Abstract
Biofilm formation and microbial adhesion are two related and complex phenomena. These phenomena are known to play an important role in microbial life and various functions with positive and negative aspects. Actinobacteria have wide distribution in aquatic and terrestrial ecosystems. This phylum is very large and diverse and contains two important genera Streptomyces and Mycobacteria. The genus Streptomyces is the most biotechnologically important, while the genus Mycobacteria contains the pathogenic species of Mycobacteriaceae. According to the literature, the majority of studies carried out on actinomycetes are focused on the detection of new molecules. Despite the well-known diversity and metabolic activities, less attention has been paid to this phylum. Research on adhesion and biofilm formation is not well developed. In the present review, an attempt has been made to review the literature available on the different aspects on biofilm formation and adhesion of Actinobacteria. We focus especially on the genus Streptomyces. Furthermore, a brief overview about the molecules and structures involved in the adhesion phenomenon in the most relevant genus is summarized. We mention the mechanisms of quorum sensing and quorum quenching because of their direct association with biofilm formation.
1. Introduction
A biofilm is a well-organized and collaborative community of microorganisms. These associated cells are differentiated from the suspended ones by reduced growth rate, gene regulation, and the secretion of an extracellular polymer matrix [1]. Biofilm formation undergoes several stages (Figure 1) and involves intracellular signaling and transcription mechanisms distinct from planktonic cells. The biofilm state plays an important role in the community's survival through the protection against various environmental stresses such as pH change, UV radiation, osmotic shock and desiccation, antimicrobial agent penetration, acquisition of new genetic traits, and nutrient availability [3]. This fundamental change in living conditions leads to constant changes in the expression of multiple genes, which causes changes in the morphological, physiological, and biochemical state of cells. Living inside a biofilm provides cells resistance to stressful conditions.
Figure 1.
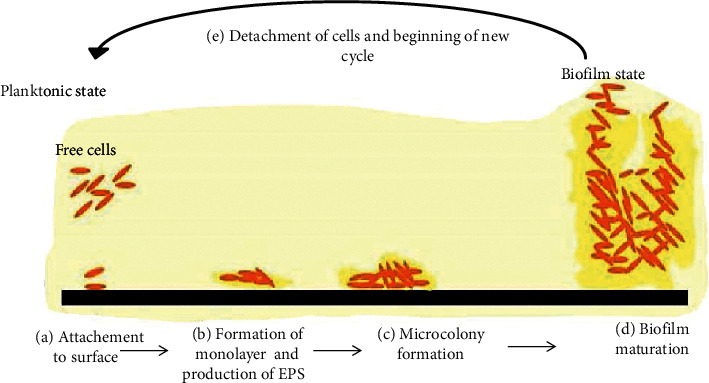
Typical illustration of biofilm formation. The process begins with reversible adhesion of the planktonic cells (brown ovals) with subsequent attachment to the surface (black) (a). The bacteria then form a monolayer and attach irreversibly, inducing an extracellular matrix formation (b). Later, a microcolony is formed where multilayers appear (c). In the following stages, the biofilm matures, forming characteristic structures due to polysaccharides existence (d). At the final step, some cells start to detach and the biofilm (yellow) disperses (e) [2].
Biofilms occur in several steps: adhesion, maturation, then detachment, and dispersion. The first and most crucial stage is microbial adhesion. Currently, research in the medical field is oriented toward the prevention of microbial adhesion [4–7] and to block quorum sensing [8].
Biofilms are a serious challenge in healthcare-associated infections, especially those involving implantation of medical devices, such as intravascular catheters, urinary catheters, and orthopedic implants [9].
The term biofilm is usually related to negative aspects of microbes association. In many industrial sectors, biofilms represent a real defiance; in addition to cleaning and disinfection problems, they cause energy losses by blocking in condenser tubes, coolers, and heat exchangers. Biofilms affect water quality in drinking water distribution systems by disseminating pathogens [10]. Biofilms can create serious problems in natural, artificial, and biomedical systems [11]. The thing that poses serious problems becomes an obvious advantage in waste treatment and biotechnology. Indeed, cells immobilized within biofilms are adequate in the white biotechnology for production of valuable molecules [12]. Cell immobilization in biofilms can provide specific metabolism in comparison with the suspended culture [13, 14]. Biofilm formation mechanisms are currently the subject of intensive studies and discussions [15].
The phylum Actinobacteria includes 23 orders, forming one of the largest phyla in the domain bacteria [16]. In December 2015, there were 342 genera with a continuous discovery of other genera. They are typically Gram positive. The life cycle of actinomycetes is unique (Figure 2).
Figure 2.
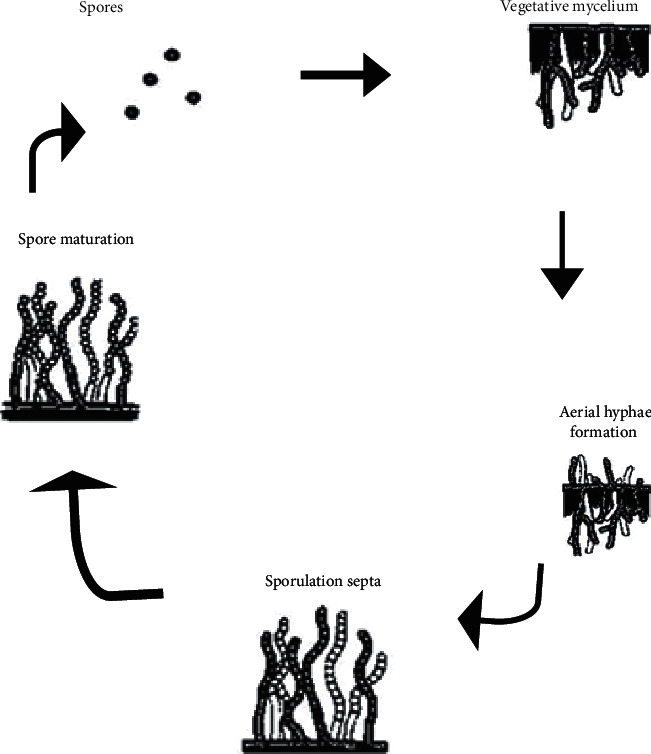
Life cycle of streptomycetes [17].
The cycle starts with the germination of “dispersal form” spores and development in branched hyphae named vegetative mycelium, which they differentiate into specialized reproductive structures called aerial mycelium [18]. Considerable effort has been devoted to attack the physiology of bacterial growth and cell division in Actinobacteria [19–22]. Despite the abundance of Actinobacteria, very few reports were available about biofilm formation in the phylum. This phylum comprises a plethora of phenotypically diverse organisms, with widespread distribution in nature and exhibiting varied oxygen, nutritional, temperature, and pH requirements for growth, making it an important phylum [23]. Their diverse physiological potential makes them a dominant role player in the biotechnology. Their applications are widespread and vary from agroindustry, pharmaceuticals, and bioremediation among numerous others [17, 24, 25]. They play a key role in natural geochemical cycles, especially through their ability to decompose organic matter. Actinobacteria are also abundant in the rhizosphere and produce a wide range of biologically active metabolites, thereby influencing plant development [26]. Many Actinobacteria are also known pathogens of plants and animals [27]. However, among the most important potential of Actinobacteria is the production of a significant number of secondary metabolites like antibiotics and other compounds of biotechnological interest that has been exploited [28].
In addition, members of the genus Streptomyces are known to produce more than 70% of commercially available antibiotics [29]. This taxon includes pathologically important strains (e.g., Mycobacterium tuberculosis and Corynebacterium diphtheriae) and biotechnologically pertinent strains (Corynebacterium glutamicum and Streptomyces griseus), so exact information on their growth and cell division dynamics is essential to design a treatment or production boosting strategies [18].
