Abstract
Squalene monooxygenase (SM, also known as squalene epoxidase) is a rate-limiting enzyme of cholesterol synthesis that converts squalene to monooxidosqualene and is oncogenic in numerous cancer types. SM is subject to feedback regulation via cholesterol-induced proteasomal degradation, which depends on its lipid-sensing N-terminal regulatory domain. We previously identified an endogenous truncated form of SM with a similar abundance to full-length SM, but whether this truncated form is functional or subject to the same regulatory mechanisms as full-length SM is not known. Here, we show that truncated SM differs from full-length SM in two major ways: it is cholesterol resistant and adopts a peripheral rather than integral association with the endoplasmic reticulum membrane. However, truncated SM retains full SM activity and is therefore constitutively active. Truncation of SM occurs during its endoplasmic reticulum–associated degradation and requires the proteasome, which partially degrades the SM N-terminus and disrupts cholesterol-sensing elements within the regulatory domain. Furthermore, truncation relies on a ubiquitin signal that is distinct from that required for cholesterol-induced degradation. Using mutagenesis, we demonstrate that partial proteasomal degradation of SM depends on both an intrinsically disordered region near the truncation site and the stability of the adjacent catalytic domain, which escapes degradation. These findings uncover an additional layer of complexity in the post-translational regulation of cholesterol synthesis and establish SM as the first eukaryotic enzyme found to undergo proteasomal truncation.
Keywords: squalene monooxygenase, cholesterol, proteasome, ubiquitylation (ubiquitination), protein degradation, endoplasmic reticulum–associated protein degradation
Abbreviations: BSA, bovine serum albumin; cDNA, complementary DNA; DMEM-HG, high-glucose Dulbecco's modified Eagle's medium; DMSO, dimethyl sulfoxide; DOS, dioxidosqualene; ER, endoplasmic reticulum; ERAD, ER-associated degradation; FCS, fetal calf serum; FCLPDS, lipoprotein-deficient FCS; GSK-3β, glycogen synthase kinase 3 beta; HA, hemagglutinin; [HA]3–SM–V5, SM fused to N-terminal (HA)3 and C-terminal V5 epitope tags; HEK293, human embryonic kidney 293; MARCHF6, membrane–associated RING-CH-type finger 6; MOS, monooxidosqualene; PBST, PBS with 0.1% (v/v) Tween-20; RIPA, radioimmunoprecipitation assay; SM, squalene monooxygenase; SM-N100, N-terminal one hundred amino acids of SM; SQLE, squalene epoxidase; trunSM, truncated SM; UNSW, University of New South Wales; VCP, valosin-containing protein
Cholesterol is a vital lipid that serves many important functions in mammalian cells, including the maintenance of membrane fluidity and integrity, assembly of cell surface microdomains for signaling and adhesion, and synthesis of steroid hormones (1). Nevertheless, excess cholesterol is cytotoxic and linked with the onset of cardiovascular disease and cancer (2, 3). It is therefore essential that cells tightly control cholesterol homeostasis by balancing its uptake, synthesis, and efflux (4).
The regulation of cholesterol synthesis is especially exquisite, given the energy-intensive and oxygen-intensive nature of the pathway. A critical point at which this regulation is exerted is squalene monooxygenase (SM, also known as squalene epoxidase or SQLE; Enzyme Commission number: 1.14.14.17), an endoplasmic reticulum (ER)–localized and rate-limiting enzyme responsible for the conversion of squalene to monooxidosqualene (MOS) and subsequently dioxidosqualene (DOS) (5). SM is positioned within the branch of the mevalonate pathway that is committed to cholesterol synthesis, contrasting it with the upstream rate-limiting enzyme and well-studied target of statins, 3-hydroxy-3-methylglutaryl-CoA reductase. Therefore, SM may be an alternative target for the treatment of hypercholesterolemia (6). Recent years have also seen increasing recognition of SM as oncogenic in a range of malignancies, including breast (7), prostate (8), and liver cancer (9). Moreover, the SM substrate squalene is implicated either as a cytotoxic intermediate (10) or as protective against cancer cell death (11), depending on the cellular context. These reports raise the interesting prospect of targeting SM therapeutically. As pharmacological inhibition of SM is toxic in mammals (12), indirect inhibition by modulating its physiological regulation may be a more viable strategy.
At the transcriptional level, SM expression is controlled by sterol regulatory element–binding proteins, the master regulators of cholesterogenic genes (13, 14). Acute regulation occurs at the post-translational level, where SM undergoes accelerated degradation in response to increased cholesterol levels (5). Reciprocally, SM is protected from degradation by the allosteric binding of squalene (15, 16). These responses depend on the N-terminal one hundred amino acids of SM (SM-N100), a regulatory domain that is both necessary and sufficient for cholesterol and squalene sensing (5, 15, 17, 18). The SM-N100 domain is absent from the yeast ortholog of SM, Erg1p, despite high sequence conservation within the SM catalytic domain (5). This suggests that the lipid-sensing capabilities of SM are unique to higher eukaryotes, in which more nuanced regulation of cholesterol synthesis is required. Cholesterol and squalene affect SM levels by modulating its ubiquitination by the E3 ubiquitin ligase membrane–associated RING-CH-type finger 6 (MARCHF6), thereby promoting or preventing its ER-associated degradation (ERAD) (15, 19). Beyond MARCHF6, the ERAD of SM involves additional effectors including the AAA+-type ATPase valosin-containing protein (VCP), which extracts client proteins from the ER membrane, the E2 ubiquitin conjugating enzyme Ube2J2, deubiquitinases, and the 26S proteasome (5, 20, 21). Plasmalogen glycerophospholipids and unsaturated fatty acids also regulate the MARCHF6-mediated degradation of SM (22, 23), implying that SM responds to other classes of lipids. However, further details of the SM ERAD mechanism remain to be elucidated.
Previously, we reported that immunoblotting of SM in human embryonic kidney 293 (HEK293) cell lysates detected full-length SM as well as a low–molecular weight and putatively truncated form of SM (15). In the present study, we characterize this SM variant and show that it arises from partial proteasomal degradation of the SM-N100 regulatory domain (referred to herein as proteasomal truncation). This has been described for only two other human proteins, NF-κB and Gli3, where it results in major changes to protein function (24, 25). In the case of SM, proteasomal truncation depends on an intrinsically disordered region adjacent to the truncation site and the stability of the C-terminal catalytic domain. Truncation yields a constitutively active form of SM that is resistant to cholesterol-induced degradation and has an altered ER membrane topology. These findings uncover an additional mode by which SM activity is regulated and establish the first known example of a proteasomally truncated eukaryotic enzyme.
Results
A truncated and cholesterol-resistant form of SM is present in a variety of cell types
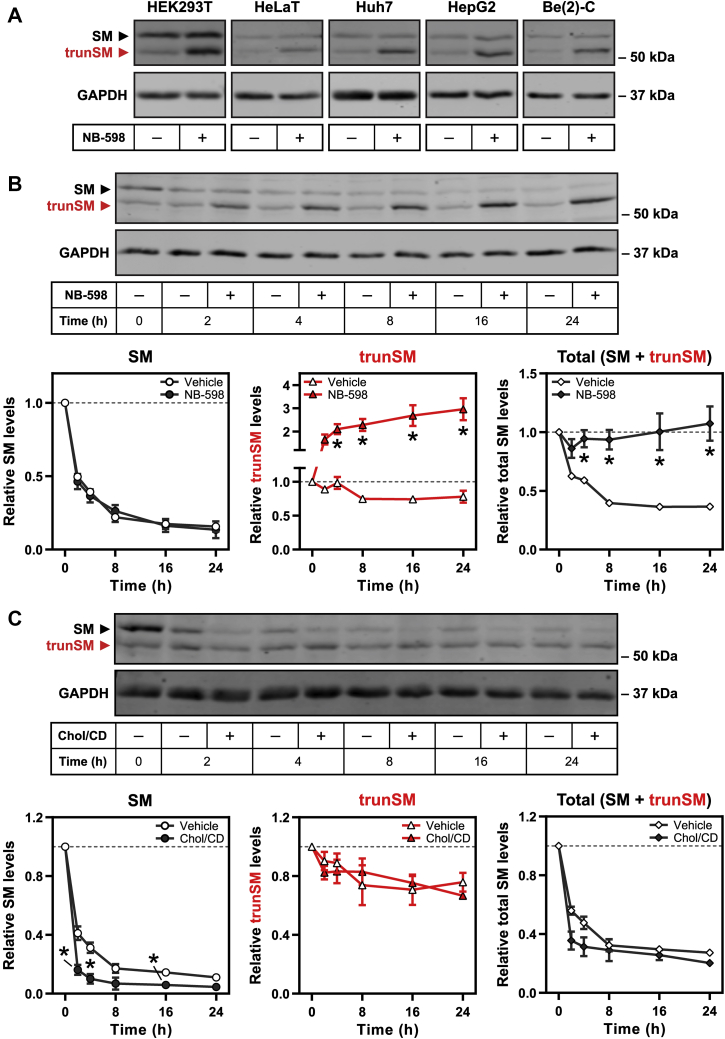
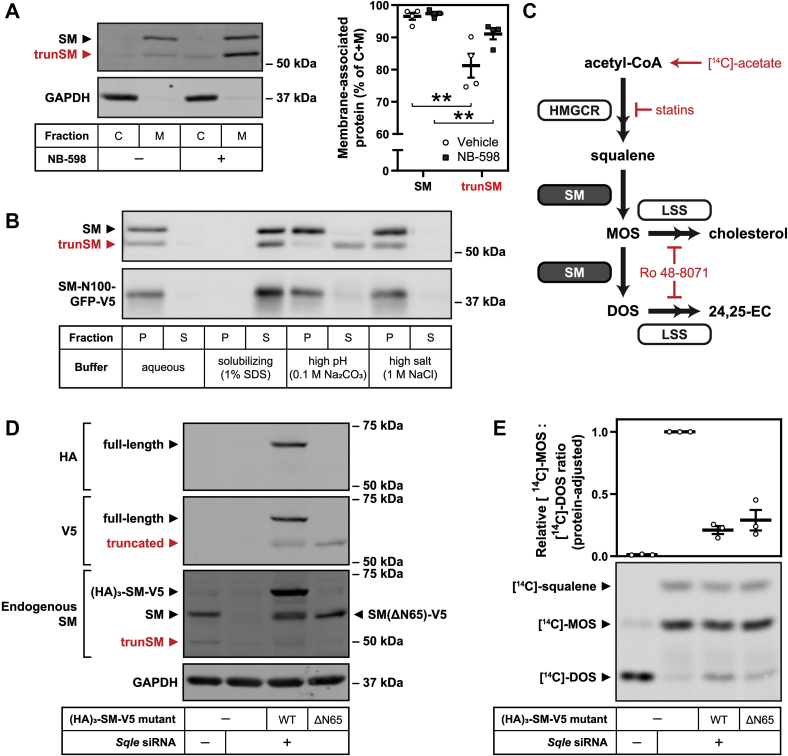
We previously reported that anti-SM immunoblotting of HEK293 cell lysates detected full-length SM (~64 kDa) as well as a low–molecular weight protein (~55 kDa) that was derived from the SQLE gene and accumulated upon treatment with the SM inhibitor NB-598 (15). This protein will henceforth be referred to as truncated SM (trunSM). Here, we observed expression and NB-598–induced accumulation of trunSM in the commonly used HEK293T and HeLaT cell lines, as well as other human cell lines derived from tissues that actively synthesize cholesterol: Huh7 (liver), HepG2 (liver), and Be(2)-C (brain) (Fig. 1A). A trunSM-like protein was also detected in the CHO subline CHO-7, where it accumulated upon prolonged NB-598 treatment (Fig. S1A). These observations confirmed that trunSM production is generalizable to a range of human cell types and the hamster ortholog of SM.
Figure 1.
A truncatedandcholesterol-resistantform of SM is present in a variety of cell types.A, the indicated cell lines were treated in the presence or the absence of 1 μM NB-598 for 8 h, and immunoblotting was performed for SM and truncated SM (trunSM; red). Immunoblots are representative of n ≥ 3 (HEK293T, HeLaT, Huh7, and HepG2) or n = 1 (Be(2)-C) independent experiments. B and C, HEK293T cells were treated with 10 μg/ml cycloheximide in the presence or the absence of (B) 1 μM NB-598 or (C) 20 μg/ml cholesterol-methyl-β-cyclodextrin complexes (Chol/CD) for the indicated times. Graphs depict densitometric quantification of SM levels (left), trunSM levels (center), or total SM levels (SM + trunSM; right) normalized to the 0 h time point, which was set to 1 (dotted line). Data presented as mean ± SEM from n ≥ 3 independent experiments (∗p ≤ 0.05; two-tailed paired t test versus vehicle condition). HEK293T, human embryonic kidney 293T; SM, squalene monooxygenase.
