Abstract
Heterosis has historically been exploited in plants; however, its underlying genetic mechanisms and molecular basis remain elusive. In recent years, due to advances in molecular biotechnology at the genome, transcriptome, proteome, and epigenome levels, the study of heterosis in vegetables has made significant progress. Here, we present an extensive literature review on the genetic and epigenetic regulation of heterosis in vegetables. We summarize six hypotheses to explain the mechanism by which genes regulate heterosis, improve upon a possible model of heterosis that is triggered by epigenetics, and analyze previous studies on quantitative trait locus effects and gene actions related to heterosis based on analyses of differential gene expression in vegetables. We also discuss the contributions of yield-related traits, including flower, fruit, and plant architecture traits, during heterosis development in vegetables (e.g., cabbage, cucumber, and tomato). More importantly, we propose a comprehensive breeding strategy based on heterosis studies in vegetables and crop plants. The description of the strategy details how to obtain F1 hybrids that exhibit heterosis based on heterosis prediction, how to obtain elite lines based on molecular biotechnology, and how to maintain heterosis by diploid seed breeding and the selection of hybrid simulation lines that are suitable for heterosis research and utilization in vegetables. Finally, we briefly provide suggestions and perspectives on the role of heterosis in the future of vegetable breeding.
Subject terms: Plant breeding, Gene regulation
Introduction
Heterosis occurs in a variety of species and has been observed and recorded in China since ancient times. For example, Jia Sixie described in “The Manual of Important Arts for the People” that interbreeding between horses and donkeys produced stronger mules, and the famous agricultural work “Tian Gong Kai Wu” also recorded crossbreeding techniques for silkworms. Heterosis has also been extensively studied in other countries. In 1763, the German scholar Koelreuter1 was the first to present concrete evidence that the growth of hybrid tobacco is superior to that of its parents. By comparing the height of hybrid and self-crossing offspring in maize, Darwin2 found that the average height of hybrid offspring was higher than that of self-crossing offspring. Beal3 found that the yield of maize hybrid offspring was greater than that of both parents. Shull4,5 observed heterosis in maize hybrid offspring and first proposed the concept of heterosis; he then formally named this phenomenon “heterosis.” Heterosis was first applied to genetic breeding in maize, and many excellent maize hybrids have been produced since the 1930s. Since 2011, the yield of maize increased by at least eightfold in America, due mostly to the cultivation of hybrids6.
As heterosis has been applied in cereal crop production, crossbreeding in vegetables has also rapidly progressed. Under natural planting conditions, 40–80% of seeds produced are usually hybrids due to fertilization competition between self-pollination and pollen from other plants7. Although the traits of randomly generated hybrid seeds are not organized at first, F1 hybrids exhibit higher yield, better adaptability, and higher stress resistance than pure line seeds under optimum production and fertilization protection management conditions. Therefore, farmers have paid much attention to the cultivation of hybrid seeds8. The first hybrid of eggplant (Solanum melongena) was released in 19249. Subsequently, hybrids of other vegetables, such as watermelon (Citrullus lanatus L.), cucumber (Cucumis sativus L.), radish (Raphanus sativus L.), tomato (Solanum lycopersicum L.), and cabbage (Brassica oleracea L.), were developed over the next 20 years7. The number of hybrid vegetable varieties is rapidly increasing, at a rate of 8–10% each year, while nonhybrid vegetable varieties are gradually being eliminated10.
The application of heterosis to vegetable cultivation was first proposed by Hayes and Jones11 using cucumbers. However, because of the high cost of producing hybrid seeds, hybrid cucumber seeds were not used until the 1930s7. Similarly, self-pollination and the occasional presence of indehiscent anthers in eggplant12 and styles that are shorter than anthers in tomato13 have resulted in a high degree of self-pollination, which in turn has limited hybrid utilization. Pearson (1933) and Jones and Clarke (1943) used the mechanisms of self-incompatibility in cabbage and cytoplasmic male sterility in onion, respectively, to produce pure line and hybrid seeds on a large scale8. To avoid undesirable selfing, various genetic and nongenetic mechanisms, including genic male sterility, cytoplasmic male sterility, self-incompatibility, gynoecious lines, auxotrophy, and the use of sex regulators and chemical hybridizing agents, have been applied to facilitate hybrid seed production in vegetables8,14. The various traits that exhibit remarkable heterosis in F1 hybrids, including yield, earliness, growth vigor, and stress tolerance15–18, have become a major area of research on vegetables. In an experiment with hybrid eggplant conducted by Balwani et al.19 and Makani et al.20 heterosis in the optimal F1 hybrid resulted in yield increases of 125.78% and 88.88%, respectively. A more productive eggplant hybrid will effectively decrease the time to first harvest18. Transgressive phenotypes have also been observed in other Solanaceae21,22, Cruciferae23,24, and Cucurbitaceae vegetables25,26.
Although heterosis in vegetables has historically been used in research and crossbreeding experiments, its genetic mechanism remains elusive. Different genetic models for heterosis have been described in various reviews27–31. However, it is apparent that the classical genetic hypothesis of heterosis cannot explain all mechanisms of heterosis. Therefore, genetic models of heterosis have been included in this review. In addition to genetic models, we also present a schematic diagram depicting the involvement of epigenetics in heterosis. Simultaneously, we discuss studies on heterosis at the molecular level based on QTL effects and differential gene expression analyses. We also describe the effects of QTL on heterosis in crop plants based on Shang et al.32 to guide future research studies on the genetic mechanisms of heterosis. We summarize recent findings on the interactions of QTL sites with regard to heterosis and discuss the contribution of various QTL effects to heterosis. Differential expression analysis of genes related to heterosis can also provide a different perspective on heterosis31. In addition, we present morphological improvement as another measure to increase yield and an important component of breeding7 and describe how to combine heterosis utilization and morphological improvement.
