Abstract
Objective
To test the hypothesis that the APOE genotype is a significant driver of heterogeneity in Alzheimer disease (AD) clinical progression, which could have important implications for clinical trial design and interpretation.
Methods
We applied novel reverse-time longitudinal models to analyze the trajectories of Clinical Dementia Rating Sum of Boxes (CDR-SOB) and Mini-Mental State Examination (MMSE) scores—2 common outcome measures in AD clinical trials—in 1,102 autopsy-proven AD cases (moderate/frequent neuritic plaques and Braak tangle stage III or greater) from the National Alzheimer's Coordinating Center Neuropathology database resembling participants with mild to moderate AD in therapeutic clinical trials.
Results
APOE ε4 carriers exhibited ≈1.5 times faster CDR-SOB increase than APOE ε3/ε3 carriers (2.12 points per year vs 1.44 points per year) and ≈1.3 times faster increase than APOE ε2 carriers (1.65 points per year), whereas APOE ε2 vs APOE ε3/ε3 difference was not statistically significant. APOE ε4 carriers had ≈1.1 times faster MMSE decline than APOE ε3/ε3 carriers (−3.45 vs −3.03 points per year) and ≈1.4 times faster decline than APOE ε2 carriers (−2.43 points per year), whereas APOE ε2 carriers had ≈1.2 times slower decline than APOE ε3/ε3 carriers (−2.43 vs −3.03 points per year). These findings remained largely unchanged after controlling for the effect of AD neuropathologic changes on the rate of cognitive decline and for the presence and severity of comorbid pathologies.
Conclusion
Compared to the APOE ε3/ε3 reference genotype, the APOE ε2 and ε4 alleles have opposite (slowing and accelerating, respectively) effects on the rate of cognitive decline, which are clinically relevant and largely independent of the differential APOE allele effects on AD and comorbid pathologies. Thus, APOE genotype contributes to the heterogeneity in rate of clinical progression in AD.
One milestone in Alzheimer disease (AD) clinical trials has been the incorporation of PET imaging and CSF biomarkers to exclude AD mimics and even design secondary prevention trials for participants with preclinical AD.1 However, trial success still depends on detecting a treatment vs placebo change in cognitive decline rate, and this is hampered by the substantial variability in rate of clinical progression among participants. One potential contributor to this heterogeneity is the APOE genotype, given the opposing effects of the APOE ε4 and ε2 alleles on AD risk, age at symptom onset, and AD neuropathologic changes (ADNC).2-5 However, prior longitudinal clinical studies after symptom onset have reported conflicting results—accelerating,6-9 neutral,10-12 and slowing effects13-15 for APOE ε4 vs slowing effects for APOE ε216-18—likely due, at least in part, to the suboptimal accuracy of a clinically based AD diagnosis.19,20 Another proposed driver is the co-occurrence of ≥2 brain pathologies,21 each of which could independently contribute to cognitive impairment and could be influenced by the APOE genotype.22-24
We tested the hypothesis that the APOE alleles differentially affect the cognitive decline rate in an autopsy-proven clinical trial–eligible AD sample. We circumvented the limitations of prior clinical studies by (1) selecting a National Alzheimer's Coordinating Center (NACC) sample with sufficient ADNC to warrant enrollment in current biomarker-based therapeutic clinical trials, (2) applying novel reverse-time longitudinal models to link autopsy findings with cognitive trajectories during life, and (3) controlling for the effects of ADNC on cognitive decline rate and for comorbid pathologies.
Methods
Standard Protocol Approvals, Registrations, and Patient Consents
As determined by the University of Washington Human Subjects Division, the NACC database itself is exempt from Institutional Review Board review and approval because it does not involve human participants, as defined by federal and state regulations. However, all contributing Alzheimer Disease Centers (ADCs) are required to obtain informed consent from their participants and to maintain their own separate Institutional Review Board review and approval from their institution before submitting data to NACC.
Inclusion and Exclusion Criteria
The NACC study is a multicenter longitudinal cohort study conducted in the ≥30 ADCs across the United States. Briefly, participants undergo baseline and annual follow-up visits with demographics and standard motor, behavioral, functional, and neuropsychological evaluations collected in a Uniform Data Set (UDS).25,26 Participants can donate their brain on death for research purposes, including a standardized neuropathologic evaluation.27 The NACC Neuropathology database was interrogated for UDS visits conducted between September 2005 and November 2018. Inclusion criteria included age at death ≥50 years and last visit <2 years from the time of death. Exclusion criteria were unknown APOE genotype, APOE ε2/ε4 genotype (due to the small number of cases and presumed cancellation of APOE ε2 and APOE ε4 opposite effects), a primary neuropathologic diagnosis other than ADNC, and cognitive impairment attributed to medical illness, medications, or alcohol. To mimic the scenario of a clinical trial with biomarker-based enrollment of participants, we excluded those with none/sparse neuritic plaques (NP) at postmortem examination because they would have had a negative baseline amyloid PET scan before enrollment.28 Similarly, to resemble a therapeutic rather than preventive design, we included only those with a Braak neurofibrillary tangle (NFT) stage III or higher because individuals with limbic (III/IV) or isocortical (V/VI) Braak NFT stages would have had tau PET scans demonstrating limbic and cortical tau deposits.29 Our prior inverse probability–weighting analyses on the NACC Neuropathology database demonstrated little impact of potential autopsy-related selection bias in clinicopathologic correlations.30
Data Collection
Data collected included (1) demographic variables (age at each visit and at death, sex, years of education, and length of follow-up), (2) APOE genotype, (3) neuropsychological scores at each visit (Clinical Dementia Rating Sum of Boxes [CDR-SOB; CDR Dementia Staging Instrument], Mini-Mental State Examination [MMSE], digit span forward and backward, Trail Making Tests A and B, Wechsler Adult Intelligence Scale Digit-Symbol Substitution Test, logical memory immediate and delayed recall, semantic fluency [animals and vegetables in 1 minute], and Boston Naming Test), and (4) autopsy neuropathologic findings (Consortium to Establish a Registry of Alzheimer’s Disease [CERAD] NP score, Braak NFT stage, presence of hippocampal sclerosis [HScl] and Lewy bodies [LB], and presence and severity of both arteriolosclerosis and cerebral amyloid angiopathy [CAA; none, mild, moderate, severe]), all of which are associated with antemortem CDR-SOB score within the AD continuum.30
Statistics
Statistical analyses were run in R software version 3.6 using R package lcmm (R Foundation for Statistical Computing).31 Outcome variables included CDR-SOB score, MMSE score, and cognitive domain–specific scores. For the last ones, the scores of the neuropsychological outcome variables at each visit were converted into z scores. Briefly, tests were grouped in 4 cognitive domains based on a validated factor structure32 as follows: z scores for logical memory immediate and delayed recall were averaged into a memory composite score; z scores for Trail Making Tests A and B and Digit-Symbol Substitution Test were averaged into an executive composite score; z scores for digits forward trials and length, and digits backward trials and length were averaged into an attention composite score; and z scores for animals and vegetables in 1 minute and Boston Naming Test were averaged into a language composite score.
To evaluate the association between APOE genotype and rate of cognitive decline as reflected in longitudinal global functional and cognitive measures (CDR-SOB and MMSE scores), we used statistical methods previously described in detail elsewhere.33 Briefly, the main specifications and advantages of this methodology are as follows:
1. Reverse time. Traditional forward time analysis precludes linking the neuropathologic measures with individual cognitive trajectories during life because neuropathologic measures are time varying and measurable only at postmortem examination. Therefore, we treated the neuropathologic variables as baseline covariates and modeled the longitudinal cognitive trajectories in reverse time, that is, beginning with neuropsychological scores at the visit closest to death (<2 years prior per inclusion criteria) and moving backward toward the scores obtained at the initial visit.
