Abstract
Plants are complex organisms that adapt to changes in their environment using an array of regulatory mechanisms that span across multiple levels of biological organization. Due to this complexity, it is difficult to predict emergent properties using conventional approaches that focus on single levels of biology such as the genome, transcriptome, or metabolome. Mathematical models of biological systems have emerged as useful tools for exploring pathways and identifying gaps in our current knowledge of biological processes. Identification of emergent properties, however, requires their vertical integration across biological scales through multiscale modeling. Multiscale models that capture and predict these emergent properties will allow us to predict how plants will respond to a changing climate and explore strategies for plant engineering. In this review, we (1) summarize the recent developments in plant multiscale modeling; (2) examine multiscale models of microbial systems that offer insight to potential future directions for the modeling of plant systems; (3) discuss computational tools and resources for developing multiscale models; and (4) examine future directions of the field.
Keywords: computational models, multiscale plant models, plant engineering
Introduction
Plants have developed complex regulatory strategies, across all levels of biological organization, to adapt to changes in their environment [1]. With the development of next-generation sequencing techniques, scientists have access to large-scale molecular phenotypes, providing information to help connect plant genomes to their phenomes. While the flow of biological information along the Central Dogma of biology (DNA → RNA → Protein) seems simple, it is exceedingly complex, making it difficult to predict plant phenotypes based solely on the genome. Advances in multiscale modeling are needed to account for this inherent complexity and to capture dynamic biological system responses to perturbations.
Plant modeling efforts range from empirical to mechanistic and vary in their predictive ability. Highly parameterized empirical models provide accurate representations of living systems but are rarely able to predict emergent properties. An exception to this is the OpenSimRoot model that predicted yield of maize as an emergent property of root architecture and water uptake [2]. Alternatively, data-driven models, such as machine learning models are entirely predictive, detecting correlations and extracting unseen patterns in the data. However, they do not reveal the underlying causal mechanisms driving the predicted phenotypes. Further, these models are unable to predict beyond the scope of the data used to train the models [3,4]. Finally, mechanistic models are mathematical representations of observed phenomena that attempt to identify causal relationships that result in an emergent phenotype, enabling the extrapolation of predictions about behaviors not present in the original data [3,4]. However, like empirical models, many mechanistic models are only predictive within the scope of the observed data, failing to predict phenotypes when new parameters are introduced [3].
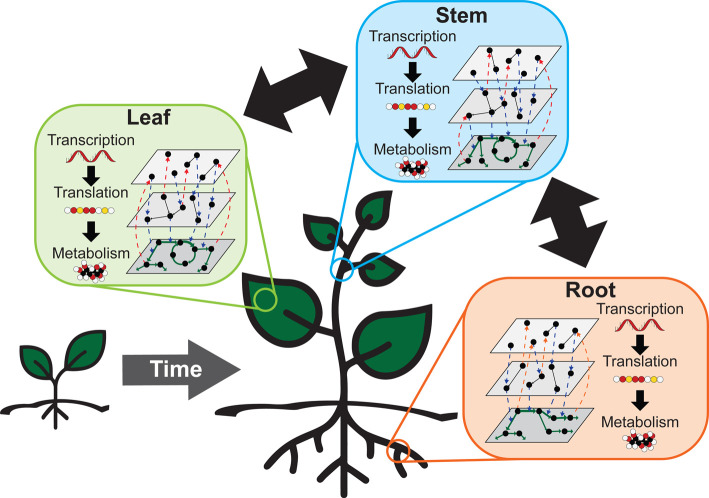
Mechanistic plant modeling has often focused on biochemical or metabolic processes like photosynthesis [5–7], other primary or secondary metabolic pathways [8–10], and genome-scale metabolic models which aim to capture all of the metabolic fluxes in an organism [11,12]. However, similar to just connecting plant genomes and phenomes, predicting emergent phenotypic responses based solely on metabolic models are difficult. Integrating empirical and mechanistic modeling approaches [3,4,13–15] and creating models that span across biological levels through integrative multiscale modeling (Figure 1) is necessary to understand how plants respond to a changing climate [16] or to explore how plants can be engineered to achieve specific phenotypic goals [17].
