8D, 9D) and lower MDA (Figures 8H, 9H), H2O2 (Figures 8I, 9I), and O2– (Figures 8J, 9J) contents than the EV lines. In order to visually show the degree of damage in soybean EV and GmDUF4228-70-OE plant leaves, we used trypan blue (Figures 8E, 9E), DAB (Figures 8F, 9F), and NBT (Figures 8G, 9G) staining to measure cell viability in soybean leaves under drought and salt stresses. No difference was observed in both the GmDUF4228-70-OE and EV soybean leaves under normal conditions based on trypan blue, DAB, or NBT staining methods. However, under the drought and salt treatments, the GmDUF4228-70-OE plant leaves were stained significantly less by all three staining methods than the EV plant leaves, suggesting that the leaves of GmDUF4228-70-OE plants suffered less damage under drought and salt treatments than the EV plants.
GmDUF4228-70-OE Plants Exhibited Increased Transcripts of Drought- and Salt-Inducible Genes
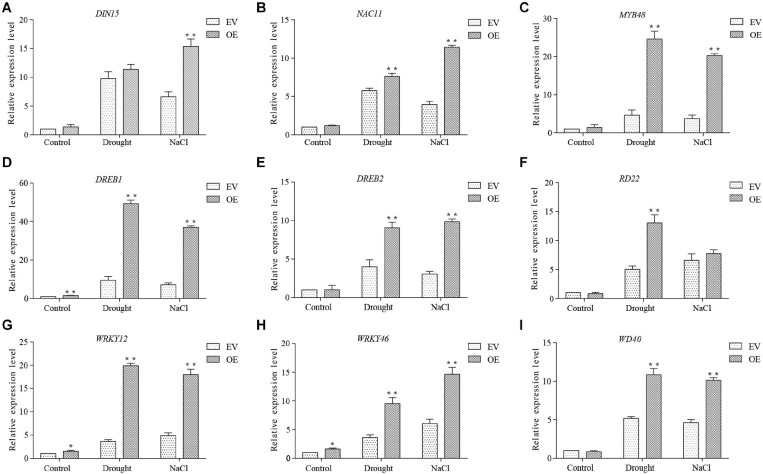
A group of 9 marker genes up-regulated under both drought and salt stresses were chosen based on previous studies to investigate the molecular mechanism of GmDUF4228-70 in response to stress, including DIN15 (Guo et al., 2019), NAC11 (Hao et al., 2011), MYB48 (Liao et al., 2008), DREB1 (Kasuga et al., 1999), DREB2 (Chen et al., 2007), RD22 (Matus et al., 2014), WRKY12 (Shi et al., 2018), WRKY46 (Chen et al., 2017), and WD40 (Mishra et al., 2014; Figure 10). The expression of these marker genes under normal and stress conditions was analyzed by qRT-PCR. Under normal and stress conditions, the transcript levels of DREB1, WRKY12, and WRKY46 in GmDUF4228-70-OE were significantly up-regulated compared with those in EV lines, while no significant difference was observed in DIN15, NAC11, MYB48, DREB2, RD22, and WD40 under normal conditions. The transcriptional levels of NAC11, MYB48, DREB2, RD22, and WD40 were significantly higher in GmDUF4228-70-OE than EV under drought treatment, while the transcriptional levels of DIN15, NAC11, MYB48, DREB2, and WD40 were significantly higher in GmDUF4228-70-OE than EV under salt treatment (Figure 10). Under drought and salt treatments, the transcriptional levels of these marker genes increased by varied degrees, indicating that these marker genes may be regulated by GmDUF4228-70. Further studies are necessary to identify the relationships between GmDUF4228-70 and stress-related marker genes in soybean.
FIGURE 10.
The expression levels of stress-related marker genes DIN15 (A), NAC11 (B), MYB48 (C), DREB1 (D), DREB2 (E), RD22 (F), WRKY12 (G), WRKY46 (H), and WD40 (I) based on qRT-PCR. The actin gene is used as an internal control. Data are shown as mean ± standard deviation (n = 3). Significant differences based on ANOVA are set at p < 0.05 (*) and p < 0.01 (**), respectively.