2. Shared latent classes. Longitudinal cognitive trajectories are truncated by events such as last visit or death, and the neuropathologic variables are ascertained at death. Therefore, any potential association between the longitudinal cognitive trajectories and these time-to-events must be accounted for. To achieve this and to control for any unmeasured (latent) features that may be associated with both, we implemented a joint latent class model for the longitudinal cognitive trajectories (mixed-effects submodel), the time-to-event analyses (death to first NACC visit, Cox proportional hazards submodel), and class membership (logistic submodel) and evaluated the number of latent classes best supported by the data with the Bayesian information criterion (BIC).
3. Change point. Longitudinal neuropsychological testing is often affected by floor or ceiling effects as the dementia advances and individuals become untestable. To account for these possible floor/ceiling effects of cognitive outcomes and to capture any change in the slope of cognitive trajectories in advanced AD, we used a piecewise linear model with 2 different linear slopes before and after a change point and determined the change point best supported by the data (2, 2.5, or 3 years before death) with the BIC. We decided not to use change point as a random variable due to the complexity of the model.
4. Right truncation adjustment by time to death. Because this is a clinicopathologic autopsy sample, time to last NACC visit is right truncated by time to death. To avoid potential bias derived from the oversampling of shorter times to death, we adjusted for right truncation of time to last visit by time to death. For a more detailed discussion of the statistical methodology, we refer the reader to our previous article.32
The covariates used in the mixed-effects submodel for the longitudinal cognitive trajectory analyses and in Cox proportional hazards submodel for the time-to-event analyses included sex, education, age at death, APOE genotype (presence vs absence of ε2 allele, and presence vs absence of ε4 allele, with e3/e3 carriers as reference group), CERAD NP score (frequent vs moderate), and Braak NFT stages (V/VI vs III/IV). After selection of the most suitable number of latent classes and change point for each cognitive variable, CERAD NP score, Braak NFT stage, and the neuropathologic comorbid pathologies (i.e., presence of HScl and LB, presence and severity of both arteriolosclerosis and CAA [none, mild, moderate, severe]) with a significant association with the antemortem cognitive variable were added to the models. To assess the differential effects of APOE genotype, CERAD NP score, and Braak NFT stage on cognitive trajectory, we allowed interaction terms between each of these 3 variables and the slope before or after the change point in the modeling. We started with simpler models with APOE genotype as predictor and each cognitive measure as outcome variable, adjusted by age, sex, education, CERAD NP score, and Braak NFT stage (model 1). To investigate whether APOE genotype effects on the rate of cognitive decline are independent from ADNC and comorbid pathologies, we then built more complex models by further adjusting for interaction terms between CERAD NP score or Braak NFT stage and the slope before or after the change point and for concurrent pathologies (model 2). To use the autopsy variables in the forward-time translation of the analyses, we assumed that amyloid plaque burden does not substantially change over the clinical course of AD and that the sequence of Braak NFT stages is preserved over the extent of longitudinal follow-up. These assumptions are well supported by prior β-amyloid (Aβ) and tau PET imaging studies.34,35
Data Availability
The NACC database is a public resource available to researchers. Data requests can be submitted online at the following NACC website: alz.washington.edu/NONMEMBER/QUERY/datareqnew.html.
Results
Characteristics of Study Participants
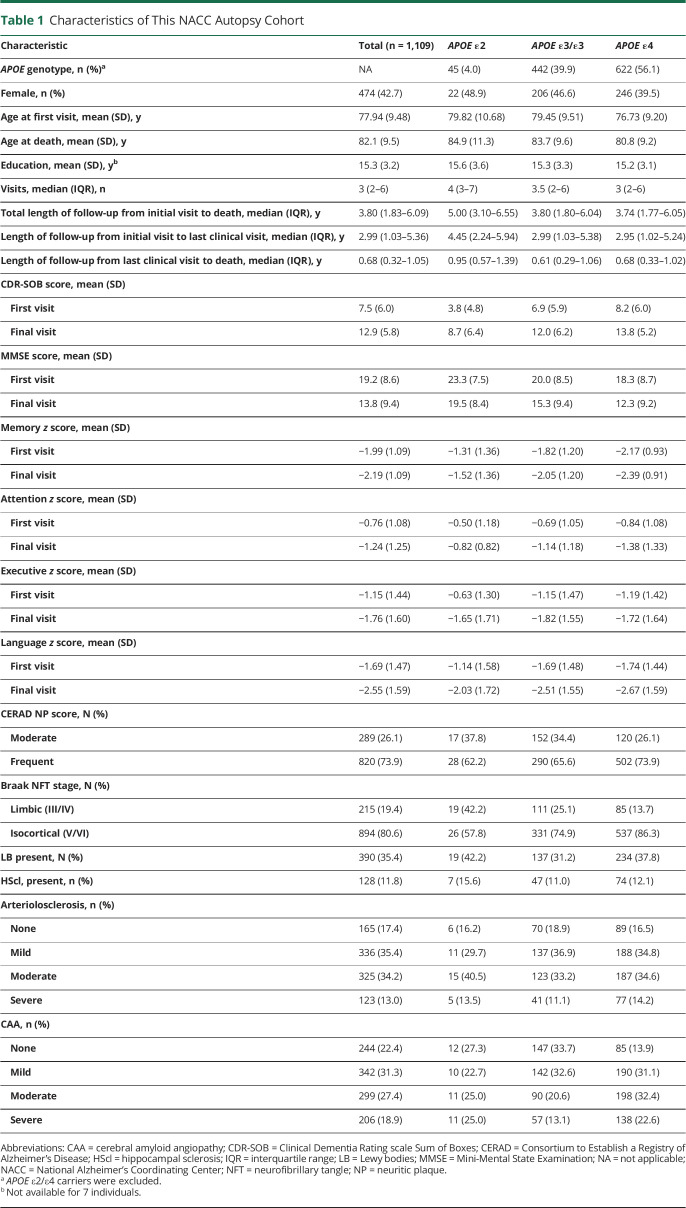
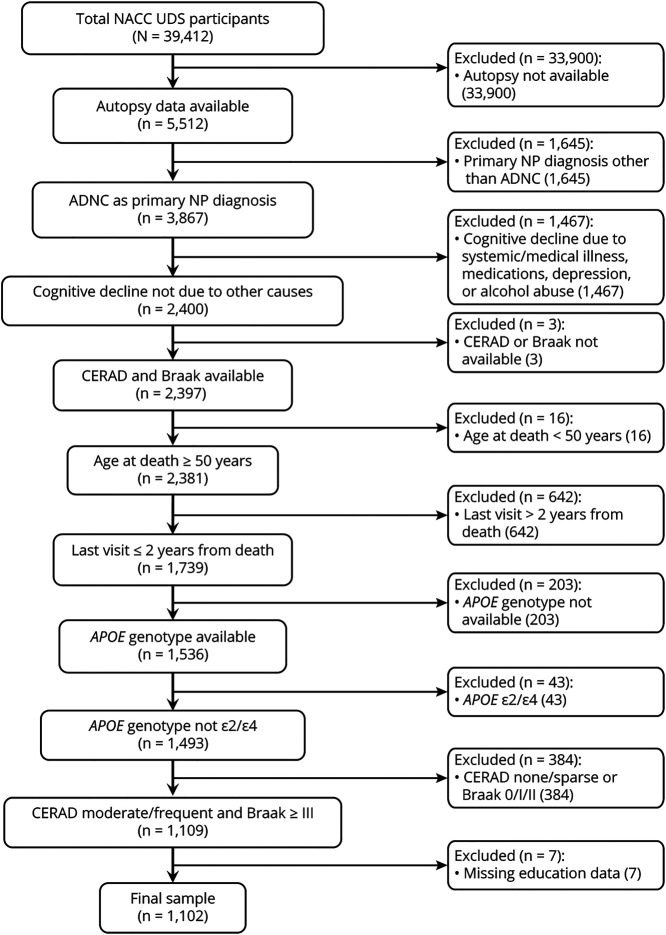
Table 1 summarizes the demographic characteristics, neuropathologic autopsy findings, and cognitive measures of the study participants at baseline and last clinical visit. The flowchart in figure 1 shows that 1,109 individuals met the inclusion criteria and none of the exclusion criteria, but 7 individuals had to be excluded due to missing education data, hence the final sample of 1,102.