Figure 1.
Multiscale plant modeling involves integrating mathematical descriptions across space, time, and biological levels of organization.
In this review we discuss the current state of the art in plant multiscale modeling, as well as recent advances in microbial multiscale modeling that offer insight to potential future directions for the multiscale modeling of plant systems. We also discuss recently developed computational tools and resources for plant multiscale modeling, and other future directions of the field.
Multiscale plant models
Many of the current plant multiscale models have been developed for the model plant Arabidopsis, which has been extensively studied over the last few decades [18,19]. One of the most complete multiscale plant models to date is a family of Arabidopsis framework models. The first version of this model integrated (1) carbon dynamic, (2) functional-structural, (3) photothermal, and (4) photoperiodism models to describe Arabidopsis rosette growth [20]. Together, these four models link genetic regulation and biochemical dynamics to organ and plant growth. Chew et al. used their multiscale model to show that increasing leaf production rate in developmentally misregulated transgenic Arabidopsis sufficiently explained the smaller leaf size phenotype of this transgenic. The authors added a clock sub-model that explicitly represented key pathways in the Arabidopsis clock gene circuit to create version 2 of the Arabidopsis framework model (FMv2) [21]. The outputs of the clock sub-model were used to regulate tissue elongations and starch metabolism. With these updates, Arabidopsis FMv2 was able to predict phenotypic responses due to altered circadian timing in clock-mutant plants [21].
Kinmonth-Schultz et al. further expanded on the Arabidopsis FMv1 model [20] by adding a mechanistic description of temperature influence on FLOWERING LOCUS T (FT) expression, a photoperiodic flowering regulator [22]. Their model also highlighted some areas for further improvement such as incorporating a more mechanistic description of the relationship between leaf developmental age and FT expression. Zardilis et al. further extended the framework models [20,21] to include reproductive growth and developed a whole life cycle model of Arabidopsis, FM-life [23]. They used FM-life to explore different life cycle strategies for two genetic inputs (low seed dormancy with low floral repression and high seed dormancy with high floral repression) in two different environments (Valencia, Spain and Oulu, Finland). Their model helps explain how genetic variances result in different plant fitness responses in different environments [23].
Genome-scale metabolic models
Another area in multiscale plant modeling has been to integrate genome-scale metabolic flux models with gene expression data [24,25], spatial data, and temporal data [25–29].
Liu et al. integrated a genome-scale metabolic flux model with condition-specific transcriptome data to explore the metabolic response of Arabidopsis under low and high CO2 conditions [24]. However, in many cases using only transcriptomic data has not resulted in the expected improvement in model predictions [30]. Possible reasons for this include that key regulation could be occurring after transcription at either proteomic [31] or metabolic [32] levels. The authors also found this to be true with their Arabidopsis model, and they used their model to explore potential areas of post-transcriptional regulation [24]. Their findings suggested that Arabidopsis adapts to low or high CO2 environments by regulating the metabolic activity after transcription [24].
Multi-tissue genome-scale metabolic models have recently been used to study the balance of resources between plant tissues and across growth stages. Scheunemann et al. used a genome-scale metabolic model to develop a model of Arabidopsis root metabolism that consisted of several coupled cell-type specific models [25]. They used this model to predict the flux of the growth hormone indole-3-actetate through the root and its developmental stages [25]. De Oliveira Dal'Molin et al. created a multi-tissue genome-scale reconstruction of Arabidopsis leaf, stem, and roots, which they used to explore carbon (C) and nitrogen (N) resource allocation between the sink and source tissues with diurnal cycle constraints [26]. Likewise, a multi-tissue dynamic genome-scale model of Arabidopsis was developed and used to explore C/N balance over different growth stages [27]. Using this model, they identified sets of growth-stage specific reactions and reactions that were present during all of the growth stages [27]. This approach has also been successful in crop plants as demonstrated by Moreira et al. who developed a soybean multi-tissue genome-scale metabolic model of cotyledon and hypocotyl/root axis tissues and used it to study how stored reserves in seeds are remobilized during seedling growth [28]. In barley, a multiscale metabolic model was built to couple organ level static FBA models of metabolism with a dynamic functional plant model to simulate the spatiotemporal metabolic behavior of barley [29]. They used this model to study the source-sink interactions during the seed developmental phase of barley plants.