Discussion
DUF4228 has been reported to function in various abiotic stress processes of plants (Wang et al., 2017). However, the role of DUF4228 genes in soybean have not been investigated. Previous studies have identified a total of 22 DUF4228 genes in Aquilegia coerulea, 25 in Arabidopsis, 29 in Medicago truncatula, and 52 in Populus trichocarpa (Yang Q. et al., 2020). Our results of phylogenetic analysis using ML method showed that the topological relationships among a total of 135 DUF4228 protein sequences were largely consistent with those reported previously by Yang Q. et al. (2020) with one exception, i.e., AT1G18290 identified in Group I of Yang Q. et al. (2020) was revealed in Group II in our study.
Profiling of GmDUF4228 gene expression showed that most of the genes were expressed low in various tissues and organs of soybean (Figure 4). Our results showed that 27 and 22% of GmDUF4228 genes were up-regulated under drought and salt stress conditions, respectively (Figure 5), indicating that most of these genes may be related to abiotic stresses. It has been shown that cis-acting elements in the promoter region are closely related to gene function (Chow et al., 2018). Consistent with their expression profiles, GmDUF4228 genes contained a variety of cis-acting elements related to abiotic stresses, including MYB, MYC, MBS, and GT-1 elements (Figure 6), which play an important role in response to drought and salt treatments (Abe et al., 1997; Li et al., 2018; Tang et al., 2019; Zhang L. et al., 2019). Studies have shown that MYC also functions in cold stress (Ohta et al., 2018) and disease resistance (Lorenzo et al., 2004). Other elements found in GmDUF4228 gene promoters are also associated with abiotic stress. For example, the ABRE element is bound by ABRE-binding proteins, involved in dehydration, salt, and ABA stresses (Wang X. et al., 2019), while both ERE and TC-rich repeats may be associated with ethylene and defense and stress response-related elements (Zhang et al., 2015; Song et al., 2019). ARE, LTR, and CGTCA-motif are associated with anaerobic inducement response (Geffers et al., 2001), low temperature stress (Maestrini et al., 2009), and methyl jasmonate response (Xu et al., 2018), respectively. These results suggest that soybean GmDUF4228 genes may be involved in abiotic stress response.
Soil drought and salinization are two major stress factors influencing plant growth and development. Plants have evolved a variety of survival mechanisms to cope with stress, including the maintenance of morphological characteristics related to plant tissues and physiological mechanisms related to the regulation of metabolism (Vijayaraghavareddy et al., 2020). Under drought stress, in order to improve water use efficiency, stomatal opening on leaves is mainly regulated to control carbon dioxide emissions and reduce transpiration (Mega et al., 2019). In response to drought stress, roots generate CLE25 peptide, which is transferred to leaves via the vascular system to activate NCED3 and promote ABA production (Takahashi et al., 2018). Under salt stress, the growth and development of plants are mainly inhibited by changes in osmotic pressure and ion imbalance. The excessive accumulation of Na+ and Cl– in the cell leads to inhibition of the activity of intracellular enzymes involved in Calvin cycle, phenylpropionic acid pathway, glycolysis, and starch synthesis, resulting in disruption of metabolism, ultimately inhibiting plant growth (Zhao et al., 2020). Under salt stress, when roots perceive the change in osmotic pressure, the signal is almost immediately transmitted to the leaves through the stem, causing a decrease in leaf stomatal opening, thus protecting the plant against water loss (Cosgrove and Hedrich, 1991; Christmann et al., 2013). Both drought and salt stresses are perceived by the root system, which transmits a signal to the leaves through a feedback mechanism, causing leaves damaged to varying degrees. Our results showed that the overexpression of GmDUF4228-70 in soybean reduced water loss and chlorophyll degradation, while improved the viability of soybean under drought and salt stresses. Similarly, the cell activity of GmDUF4228-70-OE plant leaves was stronger than that of EV leaves, suggesting that GmDUF4228 improves the survival of soybean under drought and salt stresses.
In this study, Agrobacterium rhizogenes-mediated soybean hairy root transformation technology was used to induce transgenic roots on soybeans to study the function of GmDUF4228-70 gene (Kereszt et al., 2007). Phenotypic and physiological analyses showed that overexpression of GmDUF4228-70 improved soybean tolerance to drought and salt (Figures 8, 9). While Agrobacterium rhizogenes-mediated transformation of soybean roots generated transgenic seedlings with increased expression of GmDUF4228-70 gene in roots, this gene cannot be inherited to future generations by sexual reproduction (i.e., seeds). Further transformation studies are necessary to explore the applications of GmDUF4228-70 gene in breeding of transgenic soybean lines with increased tolerance to drought and salt stresses. In summary, our study provides a scientific foundation for further analysis of the functions of GmDUF4228-70 gene in response to drought and salt stresses.