Table 1.
Characteristics of This NACC Autopsy Cohort
Figure 1. Flowchart of Study Participants.
ADNC = Alzheimer disease neuropathological changes; CERAD = Consortium to Establish a Registry of Alzheimer’s Disease; NACC = National Alzheimer’s Coordinating Center; NP = neuritic plaque; UDS = Uniform Data Set.
Selection of Neuropathologic Covariates for Longitudinal Modeling
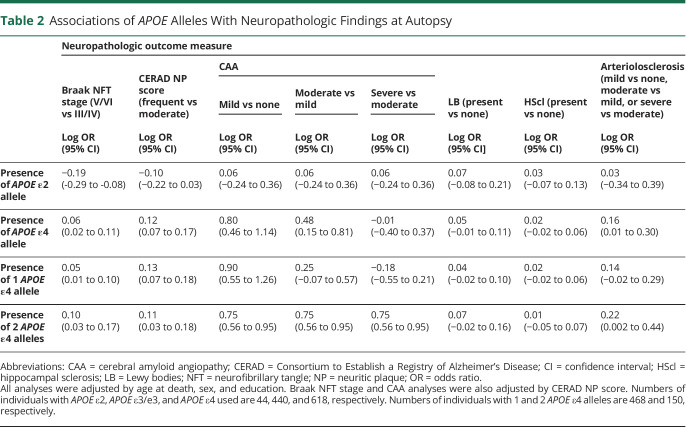
To select the neuropathologic covariates for the longitudinal models, we first investigated the effects of APOE alleles on ADNC and comorbid pathologies in this convenience sample using multivariate regression models controlling for age at death, sex, and education (table 2). The APOE ε4 allele was associated with a higher CERAD NP score (frequent vs moderate), a higher Braak stage (V/VI vs III/IV), more severe CAA (moderate vs mild and mild vs none), and more severe arteriolosclerosis than the APOE ε3/ε3 reference group. An APOE ε4 dose-dependent effect was observed for most of these associations. In contrast, the APOE ε2 allele was associated with a lower Braak stage (III/IV vs V/VI) compared to the APOE ε3/ε3 group but not with a lower CERAD NP score (log odds ratio −0.10, 95% confidence interval [CI] −0.22 to 0.03, p = 0.141). No significant effect was observed for either APOE allele on the presence of LB or HScl. These results largely agree with those from a NACC sample of cognitively impaired (CDR-SOB score >0) selected to represent the AD clinicopathologic continuum.5
Table 2.
Associations of APOE Alleles With Neuropathologic Findings at Autopsy
To further refine the selection of neuropathologic covariates, we examined the effects of concurrent pathologies (CAA, LB, HScl, and arteriolosclerosis) on antemortem global cognitive measures (CDR-SOB and MMSE scores) and domain-specific composites. With age at death, sex, education, CERAD NP score, Braak NFT stage, and APOE genotype held constant, presence vs absence of HScl was associated with worse memory (−0.294 ± 0.063, p < 0.001) and language (−0.666 ± 0.149, p < 0.001), higher CDR-SOB (2.694 ± 0.436, p < 0.001) and lower MMSE (−3.801 ± 0.752, p < 0.001) scores, presence vs absence of LB with worse executive function (−0.286 ± 0.118, p = 0.015), severe arteriolosclerosis vs none with worse attention (−0.376 ± 0.140, p = 0.007), and moderate and severe CAA vs none with higher CDR-SOB (moderate vs none 1.400 ± 0.410, p < 0.001; severe vs none 0.977 ± 0.453, p = 0.031) and lower MMSE (moderate vs none −2.478 ± 0.700, p = 0.002; severe vs none −1.665 ± 0.792, p = 0.035) scores.
Longitudinal Modeling Reveals Opposing Effects of APOE Alleles on Global Cognitive Trajectory
Overall, on the basis of the BIC, longitudinal models with a change point at 3 years before death were preferred to those with a 2- or 2.5-year change point; with a change point at 3 years, the 2-latent-class model was preferred over the 1-latent-class model for all the cognitive outcomes.
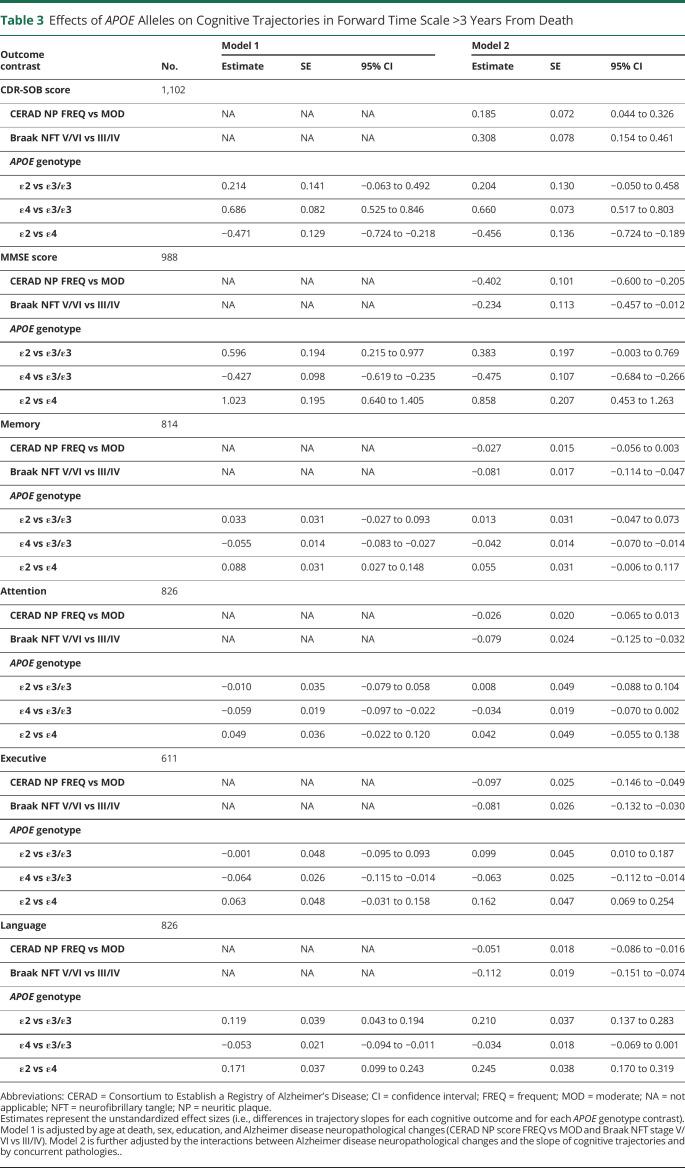
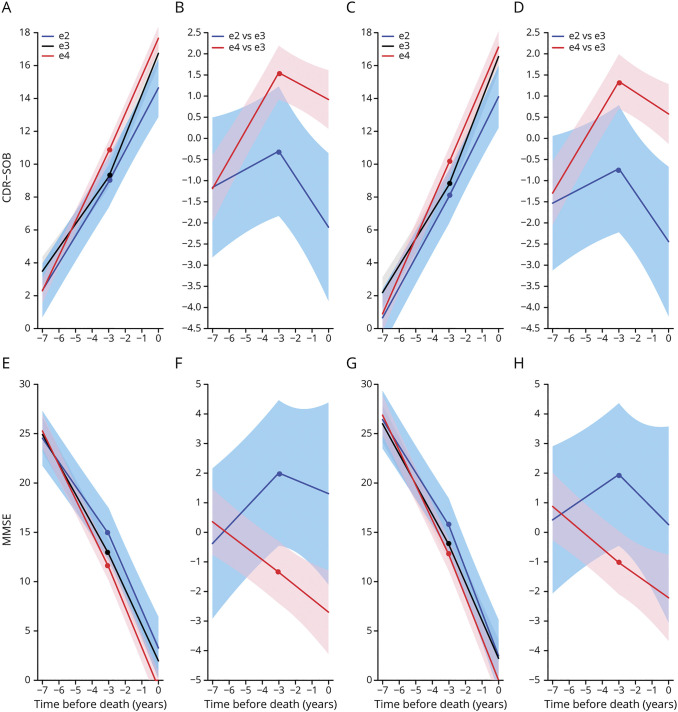