To further characterize trunSM, we examined its stability by treating HEK293T cells with the protein synthesis inhibitor cycloheximide in the presence or the absence of NB-598. The trunSM protein was remarkably long lived: in the absence of NB-598, ∼80% of its starting material remained after 24 h of cycloheximide treatment, compared with only ∼15% of full-length SM (Fig. 1B). NB-598 had no effect on the disappearance of full-length SM but rapidly induced trunSM accumulation, with total SM levels (the sum of full-length SM and trunSM) remaining constant during the treatment. This strongly suggested that trunSM is derived from full-length SM, and that NB-598 promotes this conversion. To directly verify this, we sterol-depleted cells to maximize SM expression, labeled newly synthesized protein with an azidomethionine analog, and then induced trunSM formation using NB-598 treatment. Immediately following removal of the azidomethionine label, nascent SM was mostly in its full-length form (Fig. S1B). However, the proportion of labeled trunSM increased during the subsequent NB-598 treatment, confirming that trunSM arises directly from full-length SM.
Next, we tested if trunSM undergoes the cholesterol-induced degradation characteristic of full-length SM (5) by cotreating cells with cycloheximide and exogenous cholesterol. Strikingly, cholesterol had no effect on trunSM levels, whereas accelerated degradation of SM was apparent within 2 h of cholesterol treatment (Fig. 1C). Together, these data indicated that trunSM is resistant to both basal and cholesterol-induced degradation, raising the possibility that it lacks part or all the SM-N100 regulatory domain. This was consistent with the shift in apparent molecular weight between SM and trunSM, which corresponded to a difference of ~50 to 100 amino acids.
trunSM is not produced by alternative SQLE transcripts
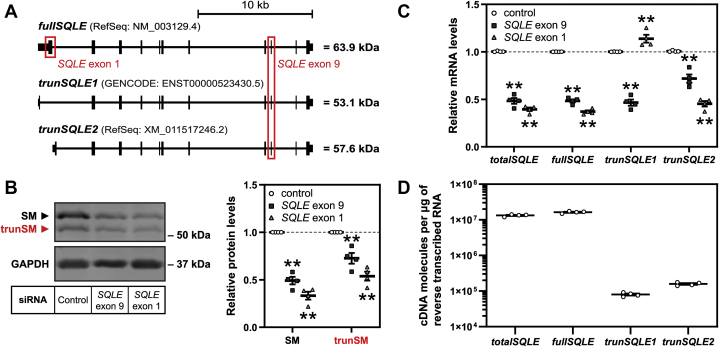
Each of the GENCODE-annotated and RefSeq-annotated human genomes predicts a different protein-coding isoform of SQLE. These isoforms utilize alternative first exons that substitute the coding sequence of the first 97 amino acids of full-length SM with a two-amino acid or 39-amino acid sequence, respectively (Fig. 2A). Given our hypothesis that trunSM lacks the SM-N100 domain, as well as the similarity between the apparent molecular weight of trunSM (~55 kDa) and the predicted molecular weights of the SQLE isoforms (53.1 kDa for the GENCODE isoform, trunSQLE1; and 57.6 kDa for the RefSeq isoform, trunSQLE2), we sought to confidently rule out the possibility that alternative SQLE transcripts also contribute to trunSM production.
Figure 2.
trunSM is not produced by alternative SQLE transcripts.A, schematic of full-length (fullSQLE) and alternative protein-coding (trunSQLE1 and trunSQLE2) SQLE transcripts. Exons and untranslated regions are indicated by black bars, and siRNA target regions are indicated by red boxes. B and C, HEK293T cells were transfected with the indicated siRNAs for 24 h and refreshed in maintenance medium for a further 24 h. B, graph depicts densitometric quantification of SM and trunSM levels normalized to the control siRNA condition, which was set to 1 (dotted line). C, SQLE transcript levels were normalized to PBGD housekeeping transcript levels and adjusted relative to the control siRNA condition, which was set to 1 (dotted line). D, absolute quantification of SQLE cDNA levels in control siRNA samples from (C). B–D, data are presented as mean ± SEM from n = 4 independent experiments, each performed in triplicate for quantitative RT-PCR analysis (∗∗p ≤ 0.01; two-tailed paired t test versus control siRNA). cDNA, complementary DNA; HEK293T, human embryonic kidney 293T; SQLE, squalene epoxidase; trunSM, truncated SM.
To this end, we transfected HEK293T cells with siRNA targeting exon 9 of SQLE, which is present in all three SQLE isoforms (quantified collectively as totalSQLE) or exon 1 of the full-length SQLE isoform only (fullSQLE; Fig. 2A). Both siRNAs reduced trunSM protein levels (Fig. 2B), whereas trunSQLE1 mRNA expression was downregulated by only exon 9 siRNA (Fig. 2C), ruling out this isoform as giving rise to trunSM. Unexpectedly, trunSQLE2 mRNA expression was downregulated by exon 1 siRNA, perhaps because of the presence of the siRNA target sequence in an unannotated 3′-untranslated region of this transcript. To determine the likelihood of trunSQLE2 accounting for trunSM formation, we next performed absolute quantification of SQLE complementary DNA (cDNA). Full-length SQLE cDNA comprised the great majority of SQLE transcripts (~1.6 × 107 cDNA copies per microgram of reverse transcribed RNA), whereas trunSQLE1 and trunSQLE2 cDNA were less abundant by over two orders of magnitude (~8.0 × 104 and ~1.5 × 105 cDNA copies, respectively) (Fig. 2D). Given that (1) trunSM protein levels are comparable to full-length SM even in the absence of NB-598 (Fig. 2B), (2) accumulation of trunSM can occur during inhibition of protein synthesis (Fig. 1B), and (3) we later found that ectopic SM also produces a trunSM-like protein (Fig. 3A), we concluded that trunSM is highly unlikely to be derived from low-abundance SQLE isoforms.
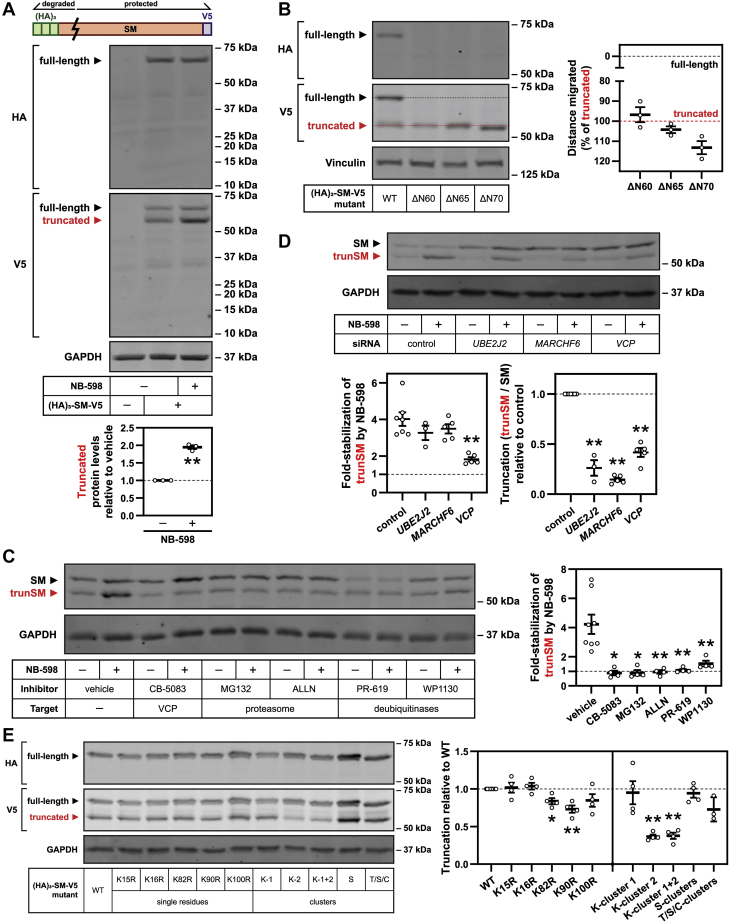
Figure 3.
trunSM arises from partial proteasomal degradation of the SM N-terminus.A, HEK293T cells were transfected with empty vector or (HA)3–SM–V5 expression vector for 24 h, refreshed in maintenance medium for 16 h, and treated in the presence or the absence of 1 μM NB-598 for 8 h. Lysates were separated by 4 to 15% gradient Tris–glycine SDS-PAGE. Graph depicts densitometric quantification of truncated protein levels normalized to the vehicle condition, which was set to 1 (dotted line). B, HEK293T cells were transfected with the indicated constructs for 24 h and refreshed in maintenance medium for 24 h. Lysates were separated by 7.5% Tris–glycine SDS-PAGE. Graph depicts quantification of construct migration relative to the separation of WT full-length and truncated (HA)3–SM–V5, which were set to 0% (gray dotted line) and 100% (red dotted line), respectively. C, HEK293T cells were treated with 5 μM CB-5083, 20 μM MG132, 25 μg/ml ALLN, 40 μM PR-619, or 10 μM WP1130, in the presence or the absence of 1 μM NB-598, for 8 h. Graph depicts densitometric quantification of trunSM stabilization by NB-598. D, HEK293T cells were transfected with the indicated siRNAs for 24 h, refreshed in maintenance medium for 16 h, and treated in the presence or the absence of 1 μM NB-598 for 8 h. Graphs depict densitometric quantification of (left) trunSM stabilization by NB-598 or (right) SM truncation normalized to the control siRNA condition, which was set to 1 (dotted line). E, HEK293T cells were transfected with the indicated constructs for 24 h and refreshed in maintenance medium for 24 h. Graph depicts densitometric quantification of (HA)3–SM–V5 truncation normalized to the WT construct, which was set to 1 (dotted line). Cluster mutations: K-cluster 1 (K15R and K16R); K-cluster 2 (K82R, K90R, and K100R); K-cluster 1 + 2 (K15R, K16R, K82R, K90R, and K100R); S-clusters (S59A, S61A, S83A, and S87A); T/S/C-clusters (T3A, T9A, T11A, S43A, C46A, S59A, S61A, S67A, S71A, S83A, and S87A). A–E, data presented as mean ± SEM from n ≥ 3 independent experiments (∗p ≤ 0.05; ∗∗p ≤ 0.01, two-tailed paired t test versus [A and B] vehicle, [C] control siRNA, or [D] WT). HEK293T, human embryonic kidney 293T; trunSM, truncated SM.
trunSM arises from partial proteasomal degradation of the SM N-terminus
To determine if trunSM is a proteolytic product of full-length SM, we transfected HEK293T cells with SM fused to N-terminal (hemagglutinin [HA])3 and C-terminal V5 epitope tags ([HA]3–SM–V5). Immunoblotting detected two C-terminally tagged proteins with molecular weights corresponding to SM and trunSM, the latter of which accumulated upon NB-598 treatment (Fig. 3A). Only the full-length protein was N-terminally tagged, confirming that the trunSM-like fragment lacks the SM N-terminus. Significantly, we were unable to recover a low–molecular weight and N-terminally tagged fragment, suggesting that the SM N-terminus undergoes complete proteolysis during truncation. To estimate the truncation site, we inserted a FLAG epitope tag at various positions within the (HA)3–SM–V5 construct and monitored for its appearance in the truncated fragment. Truncation eliminated the FLAG tag when it was inserted after SM residue 60 but not residue 70 (Fig. S2A), implying that truncation occurs between these two residues. For a more accurate estimation, we next generated N-terminally truncated SM–V5 constructs (ΔN60, ΔN65, and ΔN70) and measured their SDS-PAGE migration relative to full-length and truncated (HA)3–SM–V5. On average, the SM(ΔN60)–V5 construct migrated a shorter distance than the truncated fragment, whereas the SM(ΔN65)–V5 constructs migrated a longer distance (Fig. 3B), indicating that trunSM lacks the N-terminal 60 to 65 residues of SM. We also attempted N-terminal sequencing of truncated (HA)3–SM–V5 to precisely pinpoint the truncation site, but a sequence could not be obtained because of N-terminal blockage (data not shown). Although the reason for the blockage is unknown, we note that N-terminal glutamine residues can prevent protein sequencing (26), and the 60 to 65 region of SM contains residue Gln-62. Based on the aforementioned size estimations, we concluded that truncation of SM eliminates part of the cholesterol-sensing SM-N100 regulatory domain but retains the full C-terminal catalytic domain.