Significant information is available on the growth of actinomycetes in shaken-flask cultures and fermenters [30]. Surface growth has received less attention, although surface-associated growth is certainly a different phenotype and a critical stage in their life cycle, with putative morphological and physiological variations that could impact metabolite production [31]. Most of what is known about biofilm formation in Actinobacteria comes from the study of antibiotic production in this taxon. While it is indeed the most important phenomenon, the aim of this review is not to present an overview on the biofilm formation mechanisms. The expression of virulence determinants, production of secondary metabolites, and morphogenesis is associated with cell-cell interaction and cell-surface interactions and typically controlled by growth state [32–34]. Further exploration of novel phenotypes under biofilm formation regulations is likely to contribute to the advancement in medical, biotechnological, and ecological fields. Hence, there is a need of studying biofilm formation in Actinobacteria especially in Streptomyces genus.
In the present work, an attempt has been made to review the literature available on different aspects on biofilm formation and adhesion of Actinobacteria. We focus especially on the genus Streptomyces. In addition, the literature about the molecules and structures involved in the adhesion phenomenon in the most relevant genus is summarized. We also mention the mechanisms of quorum sensing and quorum quenching; there is a direct relation between these phenomena.
2. Biofilm Formation in the Phylum Actinobacteria
2.1. Importance of Biofilms in Biotechnology
Microorganisms forming monospecific stable biofilms are the most required for their use in industrial processes to produce valuable substances [35]. Many processes are developed to obtain the desired product in bioreactors based on microbial biofilms in the tree levels: laboratory, pilot, and industrial scale. Special designs of “biofilm” bioreactors for culturing microorganisms in biofilm form are described in reference [36]. In fact, the production of organic acids and alcohols and antimicrobial activity are more interesting when the strain is biofilm forming. Various applications in biotechnology using bacterial and fungal biofilms have been published [36, 37]. Recently, environmental biotechnology essentially based on the biodegradation of waste using microorganisms has a great tendency. It is mainly utilized in the processes of industrial wastewater treatment like detoxification and bioremediation and production of electrochemical energy. The application of biofilms for biotechnological purposes has shown great efficiency. Environmental processes based on biofilms are used in the biosorption of heavy metals from wastewater, also for obtaining electrochemical energy from aquatic environments and wastewater [38]. Moreover, in bioremediation and purification of natural environments, the creation of artificial biofilms involving microorganisms gives optimal results and contributes to the resolution of biopurification and bioremediation problems [39]. There is a higher final product yield in bioreactors where a producer is grown as biofilms compared with bioreactors using conventional culture methods; “planktonic cells” occur in the preparation of alcohols—such as ethanol and butanol—and organic acids—such as acetic, lactic, succinic, and fumaric [37, 40]. Qureshi et al. reported that, in bioreactors where Clostridium acetobutylicum is grown as a biofilm, butanol production is increased by about 40 to 50 times compared with a conventional bioreactor in a producing batch culture [37]. Moreover, the cellulose production process is reliable and low cost when Acetobacter xylinum is grown as a biofilm in a special bioreactor [41]. Acetic acid is produced based on the biofilm culture on industrial scale [36].
With Streptomycetes involvement in biofilm formation, there are different approaches. The first approach is co-culture of two organisms—one nonbiofilm former but produces the desired product, and the other is a biofilm producer. This method can optimize the whole process. The second approach is the cultivation of monobiofilms of Streptomyces producers. Each of these approaches has technological characteristics and some advantages. The most important tasks of biotechnological biofilms of Streptomyces are screening the organism that can actively form a stable biofilm, choosing the conditions of its adherence, selecting an optimal substrate for the fixation and development of biofilms, and studying the parameters of conventional culture (composition of the medium, temperature, agitation rate etc.). For example, the rough surface of the polymethylmethacrylate block as a substrate for adhesion has been found to be more suitable for the formation of streptomycetes biofilms than a smooth surface [40]. Previous studies described different models of bioreactors using biofilms of streptomycetes for laboratory and pilot scales [40]). The production of lactic acid in bioreactors involves the presence of two strains: Lactobacillus casei subsp. Rhamnosus convert glucose to lactic acid, and Streptomyces T7A form stable biofilms. The biofilms formed by Streptomyces T7A protect Lactobacillus casei subsp. rhamnosus from acidity and subsequently contribute to the stability of the process [42, 43].
2.2. Current Knowledge on Actinomycetes Adhesion
Attachment is so important in the life of the pathogens of Streptomyces. These microorganisms can grow in the liquid-air interface. They can also grow and attach to hydrophobic surfaces such as the leaves of a plant or an animal's skin. Attachment/adhesion could contribute to the effective degradation of substrates by saprophytic streptomycetes. Furthermore, the attachment of microbes to host surfaces is crucial for initiation of infections. Adherence to surfaces is required to establish infections by pathogenic streptomycetes, such as the plant pathogen Streptomyces scabies or the human pathogen Streptomyces somaliensis. Cell-surface-associated polymers involved in the adhesion of pathogens are unknown because surface polymers have multiple functions in the biology of these organisms [44].
Studying of bacteria to surfaces requires deep knowledge and characterization of both bacterial and substratum surfaces. Research on adhesion of actinomycetes has not received much attention from scientists. The lack of information about this fascinating group comes from the fact that screening for novel molecules with high value is still in progress.
The pathogenic actinomycetes adhesion on medical devices creates a real challenge, so further understanding of the adhesive behavior could contribute to prevention of biofilm formation [45, 46]. Several methods are used to study physicochemical properties and biofilm formation phenomena such as surface hydrophobicity, electron donor character, electron acceptor character, and surface free energy. In physicochemical characterization of the actinomycetes surface, the three pathogenic Streptomyces had a hydrophilic cell surface with a weak electron donor character, whereas the surface of Nocardia was hydrophobic and strongly electron donor [47].
As a contribution to future research on biofilm formation in soil, actinomycetes were isolated from the saline soil of Beni Amir, Morocco. Bacterial surfaces of actinomycetes were hydrophilic and bipolar at 1 M of salinity. These properties are affected by the salt concentration in the medium [48]. Thermobifida fusca's surface was considered hydrophobic using the MATH method; cell surface hydrophobicity has been implicated in the interactions between cellulosic surfaces. The purpose of investigation is to correlate between attachment and effective degradation of cellulose [49].
3. Extracellular Matrix of Actinobacteria
Although Actinobacteria is one of the largest groups of organisms in the bacterial field, very few reports are available about the biofilm formation in the phylum. An analysis of literature for biofilm formation in Actinobacteria revealed that only 9 actinobacterial genera show biofilm ability. Also, limited reports are dissecting the extracellular matrix in some genera [31, 35, 50].
The extracellular polymer matrix of microbial biofilms is a highly complex scaffold. It is characterized by a multitude of structurally and chemically heterogeneous microenvironments. It plays various roles in the structure and function of different biofilm communities. It can be a barrier against adverse chemical and biological influences, such as osmotic stress, acid/base challenges, oxygen, antibiotics and antiseptics, the host immune defense, and grazing protozoa. Moreover, it provides mechanical stability to the biofilm and protects the microorganisms from desiccation and contributes to the sorption and storage of nutrients and trace elements. It is the location of numerous extracellular enzymatic reactions, surface-mediated adhesion, regulatory capacity, and a cohesive network in which the cells are temporarily immobilized. If the biofilm is a microbial city, then the matrix is its infrastructure [3]. Exact and molecular interactions of the extracellular matrix in the biofilm of Actinobacteria have not been defined, and the contributions of these components to matrix integrity are poorly understood at a molecular level [51]. The composition of this matrix depends on the species. The extracellular matrix formed by Streptomyces—based on building blocks—[44] is morphologically and functionally different from other bacteria. Streptomyces form a so-called mycelium that consists of branched filaments that grow by tip extension [52]. This mycelium invades and colonizes the soil environment and secretes numerous enzymes that facilitate break down of plant material, which yields nutrients that can be used as food for the growing mycelium [53].