Conclusion
This study identified a total of 81 DUF4228 genes in the soybean genome. The expression levels of nine of these genes were up-regulated under both drought and salt stresses as identified by the RNA-seq transcriptome data and further validated by qRT-PCR. The results showed that GmDUF4228-70 was sensitive to both drought and salt stresses. Analysis of plant phenotypes and stress-related physiological indicators showed that overexpression of GmDUF4228-70 enhanced soybean tolerance to drought and salt stresses. The results of qRT-PCR showed that the transcription levels of marker genes in transgenic soybean overexpressing GmDUF4228-70 were increased significantly under drought and salt stresses. These results have advanced our understanding of the functions of GmDUF42228 family and provide a strong foundation for further investigations on the molecular mechanisms of GmDUF4228-70 in response to abiotic stresses in soybean.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Z-SX coordinated the project, conceived and designed experiments, and edited the manuscript. Z-XL performed experiments and wrote the first draft. YL revised the manuscript. Z-YC, JG, JC, and Y-BZ contributed to data analysis and managed reagents. MC, Y-ZM, and X-YC contributed with valuable discussions. All authors reviewed and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Drs. Lijuan Qiu and Tianfu Han at the Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS) for kindly providing soybean seeds, Dr. Wensheng Hou and Hui Zhang at the Institute of Crop Science, CAAS, for kindly providing vectors, and Dr. Hui Zhang at the Institute of Crop Science, CAAS, for providing the procedure for high-efficiency A. rhizogenes-mediated transformation.
Abbreviations
- CDS
Coding sequence
- FPKM
Fragments per kilobase of transcript per million mapped reads
- GSDS
Gene structure display server
- HMM
Hidden Markov model
- MDA
Malondialdehyde
- MEME
Multiple Em for motif elicitation
- MEGA
Molecular evolutionary genetics analysis
- pI
Isoelectric point
- qRT-PCR
quantitative real-time PCR
- RNA-Seq
RNA sequencing
- SMART
Simple modular architecture research tool.
Funding. This research was financially supported by the National Science Foundation of Jilin Province (20180101026JC) and the Fundamental Research Funds for Central Non-Profit of Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (Y2020PT12 and S2020ZC02).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.628299/full#supplementary-material
qRT-PCR analysis of GmDUF4228-70 expression level in EV and GmDUF4228-70-OE transgenic hairy roots.
Primers and sequences used in this study.
FPKM values for DUF4228 genes in different tissues and organs of soybean.
References
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9 1859–1868. 10.1105/tpc.9.10.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad P., Wani M. R., Azooz M. M., Tran L. S. P. (eds). (2014). Improvement of Crops in the Era of Climatic Changes Volume 2. New York: Springer. [Google Scholar]
- Bateman A., Coggill P., Finn R. D. (2010). DUFs: families in search of function. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66 1148–1152. 10.1107/S1744309110001685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J. S. (1982). Plant productivity and environment. Science 218 443–448. 10.1126/science.218.4571.443 [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen J., Nolan T. M., Ye H., Zhang M., Tong H., Xin P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 29 1425–1439. 10.1105/tpc.17.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wang Q. Y., Cheng X. G., Xu Z. S., Li L. C., Ye X. G., et al. (2007). GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353 299–305. 10.1016/j.bbrc.2006.12.027 [DOI] [PubMed] [Google Scholar]
- Chow C. N., Chiang-Hsieh Y. F., Chien C. H., Zheng H. Q., Lee T. Y., Wu N. Y., et al. (2018). Delineation of condition specific Cis- and Trans-acting elements in plant promoters under various Endo- and exogenous stimuli. BMC Genomics 19:85. 10.1186/s12864-018-4469-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A., Grill E., Huang J. (2013). Hydraulic signals in long-distance signaling. Curr. Opin. Plant Biol. 16 293–300. 10.1016/j.pbi.2013.02.011 [DOI] [PubMed] [Google Scholar]