Given that SM truncation does not yield an intact N-terminal fragment (Fig. 3A), we hypothesized that this process requires the proteasome and, by extension, the ERAD of SM. ERAD effectors involved in the proteasomal degradation of SM include the AAA+-type ATPase VCP, the E3 ubiquitin ligase MARCHF6 and its associated E2 ubiquitin–conjugating enzyme Ube2J2, and unidentified deubiquitinases (19, 21). Treating HEK293T cells with VCP, proteasome, or deubiquitinase inhibitors blocked the NB-598–induced accumulation of trunSM (Fig. 3C), confirming that a functional ERAD pathway and the proteasome are required for truncation. To corroborate this finding, we performed siRNA-mediated knockdown of ERAD effectors. Knockdown of VCP also impaired NB-598–induced accumulation of trunSM, whereas UBE2J2 or MARCHF6 knockdown had no effect (Fig. 3D, left). However, we noted that in the absence of NB-598, all three knockdowns greatly reduced the basal truncation of SM (Fig. 3D, right; expressed as the ratio between trunSM and full-length SM levels). This suggested that while Ube2J2 and MARCHF6 are the major E2 and E3 proteins required for SM truncation under normal conditions, other proteins can compensate for their absence during NB-598–stimulated truncation. This contrasted with the apparent absolute requirement for VCP, a core component of ERAD. A lysosome-dependent route for SM degradation has been proposed (9); however, inhibitors of lysosomal acidification had no effect on trunSM formation (Fig. S2B), further supporting an ERAD-dependent mechanism.
As ERAD requires substrate ubiquitination, typically at lysine residues, we next tested whether a ubiquitin signal controls truncation. We predicted that this signal would occur within the SM-N100 regulatory domain, given that SM is truncated at its N-terminus. While mutation of Lys-82 and the published ubiquitination site Lys-90 (27) slightly reduced the truncation of the (HA)3–SM–V5 construct, a more marked effect was observed upon combined mutation of the Lys-82/90/100 cluster (Fig. 3E and Fig. S2C), implying functional redundancy amongst these residues. This reduction in truncation was not compounded by additional mutation of Lys-15/16, residues located nearer to the SM N-terminus, suggesting that Lys-82, Lys-90, and Lys-100 are most critical for truncation. We previously showed that threonine, serine, and cysteine residues within SM-N100 contribute to the cholesterol-induced degradation of SM, with Ser-83 serving as a noncanonical ubiquitination site (20) (Fig. S2C). However, mutating these residues did not affect truncation (Fig. 3E). Given that lysine residues within the SM-N100 domain are not required for cholesterol-induced degradation of SM (5, 20), this indicated that proteasomal truncation of SM depends on a distinct ubiquitin signal.
SM truncation depends on an intrinsically disordered region and the stability of the catalytic domain
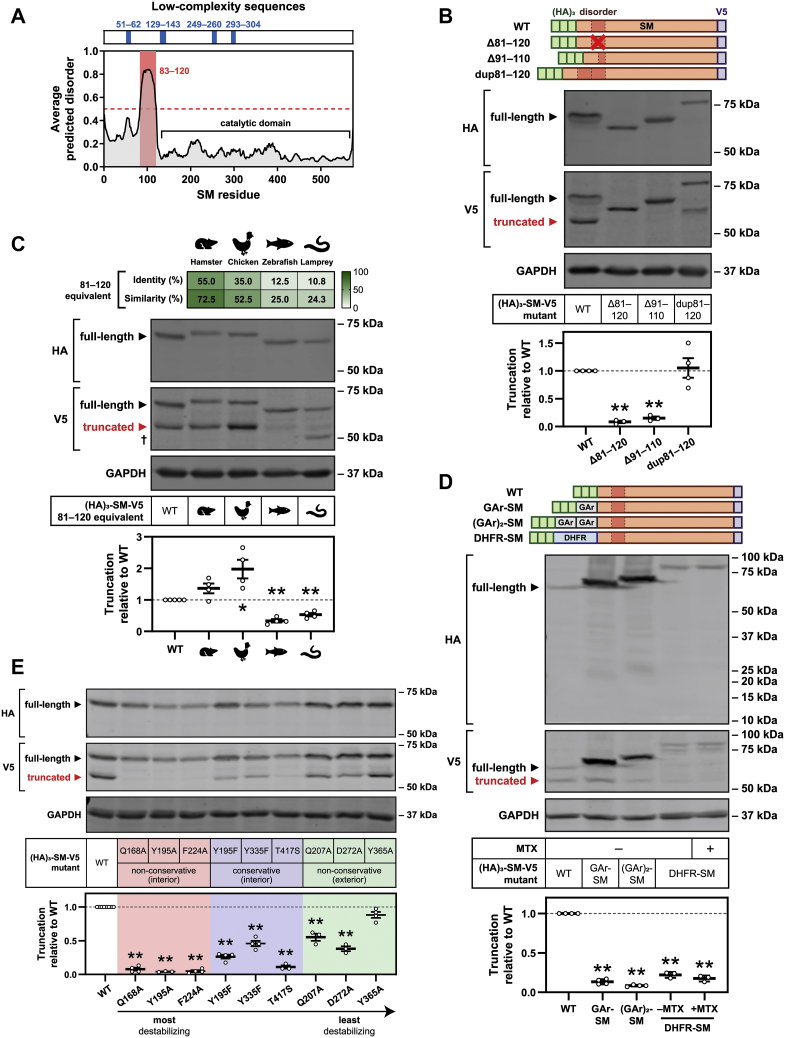
Few other substrates of partial proteasomal degradation are known. However, two features are associated with truncation: (1) a low-complexity sequence (28) or (2) high intrinsic disorder (29). In both cases, the region must be adjacent to a tightly folded domain that is resistant to proteasomal unfolding and degradation, allowing an opportunity for substrate release (28, 29, 30). To investigate whether these features could account for SM truncation, we analyzed the SM protein sequence using predictors of sequence complexity and intrinsic disorder. Four short regions of low sequence complexity were found throughout SM, including one within the SM-N100 domain (residues 51–62; Fig. 4A). Deletion of this region (Δ50–60) slightly reduced the truncation of the (HA)3–SM–V5 construct but did not alter the size of the truncated fragment (Fig. S3A), further supporting the idea that truncation occurs after residue 60. More strikingly, we identified a high intrinsically disordered region between residues 83 and 120 (Fig. 4A), adjacent to the estimated truncation site. By contrast, residues in the C-terminal direction of this region, comprising the SM catalytic domain, were highly ordered.
Figure 4.
SM truncation depends on an intrinsically disordered region and the stability of the catalytic domain.A, low-complexity regions (blue) and intrinsically disordered regions (red) within the SM protein sequence. B, C, and E, HEK293T cells were transfected with the indicated constructs for 24 h and refreshed in maintenance medium for 24 h. C, dagger indicates an additional non-trunSM fragment. D, HEK293T cells were transfected with the indicated constructs for 24 h, refreshed in maintenance medium for 16 h, and treated in the presence or the absence of 10 μM methotrexate (MTX) for 8 h. Lysates were separated by 4 to 15% gradient Tris–glycine SDS-PAGE. B–E, graphs depict densitometric quantification of (HA)3–SM–V5 truncation normalized to the WT construct, which was set to 1 (dotted line). Data are presented as mean ± SEM from n ≥ 3 independent experiments (∗p ≤ 0.05; ∗∗p ≤ 0.01, two-tailed paired t test versus WT). DHFR, dihydrofolate reductase; GAr, glycine-alanine repeat; HEK293T, human embryonic kidney 293T; SM, squalene monooxygenase; trunSM, truncated SM.
Supporting the importance of the disordered region in partial degradation of SM, its deletion (Δ81–120) abolished truncation (Fig. 4B). Halving the length of the disordered region (Δ91–110) also prevented truncation, whereas tandem duplication of the disordered region (dup81–120) had little effect, implying that a minimum length of intrinsic disorder is required. We also noted that the apparent molecular weight of the truncated fragment increased when the disordered region was duplicated, suggesting that the truncation site remained unchanged despite the extended disorder length. As the region corresponding to residues 81 to 120 is highly disordered in SM orthologs from Chinese hamster, chicken, zebrafish, and sea lamprey, despite their differing levels of sequence conservation (Fig. 4C and Fig. S3, B and C), we next tested the effect of substituting these regions into human SM. Truncation was maintained or even increased in constructs derived from Chinese hamster and chicken and approximately halved in constructs derived from zebrafish and sea lamprey SM (Fig. 4C). The persistence of truncation in all four mutant constructs indicated that the intrinsically disordered nature of the 81 to 120 region is sufficient to promote truncation, although sequence-specific features may contribute.
The proteasome typically engages and initiates degradation from intrinsically disordered regions of its substrates (31). Therefore, we considered whether residues 81 to 120 of SM are an internal proteasomal engagement site that results in preferential degradation of the N-terminus. A similar mechanism has been reported for other truncation substrates (30). To test this, we generated N-terminal fusions of SM with two proteins that impede proteasomal processivity: a 30-amino acid glycine–alanine repeat from the Epstein-Barr virus nuclear antigen-1 (32), or dihydrofolate reductase, which becomes tightly folded and resistant to degradation upon the binding of its ligand methotrexate (33). We reasoned that if degradation were initiated internally, these fusions would not prevent truncation but rather protect the N-terminus from complete degradation. However, we found that fusion with glycine–alanine repeat sequences dramatically ablated truncation, and we were unable to recover N-terminal fragments of the expected molecular weight (10–15 kDa; Fig. 4D). Fusion with dihydrofolate reductase similarly reduced truncation, and its further stabilization by methotrexate did not rescue the N-terminus from degradation. This indicated that partial proteasomal degradation of SM is initiated from the N-terminus rather than an internal site. To support this conclusion, we further manipulated the SM N-terminus by sequentially removing HA epitope tags from the (HA)3–SM–V5 construct. These tags have a propensity for intrinsic disorder (34) and may enhance proteasomal engagement at the N-terminus. As expected, their deletion led to a stepwise reduction in SM truncation (Fig. S3D), confirming that truncation proceeds from the N-terminus.
All known examples of partial proteasomal degradation require a tightly folded domain adjacent to the truncation site. Given that the SM catalytic domain has a compact structure (35) and is predicted to be highly ordered (Fig. 4A), we considered the possibility that its stability is also essential for truncation. Supporting this idea was our earlier observation that NB-598 treatment rapidly accumulates trunSM (Fig. 1B). NB-598 is a potent and tight-binding inhibitor of SM that strongly stabilizes the catalytic domain (35), likely increasing its resistance to proteasomal unfolding. To examine the inverse situation, we generated point mutations within the catalytic domain based on the crystal structure of SM (35). Our rationale was that nonconservative substitutions in the interior of the domain would be more destabilizing than conservative substitutions, which would in turn be more destabilizing than mutations on the exterior of the domain. As expected, most of the substitutions significantly reduced SM truncation, and those that were both internal and nonconservative tended to have a larger effect than those that were conservative or external (Fig. 4E). Of note, the nonconservative mutation of Tyr-195 (Y195A) ablated truncation to a greater extent than its conservative equivalent (Y195F). This strongly supported the idea that catalytic domain stability is required for partial degradation. Taken together, our data indicate that the truncation of SM depends on two major structural features: the 81 to 120 disordered region and the stability of the SM catalytic domain.
trunSM has an altered ER membrane topology and retains SM activity
We next sought to confirm that trunSM retains the ER localization of full-length SM. Fractionation of HEK293T cell lysates revealed that like full-length SM (5), trunSM is membrane associated (Fig. 5A). However, a greater proportion of trunSM was found in the cytoplasmic fraction compared with full-length SM, particularly in the absence of NB-598. This suggested that trunSM is more loosely bound to the membrane than full-length SM, possibly because of the loss of the SM-N100 re-entrant loop (residues ~15 to 40). To investigate this further, membranes were isolated and treated with aqueous buffer (control), 1% SDS (solubilizing), 0.1 M Na2CO3 (high pH), or 1 M NaCl (high salt). Solubilizing conditions disrupt the membrane association of all membrane proteins, whereas high-pH or high-salt conditions release peripheral membrane proteins (in the latter case, those associated via electrostatic interactions) (36, 37). Both full-length SM and trunSM remained membrane associated under aqueous or high-salt conditions and were released into the supernatant fraction under solubilizing conditions (Fig. 5B). A similar distribution was observed for SM-N100–GFP-V5, a fusion construct that contains the SM-N100 re-entrant loop (18). However, unlike full-length SM and SM-N100–GFP-V5, the membrane association of trunSM was readily disrupted by high-pH conditions. This suggested that the loss of the SM-N100 re-entrant loop renders trunSM a peripheral ER membrane protein.