Table 3 shows the results of these models controlled for age at death, sex, education, and ADNC severity (model 1) and with additional adjustments for the effect of ADNC on rate of cognitive decline and for presence and severity of concurrent pathologies (model 2) >3 years before death. Figure 2 illustrates these results. With only demographic and ADNC variables (model 1) held constant, APOE ε4 carriers exhibited ≈1.5 times faster progression by CDR-SOB score than APOE ε3/ε3 carriers (2.12 vs 1.44 points per year, 95% CI for the difference 0.53–0.85) and ≈1.3 times faster than APOE ε2 carriers (2.12 vs 1.65 points per year, 95% CI 0.22–0.72), but APOE ε2 carriers did not significantly differ from APOE ε3/ε3 carriers (1.65 vs 1.44 points per year, 95% CI −0.06 to 0.49) (figure 2, A and B). By MMSE score, APOE ε4 carriers had ≈1.1 times faster decline than APOE ε3/ε3 carriers (−3.45 vs −3.03 points per year, 95% CI −0.62 to −0.24) and ≈1.4 times faster decline than APOE ε2 carriers (−3.45 vs −2.43 points per year, 95% CI −1.41 to −0.64), whereas APOE ε2 carriers had ≈1.2 times slower decline than APOE ε3/ε3 carriers (−2.43 vs −3.03 points per year, 95% CI 0.22–0.98) (figure 2, E and F).
Table 3.
Effects of APOE Alleles on Cognitive Trajectories in Forward Time Scale >3 Years From Death
Figure 2. Effect of APOE Alleles on Global Functional and Cognitive Outcome Measures (CDR-SOB and MMSE) Scores.
(A and E) Model-based cognitive trajectories of Clinical Dementia Rating Sum of Boxes (CDR-SOB) (A) and Mini-Mental State Examination MMSE (E) scores by APOE allele groups (APOE ε3/ε3, APOE ε2, and APOE ε4 carriers) with the intercept at the time of death calculated for an 82-year-old woman with 15 years of education and autopsy findings of frequent Consortium to Establish a Registry of Alzheimer’s Disease (CERAD) neuritic plaque (NP) score and Braak neurofibrillary tangle (NFT) stage V/VI, but without adjustments for the effect of these neuropathologic variables on the rate of cognitive decline or for comorbid pathologies (model 1). (C and G) Model-based cognitive trajectories of CDR-SOB (C) and MMSE (G) scores by APOE group with the intercept at the time of death calculated for an 82-year-old woman with 15 years of education and autopsy findings of frequent CERAD NP score and Braak NFT stage V/VI and no comorbid pathologies and with adjustments for the effect of AD neuropathologic variables (CERAD and Braak) on the rate of cognitive decline and for comorbid pathologies (model 2). Note the nearly identical trajectories with and without controlling for neuropathology. (B, D, F, and H) Difference of model-based cognitive trajectories of CDR-SOB (B and D) and MMSE (F and H) scores between APOE allele groups (APOE ε2 and APOE ε4 vs APOE ε3/ε3 carriers) under models 1 (B and F) and 2 (D and H). Shaded areas represent the corresponding 95% confidence intervals.
Holding all demographic, ADNC, and comorbid neuropathologic covariates constant and controlling for the effect of ADNC on the slope of cognitive decline (model 2), we found that APOE ε4 carriers exhibited ≈1.6 times faster clinical progression by CDR-SOB score than APOE ε3/ε3 carriers (1.80 vs 1.14 points per year, 95% CI for the difference 0.52–0.80) and ≈1.3 times faster clinical progression than APOE ε2 carriers (1.80 vs 1.34 points per year, 95% CI 0.19–0.72). In contrast, APOE ε2 carriers did not significantly differ from APOE ε3/ε3 carriers (1.34 vs 1.14 points per year, 95% CI −0.05 to 0.46) (figure 2, C and D). By MMSE score, APOE ε4 carriers had ≈1.2 times faster decline than APOE ε3/ε3 carriers (−2.90 vs −2.43 points per year, 95% CI −0.68 to −0.27) and ≈1.4 times faster decline than APOE ε2 carriers (−2.90 vs −2.04 points per year, 95% CI −1.26 to −0.45), whereas APOE ε2 carriers had ≈1.2 times slower decline than APOE ε3/ε3 carriers (−2.04 vs −2.43 points per year, 95% CI 0.00–0.77) (figure 2, G and H).