- Cosgrove D. J., Hedrich R. (1991). Stretch-activated chloride, potassium, and calcium channels coexisting in plasma membranes of guard cells of Vicia faba L. Planta 186 143–153. 10.1007/BF00201510 [DOI] [PubMed] [Google Scholar]
- Cui X. Y., Du Y. T., Fu J. D., Yu T. F., Wang C. T., Chen M., et al. (2018). Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 18:93. 10.1186/s12870-018-1306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelon M., Khafif M., Godiard L., Barbacci A., Raffaele S. (2020). Patterns of Sequence and Expression Diversification Associate Members of the PADRE Gene Family With Response to Fungal Pathogens. Front. Genet. 11:491. 10.3389/fgene.2020.00491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y. T., Zhao M. J., Wang C. T., Gao Y., Wang Y. X., Liu Y. W., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18:320. 10.1186/s12870-018-1551-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., et al. (2008). The Pfam protein families database. Nucleic Acids Res. 36 D281–8. 10.1093/nar/gkm960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffers R., Sell S., Cerff R., Hehl R. (2001). The TATA box and a Myb binding site are essential for anaerobic expression of a maize GapC4 minimal promoter in tobacco. Biochim. Biophys. Acta 1521 120–125. 10.1016/s0167-4781(01)00302-5 [DOI] [PubMed] [Google Scholar]
- Guan R., Qu Y., Guo Y., Yu L., Liu Y., Jiang J., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80 937–950. 10.1111/tpj.12695 [DOI] [PubMed] [Google Scholar]
- Guo X., Zhang L., Wang X., Zhang M., Xi Y., Wang A., et al. (2019). Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS One 14:e0225090. 10.1371/journal.pone.0225090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Rico-Medina A., Cano-Delgado A. I. (2020). The physiology of plant responses to drought. Science 368 266–269. 10.1126/science.aaz7614 [DOI] [PubMed] [Google Scholar]
- Hamant O., Haswell E. S. (2017). Life behind the wall: sensing mechanical cues in plants. BMC Biol. 15:59. 10.1186/s12915-017-0403-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y. J., Wei W., Song Q. X., Chen H. W., Zhang Y. Q., Wang F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68 302–313. 10.1111/j.1365-313X.2011.04687.x [DOI] [PubMed] [Google Scholar]
- Hossain Z., Khatoon A., Komatsu S. (2013). Soybean proteomics for unraveling abiotic stress response mechanism. J. Proteome Res. 12 4670–4684. 10.1021/pr400604b [DOI] [PubMed] [Google Scholar]
- Jaroszewski L., Li Z., Krishna S. S., Bakolitsa C., Wooley J., Deacon A. M., et al. (2009). Exploration of uncharted regions of the protein universe. PLoS Biol. 7:e1000205. 10.1371/journal.pbio.1000205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. 10.1038/7036 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A., Li D., Indrasumunar A., Nguyen C. D., Nontachaiyapoom S., Kinkema M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2 948–952. 10.1038/nprot.2007.141 [DOI] [PubMed] [Google Scholar]
- Kim Y., Tsuda K., Igarashi D., Hillmer R. A., Sakakibara H., Myers C. L., et al. (2014). Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe 15 84–94. 10.1016/j.chom.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Wei S., Chen J., Yang F., Kong L., Chen C., et al. (2018). OsASR2 regulates the expression of a defence-related gene, Os2H16, by targeting the GT-1 cis-element. Plant Biotechnol. J. 16 771–783. 10.1111/pbi.12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Zou H. F., Wang H. W., Zhang W. K., Ma B., Zhang J. S., et al. (2008). Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18 1047–1060. 10.1038/cr.2008.280 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J. M., Sanchez-Serrano J. J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950. 10.1105/tpc.022319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Su W., Li H., Guo Z. (2009). Abscisic acid improves drought tolerance of triploid bermudagrass and involves H2O2- and NO-induced antioxidant enzyme activities. Plant Physiol. Biochem. 47 132–138. 10.1016/j.plaphy.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Ma Q., Xia Z., Cai Z., Li L., Cheng Y., Liu J., et al. (2018). GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 9:1979. 10.3389/fpls.2018.01979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestrini P., Cavallini A., Rizzo M., Giordani T., Bernardi R., Durante M., et al. (2009). Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba). J. Plant Physiol. 166 1544–1556. 10.1016/j.jplph.2009.03.014 [DOI] [PubMed] [Google Scholar]