Figure 5.
trunSM has an altered ER membrane topology and retains SM activity.A, HEK293T cells were treated in the presence or the absence of 1 μM NB-598 for 8 h, and cytosolic (C) or membrane (M) fractions were isolated. Graph depicts the proportion of overall protein (C + M) found in the membrane fraction. Data are presented as mean ± SEM from n = 4 independent experiments (∗∗p ≤ 0.01, two-tailed paired t test versus SM). B, HEK293T cells were transfected with pTK–SM-N100–GFP-V5 for 24 h and refreshed in maintenance medium for a further 24 h. Membrane fractions were isolated and treated as indicated, followed by collection of pellet (P) and supernatant (S) fractions. Immunoblot is representative of n = 3 independent experiments. C, simplified schematic of the cholesterol synthesis pathway. Acetyl-CoA is converted to squalene by enzymes including HMG-CoA reductase (HMGCR), the target of statins. SM catalyzes the epoxidation of squalene to form monooxidosqualene (MOS), which is converted to cholesterol by lanosterol synthase (LSS) and other downstream enzymes. When SM activity is high or LSS activity is inhibited by Ro 48-8071, SM converts MOS to dioxidosqualene (DOS), the precursor of a shunt pathway producing 24,25-epoxycholesterol (24,25-EC). [14C]-acetate can feed into the cholesterol synthesis pathway at the level of acetyl-CoA. D, CHO-7 cells were transfected in the presence or the absence of Sqle siRNA for 24 h, refreshed in sterol-depletion medium, and transfected with the indicated constructs for 16 h. Cells were then treated with 100 nM Ro 48-8071 for 4 h. Immunoblot is representative of n = 3 independent experiments. Note that immunoblotting for endogenous SM also detects ectopic constructs. E, CHO-7 cells were treated as described in (C) and in addition labeled with [14C]-acetate during the 4 h Ro 48-8071 treatment. Cells were then assayed for the synthesis of [14C]-squalene, [14C]-MOS, or [14C]-DOS. Graph depicts quantification of [14C]-MOS:[14C]-DOS (substrate:product) ratios normalized to the condition with the highest ratio, as we have done previously (19), and adjusted for the expression of cotransfected SM constructs. Data are presented as mean ± SEM from n = 3 independent experiments. ER, endoplasmic reticulum; HEK293T, human embryonic kidney 293T; trunSM, truncated SM.
The stability and cholesterol resistance of trunSM (Fig. 1C), along with its ER localization and the preservation of the entire SM catalytic domain following truncation (Fig. 3B), suggested that it is a constitutively active form of SM. Supporting this idea, previous studies have found that SM lacking the SM-N100 regulatory domain retains catalytic activity (35, 38). To independently verify trunSM activity, we used siRNA to deplete hamster Sqle (SM) in CHO-7 cells and transfected these cells with human full-length ([HA]3–SM–V5) or truncated (SM[ΔN65]–V5) SM constructs. We then assayed SM activity by labeling cells with [14C]-acetate, a precursor for cholesterol synthesis, and treating them with the oxidosqualene cyclase inhibitor Ro 48-8071 to block downstream conversion of SM products and enable their detection (Fig. 5C). In mock-transfected cells, endogenous SM converted labeled acetate to the first SM product, MOS, and then to DOS, which is formed from MOS by a second round of SM-catalyzed epoxidation (Fig. 5, D and E). Knockdown of Sqle ablated DOS synthesis and accumulated MOS and the SM substrate squalene, consistent with reduced SM activity. This activity was rescued by co-transfection with either (HA)3–SM–V5 or SM(ΔN65)–V5, as indicated by the reappearance of DOS. Importantly, when accounting for the different expression levels of the protein constructs (Fig. 5D), SM(ΔN65)–V5 restored activity to a similar extent as (HA)3–SM–V5. This confirmed that trunSM retains full SM activity and is therefore constitutively active.
Discussion
Feedback regulation of SM protein levels is conferred by its lipid-sensing SM-N100 domain, which contains structural elements required for cholesterol-induced degradation. In this study, we characterized a truncated form of SM that arises directly from full-length SM through partial proteolysis of the SM-N100 domain. This renders trunSM long lived, cholesterol resistant, and therefore constitutively active. Truncation requires ERAD and the proteasome and depends on two structural features of SM: intrinsic disorder within the 81 to 120 region and the stability of the adjacent catalytic domain. Furthermore, the loss of a membrane-embedded region at the N-terminus causes trunSM to adopt a peripheral association with the ER membrane. These findings establish a new mechanism controlling the abundance and activity of SM, with likely consequences for the homeostatic regulation of cholesterol synthesis.
Proteasomal truncation of SM
The SM-N100 domain contains two cholesterol-sensing elements that enable accelerated degradation: a re-entrant loop spanning residues ~15 to 40 that undergoes a conformational change in the presence of excess cholesterol (18) and a membrane-associated amphipathic helix from residues 62 to 73 that is ejected from the ER membrane under similar conditions (17). Truncation of the N-terminal 60 to 65 amino acids of SM (Fig. 3B) eliminates the re-entrant loop and is likely to disrupt or partially degrade the amphipathic helix, accounting for the longevity and cholesterol resistance of trunSM (Fig. 1C). This reinforces the importance of these two structural features for the metabolic regulation of full-length SM, as they have largely been studied only in the context of the isolated SM-N100 domain (17, 18). Loss of the membrane-embedded re-entrant loop also renders trunSM a peripheral membrane protein (Fig. 5B) bound to the ER via two C-terminal helices (35). Proteomic studies have shown that SM partitions to lipid droplets (39, 40), and it is possible that the peripheral membrane association of trunSM makes it more compatible with the lipid droplet monolayer than full-length SM. This possibility warrants further consideration given the constitutive activity of trunSM and lipid droplet localization of lanosterol synthase, the cholesterol synthesis enzyme immediately downstream of SM (39, 40).
In experiments with epitope-tagged SM, we were unable to recover fragments containing the N-terminal 60 to 65 amino acids of SM (with a predicted molecular weight of 10–11 kDa; Fig. 3A), suggesting that the region undergoes complete proteolysis during truncation. Through pharmacological and genetic approaches, we confirmed that truncation, like cholesterol-regulated degradation of SM (5), occurs through proteasomal ERAD and requires Ube2J2, MARCHF6, and VCP (Figs. 3, C and D and 6). However, the exact mechanism differs. Truncation is not stimulated by cholesterol (Fig. 1C), depends on a cluster of lysine residues (Lys-82/90/100) that are dispensable for cholesterol regulation (5, 20), and is independent of atypical cholesterol-dependent ubiquitination sites within SM-N100 (Fig. 3E) (20). Instead, truncation may occur for a subset of SM molecules undergoing a basal degradation route. This is supported by our finding that upon stabilization of SM by NB-598, complete degradation ceases and all SM molecules become truncated (Fig. 1B). Indeed, truncation may be a relatively rare event in the absence of NB-598, but the dramatically different stabilities of full-length and trunSM lead to an equilibrium where their protein levels are comparable. Combined with the saturation of ERAD machinery, this may explain why overexpressed SM-V5 is less truncated than endogenous SM (Fig. S3D) despite the two proteins having identical N termini. Along similar lines, we previously found that overexpressed SM exhibits blunted cholesterol regulation (5).
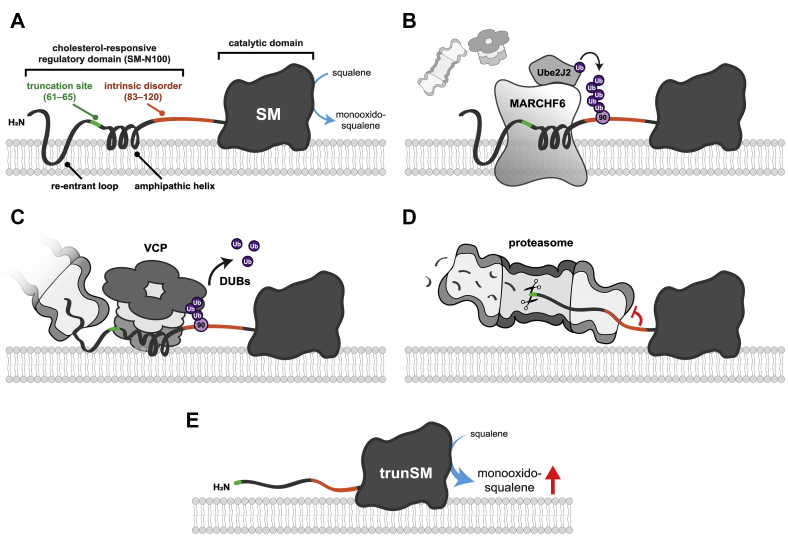
Figure 6.
Model for the mechanism of SM truncation.A, full-length SM comprises the SM-N100 regulatory domain, containing a cholesterol-sensing re-entrant loop and amphipathic helix, and the C-terminal catalytic domain that converts squalene to monooxidosqualene. B, Ube2J2 and MARCHF6 ubiquitinate the SM-N100 domain, likely at the known ubiquitination site Lys-90. C, VCP is recruited to ubiquitinated SM and extracts the SM-N100 domain from the ER membrane, allowing the proteasome to begin degrading SM from its N-terminus. Deubiquitinases (DUBs) are required for this process. D, the SM 81 to 120 disordered region impedes the unfolding of the adjacent and highly stable catalytic domain by the proteasome, preventing further degradation. E, the undegraded portion of SM (trunSM, with a new N-terminus between residues 60 and 65) is released from the proteasome. The loss of the SM-N100 re-entrant loop and disruption of the SM-N100 amphipathic helix renders trunSM resistant to cholesterol-induced degradation and therefore constitutively active. ER, endoplasmic reticulum; SM, squalene monooxygenase; VCP, valosin-containing protein.
The membrane association of trunSM (Fig. 5A) implies that the proteasome acts on SM directly at the ER without the need for cytosolic chaperones. While proteasomal recruitment to the ER has been described, this is generally in the context of interaction with the Sec61 translocon to export and degrade misfolded polypeptides (41). One notable exception is the degradation of the yeast cadmium exporter Pca1p, in which interaction between the ubiquitinated substrate and the proteasome requires Doa10p (the ortholog of MARCHF6) and is bridged by Cdc48p (the ortholog of VCP) (42). A similar pathway may be required for the truncation of SM and presumably extends to its basal or even cholesterol-induced degradation. Precedent for the latter is found in the sterol-induced degradation of Hmg2p and its mammalian equivalent HMGCR (another rate-limiting enzyme of cholesterol synthesis), which also involves direct interaction with the proteasome at the ER membrane (42, 43). In the case of SM, the AAA+ ATPase VCP likely provides the driving force to extract the membrane-associated components of the SM-N100 domain for degradation, although MARCHF6 may also contribute given the retrotranslocase function of Doa10p (44). The role of deubiquitinases in SM truncation is less clear but may involve the removal of ubiquitin chains from the Lys-82/90/100 cluster to enable processing by VCP or entry into the proteasome (45, 46). Ubiquitination of multiple residues in this cluster is seemingly required for maximal truncation (Fig. 3E), which may explain why a peptide containing Lys-90 alone was not enriched in a study that used VCP inhibition to accumulate and capture ERAD substrates (47). Interestingly, proteasomal substrates with K63-linked ubiquitin chains are degraded less efficiently than those with the more typical K48 linkages (48), but whether SM is modified in this way is unknown.