The transition from vegetative to aerial growth is characterized by a dramatic change in the surface properties of the hyphae. Whereas vegetative hyphae are hydrophilic, aerial hyphae are hydrophobic due to the assembly of an additional outer surface layer or matrix called the rodlet layer [44]. The rodlet layer has an amphipathic nature: the hydrophilic side faces the cell wall, while the hydrophobic side is exposed to the air [54]. The hydrophobic side of the rodlet layer is characterized by thin fibrils. These fibrils are formed by the assembly of so-called chaplin proteins, discovered at the first time in Streptomyces coelicolor, providing a hydrophobic character to aerial hyphae [55, 56].
Bacterial amyloids have beneficial functions, including conferring stability to biofilms, regulating development, or conferring virulence [57]. For Streptomyces coelicolor chaplin proteins are involved in the formation of aerial hyphae, they also mediate attachment to a hydrophobic surface. Chaplin proteins are associated with amyloid-type fibrils to form so-called fimbriae (Figure 3). These fimbriae are anchored to the cell wall via cellulose, providing an important new insight into the role of this polysaccharide in the bacterial domain [58]. The attachment mechanism via cellulose-anchored amyloid fimbriae is widespread in bacteria and may function in the initiation of infection and in the formation of biofilms [58]. Multicellular communities of bacteria are generally connected to each other through a variety of extracellular polymers. These polymers include amyloid fibrils, polysaccharides, lipids, and nucleic acids [59].
Figure 3.
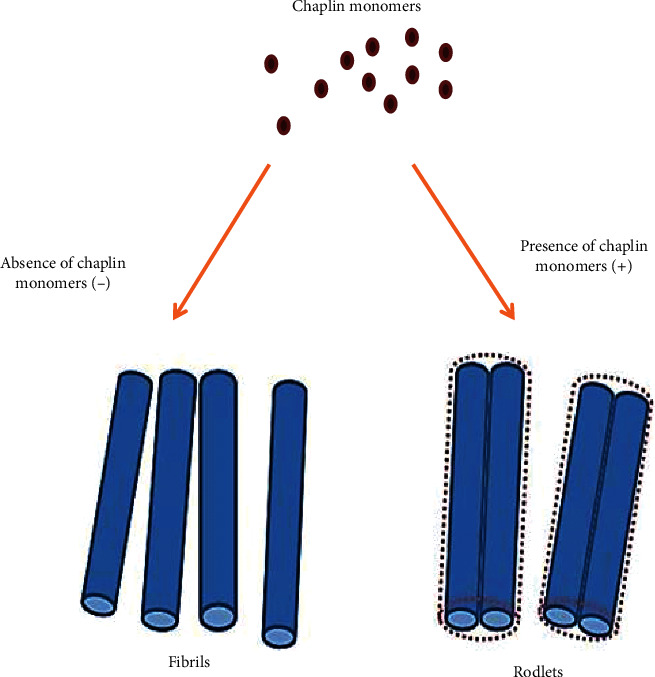
The rodlet layer formed in Streptomyces coelicolor strain; it is a result of the assembly of two types of proteins—chaplins and rodlins. In the absence of rodlins, the aggregation of chaplins provides fibrils. This illustration reproduced from Petrus and Claessen's research [44].
Industrial-scale production of important secondary metabolites, such as antibiotics, occurs in large bioreactors. Growth of streptomycetes in such conditions, as opposed to solid-growth or liquid-standing cultures, is characterized by the formation of large clumps, or pellets, consisting of interconnected hyphae [60, 61]. Formation of pellets might be caused by surface-to-surface contact between smaller particles, which as a result can become very large (several millimeters in diameter) [62]. As a consequence, growth occurs predominantly at the outer surface of pellets as oxygen and/or nutrient depletion hampers growth in the central region [60, 63].
Indeed, Manteca et al. indicated that programmed cell death (PCD)-like events occur in the central part of pellets [64, 65]. PCD gives rise to new hyphal growth and also constitutes a serious damage to the integrity of the pellets. In the presence of shear forces in bioreactors, extracellular substances probably contribute to hyphal cohesion. The extracellular DNA (eDNA), which is supposed to be released during hyphal autolysis in the central part of the granules, maintains hyphae together [66, 67]. Similarly, a role for the polysaccharide hyaluronic acid was proposed [66]. More recent evidence indicates that the CslA-produced polymer also is involved in pellet architecture [68, 69]. In contrast to the dense pellets formed in the wild-type strain, mycelium of the CslA mutant had an open structure [69]. Furthermore, the chaplin proteins were shown to be involved in control of pellet size [68]. Extracellular surface polymers are involved in shaping Streptomyces pellets. Interestingly, cell surface polymers might be interesting targets for the industrial improvement of strains, considering the fact that the morphology and size of pellets are important for productivity [61, 68].
4. Biofilm Formation by Actinobacteria
4.1. Streptomyces
The genus Streptomyces with 778 species [70] is the largest genus of Actinobacteria and is a natural inhabitant of soil and decaying vegetation. Streptomyces are widely used to produce useful enzymes and a wide variety of secondary metabolites with potential biological activities. Streptomyces produce more than half of clinically useful natural products [71]. They have an inflexible (rigid) cell wall that contains muramic acid with some containing wall teichoic acids. They are excellent producers of antimicrobial secondary metabolites and secrete numerous extracellular enzymes that decompose organic substances [26]. Streptomyces is the most studied group of Actinobacteria as it is the uncontested and reliable source of many biologically active substances, in particular, antibiotics. Furthermore, the morphological differentiation [72] and exploitation in biotechnology and therapy [73] have received much more attention [18]. However, there are few data available on Streptomyces biofilms, but biofilm lifestyle is part of the life cycle of these microorganisms. Streptomyces somaliensis and Streptomyces dangerous are two species known to be pathogenic to humans and are found in the African region [74]. It has been shown that Streptomyces found on devices used in gynecology actively form biofilms in the human body. Three isolates of actinomycetes belonging to the genus Streptomyces spp. and Nocardia sp. showed a high capacity to form biofilms. Actinomycetes biofilms on the above-mentioned devices are responsible for the development of actinomycosis [50].
Relatively, only few data on the structure of Streptomyces biofilms are available yet. In the initial studies on Streptomyces biofilms, great emphasis is placed on the ability of these organisms to adhere [58, 75]. Streptomyces biofilms are formed on the surface of ancient stone monuments, on cave walls with prehistoric drawings, on old wall paintings and frescoes in historical catacombs, and on the walls of historical and artistic building historic castles in Scotland [76, 77]. A study of 230 biofilms formed on old buildings of different types of materials (cement, lime, concrete, reinforced concrete, brick, and in six European and seven Latin American countries) showed streptomycetes cases are the main biofilm-forming microorganisms on building surfaces.
4.2. Surface-Active Proteins Involved in the Attachment of Hyphae to Surfaces and Aerial Growth
Microbial adhesion depends on surface properties of both bacteria and support [7, 78]. Here, we presented the majority of proteins expressed in the cell surface of Streptomyces, which mediate attachment. The most of what is known about surface-associated proteins in Streptomyces comes from studies completed on the most pertinent species Streptomyces coelicolor. Streptomycetes are capable of adhering to a variety of surfaces, which could be instrumental for invasive growth or for the efficient degradation of substrates, for example, dead plant material. Also, attachment could be important for establishing infections by plant pathogenic streptomycetes, such as Streptomyces ipomoeae or Streptomyces scabies. Several proteins are constituents of the cell surface of Streptomyces involved in their attachment to surfaces.