- Mahalingam R., Fedoroff N. (2010). Stress response, cell death and signalling: the many faces of reactive oxygen species. Physiol. Plant. 119 56–68. 10.1034/j.1399-3054.2003.00156.x [DOI] [Google Scholar]
- Matus J. T., Aquea F., Espinoza C., Vega A., Cavallini E., Dal Santo S., et al. (2014). Inspection of the grapevine BURP superfamily highlights an expansion of RD22 genes with distinctive expression features in berry development and ABA-mediated stress responses. PLoS One 9:e110372. 10.1371/journal.pone.0110372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mega R., Abe F., Kim J. S., Tsuboi Y., Tanaka K., Kobayashi H., et al. (2019). Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 5 153–159. 10.1038/s41477-019-0361-8 [DOI] [PubMed] [Google Scholar]
- Meher, Shivakrishna P., Ashok Reddy K., Manohar Rao D. (2018). Effect of PEG-6000 imposed drought stress on RNA content, relative water content (RWC), and chlorophyll content in peanut leaves and roots. Saudi J. Biol. Sci. 25 285–289. 10.1016/j.sjbs.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A. K., Muthamilarasan M., Khan Y., Parida S. K., Prasad M. (2014). Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS One 9:e86852. 10.1371/journal.pone.0086852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na L., Qi Y., Guojing L., Ziyi Z., Lide B., Ruigang W., et al. (2016). Cloning and expression analysis of CiDUF4228-3 from caragana intermedia. Mol. Plant Breed. 17, 2492–2501. 10.13271/j.mpb.017.002492 [DOI] [Google Scholar]
- Ohta M., Sato A., Renhu N., Yamamoto T., Oka N., Zhu J. K., et al. (2018). MYC-type transcription factors, MYC67 and MYC70, interact with ICE1 and negatively regulate cold tolerance in Arabidopsis. Sci. Rep. 8:11622. 10.1038/s41598-018-29722-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papiernik S. K., Grieve C. M., Lesch S. M., Yates S. R. (2005). Effects of Salinity, Imazethapyr, and Chlorimuron Application on Soybean Growth and Yield. Commun. Soil Sci. Plant Anal. 36 951–967. 10.1081/css-200050280 [DOI] [Google Scholar]
- Posada D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27 1164–1165. 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Xiaocui N., Ruigang W., Guojing L., Guosheng Z. (2019). Genome-wide characterization of DUF221 gene family in medicago truncatula and screening for salt response genes. Mol. Plant Breed. 16, 5255–5262 10.13271/j.mpb.017.005255 [DOI] [Google Scholar]
- Sade N., Del Mar Rubio-Wilhelmi M., Umnajkitikorn K., Blumwald E. (2018). Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 69 845–853. 10.1093/jxb/erx235 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95 5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Z., Jiang Y., Li H., Guo J., Dong M., Zhang J., et al. (2020). Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genomics 21:96. 10.1186/s12864-020-6479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W. Y., Du Y. T., Ma J., Min D. H., Jin L. G., Chen J., et al. (2018). The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 19:4087. 10.3390/ijms19124087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Pan F., Lou X., Wang D., Yang C., Zhang B., et al. (2019). Genome-wide identification and characterization of Hsp70 gene family in Nicotiana tabacum. Mol. Biol. Rep. 46 1941–1954. 10.1007/s11033-019-04644-7 [DOI] [PubMed] [Google Scholar]
- Su H. G., Li B., Song X. Y., Ma J., Chen J., Zhou Y. B., et al. (2019). Genome-Wide Analysis of the DYW Subgroup PPR Gene Family and Identification of GmPPR4 Responses to Drought Stress. Int. J. Mol. Sci. 20:5667. 10.3390/ijms20225667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H. -G., Zhang X. -H., Wang T. -T., Wei W. -L., Wang Y. -X., Chen J., et al. (2020). Genome-Wide Identification, Evolution, and Expression of GDSL-Type Esterase/Lipase Gene Family in Soybean. Front. Plant Sci. 11:726. 10.3389/fpls.2020.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Suzuki T., Osakabe Y., Betsuyaku S., Kondo Y., Dohmae N., et al. (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556 235–238. 