Multiscale models for plant engineering
A third area in plant multiscale modeling are models that were developed to explore strategies for engineering plants to achieve specific objectives. For example, a gene regulatory network model, a protein translation model, a mechanistic photosynthesis model, and a leaf-level physiological model were coupled to explore the impacts of genetic modifications to soybean photosynthesis in ambient and elevated CO2 [33]. The integrated model was used to identify gene regulatory controls of the allocation of resources from Rubisco to RuBP regeneration, which has been shown to improve photosynthesis under elevated CO2. Similarly, a multiscale model of lignin biosynthesis in the model tree Populus trichocarpa, was created by coupling a simple monolignol gene protein translation model, a mechanistic model of the monolignol biosynthetic pathway [9], and a multiple regression model to predict lignin and other wood traits under transgenic knockdowns of the monolignol genes [17]. This multiscale model was used to explore potential gene engineering strategies for producing trees with improved bioenergy traits while mitigating negative impacts on tree growth. This model was later expanded by incorporating the impact of cross-regulatory influences between the monolignol gene transcripts and proteins, capturing the effect of regulatory mechanisms that occur after transcription, such as potential post-transcriptional and post-translational modifications on predicted monolignol protein abundances [34,35].
Most of the current work in plant multiscale modeling has been to semi-integrate models, where information is passed from one layer to another, but limited feedback regulation is represented. The Arabidopsis FM models [20–23] are close to a fully-integrated framework, but also have few regulatory components modeled. As plant models become more fully-integrated, by communicating information across biological scales, they will better represent dynamic response to perturbation, which may reveal insights about system response to untested conditions. Recent works in microbial multiscale modeling have demonstrated that emergent properties can be predicted using fully-integrated models.
Learning from multiscale microbial models
Multiscale models have been developed for three different microbial organisms, Mycoplasma genitalium [36], E. coli [37–39], and S. cerevisiae [40,41].
Twenty eight submodels of diverse cellular processes were integrated to develop a whole cell multiscale model that describes the life cycle of a single cell of the human pathogen M. genitalium at the molecular level [36]. Their model provided insights into previously unobserved cellular behaviors. Through comparison of model predictions and experimental measurements they were able to detect new biological functions including a novel component of the pyruvate dehydrogenase chemical reactions [36].
A multiscale model of E. coli integrated transcriptional regulation, signal transduction, and metabolic pathways, and was used to predict E. coli growth rates in adverse conditions for single gene knockouts [37]. A multiscale model of chemotactic E. coli was also developed, which models cross-compartment mechanisms linking E. coli to its environment [38]. Further, researchers have used a large-scale integrated model of E. coli to assess the cross-consistency of millions of heterogeneous E. coli parameters collected from the literature, and used the identified inconsistencies to further improve their model [39].
In S. cerevisiae, Ye et al. followed the whole-cell modeling principles used to model M. genitalium [36,42] to develop a whole-cell multiscale model. This model bridged the gap between genotype and phenotype predictions by developing and integrating models on one second timescales of 26 cellular processes that span five areas of cell biology [40]. Using this model, they were able to study the dynamic allocation of resources during the cell cycle in real-time simulations. Ma et al. took a different approach by developing an interpretable neural network that incorporated 2526 intracellular components, processes, and functions to model the hierarchical structure and function of S. cerevisiae [41]. Their model was able to capture nearly all phenotypic variation in cellular growth, including variation due to genetic interactions under single and double gene deletions. Further, by using an interpretable neural network framework, they were able to identify previously undocumented processes related to cell growth, DNA repair, and UV sensitivity [41].
Plants are orders of magnitude more complex than microbes, so modeling whole plant systems at the same level of granularity as these microbial models may never be completely feasible. However, recently developed computational and experimental tools have the potential to make more fully-integrated plant multiscale models possible.