Streptomyces have a complex life cycle, its starts when spore senses a suitable source of nutrients. The germinating spore develops one or two tubes that elongate by apical tip extension. The hyphae form septal cross walls without compartmentation called vegetative mycelia. Aerial hyphae are formed only on solid-growth culture giving colonies their characteristic white and fluffy appearance [52]. The transition from vegetative to aerial growth coincides with a dramatic change in the cell surface properties of the hyphae; whereas vegetative hyphae are hydrophilic, aerial hyphae are characterized by their high surface hydrophobicity [79]. This hydrophobicity is due to the formation of an extracellular proteinaceous surface layer known as the rodlet layer. The rodlet layer is composed of two classes of proteins, chaplins and rodlins [79]. The chaplin proteins in Streptomyces coelicolor comprise a class of eight different members, ChpA-H, which can be divided into two groups. The long chaplins, ChpA-C, are about 225 amino acids in length, while the short chaplins, ChpD-H, are considerably smaller (approximately 63 amino acids). ChpD-H consists of a signal sequence for secretion followed by a relatively hydrophobic, so-called chaplin domain. ChpA-C contains two chaplin domains followed by a stretch of hydrophilic amino acids and a cell wall-anchoring domain [55, 56]. Two of the short chaplins, ChpE and ChpH, are secreted in the medium well before aerial growth commences [55].
These chaplins are thought to facilitate aerial growth by lowering the water surface tension, allowing hyphae to breach the medium-air interface. Subsequently, aerial hyphae secrete all chaplins (ChpA-H), which jointly assemble into small fibrils that cover the hyphal surface. Thereby, these aerial structures become hydrophobic. In most streptomycetes, these chaplin fibrils are organized by the rodlin proteins into rodlets [79]. However, this organization only appears to be critical for aerial growth under osmotic stress conditions [80]. Fibrils forming the rodlet layer are often aligned into pairs called rodlets. This process is mediated by the activity of the rodlin proteins RdlA and RdlB [75]. The chaplin protein fibrils provide not only surface hydrophobicity but also rigidity to the aerial hyphae. Notably, three chaplin proteins appear to be invariably present in streptomycetes, which are ChpC, ChpE, and ChpH [81]. ChpE and ChpH are secreted into the environment to reduce surface tension, thereby allowing the hyphae to escape from the aqueous environment into the air. These proteins therefore make it possible to overcome this high-surface-tension barrier [55]. In vitro experiments indicated that chaplins can reduce the surface tension, within minutes, from 72 to 24 mJ m−2, making them among the most potent natural surfactants known [55]. Mixtures of cell-wall-extracted chaplins can be used to modify a variety of hydrophilic and hydrophobic surfaces in vitro, thereby changing their nature. Assembly on glass leads to a protein coating that makes the surface hydrophobic. The assembly of chaplin proteins on hydrophobic surfaces make them hydrophilic [82].
In so-called liquid static cultures, strong attachment of Streptomyces coelicolor is observed coinciding with the formation of an extracellular matrix that is relatively distinct from the rodlet matrix identified on aerial hyphae [58, 83].
The attachment-associated matrix is characterized by fimbriae structures that appear to be connected to the hyphal surface via protrusions present along the adhering hyphae. Formation of fimbriae is dependent on the presence of the chaplin, inferring that fimbriae are, at least partially, composed of these proteins [58]. Indeed, without chaplins, fimbriae are no longer formed, which concomitantly also prevents hyphae from adhering to the surface.
Similar results were obtained with a mutant strain lacking the bldN gene, which encodes an extracytoplasmic function sigma factor required for the expression of the chaplin genes [84, 85]. To mediate attachment to hydrophobic surfaces, fimbriae probably require partial unfolding or unwinding to expose the hydrophobic sides of the individual chaplin fibrils, which are expected to be buried in the interior of the hydrophilic fimbriae [59]. Notably, also adhesion of streptomycetes involves a cellulose-like glycan, which is synthesized by the cellulose synthase-like protein CslA [58]. Attachment of hyphae was significantly reduced in mutant strains lacking the corresponding cslA gene. Instead, the glycan was shown to be important for proper anchoring of the fimbriae to the adhering hyphae. SapB is another macromolecule that is secreted during development [86]. This lantibiotic-like peptide [87] lowers the surface tension of the aqueous environment to enable hyphae to grow into the air [88].
4.3. γ-Butyrolactones as Inducers in Actinomycetes
Quorum sensing or cellular communication plays an important role in the attachment and biofilm formation [89–91]. Intracellular communication takes place through molecules that can be diffused in the surrounding environment. The communication system is based on signal molecules and transcription-activating proteins. In response to a cellular concentration, the signal molecules activate the target gene, which is in conjunction with the LuxR transcription activators. The signal molecules are known as acylhomoserine lactones HSL that are Gram positive. There are other types of signal molecules that are called AIP autoinducer peptides that exist in positive Gram. Streptomyces have butanolide-type molecules called gamma-butyrolactones that regulate the production of secondary metabolites such as antibiotics. Nowadays, there are no studies showing the role of autoinductors in biofilm formation in Streptomyces [23]. On the other hand, there are some reports about the involvement of signal molecules in biofilm formation in bifidobacterium. The autoinducers AI-2 are implicated in the regulation of biofilm formation in Bifidobacterium and Propionibacterium genera [92, 93]. AI-2 autoinducers are involved in the upregulation of virulence factors [92]. GBLs, MMFs, Factor-A, Factor-I, IM-2, VB, and PI factor are signal molecules regulating quorum sensing in the genus Streptomyces. These molecules control the following phenotypes: antibiotic production (Act Red, clavulanic acid, cephamycin, D-cycloserine, Kas, methylenomycin, natamycin, nikkomycin, nucleoside, pristinamycin, streptomycin, tylosin, virginiamycin), morphogenesis, and sporulation.
5. Antibiofilm Activity/Compounds from Actinobacteria
With constant rise in the number of antibiotic-resistant bacteria, there is a need to find alternative strategies to combat the phenomenon. Since pathogenicity is related to the biofilm formation ability, antibiofilm activity has become the trendiest alternative target to control their spread. Biofilms are intrinsically resistant to conventional antibiotics [94,95]. This indicates the need for new antibacterial drugs active not only against planktonic bacteria but also against drug-resistant biofilms. Various bioactive compounds have shown antibiofilm activity against Gram-positive pathogens [96–98]. The discovery of novel compounds is still very important.
Actinomycetes predominantly account for the production of the majority of bioactive compounds [99]. The antibiofilm activity of actinomycetes can be manifested by several mechanisms: by inhibiting microbial adhesion [100, 101] or by targeting quorum sensing [102, 103]. Streptomyces akiyoshinensis (A3) had shown an antibiofilm activity against S. pyogenes biofilms. Application of extracts significantly prevents biofilm formation up to 60–80%. Five extracts obtained from Streptomyces akiyoshinensis (A3) reduced the cell surface hydrophobicity, which is a crucial factor for biofilm formation in Staphylococcus pyogenes [104]. Streptomyces lunalinharesii strain 235 produces an active antimicrobial substance against sulfate-reducing bacterial biofilms, a significant problem for the oil industry. Results showed that the supra-minimal inhibitory concentration significantly reduced biofilm formation. The application of this antibiofilm substance as a potential biocide to control the growth of sulfate-reducing bacteria could be of great interest to the oil industry [105]. Streptomyces chrestomyceticus strain ADP4 inhibits biofilm formation [101]. The antibiofilm activity exerted by Streptomyces chrestomyceticus strain ADP4 against Candida spp. had a strong inhibitory effect on Candida cells adhesion and subsequent conversion to the hyphal state. These metabolites produced by Streptomyces chrestomyceticus strain ADP4 showed strong anti-Candida activity. They could be a potential drug to treat biofilm-mediated infections because they prevent cell transformation to biofilms [101] (Table 1).