Structural determinants of truncation
By establishing SM as a substrate of partial proteasomal degradation, we identify the first eukaryotic enzyme found to be truncated in this manner. Of the few other reported substrates, almost all are soluble transcription factors, and only two occur in mammalian cells: NF-κB subunit p105 (24) and the Hedgehog signaling transducer Gli3 (25, 49). Depending on the substrate, proteasomal truncation may be activating or inhibitory. In the case of NF-κB, the proteasome degrades an inhibitory domain sequestering the protein to the cytoplasm, thereby enabling its nuclear translocation (50). Conversely, the transactivation domain of Gli3 is degraded to yield a dominant-negative repressor (25). In yeast, partial degradation of Spt23p and Mga2p liberates them from the ER membrane to activate lipogenic gene expression (51). Proteasomal truncation can also have profound consequences at the organismal level: partial degradation of Gli3 or the Drosophila proteins Lola29M and Shavenbaby regulates processes including sex determination, differentiation, and stem cell maintenance (25, 29, 52, 53). As with these examples, the truncation of SM eliminates a regulatory region (cholesterol-sensing elements of the SM-N100 domain) and yields a fragment with altered properties (constitutive activity and a peripheral membrane association).
For most truncation substrates, including NF-κB (54), Gli3 (55), and Def1p (56), partial degradation requires a two-part signal: a low-complexity sequence (such as a glycine-rich or glutamine-rich region) and an adjacent tightly folded domain. The 19S regulatory particle of the proteasome is thought to poorly transduce an unfolding force when occupied by a low-complexity sequence, preventing disassembly of the folded domain and allowing substrate escape (28, 57). However, we ruled out a low-complexity sequence as the major determinant of SM truncation (Fig. S3A). Unique mechanisms have been described for the remaining substrates, with the exception of Lola29M where the structural features enabling truncation are unknown (52). Spt23p and Mga2p are truncated upon proteasomal engagement at an internal hairpin loop, which leads to preferential degradation of the C-terminus rather than the tightly folded N-terminal domain (30). Such a mechanism is unlikely to control SM truncation, which is initiated at the N-terminus (Fig. 4D). In the case of Shavenbaby, its degraded portion is a large (∼550 residue) intrinsically disordered domain juxtaposed with high predicted order in the adjacent and undegraded domain (29). This most closely resembles SM, where truncation depends on the disordered 81 to 120 region (Fig. 4B), but the much shorter length of this region clearly represents a distinct signal.
On the other hand, the presence of a tightly folded and degradation-resistant domain is essential for all known examples of partial degradation. SM is no exception, and changes to the stability of its catalytic domain (via NB-598 binding or disruptive point mutations) are closely correlated with truncation (Figs. 1B and 4E). The split-domain structure of SM, in which substrate-binding and cofactor-binding regions are interspersed throughout its primary sequence (35), may produce a compact conformation that is unusually resistant to unfolding and degradation. Proximity of the catalytic domain to the ER membrane is presumably required for its interaction with the hydrophobic substrate squalene and may sterically hinder engagement by VCP or the proteasome. Our findings thus reinforce the importance of a stable domain for disrupting proteasomal processivity, while also establishing a unique counterpart in the bipartite truncation signal: the 81 to 120 disordered region.
How does the 81 to 120 region promote truncation? The distance from the center of the proteasomal 20S core particle to the edge of the 19S regulatory particle is ∼200 Å (58), equivalent to ∼60 residues of a fully extended polypeptide chain. Therefore, as proteasomal degradation of SM reaches the predicted truncation site between residues 60 and 65, the 81 to 120 region is translocating through the regulatory particle, and the catalytic domain is on the periphery of the complex. The conspicuous spacing of these three elements suggests that the 81 to 120 region is a proteasomal ‘stop signal' that impedes further unfolding and incorporation of SM, analogous to the low-complexity sequences of other substrates (Fig. 6). This may explain why a minimum length of disorder is required for truncation (Fig. 4B), as well as the need for both the disordered region and the catalytic domain stability. Without the former, the proteasome successfully transduces the force necessary to unfold the catalytic domain, whereas without the latter, proteasomal ATPases are still capable of disassembling the catalytic domain in their inhibited state. In both cases, complete degradation is the result (Fig. 4, B and E). It is noteworthy that the SM truncation site is unchanged when residues on the N-terminal side of the disorder are removed (Fig. S3A) or when the disorder is lengthened (Fig. 4B), supporting the notion that the proteasome releases trunSM upon first encountering the 81 to 120 region.
Ubiquitination within residues 81 to 120 is unlikely to be solely responsible for its effect on truncation, given that the lowly truncated Δ91-110 mutant (Fig. 4B) retains both Lys-82 and Lys-90. Likewise, Lys-90 and Lys-100 are not conserved in highly truncated hamster and chicken orthologs (Fig. 4C). One possibility is that these sequences contain other lysine residues that serve as alternative ubiquitination sites. However, the number of lysine residues within orthologous regions (Fig. S3B) does not correlate with the extent of truncation, and partial degradation is observed for a lamprey-derived sequence containing only one lysine residue. This strongly suggests that the intrinsically disordered nature of the region promotes truncation, but further work is required to elucidate the precise mechanism. Interestingly, the partial degradation of Spt23p and NF-κB is augmented by their homodimerization with a full-length counterpart, which protects the rescued fragment from complete proteolysis (59, 60). Disordered regions often provide an interface for protein–protein interaction (61), and so a similar process may control SM truncation. It is currently unknown if SM forms dimers in vivo, but the transmembrane microprotein CASIMO1 is a confirmed interactor of SM (62), and glycogen synthase kinase 3 beta (GSK-3β) was recently reported to associate with the isolated SM-N100 domain (63). Given that GSK-3β is a soluble protein and much of SM-N100 is membrane associated, the hydrophilic 81 to 100 region is a strong candidate for a binding site. Both CASIMO1 and GSK-3β are linked with metabolism (62, 64), warranting study into whether they impact on the production of the constitutively active trunSM.
Consequences for cholesterol synthesis
trunSM retains full SM activity (Fig. 5D), and the levels of full-length SM and trunSM are similar across a range of human cell types (Fig. 1A). Therefore, two enzyme pools are maintained: one that is rapidly turned over and regulated by cholesterol availability, and another that is stable and constitutively active. As SM is a rate-limiting cholesterol synthesis enzyme, this would allow a basal level of pathway flux that can be fine tuned by metabolic demand. It is striking that amongst distant vertebrate orthologs of SM, there is strong structural conservation of the intrinsically disordered region (Fig. S3B) and sequence conservation within the catalytic domain (5), both of which are required for truncation. Indeed, truncation of SM occurs in CHO-7 cells (Fig. S1A) and is likely characteristic of higher eukaryotes possessing the SM-N100 domain. In these organisms, the steady-state cholesterol synthesis enabled by trunSM may better meet the demands of multicellular organisms.
The presence of the cholesterol-resistant trunSM may also prevent the complete ablation of SM activity under conditions of cholesterol excess, which would otherwise delay resumption of cholesterol synthesis or lead to dramatic accumulation of the substrate squalene (5). Whilst previously considered a cholesterol synthesis intermediate with few biochemical properties, squalene is protective against cell death induced by lipid peroxidation (11), yet cytotoxic when it is unable to be effectively sequestered to lipid droplets (10, 65). Increased squalene levels also accompany the dermal and gastrointestinal side effects of pharmacological SM inhibition in mammals (12). Therefore, a persistent population of trunSM may be advantageous for clearing excess squalene. Along similar lines, squalene itself stabilizes SM by binding to the SM-N100 domain and blunting its cholesterol-induced degradation (15). It remains to be determined if squalene-mediated stabilization also influences SM truncation, given that both processes depend on MARCHF6 (Fig. 3D) and the truncation-inducing NB-598 also causes dramatic squalene accumulation (15). It is also conceivable that trunSM is relevant in pathophysiological contexts, given that dysregulation of cholesterol synthesis is implicated in hepatocellular carcinoma (9) and prostate cancer (66). However, this possibility awaits future investigation.
Experimental procedures
Reagents and cell lines
Fetal calf serum (FCS), newborn calf serum, high-glucose Dulbecco's modified Eagle's medium (DMEM-HG), glutamine/methionine/cystine-free DMEM-HG, RPMI 1640, DMEM/Ham's Nutrient Mixture F-12 (DF-12), penicillin/streptomycin, Opti-MEM reduced-serum medium, RNAiMAX transfection reagent, Lipofectamine 3000 transfection reagent, TRI reagent, and the SuperScript III First-Strand Synthesis kit were from Thermo Fisher Scientific. Lipoprotein-deficient FCS (FCLPDS [30 mg/ml protein] and lipoprotein-deficient serum [50 mg/ml protein]) were prepared from FCS and newborn calf serum, respectively, as described previously (67). Primers, siRNA, and protease inhibitor cocktail were from Sigma–Aldrich. The SensiMix SYBR No-ROX kit was from Bioline. SmaI was from New England Biolabs. Tris-glycine SDS-PAGE gels were prepared inhouse. Immobilon Western chemiluminescent horseradish peroxidase substrate and nitrocellulose membranes were from Millipore. The Click-&-Go Protein Reaction Buffer Kit was from Click Chemistry Tools, and PureProteome Streptavidin Magnetic Beads were from Merck. The QIAquick PCR purification kit was from Qiagen. TransIT-2020 was from Mirus Bio. PBS was from the University of New South Wales (UNSW). Skim milk powder was from Fonterra, and bovine serum albumin (BSA) was from Bovogen Biologicals. Chemicals were from the following suppliers: ALLN (A6185; Sigma–Aldrich), ammonium chloride (31; Ajax Finechem), l-azidohomoalanine (1066; Click Chemistry Tools), bafilomycin A1 (B1793; Sigma–Aldrich), β-mercaptoethanol (M3148; Sigma–Aldrich), bromophenol blue (B0126; Sigma–Aldrich), butylated hydroxytoluene (B1378; Sigma–Aldrich), [14C]-acetate sodium salt (PerkinElmer; catalog no. NEC084H001MC), CB-5083 (16276; Cayman Chemical Company), cholesterol complexed with methyl-β-cyclodextrin (Chol/CD; C4951; Sigma–Aldrich), chloroform (102444; Supelco), cycloheximide (C7698; Sigma–Aldrich), l-cysteine (168149; Sigma–Aldrich), diethyl ether (code: EA036; Chem-Supply), dimethyl sulfoxide (DMSO) (2225; Ajax Finechem), ethanol (214; Ajax Finechem), glacial acetic acid (100063; Supelco), GlutaMAX (M4667; Thermo Fisher), glycerol (242; Ajax Finechem), glycine (1083; Ajax Finechem), Hepes (54457; Sigma–Aldrich), heptane (LC1078; RCI LabScan), hexane (104369; Supelco), hydrochloric acid (256; Ajax Finechem), IGEPAL CA-630 (I8896; Sigma–Aldrich), magnesium chloride (296; Ajax Finechem), methanol (318; Ajax Finechem), l-methionine (M9625; Sigma–Aldrich), methotrexate (13960; Cayman Chemical Company), mevalonolactone (mevalonate; M4667; Sigma–Aldrich), mevastatin (compactin; M2537; Sigma–Aldrich), MG132 (C2211; Sigma–Aldrich), NB-598 (CS-1274; Chemscene), PEG4 carboxamide-propargyl biotin (1266; Click Chemistry Tools), polyethyleneimine (03880; Sigma–Aldrich), Ponceau S solution (P7170; Sigma–Aldrich), potassium chloride (383; Ajax Finechem), potassium hydroxide (405; Ajax Finechem), PR-619 (16276; Cayman Chemical Company), Ro 48-8071 (10006415; Cayman Chemical Company), sodium carbonate (463; Ajax Finechem), sodium chloride (465; Ajax Finechem), sodium deoxycholate (D6750; Sigma–Aldrich), SDS (75746; Sigma–Aldrich), sodium EDTA (180; Ajax Finechem), sodium EGTA (E8145; Sigma–Aldrich), sodium hydroxide (482; Ajax FineChem), sodium orthovanadate (S6508; Sigma–Aldrich), sodium pyruvate (11360070; Thermo Fisher), Tris (2311; Ajax Finechem), Tween-20 (P9416; Sigma–Aldrich), and WP1130 (15277; Cayman Chemical Company).
HEK293T cells were a gift from the UNSW School of Medical Sciences, HepG2 and Huh7 cells were gifts from the Centre for Cardiovascular Research (UNSW), Be(2)-C and HeLaT cells were gifts from Drs Louise Lutze-Mann and Noel Whitaker (UNSW), and CHO-7 cells were a gift from Drs Joseph Goldstein and Michael Brown (UT Southwestern Medical Center).