The future of multiscale plant modeling
Improving and developing models of metabolic regulation across biological scales including transcriptional, post-transcriptional, translational, and post-translational regulation is one direction for the plant multiscale modeling community. Due to the advances in next-generation sequencing techniques, genomic and transcriptomic research has progressed at a much faster pace than their proteomic and metabolomic counterparts, which primarily rely on mass spectrometry technologies. However, it has become increasingly clear the importance of these omics levels when trying to scale from genotype to phenotype. Novel approaches that couple in vivo proteins with their corresponding mRNA sequences are expected to leverage next-generation sequencing capabilities for proteomics, improving the scalability, standardization, and cost of large-scale proteomic experiments [43]. Another challenge to overcome is obtaining greater resolution of gene and gene product expression over time and space, which would improve multiscale molecular models. Specifically, such spatial and temporal information would inform modeling efforts aimed at understanding development at the cellular, tissue, organ, and whole organism scales. New technologies that provide better spatial and temporal resolution include fliFISH, which allows quantification of mRNA transcripts within organs and cell types [44], and FRET sensors which are used to track dynamic protein-protein interactions, nutrient signaling, and subcellular visualization of metabolites [45].
In addition to multiscale models of metabolic regulation in plants, future directions of plant multiscale modeling will involve connecting these microscale cell-type specific, gene regulation, and metabolic models to macroscale models such as multi-tissue and organ models, plant growth models (e.g. APSIM [46], BioCro [47], DSSAT [48]), physiological models, and ecosystem models (e.g. Community Land Model [49]). In the last few years computational tools have been developed to help achieve this goal. The yggdrasil framework [50] is an Open source Python package that can couple models written in a variety of programming languages including C/C++, Python, R, Matlab, and Fortran. Kannan et al. used the yggdrasil framework to one-way couple a gene regulatory network model (R), a protein translation model (Python), and a leaf photosynthesis model (Matlab) [33]. yggdrasil is also capable of two-way coupling between models with different timescales, throughout a simulation. Another tool, Vivarium [38], is an engine that allows different models to be assembled within a hierarchy of embedded compartments and then run as integrated multiscale simulations with multiple timescales and distributed computation. Agmon et al. used this engine to develop their multiscale model of E. coli chemotaxis [38]. The Chromar modeling language [51] was used in the FM-Life Arabidopsis model [23], and is a declarative, agent-based language that allows for the compact definition of complex biological models that can evolve with new biological reactions and components throughout a simulation. Additionally, advances in high performance computing (HPC) have been developed and applied to biological problems such as parameterizing multiscale models through statistical inference [52], modeling biochemical reactions in 3D space [53], and modeling the relationships between microscale mechanisms and emergent macroscopic behavior of a gliding motility assay [54]. Multiscale models can have high computational costs, especially when scaling from the microscales to the macroscales or when creating fully-integrated models that communicate across scales. As such, HPC could be a key component in creating fully-integrated plant models similar to the fully-integrated microbial models.
There are still many processes in plants that are unknown or not well understood, and as such are difficult to model. Machine and deep learning approaches, which detect statistical correlations in data, have become popular tools to develop predictive models in many fields. These models, however, are black-boxes and do not provide information about the mechanisms behind those predictions, limiting their applicability to the data used to train them and providing little new biological insight [3,4,41,55,56]. Interpretable or knowledge-primed neural networks are a recent advancement in the deep learning field that seeks to address this problem. By forcing the neural networks to have biologically relevant structure and hierarchy, interpretable neural networks have been used to develop models that are both predictive and biologically informative. Interpretable neural networks have been developed to model transcriptional control in fruit fly embryos [55], to predict cancer and immune cell states from single-cell RNAseq data [56], and to model S. cerevisiae cellular growth [41].
Developing models that communicate across these levels of biological organization will enable us to do the following: (1) Improve sink/source dynamics in the source-driven growth models. As many plant responses to stress are sink limited [57–60], this will improve our understanding of plant responses to stress and help us to identify engineering or management strategies for mitigating yield losses under stress; (2) Study carbon and nutrient allocation and remobilization between plant organs. This would allow us to explore methods of achieving optimal allocation strategies and could lead to improved yields and the development of seeds and grains with designed nutritional profiles; and (3) Explore how different plant engineering strategies respond in differing climates. This could have possible ecological impacts through the development of plants that are engineered for higher yields or improved water and nitrogen use efficiencies.