Table 1.
Studies conducted on the antibiofilm activities of Streptomyces species.
| Strains | Molecule | Reference |
|---|---|---|
| Streptomyces akiyoshinensis (A3) | Antibiofilm activity against S. pyogenes biofilms | [104] |
| Streptomyces sp. | Bioactive compound from actinomycetes for antibiofilm activity against Gram-negative and Gram-positive bacteria | [106] |
| Streptomyces gandocaensis strain DHS334 | Cahuitamycin C showed highly effective inhibition effects on the biofilm formation of Acinetobacter baumannii | [107] |
| Streptomyces lunalinharesii strain 235 | Biofilms of sulfate-reducing bacteria | [105] |
| Streptomyces chrestomyceticus strain ADP4 | Strong anti-Candida activity | [101] |
| Streptomyces sp. SBT343 | Marine sponge-derived actinomycetes | [108] |
Balasubramanian et al. evaluated the potential of marine sponge-derived actinomycetes in inhibiting biofilm formation of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa. Results from in vitro biofilm-formation assays and scanning electron and confocal microscopy revealed that an organic extract derived from the marine sponge-associated bacterium Streptomyces sp. SBT343 significantly inhibited staphylococcal biofilm formation on polystyrene, glass, and contact lens surfaces without affecting bacterial growth [108].
6. Conclusion
Actinobacteria is a large phylum [109]. It comprises genera of particular importance in biotechnology and other pathogenic species [110]. However, few reports describe biofilm formation in this phylum [35, 50]. Great efforts are being deployed in areas related to the production of valuable compounds and antibiotics. The biotechnological processes are more efficient using biofilms than planktonic cells. The greatest metabolic and phylogenetic diversity in Actinobacteria provides an exceptional opportunity to explore their multifactorial abilities for biotechnological applications [23]. Some studies have focused on the adhesion of Streptomyces used in biotechnology. Biofilm steps are still unknown in Actinobacteria. The comprehension of biofilm formation in this bacterial group could contribute to several advances, so this issue must not be neglected. To our knowledge, the investigation of biofilm formation in Actinobacteria remains poorly explored. The mechanisms can be used in the application of biotechnology research. Future research should be oriented to the extracellular matrix, which constitutes a key factor in the integrity and stability in biofilms. Biofilm formation is an important phenomenon that is underexplored in this phylum.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Donlan R. M. Biofilms: microbial life on surfaces. Emerging Infectious Diseases. 2002;8(9) doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasudevan R. Biofilms: microbial cities of scientific significance. Journal of Microbiology & Experimentation. 2014;1(3):84–98. doi: 10.15406/jmen.2014.01.00014. [DOI] [Google Scholar]
- 3.Kokare C. R., Chakraborty S., Khopade A. N., Mahadik K. R. Biofilm : importance and applications. Indian Journal of Biotechnology. 2009;8:159–168. [Google Scholar]
- 4.Friedlander R. S., Vogel N., Aizenberg J. Role of flagella in adhesion of Escherichia coli to abiotic Surfaces. Langmuir. 2015;31(22):6137–6144. doi: 10.1021/acs.langmuir.5b00815. [DOI] [PubMed] [Google Scholar]
- 5.Garrett T. R., Bhakoo M., Zhang Z. Bacterial adhesion and biofilms on surfaces. Progress in Natural Science. 2008;18(9):1049–1056. doi: 10.1016/j.pnsc.2008.04.001. [DOI] [Google Scholar]
- 6.Pirog T. P., Konon A. D., Beregovaya K. A., Shulyakova M. A. Antiadhesive properties of the surfactants of Acinetobacter calcoaceticus IMB B-7241, Rhodococcus erythropolis IMB Ac-5017, and Nocardia vaccinii IMB B-7405. Microbiology. 2014;83(6):732–739. doi: 10.1134/S0026261714060150. [DOI] [PubMed] [Google Scholar]
- 7.Sadiki M., Elabed S., Barkai H., Balouiri M., Nasri A., Koraichi S. I. The modification of cedar wood surface properties for the prevention of fungal adhesion. International Journal of Adhesion and Adhesives. 2017;75:40–46. doi: 10.1016/j.ijadhadh.2017.01.007. [DOI] [Google Scholar]
- 8.Adak S., Upadrasta L., Kumar S. P. J., Soni R., Banerjee R. Quorum quenching–an alternative antimicrobial therapeutics. 2011.
- 9.Francolini I., Donelli G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunology & Medical Microbiology. 2010;59(3):227–238. doi: 10.1111/j.1574-695X.2010.00665.x. [DOI] [PubMed] [Google Scholar]
- 10.Percival S. L., Malic S., Cruz H., Williams D. W. Springer Series on Biofilms. Berlin, Germany: Springer; 2011. Introduction to biofilms; pp. 41–68. [DOI] [Google Scholar]
- 11.Lens P. N. L., Piet N. L. Biofilms in Medicine, Industry and Environmental Biotechnology : Characteristics, Analysis and Control. London, UK: IWA Publications; 2003. [Google Scholar]
- 12.Muffler K., Lakatos M., Schlegel C., Strieth D., Kuhne S. Application of biofilm bioreactors in white biotechnology. Advances in Biochemical Engineering / Biotechnology. 2013;146 doi: 10.1007/10_2013_267. [DOI] [PubMed] [Google Scholar]
- 13.Bryers J. D. Biofilms and the technological implications of microbial cell adhesion . Colloids and Surfaces B: Biointerfaces. 1994;2(1–3):9–23. doi: 10.1016/0927-7765(94)80013-8. [DOI] [Google Scholar]
- 14.Li X. Z., Hauer B., Rosche B. Single-species microbial biofilm screening for industrial applications. Applied Microbiology and Biotechnology. 2007;76(6):1255–1262. doi: 10.1007/s00253-007-1108-4. [DOI] [PubMed] [Google Scholar]
- 15.Zarnowski R., Westler W. M., Lacmbouh A., et al. Novel entries in a fungal biofilm matrix encyclopedia. mBio. 2014;5:1–13. doi: 10.1128/mBio.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig W., Euzéby J., Whitman W. B. Bergey’s Manual® of Systematic Bacteriology. Berlin, Germany: Springer; 2012. Taxonomic outline of the phylum Actinobacteria. [Google Scholar]
- 17.Hamedi J., Poorinmohammad N., Wink J. Biology and Biotechnology of Actinobacteria. Berlin, Germany: Springer International Publishing; 2017. The role of Actinobacteria in biotechnology; pp. 269–328. [DOI] [Google Scholar]
- 18.Wink J., Mohammadipanah F., Hamedi J. Biology and Biotechnology of Actinobacteria. Berlin, Germany: Springer International Publishing; 2017. [Google Scholar]
- 19.Arasu M. V., Duraipandiyan V., Agastian P., Ignacimuthu S. Antimicrobial activity of Streptomyces spp. ERI-26 recovered from western ghats of Tamil nadu. Journal of Medical Mycology. 2008;18:147–153. doi: 10.1016/j.mycmed.2008.07.004. [DOI] [Google Scholar]