Cell culture
Cells were maintained in a humidified incubator at 37 °C and 5% CO2 in maintenance medium (DMEM-HG [HEK293T, HepG2, Huh7, and Be(2)-C], RPMI 1640 [HeLaT] or DF-12 [CHO-7]; 10% [v/v] FCS for human cells or 5% [v/v] lipoprotein-deficient serum for CHO-7 cells; 100 U/ml penicillin; and 100 μg/ml streptomycin). To improve HEK293T and HepG2 surface adhesion, culture vessels were treated with 25 μg/ml polyethyleneimine in PBS for 15 min prior to cell seeding. Plasmid and siRNA transfections were performed in maintenance medium lacking penicillin and streptomycin. Sterol depletion of HEK293T cells was performed in medium containing 10% (v/v) FCLPDS, 5 μM compactin, and 50 μM mevalonate. Combined methionine and sterol depletion of HEK293T cells was performed in glutamine/methionine/cystine-free DMEM-HG containing 10% (v/v) FCLPDS, 100 U/ml penicillin, 100 μg/ml streptomycin, 4 mM GlutaMAX, 0.16 mM l-cysteine, 1 mM sodium pyruvate, 5 μM compactin, and 50 μM mevalonate. Sterol depletion of CHO-7 cells was performed in maintenance medium containing 5 μM compactin and 50 μM mevalonate. For all treatments, appropriate solvent controls were used (water [Chol/CD, ammonium chloride]; DMSO [cycloheximide, NB-598, l-azidohomoalanine, CB-5083, MG132, ALLN, bafilomycin A1, PR-619, WP1130, methotrexate, Ro 48-8071, compactin]; and ethanol [mevalonate]), and the final concentration of DMSO or ethanol did not exceed 0.2% (v/v) in cell culture medium. Treatments were delivered in full-medium refreshes, and all experiments were 72 h in duration.
Plasmids
A pcDNA3.1/V5-His TOPO expression vector (Invitrogen) encoding the protein-coding sequence of human SM (NM_003129.4) fused with three N-terminal HA tags, a C-terminal linker sequence, and C-terminal V5 and 6× His tags ([HA]3–SM–V5) was generated previously in our laboratory by Dr Julian Stevenson. Codon-optimized nucleotide sequences encoding orthologs of human SM-N100 were previously obtained from GenScript (17), and sequences encoding orthologs of human SM residues 101 to 120 were derived using the Integrated DNA Technologies codon optimization tool. Domain insertions and deletions within the (HA)3–SM–V5 construct were generated using the polymerase-incomplete primer extension cloning method and sequence- and ligation-independent cloning method (68, 69), and domain and nucleotide substitutions were generated using the overlap extension cloning method (70), as described previously (71). To generate standards for the absolute quantification of mRNA levels, quantitative PCR products were amplified from HEK293T cDNA and inserted into the pGL3-Basic vector (Promega) using the overlap extension cloning method (70). The identity of all plasmids was confirmed via Sanger dideoxy sequencing. The plasmids used in this study are listed in Table S1, and the primer sequences used for DNA cloning are listed in Table S2.
Protein harvest, SDS-PAGE, and immunoblotting
To quantify protein levels, cells were seeded into 12-well plates and treated as specified in the legends to the figures. Total protein was harvested in 2% SDS lysis buffer (10 mM Tris–HCl [pH 7.6], 100 mM NaCl, 2% [w/v] SDS, and 2% [v/v] protease inhibitor cocktail), passed through a 21-gauge needle until homogenous, and vortexed for 20 min. Lysate protein content was quantified using the bicinchoninic acid assay (Thermo Fisher Scientific), and sample concentrations were normalized by dilution in 2% SDS lysis buffer and 1× Laemmli buffer (50 mM Tris–HCl [pH 6.8], 2% [w/v] SDS, 5% [v/v] glycerol, 0.04% [w/v] bromophenol blue, and 1% [v/v] β-mercaptoethanol). Samples were heated at 95 °C for 5 min and separated by 10% (w/v) Tris-glycine SDS-PAGE, unless otherwise specified in the legends to the figures. Proteins were electroblotted onto nitrocellulose membranes and blocked in 5% (w/v) skim milk powder (Diploma) in PBS with 0.1% (v/v) Tween-20 (PBST) or in 5% (w/v) BSA in PBST for FLAG detection.
Immunoblotting was performed using rabbit polyclonal anti-SQLE (12544-1-AP; Proteintech; 1:2500 at 4 °C for 16 h), rabbit monoclonal anti-GAPDH (2118; Cell Signaling Technology; 1:2500 at 4 °C for 16 h), rabbit monoclonal anti-HA (3724; Cell Signaling Technology; 1:2000 at 4 °C for 16 h), mouse monoclonal anti-V5 (R960-25; Invitrogen; 1:5000 at room temperature for 1 h), rabbit monoclonal antivinculin (ab129002; Abcam; 1:2000 at room temperature for 1 h), rabbit polyclonal anti-FLAG (F7425; Millipore; 1:10,000 at 4 °C for 16 h), IRDye 680RD donkey anti-rabbit IgG (LCR-926-68073; LI-COR Biosciences; 1:5000 [SM detection] or 1:10,000 at room temperature for 1 h), IRDye 800CW donkey antimouse IgG (LCR-926-32212; LI-COR Biosciences; 1:10,000 at room temperature for 1 h), peroxidase-conjugated AffiniPure donkey anti-rabbit IgG (711-035-152; Jackson ImmunoResearch Laboratories; 1:10,000 at room temperature for 1 h), and peroxidase-conjugated AffiniPure donkey antimouse IgG (715-035-150; Jackson ImmunoResearch Laboratories; 1:10,000 at room temperature for 1 h). All antibodies were diluted in 5% (w/v) BSA in PBST, except for anti-FLAG and peroxidase-conjugated antibodies, which were diluted in 5% (w/v) skim milk in PBST. Fluorescence-based detection of SM, GAPDH, HA, V5, and vinculin was performed using an Odyssey Clx imager (LI-COR Biosciences), and enhanced chemiluminescence-based detection of FLAG was performed using Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore) and an ImageQuant LAS 500 imager (Cytiva Life Sciences). Because of low protein levels following streptavidin pulldown or differential solubilization of microsomal membranes, enhanced chemiluminescence was used to detect SM, V5, and GAPDH in these samples. Densitometry analysis of fluorescence images was performed using Image Studio Lite, version 5.2.5 (LI-COR Biosciences).
Nascent protein synthesis assay
To detect newly synthesized SM, HEK293T cells were seeded into 6 cm dishes and treated as specified in the legends to the figures. Cells were then starved in methionine-depletion and sterol-depletion medium for 1 h, followed by labeling of newly synthesized proteins with 50 μM l-azidohomoalanine (72) or treatment with 50 μM l-methionine as an unlabeled specificity control, in methionine-depletion and sterol-depletion medium for 4 h. After removal of methionine-depletion and sterol-depletion medium, cells were treated as specified in the legends to the figures in maintenance medium containing a 20-fold excess of l-methionine (2 mM) to outcompete residual l-azidohomoalanine. Cells were then lysed in click-chemistry lysis buffer (50 mM Tris–HCl [pH 8.0], 150 mM NaCl, 1% [w/v] SDS, and 2% [v/v] protease inhibitor cocktail). Lysates in dishes were incubated on ice for 30 min, scraped and passed through a 21-gauge needle until homogenous, and vortexed for 5 min. Lysates were then centrifuged at 16,000g and 4 °C for 5 min, the supernatant was collected, and its protein content was quantified using the bicinchoninic acid assay. Equal quantities of protein were taken from each sample and mixed with 1× Laemmli buffer for immunoblotting as an input control. From each sample, an additional 250 μg of protein was taken for click-chemistry conjugation of l-azidohomoalanine-labeled proteins with PEG4 carboxamide–propargyl biotin using the Click-&-Go Protein Reaction Buffer Kit (Click Chemistry Tools).
Precipitated proteins were resuspended in 500 μl radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris–HCl [pH 7.4], 150 mM NaCl, 0.1% [w/v] SDS, 1% [w/v] IGEPAL CA-630, 0.5% [w/v] sodium deoxycholate, 1 mM sodium orthovanadate, and 1 mM sodium EDTA), passed through a 23-gauge needle 50 times to shear the protein pellet, and vortexed for 30 min. After centrifugation at 16,000g for 5 min to remove the remaining insoluble protein, concentrations of solubilized protein were quantified using the bicinchoninic acid assay and normalized by dilution in RIPA buffer. Entire sample volumes were mixed with 20 μl of equilibrated PureProteome Streptavidin Magnetic Beads and rotated at 4 °C overnight. Beads were washed with RIPA buffer three times by rotating at 4 °C for 1 h, 30 min, and 15 min in succession. Bound proteins were eluted by resuspending beads in 50 μl elution buffer (0.4 vol RIPA buffer, 0.4 vol 10% SDS, 1× Laemmli buffer, and 25 mM biotin) and heating at 70 °C for 10 min, with intermittent vortexing. Entire eluates and input controls were analyzed by immunoblotting.
siRNA and plasmid transfection
To downregulate gene expression or transiently overexpress SM-derived constructs, cells were seeded into 12-well plates. The next day, cells were refreshed in antibiotic-free medium and transfected with 15 pmol siRNA using RNAiMAX (Invitrogen; 15 pmol siRNA: 2 μl reagent) or 1 μg expression vector using Lipofectamine 3000 (Invitrogen; 1 μg DNA: 2 μl reagent with 2 μl P3000 supplemental reagent), delivered in Opti-MEM. After 24 h, cells were refreshed in maintenance medium and treated as specified in the legends to the figures. To cotransfect siRNA and plasmids, cells were seeded into 6-well plates and transfected with 37.5 pmol siRNA using RNAiMAX as described previously. After 24 h, cells were refreshed in antibiotic-free medium and transfected with 2.5 μg expression vector using Lipofectamine 3000 as described previously. After 16 h, cells were refreshed in maintenance medium and treated as specified in the legends to the figures. The siRNAs used in this study are listed in Table S3.
RNA harvest, reverse transcription, and quantitative PCR
To quantify SQLE gene expression, HEK293T cells were seeded in triplicate into 12-well plates and transfected with siRNA as specified in the legends to the figures. Total RNA was harvested using TRI reagent (Sigma–Aldrich), and polyadenylated RNA was reverse transcribed using the SuperScript III First Strand Synthesis kit (Invitrogen). cDNA products were used as the template for quantitative RT-PCR using the SensiMix SYBR No-ROX kit (Bioline). For relative quantification of gene expression, mRNA levels were normalized to the porphobilinogen deaminase (PBGD) housekeeping gene using the comparative CT method (73) and adjusted relative to the control siRNA condition, as specified in the legends to the figures. For absolute quantification of SQLE expression, plasmids containing qRT-PCR amplicon sequences were linearized by digestion with SmaI for 1 h and purified using the QIAquick PCR purification kit (Qiagen). Linearized plasmids were quantified using spectrophotometry, serially diluted in nuclease-free water to concentrations of between ~5 × 102 and ~5 × 108 target copies/μl, and used as the template for qRT-PCR in triplicate as described previously. A standard curve of log(target sequence copies) versus CT value was generated and compared with CT values from cDNA samples to quantify gene expression. Data were expressed in units of cDNA molecules per microgram of reverse-transcribed RNA. The primer sequences used for qRT-PCR in this study are listed in Table S4.
Cell fractionation and differential solubilization
To examine protein membrane association, HEK293T cells were seeded into 10 cm dishes and treated as specified in the legends to the figures. Microsomal membranes were isolated as described (74) with some modifications. Briefly, cells were scraped in cold PBS, pelleted at 1000g and 4 °C for 5 min, and lysed in 500 μl buffer F1 (10 mM Hepes-KOH [pH 7.4], 10 mM KCl, 1.5 mM MgCl2, 5 mM sodium EDTA, 5 mM sodium EGTA, 250 mM sucrose, and 2% [v/v] protease inhibitor cocktail). Lysates were centrifuged at 1000g and 4 °C for 10 min, and the supernatant was centrifuged at 20,000g and 4 °C for 30 min. The 20,000g supernatant was collected and designated the cytosolic fraction. The 20,000g pellet was resuspended in 100 μl buffer F2 (10 mM Tris–HCl [pH 7.4], 100 mM NaCl, 1% [w/v] SDS, and 2% [v/v] protease inhibitor cocktail) and designated the membrane fraction. Protein content was quantified using the bicinchoninic acid assay (Thermo Fisher Scientific), and sample concentrations were normalized by dilution in buffer F1 or buffer F2, plus 1× Laemmli buffer, for immunoblotting analysis.