To date, studies on heterosis in vegetables mainly involve obtaining F1 hybrids through crossbreeding. The utilization of cucumber hybrids proposed by Hayes and Jones11 was likely the first instance of effective vegetable breeding that exploits heterosis. Kumar et al.30 introduced methods of predicting heterosis in eggplant hybrids, such as genetic distance prediction and combining ability tests, and proposed the application of a sterile line system as well as transgenic and gene editing techniques in eggplant breeding. Herath et al.33 summarized the QTL mapping of yield-related traits in chili, introduced the use of heterosis breeding to improve the economic and agronomic traits of chili, and suggested the use of genomic technology and sterile line materials in chili breeding. Mallikarjunarao et al.34 reviewed the progress of various balsam pear (bitter gourd) hybridization tests and indicated that heterosis does occur in the yield of balsam pear hybrids. However, studies on the genetic mechanisms of heterosis in vegetables are limited, which hinders the application of heterosis in vegetable breeding. Therefore, in this review, we describe the progress of research on the genetic mechanisms of heterosis, analyze the use of hybrid production systems and molecular biology technology in vegetable production, and propose a breeding strategy that can predict, obtain, and maintain heterosis. This review will provide a reference for the utilization of heterosis in vegetable breeding.
Study on the genetic mechanisms of heterosis
Genetic regulation of heterosis
Heterosis is a complex biogenetic phenomenon caused by the combination of many factors that is manifested in the performance of hybrid offspring. The classical hypotheses for the genetic mechanisms of heterosis include the dominance and overdominance hypotheses, which are based on allelic interactions, and epistasis, which is based on nonallelic interactions.
Davenport35 first proposed the dominance hypothesis (Fig. 1A), and Bruce36 and Jones37 developed it further. In the dominant hypothesis, favorable genes controlling growth and development are dominant, and unfavorable genes are recessive. In the hybrid generation, the alleles from the two parents are complementary, and the unfavorable recessive genes are suppressed by the favorable dominant genes; therefore, the hybrid generation exhibits heterobeltiosis.
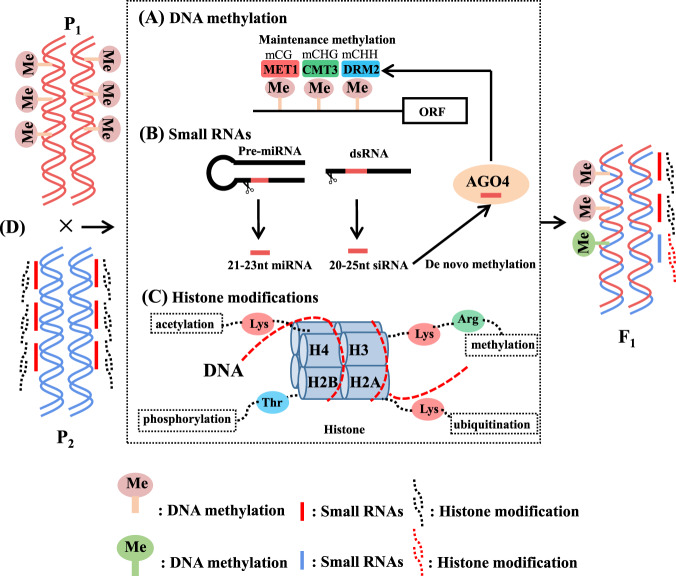
Fig. 1. There are five hypotheses to explain the mechanism of heterosis based on gene effects.
Suppose that the biomass is the sum of the genetic effects (A, B, C) and that the biomass of an organism is represented by the circular area. A Dominance effect: the dominant allele (A) inhibits the recessive allele (a); (B) overdominance effect: a single heterozygous allele (B/B−) promotes the development of heterosis; (C) Epistasis effect: nonallelic (A1/B1) interactions in the parents promote the development of heterosis; (D) active gene effect: genes from parents (C) promote heterosis when heterozygous and produce genome imprinting when homozygous, which inhibits the occurrence of heterosis; (E) gene network system: genes from parents (A, B, C) are combined into a coordinated gene network system that enables F1 to develop heterosis; (F) single-cross hybrids P1 (AB) and P2 (CD) produced from four homozygous inbred tetraploids (with genotypes A, B, C, and D) are crossed to produce F1 (ABCD), a double-cross tetraploid hybrid
The overdominance hypothesis (Fig. 1B) was originally proposed by Shull4 and East38 as the opposite of the dominance hypothesis. This hypothesis denies that there is dominant-recessive relationship between alleles and suggests that the main cause of heterosis is the interaction of heterogeneous alleles from parents. Heterozygous alleles interact more strongly than homozygous alleles; thus, the hybrids exhibit heterobeltiosis. Using the isozyme technique, Dranginis39 found that the enzymes in heterozygotes exhibit many unique conformations of hybrid enzymes. For example, the regulatory proteins of heterozygotes often present as polymers that regulate genes, and different heterozygous and homozygous proteins consistently show different activity characteristics. In addition, the anthocyanin content heterobeltiosis that occurs due to the heterozygosity of a single locus (pl) in maize40 and the yield heterosis induced by the heterozygosity of a single locus (sft) in tomato15 also provide experimental evidence for the overdominance hypothesis. However, the interaction of closely linked alleles can also result in an overdominance effect that is known as pseudo-overdominance41.
The dominance and overdominance hypotheses for the heterosis phenomenon both suggest that heterosis is caused by individual allele loci. However, several reports have shown that plant traits such as yield and growth vigor are complex quantitative traits42. Wright43 visualized the network structure of population genotypes, i.e., multiple loci control the variations in most traits; in such networks, the replacement of anu gene may affect multiple traits. Based on this perspective, Sheridan44 proposed the concept of epistasis. He believed that heterosis may arise from interactions between nonalleles. In genetics, the phenomenon in which the genetic effect of a nonallele deviates from its additive effect is called epistasis (Fig. 1C). The significant special combining ability (SCA) effects in the hybridization experiment of Sao and Mehta indicated that epistasis plays a predominant role in the genetic control of eggplant heterosis45. Using a genetic map that covered the whole rice (Oryza sativa) genome, QTL mapping for yield-related traits was conducted in 250 F2:3 lines. The results showed that the correlation between marker heterozygosity and yield-related traits was low and that the interaction between most genes could not be detected on the basis of single-gene loci; the interactions were classified as dominance by dominance, additive by dominance, and additive by additive46. Therefore, Yu et al.46 also believed that epistasis is an important genetic basis for the development of heterosis.