- 20.Ningthouja D. S., Ningthoujam S. S. Screening of actinomycete isolates from niche habitats in Manipur for antibiotic activity. American Journal of Biochemistry and Biotechnology. 2009;5(4):221–225. doi: 10.3844/ajbbsp.2009.221.225. [DOI] [Google Scholar]
- 21.Kavitha A., Vijayalakshmi M., Sudhakar P., Narasimha G. Screening of Actinomycete strains for the production of antifungal metabolites. African Journal of Microbiology Research. 2010;4:27–032. [Google Scholar]
- 22.Sanchez S. F., Karl-Heinz Schleifer E. S. A Handbook on the Biology of Bacteria. 3rd. Berlin, Germany: Springer; 2006. The prokaryotes. [Google Scholar]
- 23.Polkade A. V., Mantri S. S., Patwekar U. J., Jangid K. Quorum sensing: an under-explored phenomenon in the phylum Actinobacteria. Frontiers in Microbiology. 2016;7:1–13. doi: 10.3389/fmicb.2016.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harir M., Bellahcene M., Baratto M. C., et al. Isolation and characterization of a novel tyrosinase produced by Sahara soil Actinobacteria and immobilization on nylon nanofiber membranes. Journal of Biotechnology. 2018;265:54–64. doi: 10.1016/j.jbiotec.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., Wang J.-D., Liu C.-X., Yuan J.-H., Wang X.-J., Xiang W.-S. A new prenylated indole derivative from endophytic actinobacteria Streptomyces sp. neau-D50. Natural Product Research. 2014;28(7):431–437. doi: 10.1080/14786419.2013.871546. [DOI] [PubMed] [Google Scholar]
- 26.Dharmaraj S., Dhevendaran K. Evaluation of Streptomyces as a probiotic feed for the growth of ornamental fish Xiphophorus helleri. Food Technol Biotechnol. 2010;48:497–504. [Google Scholar]
- 27.Miao V., Davies J. Actinobacteria: the good, the bad, and the ugly. Antonie van Leeuwenhoek. 2010;98(2):143–150. doi: 10.1007/s10482-010-9440-6. [DOI] [PubMed] [Google Scholar]
- 28.Singh R. P., Kumari P., Reddy C. R. K. Antimicrobial compounds from seaweeds-associated bacteria and fungi. Applied Microbiology and Biotechnology. 2015;99(4):1571–1586. doi: 10.1007/s00253-014-6334-y. [DOI] [PubMed] [Google Scholar]
- 29.Weber W., Schoenmakers R., Spielmann M., et al. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Research. 2003;31(14):71e–71. doi: 10.1093/nar/gng071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitaker A. Actinomycetes in submerged culture. Applied Biochemistry and Biotechnology. 1992;32(1-3):23–35. doi: 10.1007/BF02922146. [DOI] [PubMed] [Google Scholar]
- 31.Alonso A. N., Pomposiello P. J., Leschine S. B. Biofilm formation in the life cycle of the cellulolytic actinomycete Thermobifida fusca. Biofilms. 2008:1–11. doi: 10.1017/S1479050508002238. [DOI] [Google Scholar]
- 32.Brescia F., Marchetti-Deschmann M., Musetti R., Perazzolli M., Pertot I., Puopolo G. The rhizosphere signature on the cell motility, biofilm formation and secondary metabolite production of a plant-associated Lysobacter strain. Microbiological Research. 2020;234 doi: 10.1016/j.micres.2020.126424.126424 [DOI] [PubMed] [Google Scholar]
- 33.Krzyściak W., Jurczak A., Kościelniak D., Bystrowska B., Skalniak A. The virulence of Streptococcus mutans and the ability to form biofilms. European Journal of Clinical Microbiology & Infectious diseases: Official Publication of the European Society of Clinical Microbiology. 2014;33:499–515. doi: 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martins M., Henriques M., Azeredo J., Rocha S. M., Coimbra M. A., Oliveira R. Morphogenesis control in Candida albicans and Candida dubliniensis through signaling molecules produced by planktonic and biofilm cells. Eukaryotic Cell. 2007;6(12):2429–2436. doi: 10.1128/EC.00252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winn M., Casey E., Habimana O., Murphy C. D. Characteristics ofStreptomyces griseusbiofilms in continuous flow tubular reactors. FEMS Microbiology Letters. 2014;352(2):157–164. doi: 10.1111/1574-6968.12378. [DOI] [PubMed] [Google Scholar]
- 36.Maksimova Y. G. Microbial biofilms in biotechnological processes. Applied Biochemistry and Microbiology. 2014;50(8):750–760. doi: 10.1134/S0003683814080043. [DOI] [Google Scholar]
- 37.Qureshi N., Annous B. A., Ezeji T. C., Karcher P., Maddox I. S. Biofilm reactors for industrial bioconversion processes : employing potential of enhanced reaction rates. Microbial Cell Factories. 2005;4(1):24–21. doi: 10.1186/1475-2859-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandecandelaere I., Nercessian O., Segaert E., et al. Alteromonas genovensis sp. nov., isolated from a marine electroactive biofilm and emended description of Alteromonas macleodii Baumann et al. 1972 (Approved Lists 1980) International Journal of Systematic and Evolutionary Microbiology. 2008;58(11):2589–2596. doi: 10.1099/ijs.0.65691-0. [DOI] [PubMed] [Google Scholar]
- 39.Elsayed A., Shatoury E., Tolba S. Hexavalent chromate reduction in soil microcosm using bacterial consortium. The Egyptian Journal of Experimental Biology (Botany) 2019;15(2):1–313. doi: 10.5455/egyjebb.20190818073117. [DOI] [Google Scholar]
- 40.Sarkar S., Roy D., Mukherjee J. Production of a potentially novel antimicrobial compound by a biofilm-forming marine Streptomyces sp. in a niche-mimic rotating disk bioreactor. Bioprocess and Biosystems Engineering. 2010;33(2):207–217. doi: 10.1007/s00449-009-0314-0. [DOI] [PubMed] [Google Scholar]
- 41.Cheng A.-L., Kang Y.-K., Chen Z., et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. The Lancet Oncology. 2009;10(1):25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 42.Demirci A., Pometto A. L., Johnson K. E. Evaluation of biofilm reactor solid support for mixed-culture lactic acid production. Applied Microbiology and Biotechnology. 1993;38(6):728–733. doi: 10.1007/BF00167135. [DOI] [Google Scholar]
- 43.Demirci A., Pometto A. L., III, Johnson K. E. Lactic acid production in a mixed-culture biofilm reactor †. Applied and Environmental Microbiology. 1993;59(1):203–207. doi: 10.1128/aem.59.1.203-207.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrus M. L. C., Claessen D. Pivotal roles for Streptomyces cell surface polymers in morphological differentiation, attachment and mycelial architecture. Antonie van Leeuwenhoek. 2014;106(1):127–139. doi: 10.1007/s10482-014-0157-9. [DOI] [PubMed] [Google Scholar]