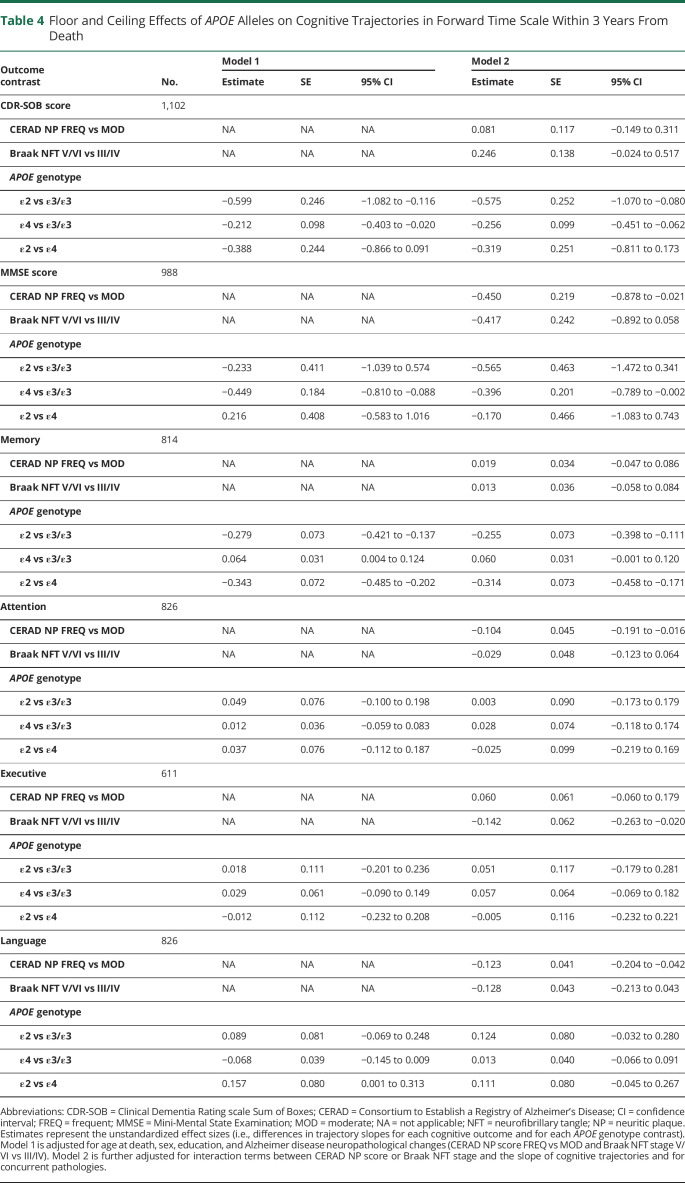
As expected, the 3-year change point revealed and isolated ceiling effects of CDR-SOB score and floor effects of MMSE score within 3 years before death (table 4 and figure 2). In this time frame, the APOE ε2 carriers exhibited a significantly slower increase in CDR-SOB score compared to the APOE ε3/ε3 group (1.70 vs 2.28 points per year, 95% CI −1.07 to −0.08), but a ceiling effect of the CDR-SOB score in the APOE ε4 group is apparent, with a slower decline relative to the APOE ε3/ε3 group (2.02 vs 2.28 points per year, 95% CI −0.06 to −0.45). Indeed, 29% APOE ε4 carriers had already reached the maximum CDR-SOB score of 18 at the 3-year change time point compared to 19% of APOE ε3/ε3 carriers. Similarly, the APOE ε4 carriers showed a significantly faster decline in MMSE score than the APOE ε3/ε3 group (−3.37 vs −2.97 points per year, 95% CI[−0.79 to −0.01), but a floor effect of the MMSE score became apparent in the APOE ε3/ε3 group, causing APOE ε2 carriers to show a nonsignificant trend toward an apparent faster decline (−3.54 vs −2.97 points per year, 95% CI −1.47 to 0.34). As an example, 8.2% of APOE ε3/ε3 carriers had reached an MMSE score ≤3 at the 3-year change time point vs none of the APOE ε2 carriers.
Table 4.
Floor and Ceiling Effects of APOE Alleles on Cognitive Trajectories in Forward Time Scale Within 3 Years From Death
Longitudinal Modeling Reveals Opposing Effects of APOE Alleles on Specific Cognitive Domains
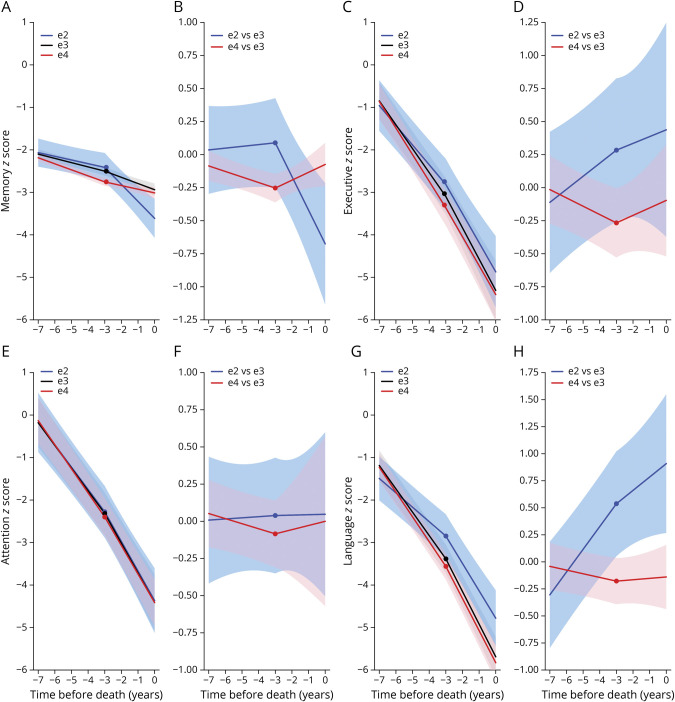
Without controlling for the effect of ADNC on rate of cognitive decline or for concurrent pathologies, APOE ε4 carriers had a significantly faster decline in all 4 domains analyzed (memory, executive, attention, language) compared to APOE ε3/ε3 carriers, whereas APOE ε2 carriers had a significantly slower decline in language compared to APOE ε3/ε3 carriers and in both memory and language compared to APOE ε4 carriers (table 3, figure 3, and figure e-1 available from Dryad: doi.org/10.5061/dryad.w0vt4b8qk). The models controlling for ADNC effects on rate of cognitive decline and these comorbid pathologies rendered somewhat different results, likely due to the confounding effects of the latter on different cognitive domains. Possession of an APOE ε4 allele was associated with a significantly faster decline compared to the APOE ε3/ε3 reference group in only the memory and executive domains, whereas APOE ε2 carriers had a slower decline in language and executive functions than both APOE ε3/ε3 carriers and APOE ε4 carriers (table 3, figure 3, and figure e-1 available from Dryad). On the other hand, within 3 years before death, the APOE ε2 carriers declined significantly faster than the APOE ε4 and APOE ε3/ε3 carriers in the memory domain (table 4, figure 3, and figure e-1 available from Dryad), likely reflecting a floor effect of the memory composite z score in the last 2 groups at advanced dementia stages.
Figure 3. Effect of APOE Alleles on Cognitive Domain–Specific Composite Measures.
(A, C, E, and G) Model-based cognitive trajectories of (A) memory, (C) executive, (E) attention, and (G) language composite-measures z scores by APOE allele with the intercept at the time of death calculated for an 82-year-old woman with 15 years of education and autopsy findings of frequent Consortium to Establish a Registry of Alzheimer’s Disease (CERAD) neuritic plaque score and Braak neurofibrillary tangle stage V/VI and no comorbid pathologies and with adjustments for the effect of Alzheimer disease neuropathologic variables (CERAD and Braak) on the rate of cognitive decline and for comorbid pathologies (model 2). (B, D, F, and H) Difference of model-based cognitive trajectories of (B) memory, (D) executive, (F) attention, and (H) language composite-measures z scores between APOE allele groups (APOE ε2 and APOE ε4 vs APOE ε3/ε3 carriers) under model 2. Shaded areas represent the corresponding 95% confidence intervals.
The goodness of fit of the proposed models for global cognitive measures (CDR-SOB and MMSE scores) and domain-specific composites was checked with the use of goodness-of-fit diagnostic graphs. Overall, we found that (1) the participant-specific residuals were approximately symmetric around zero; (2) the normal QQ plots of participant-specific residuals suggested that the residuals are normally distributed in most of their quantile range, and (3) in comparisons of the weighted predicted transformed cognitive outcome values averaged by time intervals along with the weighted average transformed observations,31 the individual predictions were close enough to the observations in the mixed-effects submodel (data not shown).