Other ideas in addition to the classical hypotheses have been proposed. Zhong47 proposed the active gene effect hypothesis (Fig. 1D) by comparing the relationship between genomic imprinting and heterosis; this hypothesis suggests that heterosis is caused by additive effects between the active genes. When alleles are homozygous, only one of them is active. When genes are heterozygous, genomic imprinting does not occur, and all genes are active, showing all effects. The interaction between active genes increases the overall effect of gene expression; as a result, the hybrid exhibits heterosis. For example, in maize, the red1 (r1) gene, when inherited from both parents, causes different colors in corn kernels48. Genomic imprinting affects the differential expression of genes by affecting DNA methylation and histone modification49. Bao50 suggested that individuals have a specific set of genetic information that controls their growth. Genetic information is expressed as different coding genes in organisms; these genes form an orderly network of expression, and the activities of each gene are related to each other. An alteration in a single gene may cause changes in the entire network. The network of F1 hybrids is a new gene network system that is formed from the two different gene networks of the parents. If the interactions between alleles bring the whole genetic network system to an optimal state, the F1 hybrid exhibits heterosis; otherwise, it remains typical (Fig. 1E). In addition, the effects caused by genomic imprinting or active gene effects may be components of genomic dosage effects51; the other part of genomic dosage effects usually caused by polyploidy, which is a specific phenomenon in polyploid plants called progressive heterosis (Fig. 1F)52,53. The genomic dosage effects produced by allopolyploids are usually stronger than those produced by homologous polyploids38,51,54,55. The formation of polyploids is accompanied by extensive genetic and epigenetic changes56, which may provide the molecular basis for the development of heterosis.
Epigenetics is involved in the development of heterosis
Although many hypotheses have been proposed to explain the mechanisms of plant heterosis at the genetic level, studies have shown that the genetic mechanisms of heterosis cannot be fully explained by one or even several hypotheses at the genetic level. Through the intensive study of epigenetics, epigenetic factors such as DNA methylation, small RNAs, and histone modifications have been found to be involved in the development of heterosis in plants57–62.
Epigenetic modifications play an important role in the formation of plant phenotypes by regulating gene transcription and gene expression63–65. Alleles of known phenotypes have been studied more extensively in the context of DNA methylation than in the context of other epigenetic modifications63. RNA-directed de novo methylation (RdDM) is one of the pathways that triggers DNA methylation by 24 nt-siRNA, which is regulated by two key genes, namely, NRPD1 and NRPE166 (Fig. 2A, B). A silent epigenetic variant caused by differentially methylated regions (DMRs) in the promoter, sulfurea (sulf/+), can result in homozygous lethal tomato plants that exhibit only chlorotic leaf sectors64,65. This may occur due to the random combination of genetic information from the parents of the F1 hybrids because their genotypes are more prone to heterozygosity at the DNA methylation level; this is in line with the findings of Shen et al.59. The gene effect caused by such heterozygosity may enable F1 hybrids to avoid producing common phenotypes or hybrid weakness, thus achieving heterobeltiosis. Using experiments involving heterograft eggplants, Cerruti et al.62 found that scion vigor is related to DNA methylation and that the reduction in methylation in the CHH context promotes scion vigor. Tomato grafting experiments revealed that RdDM can cause a heritable enhancement-through-grafting phenotype67,68.
Fig. 2. Putative model of heterosis triggered by epigenetics.
A DNA methylation: De novo methylation was catalyzed by DRM2, a homologous enzyme of DNMT3. In maintenance methylation, CG is catalyzed by MET1, a homologous enzyme of DNMT1; CHG is catalyzed by CMT3; and CHH is still catalyzed by DRM2. B Small RNA: Includes the miRNA produced by premiRNA and the siRNA produced by dsRNA. In general, 24 nt-siRNA mediates de novo DNA methylation catalyzed by the AGO4 protein. C Histone modifications: The modifications of histone amino acid residue includes acetylation, phosphorylation, methylation, and ubiquitination processes. Epigenetic modifications are produced by the parents. New epigenetic modifications may occur in F1 hybrids. D Epigenetic modification status of the parents and F1 hybrid: the increase and decrease in or recombination of epigenetic modifications induces the F1 hybrid to exhibit heterosis
Because de novo DNA methylation is mediated by siRNAs (Fig. 2B), siRNAs may also be involved in the regulation of heterosis. The level of siRNAs decreased in different genome regions between parents and hybrids, but this phenomenon was limited to 24 nt-siRNAs; in contrast, the levels of siRNAs of other sizes did not decrease67. Noncoding small RNAs can be used as signaling molecules in plants67. Shivaprasad et al.61 observed that miR395 is differentially expressed, mediates transgressive phenotypes in the hybrid progeny of tomato and is associated with suppression of the corresponding target genes, which indicates that the combination of parental genetic information can cause differences in miR395 abundance in the progeny. Simultaneously, 21–24 nt small RNAs can move through the intercellular filaments and phloem of the graft site69, and 24 nt sRNAs can guide genomic DNA methylation in recipient cells70; this information provides a theoretical basis for guiding grafting. In addition, sRNAs in plants usually play a major role in inducing gene expression silencing and gene posttranscriptional silencing71,72. This may be due to the downregulation of sRNA levels in hybrids, which lifts the silencing of some favorable genes and thus allows hybrids to exhibit heterobeltiosis71,72.