To determine the peripheral or integral nature of protein membrane association, differential solubilization of microsomal membranes was performed as described (74) with some modifications. Briefly, cells were seeded into 14.5 cm dishes and transfected with 40 μg pTK–SM-N100–GFP-V5 expression vector using TransIT-2020 (Mirus Bio; 1 μg DNA: 2 μl reagent), delivered in Opti-MEM. After 24 h, cells were refreshed in maintenance medium for a further 24 h, and microsomal membranes were isolated as described previously. Equivalent volumes of membrane preparations (20 μl) were treated with 200 μl buffer F1, 1% (w/v) SDS (with 10 mM Tris–HCl [pH 7.4]), 0.1 M Na2CO3 (pH 11.5), or 1 M NaCl (with 10 mM Tris–HCl [pH 7.4]), and incubated at 4 °C with end-over-end mixing for 30 min. Mixtures were then centrifuged at 20,000g and 4 °C for 30 min. The soluble supernatant fraction was collected, and the insoluble pellet fraction was resuspended in 200 μl buffer F3 (buffer F1 containing 100 mM NaCl). Equal volumes of supernatant and pellet fractions were mixed with 1× Laemmli buffer for immunoblotting analysis.
SM activity assay
To assay SM activity, CHO-7 cells were seeded into 6-well plates and treated as specified in the legends to the figures. Activity was determined by thin layer chromatography as described previously (5, 75), with minor modifications. Briefly, cells were metabolically labeled for 4 h in maintenance medium containing 1 μCi/well [14C]-acetic acid sodium salt and 100 nM Ro 48-8071. Cells were lysed in 1 ml 0.05 M sodium hydroxide, and protein concentrations were quantified using the bicinchoninic acid assay, and then normalized by dilution in 0.05 M sodium hydroxide. Lysates were saponified in 1 ml 100% (v/v) ethanol, 500 μl 75% (w/v) potassium hydroxide, 1 μl 20 mM butylated hydroxytoluene, and 20 μl 20 mM EDTA, at 70 °C for 1 h. Once cooled to room temperature, lipids were extracted from saponified lysates by mixing with 1 ml 100% (v/v) ethanol and 2.5 ml of hexane, centrifuging at 2000g for 5 min, and evaporating 2 ml of the upper organic phase to dryness. Lipids were redissolved in 50 μl hexane and separated via thin layer chromatography on TLC Silica gel 60 F254 plates using a heptane: diethyl ether: glacial acetic acid (60:40:1, v/v) mobile phase. Silica plates were dried, exposed to phosphor screens for 5 to 7 days, and imaged using the Typhoon FLA 9500 (GE Healthcare). Densitometry analysis was performed using Image Studio Lite, version 5.2.5.
Sequences and alignments
DNA sequences of protein-coding SQLE isoforms (fullSQLE, NM_003129.4; trunSQLE1, ENST00000523430.5; trunSQLE2, XM_011517246.2) were obtained from the RefSeq-annotated (GRCh38.p13 109.20200228) and GENCODE-annotated (GRCh38.p13 GCA_000001405.28) human genomes. Protein sequences of human SM (Homo sapiens, Q14534), Chinese hamster SM (Cricetulus griseus, A0A3L7IPT3), chicken SM (Gallus gallus, A0A1D5NWK3), zebrafish SM (Danio rerio, F1QDN5), sea lamprey SM (Petromyzon marinus, S4R6S3), and yeast Erg1p (Saccharomyces cerevisiae, P32476) were obtained from the UniProt database (76). Protein sequence complexity was predicted using the SEG (77), CAST (78), and fLPS (79) algorithms, and regions identified by all three tools were defined as low-complexity sequences. Protein intrinsic disorder was predicted using the online tools SPOT-dis2 (80), MFDp2 (81), AUCpreD (82), IUPred2A (83), DISOPRED3 (84), PrDOS (85), and DisProt (VL2E) (86), and residues with an average intrinsic disorder probability of >0.5 were defined as intrinsically disordered. Protein sequence alignments were generated using Geneious Basic, version 2020.1 (Biomatters Ltd) with a BLOSUM62 cost matrix.
Data analysis and presentation
Data were normalized as described in the legends to the figures. Data visualization and statistical testing were performed using GraphPad Prism, version 8.4 (GraphPad Software, Inc) as specified in the legends to the figures. Thresholds for statistical significance were defined as ∗p ≤ 0.05 and ∗∗p ≤ 0.01. Schematics and figures were assembled using Adobe Illustrator, version 24.1 (Adobe, Inc).
Data availability
All described data are contained within this article.
Supporting information
This article contains supporting information (5, 17, 20, 27, 68, 69, 70).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Julian Stevenson for generating the pCMV–(HA)3–SM–V5 plasmid used in this study, Dr Ngee Kiat (Jake) Chua for insightful discussions, and the members of the Brown laboratory for critically reviewing this article.
This work was supported by Australian Research Council grant DP170101178 and a NSW Health Investigator Development grant.
Author contributions
H. W. C. and A. J. B. conceptualization; H. W. C., I. M. C.-H., and A. J. B. methodology; H. W. C. and I. M. C.-H. investigation; H. W. C. and I. M. C.-H. writing—original draft; H. W. C., I. M. C.-H., and A. J. B. writing—review and editing; A. J. B. supervision; A. J. B. project administration; and A. J. B. funding acquisition.
Funding and additional information
H. W. C. is a recipient of an Australian Research Training Program scholarship, and I. M. C.-H. is a recipient of a UNSW Sydney Scientia PhD Scholarship.
Edited by George DeMartino
Supporting information
References
- 1.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 2.Prospective Studies Collaboration Blood cholesterol and vascular mortality by age, sex, and blood pressure: A meta-analysis of individual data from 61 prospective studies with 55 000 vascular deaths. Lancet. 2007;370:1829–1839. doi: 10.1016/S0140-6736(07)61778-4. [DOI] [PubMed] [Google Scholar]
- 3.Kuzu O.F., Noory M.A., Robertson G.P. The role of cholesterol in cancer. Cancer Res. 2016;76:2063–2070. doi: 10.1158/0008-5472.CAN-15-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe V., Sharpe L.J., Alexopoulos S.J., Kunze S.V., Chua N.K., Li D., Brown A.J. Cholesterol homeostasis: How do cells sense sterol excess? Chem. Phys. Lipids. 2016;199:170–178. doi: 10.1016/j.chemphyslip.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Gill S., Stevenson J., Kristiana I., Brown A.J. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011;13:260–273. doi: 10.1016/j.cmet.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Belter A., Skupinska M., Giel-Pietraszuk M., Grabarkiewicz T., Rychlewski L., Barciszewski J. Squalene monooxygenase - a target for hypercholesterolemic therapy. Biol. Chem. 2011;392:1053–1075. doi: 10.1515/BC.2011.195. [DOI] [PubMed] [Google Scholar]
- 7.Brown D.N., Caffa I., Cirmena G., Piras D., Garuti A., Gallo M., Alberti S., Nencioni A., Ballestrero A., Zoppoli G. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci. Rep. 2016;6:19435. doi: 10.1038/srep19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stopsack K.H., Gerke T.A., Sinnott J.A., Penney K.L., Tyekucheva S., Sesso H.D., Andersson S.-O.O., Andrén O., Cerhan J.R., Giovannucci E.L., Mucci L.A., Rider J.R. Cholesterol metabolism and prostate cancer lethality. Cancer Res. 2016;76:4785–4790. doi: 10.1158/0008-5472.CAN-16-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D., Wong C.C., Fu L., Chen H., Zhao L., Li C., Zhou Y., Zhang Y., Xu W., Yang Y., Wu B., Cheng G., Lai P.B.-S., Wong N., Sung J.J. Squalene epoxidase drives NAFLD-induced hepatocellular carcinoma and is a direct pharmaceutical target. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aap9840. [DOI] [PubMed] [Google Scholar]
- 10.Mahoney C.E., Pirman D., Chubukov V., Sleger T., Hayes S., Fan Z.P., Allen E.L., Chen Y., Huang L., Liu M., Zhang Y., McDonald G., Narayanaswamy R., Choe S., Chen Y. A chemical biology screen identifies a vulnerability of neuroendocrine cancer cells to SQLE inhibition. Nat. Commun. 2019;10:96. doi: 10.1038/s41467-018-07959-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Bermudez J., Baudrier L., Bayraktar E.C., Shen Y., La K., Guarecuco R., Yucel B., Fiore D., Tavora B., Freinkman E., Chan S.H., Lewis C., Min W., Inghirami G., Sabatini D.M. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature. 2019;567:118–122. doi: 10.1038/s41586-019-0945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagaraja R., Olaharski A., Narayanaswamy R., Mahoney C., Pirman D., Gross S., Roddy T.P., Popovici-Muller J., Smolen G.A., Silverman L. Preclinical toxicology profile of squalene epoxidase inhibitors. Toxicol. Appl. Pharmacol. 2020;401:115103. doi: 10.1016/j.taap.2020.115103. [DOI] [PubMed] [Google Scholar]
- 13.Horton J.D., Shah N.A., Warrington J.A., Anderson N.N., Park S.W., Brown M.S., Goldstein J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howe V., Sharpe L.J., Prabhu A.V., Brown A.J. New insights into cellular cholesterol acquisition: Promoter analysis of human HMGCR and SQLE, two key control enzymes in cholesterol synthesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:647–657. doi: 10.1016/j.bbalip.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Yoshioka H., Coates H.W., Chua N.K., Hashimoto Y., Brown A.J., Ohgane K. A key mammalian cholesterol synthesis enzyme, squalene monooxygenase, is allosterically stabilized by its substrate. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7150–7158. doi: 10.1073/pnas.1915923117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nathan J.A. Squalene and cholesterol in the balance at the ER membrane. Proc. Natl. Acad. Sci. U. S. A. 2020;117:8228–8230. doi: 10.1073/pnas.2003388117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chua N.K., Howe V., Jatana N., Thukral L., Brown A.J. A conserved degron containing an amphipathic helix regulates the cholesterol-mediated turnover of human squalene monooxygenase, a rate-limiting enzyme in cholesterol synthesis. J. Biol. Chem. 2017;292:19959–19973. doi: 10.1074/jbc.M117.794230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe V., Chua N.K., Stevenson J., Brown A.J. The regulatory domain of squalene monooxygenase contains a re-entrant loop and senses cholesterol via a conformational change. J. Biol. Chem. 2015;290:27533–27544. doi: 10.1074/jbc.M115.675181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelcer N., Sharpe L.J., Loregger A., Kristiana I., Cook E.C., Phan L., Stevenson J., Brown A.J.A.J. The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme A reductase and the cholesterol synthesis pathway. Mol. Cell. Biol. 2014;34:1262–1270. doi: 10.1128/MCB.01140-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua N.K., Hart-Smith G., Brown A.J. Non-canonical ubiquitination of the cholesterol-regulated degron of squalene monooxygenase. J. Biol. Chem. 2019;294:8134–8147. doi: 10.1074/jbc.RA119.007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chua N.K., Scott N.A., Brown A.J. Valosin-containing protein mediates the ERAD of squalene monooxygenase and its cholesterol-responsive degron. Biochem. J. 2019;476:2545–2560. doi: 10.1042/BCJ20190418. [DOI] [PubMed] [Google Scholar]
- 22.Stevenson J., Luu W., Kristiana I., Brown A.J. Squalene mono-oxygenase, a key enzyme in cholesterol synthesis, is stabilized by unsaturated fatty acids. Biochem. J. 2014;461:435–442. doi: 10.1042/BJ20131404. [DOI] [PubMed] [Google Scholar]
- 23.Honsho M., Dorninger F., Abe Y., Setoyama D., Ohgi R., Uchiumi T., Kang D., Berger J., Fujiki Y. Impaired plasmalogen synthesis dysregulates liver X receptor-dependent transcription in cerebellum. J. Biochem. 2019;166:353–361. doi: 10.1093/jb/mvz043. [DOI] [PubMed] [Google Scholar]
- 24.Fan C.-M., Maniatis T. Generation of p50 subunit of NF-kB by processing of p105 through an ATP-dependent pathway. Nature. 1991;354:395–398. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- 25.Wang B., Fallon J.F., Beachy P.A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 26.Mozdzanowski J. Deblocking of proteins containing N-terminal pyroglutamic acid. In: Smith B.J., editor. Protein Sequencing Protocols. Humana Press; Totowa, NJ: 2003. pp. 365–369. [PubMed] [Google Scholar]