Discussion
In this large, national, clinicopathologic sample, selected to be representative of participants in clinical trials with biomarker-based eligibility criteria, we found a statistically significant difference in cognitive trajectory across APOE genotypes. In general, APOE ε2 carriers exhibited a slower decline and APOE ε4 carriers a faster decline than APOE ε3/ε3 carriers.
Our reverse longitudinal modeling approach enabled us to use information from patients with definite AD, to control for neuropathologic comorbidities that may affect rate of progression, and to focus on the effect of APOE genotype. Previous disparate results on the cognitive impact of the APOE genotype may have reflected the lack of autopsy confirmation and noise introduced by variation in the extent and severity of ADNC and concurrent pathologies.6-15 Moreover, the 3-year change point is consistent with a prior report on the Religious Orders Study and Rush Memory and Aging Project (ROSMAP) cohort36 and allowed us to identify and remove the expected ceiling/floor effects of cognitive outcome measures in advanced AD stages, providing data most relevant to mild and moderate dementia stages. Thus, our findings are consistent with a scenario in which the APOE ε4 allele not only anticipates the onset of cognitive decline but also accelerates its progression later in the disease course, with the APOE ε2 allele having opposite effects.
The magnitude of these differences approached clinical relevance even after controlling for the presence and severity of ADNC and concurrent pathologies (i.e., ≈0.7 points of CDR-SOB score per year and ≈0.5 points of MMSE score per year in APOE ε4 carriers vs APOE ε3/ε3 carriers). Thus, our current results may help inform clinical trial design. While randomization would ensure matching of treatment and placebo groups by APOE genotype and APOE-based post hoc analyses are usual practice,37 stratification at enrollment by APOE genotype might be considered regardless of expected APOE-driven differences in drug response or frequency of drug adverse effects. Some clinical trials have specified alternative protocols for APOE ε4 carriers38,39 because APOE ε4 increases the risk of blood-brain barrier disruption caused by monoclonal anti–Aβ antibodies leading to amyloid-related imaging abnormalities,40 which could in addition affect the relative rates of progression among treatment groups. Moreover, although some of the concurrent pathologies controlled for here cannot be accurately ascertained antemortem, the data illustrate the importance of using available biomarkers to account for as many variables as possible in a clinical trial setting.
Of note, our results are at odds with the hypothesis that the APOE genotype drives different AD clinical phenotypes, that is, an amnestic/temporo-limbic presentation in APOE ε4 carriers vs a dysexecutive/frontoparietal in APOE ε4 noncarriers.41-43 APOE ε4 carriers had a significantly faster decline in all cognitive domains examined (memory, attention, executive, and language) compared to APOE ε3/ε3 carriers in models not adjusted by ADNC effects on rate of cognitive decline and concurrent pathologies, and in both memory and executive function in adjusted models. Larger clinical and multimodal imaging studies including higher numbers of APOE ε2 carriers across AD preclinical and clinical stages are needed to confirm this hypothesis.
Our findings also provide important pathophysiologic insights. We were able to compare estimates between models with and without adjustment for the impact of ADNC on rate of cognitive decline and for comorbid pathologies. Overall, the results from both models were largely comparable: adjusting for interactions between ADNC and the slope of cognitive decline and for concurrent pathologies (model 2) rendered statistically significant point slope estimates that were only 3% to 24% smaller than those obtained without including these neuropathologic covariates (model 1), and some of the estimates for APOE ε2 allele in model 2 were even larger than their counterparts in model 1 (table 3). Therefore, these results suggest that, relative to the APOE ε3 allele, the APOE ε2 allele confers protection against cognitive decline beyond its known protective effects against ADNC, whereas the APOE ε4 allele accelerates cognitive decline beyond its known promoting effects of ADNC and concurrent pathologies, that is, cerebrovascular disease,22 LB disease,23 and transactive response DNA-binding protein 43kDa (TDP-43) proteinopathy24 (controlled here by the presence/absence of HScl). The cognitive protective effects of the APOE ε2 allele observed in this moderate/high ADNC sample are reminiscent of the cognitive resilience to ADNC recently reported for both the Christchurch homozygous mutation in the APOE gene44 and APOE ε2 homozygosity.45 Multiple Aβ-dependent and -independent mechanisms could explain these APOE-mediated differences in cognitive decline rate: impaired glucose utilization46; stabilization of synaptotoxic Aβ oligomers47; increased colocalization of Aβ oligomers with synapses48; altered synaptic pruning49; exacerbated microglial inflammation, tau spreading, and neurodegeneration9,50; and impaired neuroprotective mechanisms.51
Some limitations of our study pertinent to available or collected data should be acknowledged. This moderate/high ADNC sample is not suited to capture possible differences in rate of cognitive decline or phenotypic presentation differences across APOE genotypes at the earliest clinical stages. Thus, our study participants do not resemble those enrolled in current therapeutic clinical trials targeting subjective or very mild cognitive impairment or in secondary prevention trials (i.e., cognitively intact individuals with positive AD biomarkers). The underrepresentation of APOE ε2 carriers with substantial ADNC (n = 44) is expected given the protective effect of this allele against AD; APOE ε4 carriers would also be underrepresented in a sample of cognitively intact individuals with low ADNC.5,45 In addition, the effects of APOE genotype on the visuospatial/perceptive domain could not be studied due to insufficient specific neuropsychological tests for these skills in the first 2 NACC UDS iterations,26 and we had to use the presence of HScl as an imperfect surrogate of TDP-43 pathology because this has been recorded only since January 2014.52 Other limitations concern the statistical modeling. To be able to use the autopsy variables as baseline covariates and to apply reverse longitudinal modeling, we assumed that amyloid plaque burden plateaus early in the clinical course of AD and that the sequence of Braak NFT stages from limbic (III/IV) to neocortical (V/VI) is valid over the extent of the follow-up; these assumptions are well supported by prior Aβ and tau PET imaging studies.34,35 The complexity of our modeling strategy prevented us from including some terms in the models due to risk of overfitting such as a participant-specific change point as a random variable and interaction terms between APOE genotype and comorbid pathologies or between comorbid pathologies and slope of cognitive decline.
The APOE ε2 and APOE ε4 alleles have opposing effects on the rate of cognitive decline compared to the most common APOE ε3/ε3 genotype. These effects are clinically relevant, detectable in samples comparable in size and demographics to those enrolled in prototypical clinical trials, and largely independent of their known effects on measured ADNC and comorbid pathologies. Thus, besides neuropathology, other APOE-related phenotypes —perhaps microglial and astrocytic reactions9,49,50—might drive AD clinical progression. Further research to understand this APOE ε2–mediated resilience and APOE ε4–linked adversity is warranted.