Different modifications, such as acetylation, phosphorylation, methylation, and ubiquitination, occur at the amino terminus of histones (Fig. 2C). These histone modifications can affect the binding of related proteins to chromatin and thereby affect the transcriptional activity of genes. At the same time, the combination of modifications of the amino terminus of histones expands the genetic information for and changes the phenotype of an individual73. Histone modifications are related to the stability of heterosis. Studies have shown that histone deacetylases cause the nonadditive expression of some genes in hybrids58. In addition, histone acetylation and methylation are related to the activation of regulatory (circadian-regulated) genes in F1 hybrids73. The biological clock controls the physiological activities of plants, including the synthesis of physiological and biochemical substances. Therefore, histone modifications can influence plant biomass heterosis.
The recombination of genetic information from parents may lead to new combinations of epigenetic modifications in the F1 generation (Fig. 2D). Epigenetic modifications essentially affect the expression of genes, causing them to be overexpressed or silenced. Therefore, epigenetic modifications may indirectly influence the development of heterosis in F1 by affecting the expression pattern of genes.
Study on heterosis at the molecular level
Progress in heterosis research based on QTL analysis
The genome contains all the genetic information of a species and determines whether an individual gene is expressed as well as its degree of expression. Heterosis is usually indicated if the hybrid generation is superior to the parents in terms of quantitative traits. Thus, it is essential to conduct a genetic analysis of heterosis from the perspective of the whole genome. With the rapid development of genome sequencing technology, it has become possible to identify gene loci related to heterosis by genome-wide association studies74, which lay a foundation for the study of individual phenotypic differences. This review summarizes the QTL effects on heterosis based on 35 studies that mainly addressed 6 crops and vegetables, i.e., rice (Oryza sativa), maize (Zea mays), cotton (Gossypium hirsutum), oilseed rape (Brassica campestris), sorghum (Sorghum vulgare), and tomato (Solanum lycopersicum) (Table S1). Among the six types of QTL effects, dominance and epistasis had equal proportions (19%, 23%, Fig. 3). Interestingly, the overdominance effect accounted for the largest proportion of all the effects (42%, Fig. 3). This means that although there are many gene loci in the plant genome, these interacted to produce different, complex, hard-to-imitate effects and resulted in heterosis; among these effects, overdominance effects occurred consistently and contributed significantly to heterosis. In addition, the overdominance effect can be conveniently used for artificial breeding, which has been well demonstrated in tomato15. However, efficiently and accurately locating the gene loci that impart the overdominance effect is necessary to make use of this effect. Heterosis may be the result of many traits. In addition, the results of QTL mapping differ among species and even within different groups of the same species75–77. Therefore, it is necessary to select a suitable genetic population based on the genetic background of the plants exhibiting heterosis.
Fig. 3.
Statistical analysis of the effect of quantitative trait loci on crop heterosis. A In the statistical analysis of the effect of quantitative trait loci on crop heterosis, the species and frequency of each species were studied; (B) in the statistical analysis of the effect of quantitative trait loci on crop heterosis, the quantitative trait locus effect on each species and the proportion of each type of effect were analyzed
Advances in gene action related to heterosis based on differential expression analysis of genes
The genome controls the formation of a biological phenotype by regulating the differential expression of genes78,79. Molecular-based expression analyses, such as allele-specific expression, DNA microarray, expression quantitative trait loci, RNA-seq, quantitative SNP-based Sequenom technology, and allele-specific RT-PCR, have made it possible to detect differential gene expression.
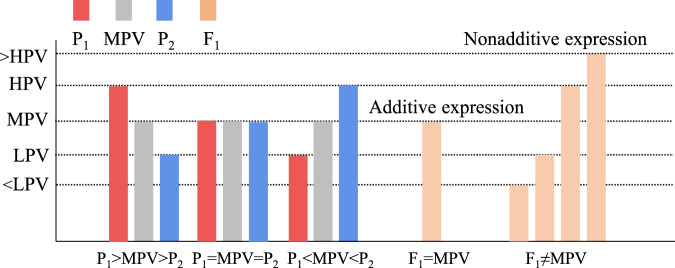
Yield and biomass heterosis in F1 hybrids may occur due to the altered expression patterns of genes that control biological functions such as carbon fixation, glucose metabolism, and circadian rhythm80. Gene Ontology (GO) analysis of pakchoi line parents and hybrids indicated that most of the differentially expressed genes between parents and hybrids enriched the photosynthetic pathway and that the enhancement of the photosynthetic capacity of the hybrids was related mainly to an increase in the number of thylakoids17. In addition, the increase in the number of thylakoids also promoted the enhancement of the carbon fixation capacity in the hybrids17; this is similar to the finding that differentially expressed genes that significantly enrich the optical signaling pathway occur between F1 and their parents in broccoli24. The same results were also found in other plants79,81. Transcriptome and differential gene expression analyses revealed that the modes of action of heterosis genes were mainly additive (F1 = MPV), overdominance (F1 > HPV), and underdominance (F1 < LPV)82 (Fig. 4). When the expression value of a differentially expressed gene in the hybrid line was higher or lower than that of the parent, the gene action patterns were classified as high-parent dominance (F1 ≈ HPV) and low-parent dominance (F1 ≈ LPV), respectively82 (Fig. 4). Li et al.24 reported that most genes exhibited additive expression patterns in hybrid broccoli and that nonadditive action was involved mainly in light and hormone signal pathways related to heterosis; a similar finding was reported in Chinese cabbage (Brassica campestris ssp. pekinensis cv. “spring flavor”)23. These gene expression patterns may have occurred due to selective inhibition or activation by the epigenetic modification of hybrid F1 genes83,84; the genes from inactive inbred lines can be activated by genes or regulatory factors of active inbred lines85,86. Epigenetic modifications and the interactions of heterogeneous factors occur in only a few genes, and the genome that produces differential expression in F1 hybrids and parents accounts for only a small part of the total genome87. Moreover, Springer and Stupar88 have shown that additive gene expression accounts for the majority of gene expression, while nonadditive gene expression is responsible for a small proportion of gene expression. These findings suggest that nonadditive expression of this fraction facilitates the development of heterosis.