- 27.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian L., Holmgren R.A., Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-κB. Nat. Struct. Mol. Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- 29.Zanet J., Benrabah E., Li T., Pélissier-Monier A., Chanut-Delalande H., Ronsin B., Bellen H.J., Payre F., Plaza S. Pri sORF peptides induce selective proteasome-mediated protein processing. Science. 2015;349:1356–1358. doi: 10.1126/science.aac5677. [DOI] [PubMed] [Google Scholar]
- 30.Piwko W., Jentsch S. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat. Struct. Mol. Biol. 2006;13:691–697. doi: 10.1038/nsmb1122. [DOI] [PubMed] [Google Scholar]
- 31.Prakash S., Tian L., Ratliff K.S., Lehotzky R.E., Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 32.Levitskaya J., Sharipo A., Leonchiks A., Ciechanover A., Masucci M.G. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston J.A., Johnson E.S., Waller P.R.H., Varshavsky A. Methotrexate inhibits proteolysis of dihydrofolate reductase by the N-end rule pathway. J. Biol. Chem. 1995;270:8172–8178. doi: 10.1074/jbc.270.14.8172. [DOI] [PubMed] [Google Scholar]
- 34.Georgieva M.V., Yahya G., Codó L., Ortiz R., Teixidó L., Claros J., Jara R., Jara M., Iborra A., Gelpí J.L., Gallego C., Orozco M., Aldea M. Inntags: Small self-structured epitopes for innocuous protein tagging. Nat. Methods. 2015;12:955–958. doi: 10.1038/nmeth.3556. [DOI] [PubMed] [Google Scholar]
- 35.Padyana A.K., Gross S., Jin L., Cianchetta G., Narayanaswamy R., Wang F., Wang R., Fang C., Lv X., Biller S.A., Dang L., Mahoney C.E., Nagaraja N., Pirman D., Sui Z. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat. Commun. 2019;10:97. doi: 10.1038/s41467-018-07928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feramisco J.D., Goldstein J.L., Brown M.S. Membrane topology of human Insig-1, a protein regulator of lipid synthesis. J. Biol. Chem. 2004;279:8487–8496. doi: 10.1074/jbc.M312623200. [DOI] [PubMed] [Google Scholar]
- 37.Fujiki Y., Hubbard L., Fowler S., Lazarow P.B. Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laden B.P., Tang Y., Porter T.D. Cloning, heterologous expression, and enzymological characterization of human squalene monooxygenase. Arch. Biochem. Biophys. 2000;374:381–388. doi: 10.1006/abbi.1999.1629. [DOI] [PubMed] [Google Scholar]
- 39.Pataki C.I., Rodrigues J., Zhang L., Qian J., Efron B., Hastie T., Elias J.E., Levitt M., Kopito R.R. Proteomic analysis of monolayer-integrated proteins on lipid droplets identifies amphipathic interfacial α-helical membrane anchors. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E8172–E8180. doi: 10.1073/pnas.1807981115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bersuker K., Peterson C.W.H.W.H., To M., Sahl S.J.J., Savikhin V., Grossman E.A.A., Nomura D.K.K., Olzmann J.A.A. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev. Cell. 2018;44:97–112. doi: 10.1016/j.devcel.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng W., Sergeyenko T., Zeng N., Brown J.D., Römisch K. Characterization of the proteasome interaction with the Sec61 channel in the endoplasmic reticulum. J. Cell Sci. 2007;120:682–691. doi: 10.1242/jcs.03351. [DOI] [PubMed] [Google Scholar]
- 42.Smith N., Adle D.J., Zhao M., Qin X., Kim H., Lee J. Endoplasmic reticulum-associated degradation of Pca1p, a polytopic protein, via interaction with the proteasome at the membrane. J. Biol. Chem. 2016;291:15082–15092. doi: 10.1074/jbc.M116.726265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris L.L., Hartman I.Z., Jun D.J., Seemann J., A. DeBose-Boyd R. Sequential actions of the AAA-ATPase valosin-containing protein (VCP)/p97 and the proteasome 19S regulatory particle in sterol-accelerated, endoplasmic reticulum (ER)- associated degradation of 3-hydroxy-3-methylglutarylcoenzyme A reductase. J. Biol. Chem. 2014;289:19053–19066. doi: 10.1074/jbc.M114.576652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt C.C., Vasic V., Stein A. Doa10 is a membrane protein retrotranslocase in ER-associated protein degradation. Elife. 2020;9 doi: 10.7554/eLife.56945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst R., Mueller B., Ploegh H.L., Schlieker C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao T., Cohen R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 47.Huang E.Y., To M., Tran E., Dionisio L.T.A., Cho H.J., Baney K.L.M., Pataki C.I., Olzmann J.A. A VCP inhibitor substrate trapping approach (VISTA) enables proteomic profiling of endogenous ERAD substrates. Mol. Biol. Cell. 2018;29:1021–1030. doi: 10.1091/mbc.E17-08-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reichard E.L., Chirico G.G., Dewey W.J., Nassif N.D., Bard K.E., Millas N.E., Kraut X.D.A. Substrate ubiquitination controls the unfolding ability of the proteasome. J. Biol. Chem. 2016;291:18547–18561. doi: 10.1074/jbc.M116.720151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C.H., Von Kessler D.P., Park W., Wang B., Ma Y., Beachy P.A. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- 50.Blank V., Kourilsky P., Israël A. Cytoplasmic retention, DNA binding and processing of the NF-kappa B p50 precursor are controlled by a small region in its C-terminus. EMBO J. 1991;10:4159–4167. doi: 10.1002/j.1460-2075.1991.tb04994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H.D., Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 52.Sato K., Ito H., Yokoyama A., Toba G., Yamamoto D. Partial proteasomal degradation of Lola triggers the male-to-female switch of a dimorphic courtship circuit. Nat. Commun. 2019;10:166. doi: 10.1038/s41467-018-08146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bohère J., Mancheno-Ferris A., Al Hayek S., Zanet J., Valenti P., Akino K., Yamabe Y., Inagaki S., Chanut-Delalande H., Plaza S., Kageyama Y., Osman D., Polesello C., Payre F. Shavenbaby and Yorkie mediate Hippo signaling to protect adult stem cells from apoptosis. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-018-07569-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin L., Ghosh S. A glycine-rich region in NF-kappaB p105 functions as a processing signal for the generation of the p50 subunit. Mol. Cell. Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan Y., Wang B. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J. Biol. Chem. 2007;282:10846–10852. doi: 10.1074/jbc.M608599200. [DOI] [PubMed] [Google Scholar]
- 56.Wilson M.D., Harreman M., Taschner M., Reid J., Walker J., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell. 2013;154:983–995. doi: 10.1016/j.cell.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nassif N.D., Cambray S.E., Kraut D.A. Slipping up: Partial substrate degradation by ATP-dependent proteases. IUBMB Life. 2014;66:309–317. doi: 10.1002/iub.1271. [DOI] [PubMed] [Google Scholar]
- 58.Huang X., Luan B., Wu J., Shi Y. An atomic structure of the human 26S proteasome. Nat. Struct. Mol. Biol. 2016;23:778–785. doi: 10.1038/nsmb.3273. [DOI] [PubMed] [Google Scholar]
- 59.Lin L., DeMartino G.N., Greene W.C. Cotranslational dimerization of the Rel homology domain of NF-kappaB1 generates p50-p105 heterodimers and is required for effective p50 production. EMBO J. 2000;19:4712–4722. doi: 10.1093/emboj/19.17.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48UFD1/NPL4, a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 61.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polycarpou-Schwarz M., Groß M., Mestdagh P., Schott J., Grund S.E., Hildenbrand C., Rom J., Aulmann S., Sinn H.P., Vandesompele J., Diederichs S., Jo H.-P.S., Vandesompele J., Diederichs S. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene. 2018;37:4750–4768. doi: 10.1038/s41388-018-0281-5. [DOI] [PubMed] [Google Scholar]
- 63.Jun S.Y., Brown A.J., Chua N.K., Yoon J.-Y., Lee J.-J., Yang J.O.K., Jang I., Jeon S.-J., Choi T.-I., Kim C.-H., Kim N.-S. Reduction of squalene epoxidase by cholesterol accumulation accelerates colorectal cancer progression and metastasis. Gastroenterology. 2021;160:1194–1207.e28. doi: 10.1053/j.gastro.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 64.Patel P., Woodgett J.R. Glycogen synthase kinase 3: A kinase for all pathways? Curr. Top. Dev. Biol. 2017;123:277–302. doi: 10.1016/bs.ctdb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 65.Valachovic M., Garaiova M., Holic R., Hapala I. Squalene is lipotoxic to yeast cells defective in lipid droplet biogenesis. Biochem. Biophys. Res. Commun. 2016;469:1123–1128. doi: 10.1016/j.bbrc.2015.12.050. [DOI] [PubMed] [Google Scholar]
- 66.Stopsack K.H., Gerke T.A., Andrén O., Andersson S.O., Giovannucci E.L., Mucci L.A., Rider J.R. Cholesterol uptake and regulation in high-grade and lethal prostate cancers. Carcinogenesis. 2017;38:806–811. doi: 10.1093/carcin/bgx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goldstein J.L., Basu S.K., Brown M.S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 68.Klock H.E., Lesley S.A. The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. In: Doyle S.A., editor. Vol 498. Humana Press; Totowa, NJ: 2009. pp. 91–103. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 69.Li M.Z., Elledge S.J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 70.Bryksin A.V., Matsumura I. Overlap extension PCR cloning: A simple and reliable way to create recombinant plasmids. Biotechniques. 2010;48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stevenson J., Krycer J.R., Phan L., Brown A.J. A practical comparison of ligation-independent cloning techniques. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dieterich D.C., Link A.J., Graumann J., Tirrell D.A., Schuman E.M. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc. Natl. Acad. Sci. U. S. A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 74.Zerenturk E.J., Sharpe L.J., Brown A.J. DHCR24 associates strongly with the endoplasmic reticulum beyond predicted membrane domains: Implications for the activities of this multi-functional enzyme. Biosci. Rep. 2014;34 doi: 10.1042/BSR20130127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong J., Quinn C.M., Gelissen I.C., Brown A.J. Endogenous 24(S),25-epoxycholesterol fine-tunes acute control of cellular cholesterol homeostasis. J. Biol. Chem. 2008;283:700–707. doi: 10.1074/jbc.M706416200. [DOI] [PubMed] [Google Scholar]
- 76.The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wootton J.C., Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 78.Promponas V.J., Enright A.J., Tsoka S., Kreil D.P., Leroy C., Hamodrakas S., Sander C., Ouzounis C.A. CAST: An iterative algorithm for the complexity analysis of sequence tracts. Bioinformatics. 2000;16:915–922. doi: 10.1093/bioinformatics/16.10.915. [DOI] [PubMed] [Google Scholar]
- 79.Harrison P.M. fLPS: Fast discovery of compositional biases for the protein universe. BMC Bioinformatics. 2017;18:476. doi: 10.1186/s12859-017-1906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hanson J., Yang Y., Paliwal K., Zhou Y. Improving protein disorder prediction by deep bidirectional long short-term memory recurrent neural networks. Bioinformatics. 2017;33:685–692. doi: 10.1093/bioinformatics/btw678. [DOI] [PubMed] [Google Scholar]
- 81.Mizianty M., Peng Z., Kurgan L. MFDp2: Accurate predictor of disorder in proteins by fusion of disorder probabilities, content and profiles. Intrinsically Disord. Proteins. 2013;1 doi: 10.4161/idp.24428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang S., Ma J., Xu J. AUCpreD: Proteome-level protein disorder prediction by AUC-maximized deep convolutional neural fields. Bioinformatics. 2016;32:i672–i679. doi: 10.1093/bioinformatics/btw446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mészáros B., Erdös G., Dosztányi Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones D.T., Cozzetto D. DISOPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ishida T., Kinoshita K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peng K., Radivojac P., Vucetic S., Dunker A.K., Obradovic Z. Length-dependent prediction of protein in intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All described data are contained within this article.