Glossary
- Aβ
β-amyloid
- AD
Alzheimer disease
- ADC
Alzheimer Disease Center
- ADNC
Alzheimer disease neuropathological changes
- BIC
Bayesian information criterion
- CAA
cerebral amyloid angiopathy
- CDR-SOB
Clinical Dementia Rating Sum of Boxes
- CERAD
Consortium to Establish a Registry of Alzheimer’s Disease
- CI
confidence interval
- HScl
hippocampal sclerosis
- LB
Lewy bodies
- MMSE
Mini-Mental State Examination
- NACC
National Alzheimer’s Coordinating Center
- NFT
neurofibrillary tangle
- NP
neuritic plaque
- ROSMAP
Religious Orders Study and Rush Memory and Aging Project
- TDP-43
transactive response DNA-binding protein 43kDa
- UDS
Uniform Data Set
Appendix. Authors

Study Funding
J. Qian received research support from the National Institute of Neurological Disorders and Stroke (NINDS) (R01NS094610) and National Institute on Aging (NIA) (R21AG053695). R.A. Betensky received research support from NINDS (R01NS094610) and NIA (P30AG066512-01 and R21AG053695). B.T. Hyman received research support from NIA (P30 AG062421-01). A. Serrano-Pozo received research support from NINDS (R25NS065743), NIA (K08AG064039), and the Alzheimer's Association (AACF-17-524184). The NACC database is funded by NIA/NIH grant U01 AG016976. NACC data are contributed by the NIA-funded ADCs: P30 AG019610 (principal investigator [PI] Eric Reiman, MD), P30 AG013846 (PI Neil Kowall, MD), P30 AG062428-01 (PI James Leverenz, MD), P50 AG008702 (PI Scott Small, MD), P50 AG025688 (PI Allan Levey, MD, PhD), P50 AG047266 (PI Todd Golde, MD, PhD), P30 AG010133 (PI Andrew Saykin, PsyD), P50 AG005146 (PI Marilyn Albert, PhD), P30 AG062421-01 (PI Bradley Hyman, MD, PhD), P30 AG062422-01 (PI Ronald Petersen, MD, PhD), P50 AG005138 (PI Mary Sano, PhD), P30 AG008051 (PI Thomas Wisniewski, MD), P30 AG013854 (PI Robert Vassar, PhD), P30 AG008017 (PI Jeffrey Kaye, MD), P30 AG010161 (PI David Bennett, MD), P50 AG047366 (PI Victor Henderson, MD, MS), P30 AG010129 (PI Charles DeCarli, MD), P50 AG016573 (PI Frank LaFerla, PhD), P30 AG062429-01 (PI James Brewer, MD, PhD), P50 AG023501 (PI Bruce Miller, MD), P30 AG035982 (PI Russell Swerdlow, MD), P30 AG028383 (PI Linda Van Eldik, PhD), P30 AG053760 (PI Henry Paulson, MD, PhD), P30 AG010124 (PI John Trojanowski, MD, PhD), P50 AG005133 (PI Oscar Lopez, MD), P50 AG005142 (PI Helena Chui, MD), P30 AG012300 (PI Roger Rosenberg, MD), P30 AG049638 (PI Suzanne Craft, PhD), P50 AG005136 (PI Thomas Grabowski, MD), P30 AG062715-01 (PI Sanjay Asthana, MD, FRCP), P50 AG005681 (PI John Morris, MD), and P50 AG047270 (PI Stephen Strittmatter, MD PhD). The authors express their gratitude to the participants and informants involved in research at the US Alzheimer Disease Research Centers.
Disclosure
B.T. Hyman has a family member who works at Novartis and owns stock in Novartis; he serves on the scientific advisory board of Dewpoint and owns stock; he serves on a scientific advisory board or is a consultant for Abbvie, Arvinas, Biogen, Novartis, Cell Signaling, the US Department of Justice, Takeda, Vigil, W20 group, and Seer; and his laboratory is supported by sponsored research agreements with Abbvie, Amgen, Arvinas, Biogen, Denali, Dewpoint, Fidelity Biosciences, F Prime, General Electric, Eli Lilly, Merck, Sangamo, and Spark, as well as research grants from the NIH (PI), Cure Alzheimer's Fund (PI), Tau Consortium (PI), the JPB Foundation (PI), Alzheimer's Association (mentor), and Brightfocus (mentor). R.A. Betensky serves on the National Cancer Institute Board of Scientific Counselors for Clinical Sciences and Epidemiology, Biogen, Alexion, Reata, and PTC Therapeutics; she is consultant for Cowen Inc and has served as expert witness for Amarin, Actavis, Amazon, and Apotex and as consultant for Pfizer and Amgen. Drs. Qian and Serrano-Pozo have no disclosures. Go to Neurology.org/N for full disclosures.
References
- 1.Sperling RA, Rentz DM, Johnson KA, et al. . The A4 Study: stopping AD before symptoms begin? Sci Transl Med 2014;6:228fs13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corder EH, Saunders AM, Strittmatter WJ, et al. . Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993;261:921–923. [DOI] [PubMed] [Google Scholar]
- 3.Corder EH, Saunders AM, Risch NJ, et al. . Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 1994;7:180–184. [DOI] [PubMed] [Google Scholar]
- 4.Bennett DA, Wilson RS, Schneider JA, et al. . Apolipoprotein E epsilon4 allele, AD pathology, and the clinical expression of Alzheimer's disease. Neurology 2003;60:246–252. [DOI] [PubMed] [Google Scholar]
- 5.Serrano-Pozo A, Qian J, Monsell SE, Betensky RA, Hyman BT. APOEε2 is associated with milder clinical and pathological Alzheimer disease. Ann Neurol 2015;77:917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson RS, Schneider JA, Barnes LL, et al. . The apolipoprotein E epsilon 4 allele and decline in different cognitive systems during a 6-year period. Arch Neurol 2002;59:1154–1160. [DOI] [PubMed] [Google Scholar]
- 7.Martins CAR, Oulhaj A, de Jager CA, Williams JH. APOE alleles predict the rate of cognitive decline in Alzheimer disease: a nonlinear model. Neurology 2005;65:1888–1893. [DOI] [PubMed] [Google Scholar]
- 8.Cosentino S, Scarmeas N, Helzner E, et al. . APOE epsilon 4 allele predicts faster cognitive decline in mild Alzheimer disease. Neurology 2008;70:1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y, Yamada K, Liddelow SA, et al. . ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 2017;549:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Isla T, West HL, Rebeck GW, et al. . Clinical and pathological correlates of apolipoprotein E epsilon 4 in Alzheimer's disease. Ann Neurol 1996;39:62–70. [DOI] [PubMed] [Google Scholar]
- 11.Dal Forno G, Rasmusson DX, Brandt J, et al. . Apolipoprotein E genotype and rate of decline in probable Alzheimer's disease. Arch Neurol 1996;53:345–350. [DOI] [PubMed] [Google Scholar]
- 12.Bunce D, Fratiglioni L, Small BJ, Winblad B, Bäckman L. APOE and cognitive decline in preclinical Alzheimer disease and non-demented aging. Neurology 2004;63:816–821. [DOI] [PubMed] [Google Scholar]
- 13.Stern Y, Brandt J, Albert M, et al. . The absence of an apolipoprotein epsilon4 allele is associated with a more aggressive form of Alzheimer's disease. Ann Neurol 1997;41:615–620. [DOI] [PubMed] [Google Scholar]
- 14.Hoyt BD, Massman PJ, Schatschneider C, Cooke N, Doody RS. Individual growth curve analysis of APOE epsilon 4-associated cognitive decline in Alzheimer disease. Arch Neurol 2005;62:454–459. [DOI] [PubMed] [Google Scholar]
- 15.van der Vlies AE, Koedam ELGE, Pijnenburg YAL, Twisk JWR, Scheltens P, van der Flier WM. Most rapid cognitive decline in APOE epsilon4 negative Alzheimer's disease with early onset. Psychol Med 2009;39:1907–1911. [DOI] [PubMed] [Google Scholar]