Fig. 4. By comparing the gene expression of the F1 hybrid and its parents, the gene expression patterns of F1 were divided into additive gene expression patterns and nonadditive gene expression patterns.
Midparent value [MPV = (HPV + LPV)/2]; High-parent value (HPV); low-parent value (LPV)
Traits contributing to yield heterosis in vegetables
Traits related to yield heterosis
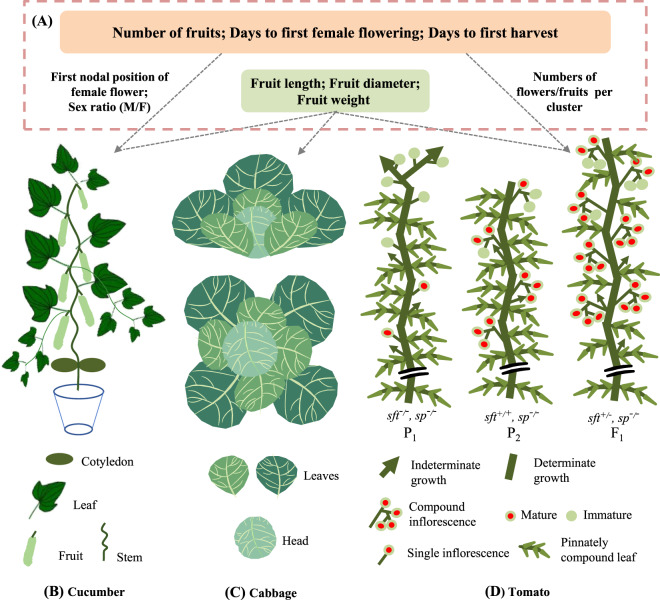
Hybrids that exhibit heterosis show significant heterobeltiosis in yield, which is a complex trait that is usually measured by weight. To clearly study the mechanisms of yield increase in hybrids, it is essential to divide yield into other, simpler traits. This review describes the traits that contribute to vegetable yields. Fruits are the source of the yield of most plants; the yield contributing traits related to fruits usually include the fruit number, fruit size and fruit weight; earliness is usually also taken into account. Cabbage is a typical leafy, head-forming vegetable in Cruciferae, so its main yield contributing traits are head weight and head size (Fig. 5A, C). Similar to that of cabbage, the yield of radish is determined by its taproot. For leafy vegetables that do not form heads, the main yield heading traits are the number and size of the leaves. Unlike cruciferous vegetables, Cucurbitaceae and Solanaceae vegetables are produce multiple harvests and multiple fruits per plant (Fig. 5B, D), so the average single fruit weight and fruit yield per plant should be taken into account. In addition, Solanaceae vegetable flowers consist mostly of compound inflorescences89, so the numbers of flowers per cluster and fruits per cluster contribute greatly to production. Cucurbitaceae are single-inflorescence vegetables; only the fruits on the main vine are harvested in production, and the first nodal position of female flowers and sex ratio (M/F) affect the days to first harvest and the number of fruits per plant, respectively. Regardless of the trait considered, the total yield can be affected only by changes in yield-related traits. Therefore, it is necessary to analyze the mechanisms that regulate yield-related traits.
Fig. 5.
Contributing traits of yield heterosis in cucumber, cabbage and tomato. A Traits contributing to yield heterosis in cucumber, cabbage, and tomato: cucumber yield contributing traits include the number of fruits, days to first female flowering, days to first harvest, first nodal position of female flower, sex ratio (M/F), fruit length, fruit diameter, and fruit weight; cabbage yield contributing traits include fruit length, fruit diameter, and fruit weight; tomato yield contributing traits include number of fruits, days to first female flowering, days to first harvest, number of flowers/fruits per cluster, fruit length, fruit diameter, and fruit weight. B Cucumber: cucumber model in production, gynoecious line with a small number of branches. C Cabbage: an aerial and cross-sectional model of cabbage consisting of leaves and heads. D Tomato: a tomato with single inflorescences and indeterminate growth is crossbred with a tomato with compound inflorescences and determinate growth to produce the hybrid F1 with earlier fruiting, more compound inflorescences, and determinate growth
Relationship between yield heterosis and plant architecture
Since the “green revolution”, interest in breeding for specific plant architecture has significantly increased, and the idea of combining heterosis breeding with plant architecture breeding has been proposed90. Donald91 conducted research on half-dwarf plant architecture, which gradually turned into the concept of the ideotype. Donald introduced the ideotype concept, which refers to the plant architecture form that results in the minimum competitive intensity in population breeding. Although this definition is no longer used, the concept of an ideal plant architecture has played a major role in promoting plant breeding for high yields. Research on ideotypes first made progress in rice. It is worth mentioning that a key gene regulating ideotype, IPA1, was proven by Huang et al.75 to influence genes that are important in heterosis by using the indica-japonica hybrid rice group. Studies of heterozygosity and ideotype were also combined effectively in tomato. The self-pruning (sp) gene promotes indeterminate growth in tomato, while the sft gene changes indeterminate growth into determinate growth by inhibiting the sp gene92. The sft gene results in the development of heterosis in tomatoes through the heterozygosity of a single gene15 and induces changes in plant architecture on the ground, causing tomato to produce compound inflorescences rather than single inflorescences93. The earliness of F1 was also higher than that of its parent (Fig. 5D), which increased tomato yield. Other vegetables in addition to tomato may also have ideotypes, and the key genes controlling plant architecture may also be important genes that are involved in the development of heterosis. Therefore, it is particularly important to study the genetic mechanisms of heterosis. By identifying the important genes involved in heterosis, the key genes that control plant ideotypes can be characterized.