Author contributions
M.L.M. and A.M.C. were both responsible for drafting and revising this article.
Funding
Research reported in the publication was supported by the Foundation for Food and Agriculture Research under award number — Grant ID: 602757. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the Foundation for Food and Agriculture Research.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Haak, D.C., Fukao, T., Grene, R., Hua, Z., Ivanov, R., Perrella, G.et al. (2017) Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 8, 1564 10.3389/fpls.2017.01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.York, L.M., Galindo-Castañeda, T., Schussler, J.R. and Lynch, J.P. (2015) Evolution of US maize (Zea mays L.) root architectural and anatomical phenes over the past 100 years corresponds to increased tolerance of nitrogen stress. J. Exp. Bot. 66, 2347–2358 10.1093/jxb/erv074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, R.E., Peña, J.-M., Jayamohan, J. and Jérusalem, A. (2018) Mechanistic models versus machine learning, a fight worth fighting for the biological community? Biol. Lett. 14, 20170660 10.1098/rsbl.2017.0660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alber, M., Buganza Tepole, A., Cannon, W.R., De, S., Dura-Bernal, S., Garikipati, K.et al. (2019) Integrating machine learning and multiscale modeling—perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. npj Digit. Med. 2, 1–11 10.1038/s41746-019-0193-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farquhar, G.D., von Caemmerer, S. and Berry, J.A. (2001) Models of photosynthesis. Plant Physiol. 125, 42–45 10.1104/pp.125.1.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu, X.-G., Wang, Y., Ort, D.R. and Long, S.P. (2013) e-photosynthesis: a comprehensive dynamic mechanistic model of C3 photosynthesis: from light capture to sucrose synthesis. Plant Cell Environ. 36, 1711–1727 10.1111/pce.12025 [DOI] [PubMed] [Google Scholar]
- 7.Morales, A., Yin, X., Harbinson, J., Driever, S.M., Molenaar, J., Kramer, D.M.et al. (2018) In silico analysis of the regulation of the photosynthetic electron transport chain in C3 plants. Plant Physiol. 176, 1247–1261 10.1104/pp.17.00779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakhartsev, M., Medvedeva, I., Orlov, Y., Akberdin, I., Krebs, O. and Schulze, W.X. (2016) Metabolic model of central carbon and energy metabolisms of growing arabidopsis thaliana in relation to sucrose translocation. BMC Plant Biol. 16, 262 10.1186/s12870-016-0868-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, J.P., Naik, P.P., Chen, H.-C., Shi, R., Lin, C.-Y., Liu, J.et al. (2014) Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26, 894–914 10.1105/tpc.113.120881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shameer, S., Ratcliffe, R.G. and Sweetlove, L.J. (2019) Leaf energy balance requires mitochondrial respiration and export of chloroplast NADPH in the light. Plant Physiol. 180, 1947–1961 10.1104/pp.19.00624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams, T.C.R., Poolman, M.G., Howden, A.J.M., Schwarzlander, M., Fell, D.A., Ratcliffe, R.G.et al. (2010) A genome-scale metabolic model accurately predicts fluxes in central carbon metabolism under stress conditions. Plant Physiol. 154, 311–323 10.1104/pp.110.158535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poolman, M.G., Kundu, S., Shaw, R. and Fell, D.A. (2014) Metabolic trade-offs between biomass synthesis and photosynthate export at different light intensities in a genome–scale metabolic model of rice. Front. Plant. Sci. 5, 656 10.3389/fpls.2014.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazestani, V.H. and Lewis, N.E. (2019) From genotype to phenotype: augmenting deep learning with networks and systems biology. Curr. Opin. Syst. Biol. 15, 68–73 10.1016/j.coisb.