- 45.Könönen E., Wade W. G. Actinomyces and related organisms in human infections. Clinical Microbiology Reviews. 2015;28(2):419–442. doi: 10.1128/CMR.00100-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westhoff C. IUDs and colonization or infection with Actinomyces. Contraception. 2007;75(6):S48–S50. doi: 10.1016/j.contraception.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Maataoui H., Barkai H., Sadiki M., Haggoud A., Ibnsouda Koraichi S., Elabed S. Physicochemical characterization of actinomycetes isolated from decayed cedar wood: contact angle measurement. Journal of Adhesion Science and Technology. 2014;28(20):2046–2053. doi: 10.1080/01694243.2014.943341. [DOI] [Google Scholar]
- 48.Zahir H., Hamadi F., Mallouki B., Imziln B., Latrache H. Effect of salinity on the adhesive power actinomycetes in soil. Journal of Materials and Environmental Science. 2016;7:3327–3333. [Google Scholar]
- 49.Alonso A. N. Hydrophobic nature and effects of culture conditions on biofilm formation by the cellulolytic actinomycete Thermobifida fusca. AIMS Microbiology. 2015;1:1–10. doi: 10.3934/microbiol.2015.1.1. [DOI] [Google Scholar]
- 50.Shanmughapriya S., Lency Francis A., Kavitha S. In vitro actinomycete biofilm development and inhibition by the polyene antibiotic, nystatin, on IUD copper surfaces. Biofouling. 2012;28(9):929–935. doi: 10.1080/08927014.2012.717616. [DOI] [PubMed] [Google Scholar]
- 51.Flemming H.-C., Wingender J. The biofilm matrix. Nature Reviews Microbiology. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 52.Flärdh K., Buttner M. J. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nature Reviews Microbiology. 2009;7(1):36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 53.Chater K. F., Biró S., Lee K. J., Palmer T., Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiology Reviews. 2010;34(2):171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- 54.Elliot M. A., Talbot N. J. Building filaments in the air: aerial morphogenesis in bacteria and fungi. Current Opinion in Microbiology. 2004;7(6):594–601. doi: 10.1016/j.mib.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Claessen D., Rink R., De Jong W., et al. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes & Development. 2003;17(14):1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elliot M. A., Karoonuthaisiri N., Huang J., et al. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes & Development. 2003;17(14):1727–1740. doi: 10.1101/gad.264403.Filamentous. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawyer E. B., Claessen D., Gras S. L., Perrett S. Exploiting amyloid: how and why bacteria use cross-β fibrils. Biochemical Society Transactions. 2012;40(4):728–734. doi: 10.1042/BST20120013. [DOI] [PubMed] [Google Scholar]
- 58.de Jong W., Wösten H. A. B., Dijkhuizen L., Claessen D. Attachment of Streptomyces coelicoloris mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Molecular Microbiology. 2009;73(6):1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- 59.Petrus M. L. C., van Veluw G. J., Wösten H. A. B., Claessen D. Sorting of streptomyces cell pellets using a complex object parametric analyzer and sorter. Journal of Visualized Experiments. 2014;84(84):p. 51178. doi: 10.3791/51178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Celler K., Picioreanu C., Van Loosdrecht M. C. M., Van Wezel G. P. Structured morphological modeling as a framework for rational strain design of Streptomyces species. Antonie Van Leeuwenhoek. 2012;102(3):409–423. doi: 10.1007/s10482-012-9760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Wezel G. P., Krabben P., Traag B. A., et al. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Applied and Environmental Microbiology. 2006;72(8):5283–5288. doi: 10.1128/AEM.00808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vecht-lifshitz S. E., Almas K. A., Zomer E., Vecht-lgshitz S. E. Microbial growth on peptones from fish industrial wastes. Letters in Applied Microbiology. 1990;10(4):183–186. doi: 10.1111/j.1472-765x.1990.tb00110.x. [DOI] [Google Scholar]
- 63.Nieminen L., Webb S., Smith M. C. M., Hoskisson P. A. A flexible mathematical model platform for Studying branching networks: experimentally validated using the model actinomycete, Streptomyces coelicolor. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0054316.e54316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manteca A., Alvarez R., Salazar N., Yagüe P. Mycelium differentiation and antibiotic production in Submerged cultures of Streptomyces coelicolor. Applied and Environmental Microbiology. 2008;74(12):3877–3886. doi: 10.1128/AEM.02715-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rioseras B., López-García M. T., Yagüe P., Sánchez J., Manteca Á. Mycelium differentiation and development of Streptomyces coelicolor in lab-scale bioreactors: programmed cell death, differentiation, and lysis are closely linked to undecylprodigiosin and actinorhodin production. Bioresource Technology. 2014;151:191–198. doi: 10.1016/j.biortech.2013.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim Y. M., Kim J. H. Formation and dispersion of mycelial pellets of Streptomyces coelicolor A3 (2) Journal of Microbiology (Seoul, Korea) 2004;42:64–67. [PubMed] [Google Scholar]
- 67.Whitchurch C. B., Tolker-nielsen T., Ragas P. C., Mattick J. S. Extracellular DNA required for bacterial biofilm formation. Science. 2002;80 doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 68.Van Veluw G. J., Petrus M. L. C., Gubbens J., et al. Analysis of two distinct mycelial populations in liquid-grown Streptomyces cultures using a flow cytometry-based proteomics approach. Applied Microbiology and Biotechnology. 2012;96(5):1301–1312. doi: 10.1007/s00253-012-4490-5. [DOI] [PubMed] [Google Scholar]
- 69.Xu H., Chater K. F., Deng Z., Tao M. A cellulose Synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. Journal of Bacteriology. 2008;190(14):4971–4978. doi: 10.1128/JB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parte A. C., Road W. LPSN-list of prokaryotic names with standing in nomenclature. Nucleic Acids Research. 2014;42(D1):D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaudhary H. S., Soni B., Shrivastava A. R., Shrivastava S. Diversity and versatility of actinomycetes and its role in antibiotic production. Journal of Applied Pharmaceutical Science. 2013;3(8) doi: 10.7324/JAPS.2013.38.S14. [DOI] [Google Scholar]
- 72.Li Q., Chen X., Jiang Y., Jiang C. Actinobacteria-Basics and Biotechnological Applications. London, UK: InTech; 2016. Morphological identification of Actinobacteria. [Google Scholar]
- 73.Jiang Y. Actinobacteria-basics and biotechnological applications. 2016.