Advances in heterosis utilization and biotechnology in vegetables
Breeding for heterosis has been extensively studied in plants, and research on the heterobeltiosis of hybrid offspring in vegetables has focused mainly on yield94 and disease resistance29. Wellington95 and Tschermak96 showed that tomato hybrids exhibit heterosis in early maturity and during yield production. Krieger et al.15 cloned the single-gene sft that affects the female flower fertility rate in tomato by infiltrating the IL and TC populations. When the sft gene exhibited heterozygosity, the tomato yield exhibited heterosis. According to this study, tomato plants that showed yield heterosis also showed resistance to both biological and abiotic stresses. The heterozygous state of the Tm and Tm22 genes contributes to tobacco mosaic virus resistance97,98 and high-temperature stress tolerance99,100. Naresh et al.101 suggested that heterosis is the result of nonadditive gene effects and that it also plays an important role in improving Cercospora leaf spot resistance in eggplant in the field. Similar to studies on other vegetables, studies on heterosis in Cucurbitaceae vegetables have also focused mainly on yield and disease resistance. Pandey et al.102 used 77 cucumber hybrid generations and their parents to study the yield heterosis and contributing traits of different cucumber hybrid varieties and found that DC–1 × B–159 and VRC–11–2 × Bihar–10 were the best hybrid combinations for yield and prematurity. Using 48 F1 hybrids and their parents, the gene effects caused by diseases and insect pests under natural conditions29 were investigated. The results indicated that nonadditive gene effects had a significant regulatory effect on other traits in cucumber (except morbidity caused by Drosophila), demonstrating the importance of heterosis in cucumber breeding for disease resistance.
Different molecular markers, such as simple sequence repeats (SSRs), inter-simple sequence repeats (ISSRs), amplified fragment length polymorphisms (AFLPs), random amplified polymorphic DNAs (RAPDs), and sequence-related amplified polymorphisms (SRAPS), have provided the molecular basis for the construction of genetic maps and the mapping of important trait genes (Table 1). Whole-genome sequencing has been conducted for a variety of vegetables (Table 1), which has provided a basis for whole-genome strategies. Whole-genome approaches can help obtain complete sequences of germplasm resources, increase the coverage of molecular markers, and increase the accuracy of genetic maps103. Molecular markers are often used for the determination of genetic distance and the classification of heterotic groups. To elucidate the breeding processes and to improve the efficiency of breeding techniques in cabbage, heterotic cabbages are usually divided into two groups: The round head type and the flat head type. Xing et al.104 further divided 21 flat cabbage inbred lines into three heterotic groups and divided 42 round cabbage inbred lines into five heterotic groups in order to provide a more definite direction for the preparation of hybrid combinations of cabbage. The method of dividing heterotic groups by molecular markers and genetic distance is widely used in vegetable breeding (Table 1).
Table 1.
Research progress in vegetable breeding
| Vegetable variety | Identification of heterotic groups | Maintenance of heterozygosity during production | Whole-genome sequencing | Regeneration system |
|---|---|---|---|---|
| Chinese cabbage | SSR, GD, ISSR, genealogical information | Polima CMS (MsMs) | Yes (Wang et al.186) | Cotyledon-petioles (Li et al.187) |
| Cabbage | SSR, microsatellites | TGMS (Ms); PGMS | Yes (Liu et al.188) | Hypocotyl (Ma et al.189) |
| Radish | AFLP, ISSR, SSR | Male sterility; self-incompatibility | Yes (Kitashiba et al.190) | Microspore (Kim et al.191) |
| Tomato | RAPD, Indel, SSR, AFLP, SNP | TGMS: Sterile pollen (ms); lack of stamens (sl–1, sl–2); positional sterility (ps); functional sterility (ps–2) | Yes (Consortium192) | Hypocotyl and cotyledon (Lee et al.193) |
| Potato | GD, SNP, RAPD | Vegetative propagation | Yes (Consortium194) | Stem segments (Zhang et al.195) |
| Pepper | GD, AFLP, SSR, RAPD | TGMS (ms–10, ms–12); TCMS | Yes (Kim et al. 2014; Qin et al.196,197) | Hypocotyl (Chee et al.198) |
| Eggplant | GD, AFLP, SSR, combining ability test | GMS; functional male sterility (fms) | Yes (Hirakawa et al.199) | Leaf (Khan200) |
| Cucumber | SSR, GD, AFLP, combining ability test | GMS (ms–3); gynoecious | Yes (Huang et al.201) | Cotyledon (Du et al.202) |
| Melon | SSR, GD, combining ability test | GMS (ms–3, ms–4, ms–5); gynoecious | Yes (Garcia–Mas et al.203) | Cotyledon (Chenarani et al.204) |
| Watermelon | RAPD, SSR | GMS (gms); gynoecious | Yes (Guo et al.205) | Cotyledon (Vasudevan et al.206) |
| Pumpkin | RAPD, SRAP | Gynoecious | Yes (Montero–Pau et al.; Sun et al.207,208) | Cotyledon (Guo et al.209) |
Chen83 proposed that determining how to obtain hybrid seeds is the key to the utilization of heterosis. The purpose of obtaining hybrid seeds is to make heterosis in the offspring permanent. The sporophyte of cruciferous vegetables is a self-incompatible system105 that can prevent self-pollination and produce normal seeds through cross-pollination. Hence, this system is convenient for the generation of hybrid seeds. In cabbage106,107 and Chinese cabbage108, hybrids are usually obtained using self-incompatible and male-sterile lines. To produce hybrid tomato seeds, pollen-abortive type and functionally sterile lines are often used109–111. Cytoplasmic male sterility occurs in eggplant112,113 and pepper114,115. Gynoecious lines tend to exist in Cucurbitaceae116. A new male-sterile system in tomato was developed by Du et al.117. Plant growth regulators such as ethylene, auxins, and brassinosteroids118,119 can increase the number of female flowers in Cucurbitaceae; this effect and male sterility are both convenient for hybrid seed production.