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costello, Z. and Martin, H.G. (2018) A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. npj Syst. Biol. Appl. 4, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, J., Petersen, S.D., Radivojevic, T., Ramirez, A., Pérez-Manríquez, A., Abeliuk, E.et al. (2020) Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism. Nat. Commun. 11, 4880 10.1038/s41467-020-17910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baysal, C., He, W., Drapal, M., Villorbina, G., Medina, V., Capell, T.et al. (2020) Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. U.S.A. 117, 26503–26512 10.1073/pnas.2014860117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, J.P., Matthews, M.L., Williams, C.M., Shi, R., Yang, C., Tunlaya-anukit, S.et al. (2018) Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 9, 1579 10.1038/s41467-018-03863-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koornneef, M. and Meinke, D. (2010) The development of Arabidopsis as a model plant. Plant J. 61, 909–921 10.1111/j.1365-313X.2009.04086.x [DOI] [PubMed] [Google Scholar]
- 19.Berardini, T.Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E.et al. (2015) The arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485 10.1002/dvg.22877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chew, Y.H., Wenden, B., Flis, A., Mengin, V., Taylor, J., Davey, C.L.et al. (2014) Multiscale digital Arabidopsis predicts individual organ and whole-organism growth. Proc. Natl. Acad. Sci. U.S.A. 111, E4127–E4136 10.1073/pnas.1410238111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chew, Y.H., Seaton, D.D., Mengin, V., Flis, A., Mugford, S.T., Smith, A.M.et al. (2017) Linking circadian time to growth rate quantitatively via carbon metabolism. bioRxiv 105437 10.1101/105437 [DOI] [Google Scholar]
- 22.Kinmonth-Schultz, H.A., MacEwen, M.J.S., Seaton, D.D., Millar, A.J., Imaizumi, T. and Kim, S.-H. (2019) An explanatory model of temperature influence on flowering through whole-plant accumulation of FLOWERING LOCUS T in Arabidopsis thaliana. in silico Plants 1, diz006 10.1093/insilicoplants/diz006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zardilis, A., Hume, A. and Millar, A.J. (2019) A multi-model framework for the Arabidopsis life cycle. J. Exp. Bot. 70, 2463–2477 10.1093/jxb/ery394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, L., Shen, F., Xin, C. and Wang, Z. (2016) Multi-scale modeling of Arabidopsis thaliana response to different CO2 conditions: from gene expression to metabolic flux. J. Integr. Plant Biol. 58, 2–11 10.1111/jipb.12370 [DOI] [PubMed] [Google Scholar]
- 25.Scheunemann, M., Brady, S.M. and Nikoloski, Z. (2018) Integration of large-scale data for extraction of integrated Arabidopsis root cell-type specific models. Sci. Rep. 8, 7919 10.1038/s41598-018-26232-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomes de Oliveira Dal'Molin, C., Quek, L.-E., Saa, P.A., Nielsen, L.K. and Front Plant, S. (2015) A multi-tissue genome-scale metabolic modeling framework for the analysis of whole plant systems. Front. Plant Sci. 6, 4 10.3389/fpls.2015.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw, R. and Cheung, C.Y.M. (2018) A dynamic multi-tissue flux balance model captures carbon and nitrogen metabolism and optimal resource partitioning during arabidopsis growth. Front. Plant Sci. 9, 884 10.3389/fpls.2018.00884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreira, T.B., Shaw, R., Luo, X., Ganguly, O., Kim, H.-S., Coelho, L.G.F.et al. (2019) A genome-scale metabolic model of soybean (Glycine max) highlights metabolic fluxes in seedlings. Plant Physiol. 180, 1912–1929 10.1104/pp.19.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grafahrend-Belau, E., Junker, A., Eschenröder, A., Müller, J., Schreiber, F. and Junker, B.H. (2013) Multiscale metabolic modeling: dynamic flux balance analysis on a whole-plant scale. Plant Physiol. 