- 16.Wilson RS, Bienias JL, Berry-Kravis E, Evans DA, Bennett DA. The apolipoprotein E epsilon 2 allele and decline in episodic memory. J Neurol Neurosurg Psychiatry 2002;73:672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonner-Jackson A, Okonkwo O, Tremont G; Alzheimerʼs Disease Neuroimaging Initiative. Apolipoprotein E ε2 and functional decline in amnestic mild cognitive impairment and Alzheimer disease. Am J Geriatr Psychiatry 2012;20:584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinohara M, Kanekiyo T, Yang L, et al. . APOE2 eases cognitive decline during aging: clinical and preclinical evaluations. Ann Neurol 2016;79:758–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beach TG, Monsell SE, Phillips LE, Kukull W. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005-2010. J Neuropathol Exp Neurol 2012;71:266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano-Pozo A, Qian J, Monsell SE, et al. . Mild to moderate Alzheimer dementia with insufficient neuropathological changes. Ann Neurol 2014;75:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyle PA, Yu L, Leurgans SE, et al. . Attributable risk of Alzheimer's dementia attributed to age-related neuropathologies. Ann Neurol 2019;85:114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montagne A, Nation DA, Sagare AP, et al. . APOE4 leads to blood-brain barrier dysfunction predicting cognitive decline. Nature 2020;581:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson DW, Heckman MG, Murray ME, et al. . APOE ε4 is associated with severity of Lewy body pathology independent of Alzheimer pathology. Neurology 2018;91:e1182–e1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang HS, Yu L, White CC, et al. . Evaluation of TDP-43 proteinopathy and hippocampal sclerosis in relation to APOE ε4 haplotype status: a community-based cohort study. Lancet Neurol 2018;17:773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besser L, Kukull W, Knopman DS, et al. . Version 3 of the National Alzheimer's Coordinating Center's Uniform Data Set. Alzheimer Dis Assoc Disord 2018;32:351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weintraub S, Besser L, Dodge HH, et al. . Version 3 of the Alzheimer Disease Centers' neuropsychological test battery in the Uniform Data Set (UDS). Alzheimer Dis Assoc Disord 2018;32:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montine TJ, Phelps CH, Beach TG, et al. . National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer's disease: a practical approach. Acta Neuropathol (Berl) 2012;123:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark CM, Pontecorvo MJ, Beach TG, et al. . Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-β plaques: a prospective cohort study. Lancet Neurol 2012;11:669–678. [DOI] [PubMed] [Google Scholar]
- 29.Marquié M, Siao Tick Chong M, Antón-Fernández A, et al. . [F-18]-AV-1451 binding correlates with postmortem neurofibrillary tangle Braak staging. Acta Neuropathol (Berl) 2017;134:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serrano-Pozo A, Qian J, Monsell SE, Frosch MP, Betensky RA, Hyman BT. Examination of the clinicopathologic continuum of Alzheimer disease in the autopsy cohort of the National Alzheimer Coordinating Center. J Neuropathol Exp Neurol 2013;72:1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proust-Lima C, Philipps V, Liquet B. Estimation of extended mixed models using latent classes and latent processes: the R package lcmm. J Stat Softw 2017;78.Available at jstatsoft.org/v78/i02/. Accessed December 21, 2020. [Google Scholar]
- 32.Hayden KM, Jones RN, Zimmer C, et al. . Factor structure of the National Alzheimer's Coordinating Centers Uniform Dataset Neuropsychological Battery: an evaluation of invariance between and within groups over time. Alzheimer Dis Assoc Disord 2011;25:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian J, Hyman BT, Betensky RA. Neurofibrillary tangle stage and the rate of progression of Alzheimer symptoms: modeling using an autopsy cohort and application to clinical trial design. JAMA Neurol 2017;74:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jack CR, Wiste HJ, Lesnick TG, et al. . Brain β-amyloid load approaches a plateau. Neurology 2013;80:890–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schöll M, Lockhart SN, Schonhaut DR, et al. . PET imaging of tau deposition in the aging human brain. Neuron 2016;89:971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu L, Boyle P, Schneider JA, et al. . APOE ε4, Alzheimer's disease pathology, cerebrovascular disease, and cognitive change over the years prior to death. Psychol Aging 2013;28:1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy RE, Cutter GR, Schneider LS. Effect of APOE genotype status on targeted clinical trials outcomes and efficiency in dementia and mild cognitive impairment resulting from Alzheimer's disease. Alzheimers Dement 2014;10:349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salloway S, Sperling R, Fox NC, et al. . Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer's disease. N Engl J Med 2014;370:322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sevigny J, Chiao P, Bussière T, et al. . The antibody aducanumab reduces Aβ plaques in Alzheimer's disease. Nature 2016;537:50–56. [DOI] [PubMed] [Google Scholar]
- 40.Sperling RA, Jack CR, Black SE, et al. . Amyloid-related imaging abnormalities in amyloid-modifying therapeutic trials: recommendations from the Alzheimer's Association Research Roundtable Workgroup. Alzheimers Dement 2011;7:367–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Flier WM, Schoonenboom SNM, Pijnenburg YAL, Fox NC, Scheltens P. The effect of APOE genotype on clinical phenotype in Alzheimer disease. Neurology 2006;67:526–527. [DOI] [PubMed] [Google Scholar]
- 42.Wolk DA, Dickerson BC. Alzheimer’s Disease Neuroimaging Initiative. Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional-executive network function in Alzheimer's disease. Proc Natl Acad Sci USA 2010;107:10256–10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dickerson BC, Wolk DA. Alzheimer’s Disease Neuroimaging Initiative. Dysexecutive versus amnesic phenotypes of very mild Alzheimer's disease are associated with distinct clinical, genetic and cortical thinning characteristics. J Neurol Neurosurg Psychiatry 2011;82:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arboleda-Velasquez JF, Lopera F, O'Hare M, et al. . Resistance to autosomal dominant Alzheimer's disease in an APOE3 Christchurch homozygote: a case report. Nat Med 2019;25:1680–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiman EM, Arboleda-Velasquez JF, Quiroz YT, et al. . Exceptionally low likelihood of Alzheimer's dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun 2020;11:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosconi L, Nacmias B, Sorbi S, et al. . Brain metabolic decreases related to the dose of the ApoE e4 allele in Alzheimer's disease. J Neurol Neurosurg Psychiatry 2004;75:370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashimoto T, Serrano-Pozo A, Hori Y, et al. . Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid β peptide. J Neurosci 2012;32:15181–15192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koffie RM, Hashimoto T, Tai HC, et al. . Apolipoprotein E4 effects in Alzheimer's disease are mediated by synaptotoxic oligomeric amyloid-β. Brain J Neurol 2012;135:2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung WS, Verghese PB, Chakraborty C, et al. . Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proc Natl Acad Sci USA 2016;113:10186–10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin YT, Seo J, Gao F, et al. . APOE4 causes widespread molecular and cellular alterations associated with Alzheimer's disease phenotypes in human iPSC-derived brain cell types. Neuron 2018;98:1141–1154.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao J, Davis MD, Martens YA, et al. . APOE ε4/ε4 diminishes neurotrophic function of human iPSC-derived astrocytes. Hum Mol Genet 2017;26:2690–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Besser LM, Kukull WA, Teylan MA, et al. . The revised National Alzheimer's Coordinating Center's Neuropathology form: available data and new analyses. J Neuropathol Exp Neurol 2018;77:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The NACC database is a public resource available to researchers. Data requests can be submitted online at the following NACC website: alz.washington.edu/NONMEMBER/QUERY/datareqnew.html.