Strategies for heterosis breeding in vegetables (with tomato as an example)
Obtaining F1 hybrids that exhibit heterosis based on heterosis prediction
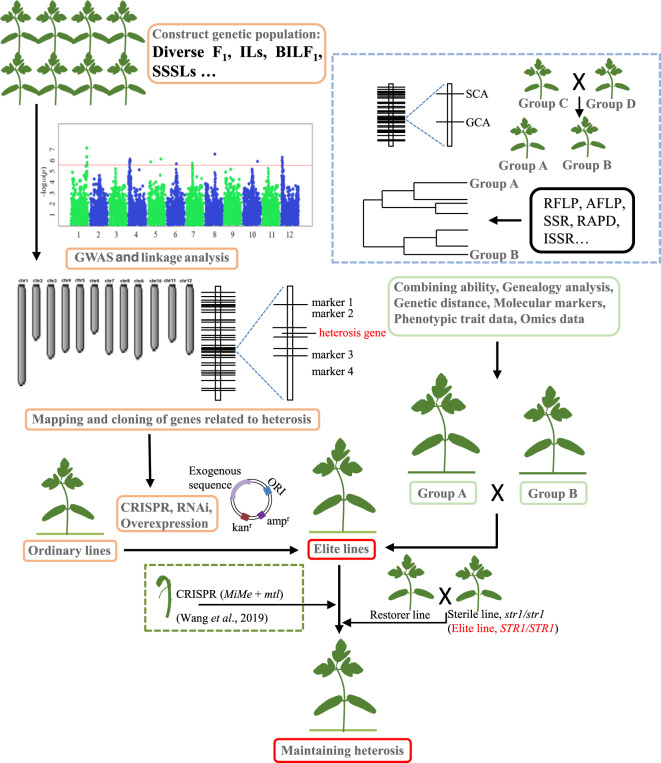
It is not advisable to conduct extensive hybridization tests to obtain hybrid F1 lines that exhibit heterosis, as this approach requires considerable resources and time and produces unreliable results13. Melchinger and Gumber120 proposed that heterotic groups should be used as the basis for crossbreeding. The heterotic group is the population that is classified according to breeding requirements, with abundant genetic variation and high combining ability. Chen et al.121 carried out a genome-wide association study (GWAS) on the yield traits, general combining ability (GCA), and SCA of rice. The study provided strong evidence for the use of combining ability to classify heterotic groups and provided a reference for studies on combining ability in vegetables (Fig. 6). Other studies have also shown that combining ability, genetic distance, and molecular markers can provide the basis for evaluating parental inbred lines and predicting F1 hybrid heterosis in vegetables122–125.
Fig. 6. There are two key factors involved in applying heterosis breeding strategies: obtaining heterotic lines and maintaining heterosis in the elite lines in the offspring.
There are two strategies for obtaining heterotic lines in crop breeding. The first is the use of crossbreeding or molecular biotechnology. Genealogical analysis, molecular markers, combining ability, and genetic distance can usually predict heterosis development, so they are often used to classify heterotic groups. The inbred lines from different heterotic groups can be crossed with each other to obtain elite lines that exhibit heterosis. The second strategy is to use modern molecular biotechnology. Elite lines were obtained based on GWAS and linkage analysis, mapping and cloning genes related to heterosis, gene editing, and gene transformation
The GCA characterizes the average performance of a set of hybrid combinations and is mainly the consequence of additive gene effects and additive × additive interactions; SCA evaluates the average performance of certain hybrid combinations compared to the parental lines and is the result of dominance, epistatic deviation and genotype × environmental interactions126. Parents with a high GCA effect have higher adaptability and fewer environmental effects127. Parents with superior traits do not always pass on their traits to offspring126; hence, the evaluation of combining ability is more reliable than the performance of the lines per se. Many types of combining ability tests can be used to identify superior parental lines for developing heterotic hybrids, including line × tester analysis, topcross tests, single-cross tests, poly-cross tests, and diallel mating128. Singh et al.129 conducted a complete diallel cross test on seven diverse bitter gourd lines and found that combinations with high × high GCA usually produced high SCA effects and could therefore be considered for use in developing superior variants through the pedigree method. High/low × low GCA combinations can also achieve high but unstable SCA effects that are suitable for heterosis breeding and are in line with the results of Kenga et al.130 in sweet sorghum and Franco et al.131 in common bean.
In addition to combining ability, heterotic groups are often classified by genealogical information132. For parents with known genealogical relationships, heterosis in hybrids can usually be predicted according to these genealogical relationships. Genetic distance is a quantitative description of the genetic differences that provide the genetic basis for the development of heterosis in offspring133,134. Parental lines with a longer genetic distance are more likely to produce hybrids with strong predominance135,136. Molecular markers can also be used to directly or indirectly classify heterotic groups by assessing their genetic distance125,137,138. RAPD and AFLP have been successfully used to detect the genetic distance between tested lines, and the yield of carrots was found to be significantly correlated with genetic distance125. Genetic distance has also been applied to predict hybrid pepper fruit diameter139 and hybrid melon (Cucumis melo L.) fruit shape diameter140. The scientific classification of heterotic groups improves the efficiency of selecting hybrid combinations of superior parents and utilizing heterosis (Fig. 6).
In addition, some omics approaches, such as genomics, transcriptomics, and metabolomics, have become tools for predicting hybrid yield in rice141. Xu et al.141 analyzed metabolomic and genomic data from 21,445 hybrids developed by 210 recombinant inbred lines and found that metabolomic data were more effective than genomic data in predicting hybrid yield. Research on the prediction of heterosis in vegetables with omics data has not been published. However, the genome or epigenome is the most fundamental source of the plant phenotype, and the transcriptome, proteome, and metabolism are the direct sources of plant phenotypes. Therefore, omics data could represent a more accurate way to predict vegetable hybrid heterosis, and studies of crop hybrid yields can provide a reference for predicting heterosis in vegetables.