163, 637–647 10.1104/pp.113.224006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machado, D. and Herrgård, M. (2014) Systematic evaluation of methods for integration of transcriptomic data into constraint-based models of metabolism. PLOS Comput. Biol. 10, e1003580 10.1371/journal.pcbi.1003580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel, C. and Marcotte, E.M. (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 10.1038/nrg3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackett, S.R., Zanotelli, V.R.T., Xu, W., Goya, J., Park, J.O., Perlman, D.H.et al. (2016) Systems-level analysis of mechanisms regulating yeast metabolic flux. Science 354, aaf2786 10.1126/science.aaf2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kannan, K., Wang, Y., Lang, M., Challa, G.S., Long, S.P. and Marshall-Colon, A. (2019) Combining gene network, metabolic and leaf-level models shows means to future-proof soybean photosynthesis under rising CO2. in silico Plants 1, diz008 10.1093/insilicoplants/diz008 [DOI] [Google Scholar]
- 34.Matthews, M.L., Wang, J.P., Sederoff, R., Chiang, V.L. and Williams, C.M. (2020) Modeling cross-regulatory influences on monolignol transcripts and proteins under single and combinatorial gene knockdowns in Populus trichocarpa. PLOS Comput. Biol. 16, e1007197 10.1371/journal.pcbi.1007197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews, M.L., Wang, J.P., Sederoff, R., Chiang, V.L. and Williams, C.M. (2021) A multiscale model of lignin biosynthesis for predicting bioenergy traits in Populus trichocarpa. Comput. Struct. Biotechnol. J. 19, 168–182 10.1016/j.csbj.2020.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karr, J.R., Sanghvi, J.C., Macklin, D.N., Gutschow, M.V., Jacobs, J.M., Bolival, B.et al. (2012) A whole-cell computational model predicts phenotype from genotype. Cell 150, 389–401 10.1016/j.cell.2012.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrera, J., Estrela, R., Luo, J., Rai, N., Tsoukalas, A. and Tagkopoulos, I. (2014) An integrative, multi-scale, genome-wide model reveals the phenotypic landscape of Escherichia coli. Mol. Syst. Biol. 10, 735 10.15252/msb.20145108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agmon, E. and Spangler, R.K. (2020) A multi-scale approach to modeling E. coli chemotaxis. Entropy 22, 1101 10.3390/e22101101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macklin, D.N., Ahn-Horst, T.A., Choi, H., Ruggero, N.A., Carrera, J., Mason, J.C.et al. (2020) Simultaneous cross-evaluation of heterogeneous E. coli datasets via mechanistic simulation. Science 369, eaav3751. 10.1126/science.aav3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye, C., Xu, N., Gao, C., Liu, G., Xu, J., Zhang, W.et al. (2020) Comprehensive understanding of Saccharomyces cerevisiae phenotypes with whole-cell model WM_S288C. Biotechnol. Bioeng. 117, 1562–1574 10.1002/bit.27298 [DOI] [PubMed] [Google Scholar]
- 41.Ma, J., Yu, M.K., Fong, S., Ono, K., Sage, E., Demchak, B.et al. (2018) Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods 15, 290–298 10.1038/nmeth.4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karr, J.R., Takahashi, K. and Funahashi, A. (2015) The principles of whole-cell modeling. Curr. Opin. Microbiol. 27, 18–24 10.1016/j.mib.2015.06.004 [DOI] [PubMed] [Google Scholar]
- 43.Oikonomou, P., Salatino, R. and Tavazoie, S. (2020) In vivo mRNA display enables large-scale proteomics by next generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 117, 26710–26718 10.1073/pnas.2002650117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui, Y., Hu, D., Markillie, L.M., Chrisler, W.B., Gaffrey, M.J., Ansong, C.et al. (2018) Fluctuation localization imaging-based fluorescence in situ hybridization (fliFISH) for accurate detection and counting of RNA copies in single cells. Nucleic Acids Res. 46, e7 10.1093/nar/gkx874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Littlejohn, G.R., Meckel, T., Schwarzländer, M. and Costa, A. (2014) Functional imaging in living plants—cell biology meets physiology. Front. Plant Sci. 5, 740 10.3389/fpls.2014.00740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holzworth, D.P., Huth, N.I., deVoil, P.G., Zurcher, E.J., Herrmann, N.I., McLean, G.et al. (2014) APSIM – evolution towards a new generation of agricultural systems simulation. Environ. Model. Softw. 62, 327–350 10.1016/j.envsoft.2014.07.009 [DOI] [Google Scholar]