- 74.Kirby R., Sangal V., Tucker N. P., et al. Draft genome sequence of the human pathogen Streptomyces somaliensis, a significant cause of actinomycetoma. Journal of Bacteriology. 2012;194(13) doi: 10.1128/JB.00534-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Claessen D., Wösten H. A. B., Keulen G. v., et al. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Molecular Microbiology. 2002;44(6):1483–1492. doi: 10.1046/j.1365-2958.2002.02980.x. [DOI] [PubMed] [Google Scholar]
- 76.Lupan I., Popescu O. Metagenomics and future perspectives for biodeterioration and biodegradation studies. Annals of the Romanian Society for Cell Biology. 2012;17(2) [Google Scholar]
- 77.Scheerer S., Ortega-morales O., Gaylarde C. Chapter 5-Microbial Deterioration of Stone Monuments-An Updated Overview. 1st. Amsterdam, Netherlands: Elesvier Inc; 2009. [DOI] [PubMed] [Google Scholar]
- 78.Soumya E., Saad I. K., Abdellah H., Hassan L. Experimental and theoretical investigations of the adhesion time of Penicillium spores to cedar wood surface. Materials Science and Engineering: C. 2013;33(3):1276–1281. doi: 10.1016/j.msec.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 79.Claessen D., Stokroos I., Deelstra H. J., et al. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Molecular Microbiology. 2004;53(2):433–443. doi: 10.1111/j.1365-2958.2004.04143.x. [DOI] [PubMed] [Google Scholar]
- 80.De Jong I. P., Claessen D. A sandwich-culture technique for controlling antibiotic production and morphological development in Streptomyces coelicolor. Journal of Microbiological Methods. 2012;91(2):318–320. doi: 10.1016/j.mimet.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 81.Di Berardo C., Capstick D. S., Bibb M. J., Findlay K. C., Buttner M. J., Elliot M. A. Function and redundancy of the chaplin cell surface proteins in aerial hypha formation, rodlet assembly, and viability in Streptomyces coelicolor. Journal of Bacteriology. 2008;190(17):5879–5889. doi: 10.1128/JB.00685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ekkers D. M., Claessen D., Galli F., Stamhuis E. Surface modification using interfacial assembly of the Streptomyces chaplin proteins. Applied Microbiology and Biotechnology. 2014;98(10):4491–4501. doi: 10.1007/s00253-013-5463-z. [DOI] [PubMed] [Google Scholar]
- 83.de Jong W., Manteca A., Sanchez J., et al. NepA is a structural cell wall protein involved in maintenance of spore dormancy in Streptomyces coelicolor. Molecular Microbiology. 2009;71(6):1591–1603. doi: 10.1111/j.1365-2958.2009.06633.x. [DOI] [PubMed] [Google Scholar]
- 84.Bibb M. J., Domonkos Á., Chandra G., Buttner M. J. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σBldN and a cognate anti-sigma factor, RsbN. Molecular Microbiology. 2012;84(6):1033–1049. doi: 10.1111/j.1365-2958.2012.08070.x. [DOI] [PubMed] [Google Scholar]
- 85.Elliot M. A., Locke T. R., Galibois C. M., Leskiw B. K. BldD fromStreptomyces coelicoloris a non-essential global regulator that binds its own promoter as a dimer. FEMS Microbiology Letters. 2003;225(1):35–40. doi: 10.1016/S0378-1097(03)00474-9. [DOI] [PubMed] [Google Scholar]
- 86.Capstick D. S., Willey J. M., Buttner M. J., Elliot M. A. SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Molecular Microbiology. 2007;64(3):602–613. doi: 10.1111/j.1365-2958.2007.05674.x. [DOI] [PubMed] [Google Scholar]
- 87.Kodani S., Lodato M. A., Durrant M. C., Picart F., Willey J. M. SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Molecular Microbiology. 2005;58(5):1368–1380. doi: 10.1111/j.1365-2958.2005.04921.x. [DOI] [PubMed] [Google Scholar]
- 88.Tillotson R. D., Wösten H. A. B., Richter M., Willey J. M. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Molecular Microbiology. 1998;30(3):595–602. doi: 10.1046/j.1365-2958.1998.01093.x. [DOI] [PubMed] [Google Scholar]
- 89.Dobretsov S., Dahms H.-U., YiLi H., Wahl M., Qian P.-Y. The effect of quorum-sensing blockers on the formation of marine microbial communities and larval attachment. FEMS Microbiology Ecology. 2007;60(2):177–188. doi: 10.1111/j.1574-6941.2007.00285.x. [DOI] [PubMed] [Google Scholar]
- 90.Kjelleberg S., Molin S. Is there a role for quorum sensing signals in bacterial biofilms? Current Opinion in Microbiology. 2002;5(3):254–258. doi: 10.1016/s1369-5274(02)00325-9. [DOI] [PubMed] [Google Scholar]
- 91.Tomlin K. L., Malott R. J., Ramage G., Storey D. G., Sokol P. A., Ceri H. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Applied and Environmental Microbiology. 2005;71(9):5208–5218. doi: 10.1128/AEM.71.9.5208-5218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Santos C. L., Correia-Neves M., Moradas-Ferreira P., Mendes M. V. A walk into the LuxR regulators of Actinobacteria: phylogenomic distribution and functional diversity. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046758.e46758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun Z., He X., Brancaccio V. F., Yuan J., Riedel C. U. Bifidobacteria exhibit LuxS-dependent autoinducer 2 activity and biofilm formation. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0088260.e88260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Corona F., Martinez J. Phenotypic resistance to antibiotics. Antibiotics. 2013;2(2):237–255. doi: 10.3390/antibiotics2020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial biofilms. International Journal of Antimicrobial Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 96.Correa M. S., Schwambach J., Mann M. B., Frazzon J., Frazzon A. P. G. Antimicrobial and antibiofilm activity of the essential oil from dried leaves of Eucalyptus staigeriana. Arquivos do Instituto Biológico. 2019;86 doi: 10.1590/1808-1657000202018. [DOI] [Google Scholar]
- 97.Fujimura S., Sato T., Kikuchi T., Zaini J., Gomi K., Watanabe A. Efficacy of clarithromycin plus vancomycin in mice with implant-related infection caused by biofilm-forming Staphylococcus aureus. Journal of Orthopaedic Science. 2009;14(5):658–661. doi: 10.1007/s00776-009-1366-3. [DOI] [PubMed] [Google Scholar]
- 98.Miquel S., Lagrafeuille R., Souweine B., Forestier C. Anti-biofilm activity as a health issue. Frontiers of Microbiology. 2016;7:p. 592. doi: 10.3389/fmicb.2016.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hopwood D. A. Streptomyces in nature and medicine. 2015.
- 100.Siddharthan S., Rajamohamed B. S., Gopal V. Streptomyces diastaticus isolated from the marine crustacean Portunus sanguinolentus with potential antibiofilm activity against Candida albicans. Archives of Microbiology. 2020;202(7):1977–1984. doi: 10.1007/s00203-020-01918-8. [DOI] [PubMed] [Google Scholar]
- 101.Srivastava V., Dubey A. K. Anti-biofilm activity of the metabolites of Streptomyces chrestomyceticus strain ADP4 against Candida albicans. Journal of Bioscience and Bioengineering. 2016;122(4):434–440. doi: 10.1016/j.jbiosc.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 102.Miao L., Xu J., Yao Z., et al. The anti-quorum sensing activity and bioactive substance of a marine derived Streptomyces. Biotechnology & Biotechnological Equipment. 2017;31(5):1007–1015. doi: 10.1080/13102818.2017.1348253. [DOI] [Google Scholar]
- 103.Younis K. M., Usup G., Ahmad A. Secondary metabolites produced by marine streptomyces as antibiofilm and quorum-sensing inhibitor of uropathogen Proteus mirabilis. Environmental Science and Pollution Research. 2016;23(5):4756–4767. doi: 10.1007/s11356-015-5687-9. [DOI] [PubMed] [Google Scholar]
- 104.Nithyanand P., Thenmozhi R. Inhibition of Streptococcus pyogenes biofilm formation by coral-associated actinomycetes. Current Microbiology. 2010;60(6):454–460. doi: 10.1007/s00284-009-9564-y.460 [DOI] [PubMed] [Google Scholar]
- 105.Pacheco J., Gonc S. R., Marques J. M., Seldin L., Reed R., Coelho R. Streptomyces lunalinharesii 235 prevents the formation of a sulfate-reducing bacterial biofilm. Brazilian Journal of Microbiology. 2016;7:603–609. doi: 10.1016/j.bjm.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Waturangi D. E., Yolanda K., Lalu M. B. S. R., Mulyono N. Characterization of bioactive compound from actinomycetes for antibiofilm activity against gram-negative and gram-positive bacteria. Malaysian Journal of Microbiology. 2016;12:291–299. [Google Scholar]
- 107.Park S. R., Tripathi A., Wu J., et al. Discovery of cahuitamycins as biofilm inhibitors derived from a convergent biosynthetic pathway. Nature Communications. 2016;7(1):1–11. doi: 10.1038/ncomms10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Balasubramanian S., Othman E. M., Kampik D., et al. Marine sponge-derived Streptomyces sp. SBT343 extract inhibits staphylococcal biofilm formation. Frontiers in Microbiology. 2017;8 doi: 10.3389/fmicb.2017.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nouioui I., Carro L., García-López M., et al. Genome-based taxonomic classification of the phylum Actinobacteria. Frontiers in Microbiology. 2018;9:p. 2007. doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wolfgang L., EuJeanzéby j., Schumann P., et al. Bergey’s Manual® of Systematic Bacteriology. Berlin, Germany: Springer; 2012. Road map of the phylum Actinobacteria; pp. 1–28. [Google Scholar]