10.1038/s41586-018-0009-2 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Tanaka R. (2006). Chlorophyll metabolism. Curr. Opin. Plant Biol. 9 248–255. 10.1016/j.pbi.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Tang Y., Bao X., Zhi Y., Wu Q., Guo Y., Yin X., et al. (2019). Overexpression of a MYB Family Gene, OsMYB6, Increases Drought and Salinity Stress Tolerance in Transgenic Rice. Front. Plant Sci. 10:168. 10.3389/fpls.2019.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavareddy P., Vemanna R. S., Yin X., Struik P. C., Makarla U., Sreeman S. (2020). Acquired Traits Contribute More to Drought Tolerance in Wheat Than in Rice. Plant Phenomics 2020 1–16. 10.34133/2020/5905371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liu Y. X., Yu Q., Zhao S. P., Zhao J. Y., Ru J. N., et al. (2019). Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses. Int. J. Mol. Sci. 20:5909. 10.3390/ijms20235909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Komatsu S. (2018). Proteomic approaches to uncover the flooding and drought stress response mechanisms in soybean. J. Proteomics 172 201–215. 10.1016/j.jprot.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Wang X., Guo C., Peng J., Li C., Wan F., Zhang S., et al. (2019). ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 221 341–355. 10.1111/nph.15345 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Z., Liu H., An Y., Han B., Wu Y., et al. (2017). Overexpression of an alfalfa (Medicago sativa) gene, MsDUF, negatively impacted seed germination and response to osmotic stress in transgenic tobacco. Plant Cell Tissue Organ. Cult. 132 525–534. 10.1007/s11240-017-1348-7 [DOI] [Google Scholar]
- Xu Z., Sun M., Jiang X., Sun H., Dang X., Cong H., et al. (2018). Glycinebetaine Biosynthesis in Response to Osmotic Stress Depends on Jasmonate Signaling in Watermelon Suspension Cells. Front. Plant Sci. 9:1469. 10.3389/fpls.2018.01469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Niu X., Tian X., Zhang X., Cong J., Wang R., et al. (2020). Comprehensive genomic analysis of the DUF4228 gene family in land plants and expression profiling of ATDUF4228 under abiotic stresses. BMC Genomics 21:12. 10.1186/s12864-019-6389-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yu T. F., Ma J., Chen J., Zhou Y. B., Chen M., et al. (2020). The Soybean bZIP Transcription Factor Gene GmbZIP2 Confers Drought and Salt Resistances in Transgenic Plants. Int. J. Mol. Sci. 21:670. 10.3390/ijms21020670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Song Z., Li F., Li X., Ji H., Yang S. (2019). The specific MYB binding sites bound by TaMYB in the GAPCp2/3 promoters are involved in the drought stress response in wheat. BMC Plant Biol. 19:366. 10.1186/s12870-019-1948-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang X. Z., Zheng W. J., Cao X. Y., Cui X. Y., Zhao S. P., Yu T. F., et al. (2019). Genomic Analysis of Stress Associated Proteins in Soybean and the Role of GmSAP16 in Abiotic Stress Responses in Arabidopsis and Soybean. Front. Plant Sci. 10:1453. 10.3389/fpls.2019.01453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gao M., Wang S., Chen F., Cui D. (2015). Allelic variation at the vernalization and photoperiod sensitivity loci in Chinese winter wheat cultivars (Triticum aestivum L.). Front. Plant Sci. 6:470. 10.3389/fpls.2015.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang H., Song C., Zhu J.-K., Shabala S. (2020). Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 1:100017. 10.1016/j.xinn.2020.100017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. P., Lu D., Yu T. F., Ji Y. J., Zheng W. J., Zhang S. X., et al. (2017). Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. 119 132–146. 10.1016/j.plaphy.2017.08.026 [DOI] [PubMed] [Google Scholar]
- Zong W., Tang N., Yang J., Peng L., Ma S., Xu Y., et al. (2016). Feedback Regulation of ABA Signaling and Biosynthesis by a bZIP Transcription Factor Targets Drought-Resistance-Related Genes. Plant Physiol. 171 2810–2825. 10.1104/pp.16.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR analysis of GmDUF4228-70 expression level in EV and GmDUF4228-70-OE transgenic hairy roots.
Primers and sequences used in this study.
FPKM values for DUF4228 genes in different tissues and organs of soybean.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.