- 47.Miguez, F.E., Zhu, X., Humphries, S., Bollero, G.A. and Long, S.P. (2009) A semimechanistic model predicting the growth and production of the bioenergy crop Miscanthus×giganteus: description, parameterization and validation. GCB Bioenergy 1, 282–296 10.1111/j.1757-1707.2009.01019.x [DOI] [Google Scholar]
- 48.Hoogenboom, G., Porter, C.H., Boote, K.J., Shelia, V., Wilkens, P.W., Singh, U.et al. (2019) The DSSAT crop modeling ecosystem. In: p.173–216 (K.J. Boote, ed.) Advances in Crop Modeling for a Sustainable Agriculture. Burleigh Dodds Science Publishing, Cambridge, United Kingdom: 10.19103/AS.2019.0061.10 [DOI] [Google Scholar]
- 49.Lawrence, D.M., Fisher, R.A., Koven, C.D., Oleson, K.W., Swenson, S.C., Bonan, G.et al. (2019) The community land model version 5: description of new features, benchmarking, and impact of forcing uncertainty. J. Adv. Model. Earth Syst. 11, 4245–4287 10.1029/2018MS001583 [DOI] [Google Scholar]
- 50.Lang, M. (2019) Yggdrasil: a python package for integrating computational models across languages and scales. in silico Plants 1, diz001 10.1093/insilicoplants/diz001 [DOI] [Google Scholar]
- 51.Honorato-Zimmer, R., Millar, A.J., Plotkin, G.D. and Zardilis, A. (2019) Chromar, a language of parameterised agents. Theoretical. Comput. Sci. 765, 97–119 10.1016/j.tcs.2017.07.034 [DOI] [Google Scholar]
- 52.Jagiella, N., Rickert, D., Theis, F.J. and Hasenauer, J. (2017) Parallelization and high-performance computing enables automated statistical inference of multi-scale models. Cell Syst. 4, 194–206.e9 10.1016/j.cels.2016.12.002 [DOI] [PubMed] [Google Scholar]
- 53.Ganesan, N., Li, J., Sharma, V., Jiang, H. and Compagnoni, A. (2016) Process simulation of complex biological pathways in physical reactive space and reformulated for massively parallel computing platforms. IEEE/ACM Trans. Comput. Biol. Bioinform. 13, 365–379 10.1109/TCBB.2015.2443784 [DOI] [PubMed] [Google Scholar]
- 54.Cook S. and Shinar T. Enabling Simulation of High-Dimensional Micro-Macro Biophysical Models through Hybrid CPU and Multi-GPU Parallelism. arXiv:190804279 [physics, q-bio]. 2019 Aug 12.
- 55.Liu, Y., Barr, K. and Reinitz, J. (2020) Fully interpretable deep learning model of transcriptional control. Bioinformatics 36, i499–i507 10.1093/bioinformatics/btaa506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fortelny, N. and Bock, C. (2020) Knowledge-primed neural networks enable biologically interpretable deep learning on single-cell sequencing data. Genome Biol. 21, 190 10.1186/s13059-020-02100-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fatichi, S., Leuzinger, S. and Körner, C. (2014) Moving beyond photosynthesis: from carbon source to sink-driven vegetation modeling. New Phytol. 201, 1086–1095 10.1111/nph.12614 [DOI] [PubMed] [Google Scholar]
- 58.White, A.C., Rogers, A., Rees, M. and Osborne, C.P. (2016) How can we make plants grow faster? A source–sink perspective on growth rate. J. Exp. Bot. 67, 31–45 10.1093/jxb/erv447 [DOI] [PubMed] [Google Scholar]
- 59.Wingler, A. (2015) Comparison of signaling interactions determining annual and perennial plant growth in response to low temperature. Front. Plant Sci. 5, 794 10.3389/fpls.2014.00794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonnewald, U. and Fernie, A.R. (2018) Next-generation strategies for understanding and influencing source–sink relations in crop plants. Curr. Opin. Plant Biol. 43, 63–70 10.1016/j.pbi.2018.01.004 [DOI] [PubMed] [Google Scholar]