Obtaining elite lines based on molecular biotechnology
GWAS is a method used to identify the gene loci that control certain traits in a population by combining phenotypes with genotypes. GWAS is often used to identify certain traits, such as green flesh color or thermotolerance, in cucumber142,143 but can also be used to analyze complex traits, such as yield and biomass144–156. In addition, whole-genome sequencing of various vegetables provides a basis for GWAS (Table 1). Due to the unique phenotype of heterosis and its genetic background sources, a genetic population can be composed of different populations or ecotype hybrid populations. A segregated F2 population that was produced by a strongly predominant F1 population is regarded as the best population for studying heterosis27. Such an F2 population not only has a reasonable proportion of lines with heterozygous genotypes and homozygous genotypes but also has allele combinations that are distributed evenly at each site27.
DeVicente and Tanksley157 randomly paired an RIL population obtained by strong F1 self-crossing to produce a new population. This population not only preserves the genotype of the RIL population but also reproduces the F2 population; thus, it is called an IF2 population. At present, IF2 populations have been established in rice158–161, maize150,162–169, cotton170, and other crops. In addition, there are also diverse F1156, IL171–175, BILF1176,177, and SSSL178 populations that can be used to study heterosis. Except for two studies on tomato, there are few relevant studies on heterosis in vegetables using such populations that would provide a reference for conducting heterosis-related studies in other vegetables.
Using genome editing techniques to knockout adverse genes or overexpress favorable genes can transform ordinary lines into strong predominance lines. For example, biomass, plant height, and leaf photosynthetic pigment contents increased in rice expressing maize GLK genes compared with those in wild-type rice;179 such results may cause researchers to think about studying mutual heterosis promotion among different vegetables. Dominance and overdominance effects account for a large proportion of the effects that produce heterosis and are easy to mimic (Fig. 3B). Understanding the mechanisms of heterosis helps breeders to improve current varieties and generate novel cultivars27 (Fig. 6).
Maintaining heterosis
The hybridization of the selfing line of two heterotic groups can generate hybrid offspring that exhibit heterosis. Through hybrid seed production, self-incompatibility and male-sterile line technology can be used to maintain the hybrid vigor of the hybrid F1 line. Some of the characteristics of the vegetables themselves, such as the gynoecious characteristic of Cucurbitaceae116 and asexual reproduction in potato (Solanum tuberosum L.)180, are convenient for hybrid seed production or heterosis maintenance. In addition, some plant hormones or chemical reagents can also be used for plant sex regulation14. However, exogenous regulation is often not completely effective14, which may affect the purity of hybrid seeds. Therefore, it is necessary to study hybrid systems of vegetables for hybrid seed production.
Du et al.117 used gene editing technology (Cas9) to knock out the male-specific gene SlSTR1 in tomato to obtain a sterile line and generated a maintainer line by transferring a fertility-restoration gene to the sterile line; it was easy to distinguish whether offspring of crosses between the maintainer and male-sterile lines were male-fertile maintainer plants because a seedling-color gene was linked to the fertility-restoration gene. This system combined tomato sterile lines and gene editing technology and represents a highly practical potential approach to hybrid seed production in tomatoes. Moreover, it may serve as an important reference for the use of gene editing technology for hybrid seed production in other vegetables.
Khanday et al.181 and Wang et al.182 found that genome editing can cause mitosis to replace meiosis in rice such that diploid clonal seeds have the original F1 gene heterozygosity and maintain F1 traits (Fig. 6). Unlike with knocking out the infertility gene using gene editing technology, with this method, fertilization and cell division are necessary for hybridization. Some vegetables do not have sterile line material. Therefore, this method, in which plant fertilization involves only mitosis and not meiosis, will be more widely applicable.
In addition, by repeatedly screening the F2 lines that were close to the F1 phenotype, Wang et al.85 obtained pure F5/F6 lines that were close to the F1 phenotype; these were called hybrid simulation lines, indicating that the phenotype of the F1 hybrids was fixed in this line. This method has also been used to maintain F1 heterosis in other vegetables, such as tomatoes183 and peas (Pisum sativum L.)184. Therefore, the heterosis of hybrid F1 vegetables produced by hybridization or molecular biotechnology can be maintained by diploid seed breeding and selection for hybrid simulation lines in the future (Fig. 6).
Conclusions and future perspectives
Research on vegetable heterosis has focused mainly on its applications in heterosis breeding. Studies on its genetic mechanism are limited, which hinders its utilization. Extensive progress has been made in the study of heterosis in cereal crops such as rice and maize. In vegetables, both hybrid production systems (male sterility lines, self-incompatibility lines, and gynoecious lines) and molecular biological techniques (gene editing, transgenosis, and asexual reproduction) have been used. Therefore, the methods and strategies proposed by this paper for studying the genetic mechanisms of heterosis can be applied to vegetable breeding. In the near future, we will identify certain heterosis-related gene loci in vegetables to understand the molecular genetics and mechanism of heterosis formation in vegetables and to make new breakthroughs in improving the yield, quality, and safety of vegetables. This review emphasizes the following points: (1) The application of heterosis in vegetable crops allows improvements in yield and quality and enhances plant resistance to biological and environmental stresses. (2) In the future, more attention should be paid to the study of the genetic mechanisms of vegetable heterosis to identify the important genes involved in the development of heterosis and to understand the regulation and activity modes of the key genes affecting vegetable heterosis. (3) By fully referencing and adapting the strategies used in cereal crop heterosis studies, exogenous genes can be applied to produce the same function in different species179. Therefore, transgenic and genomic editing technologies can significantly improve the efficiency of research on heterosis gene identification in vegetables. (4) Although a certain basic molecular knowledge of vegetable heterosis has been obtained, applying the knowledge acquired from cereal crops to vegetables will improve vegetable production and quality. It will also be useful to compare sterile line seed production with optimized transgenic systems to achieve more breakthroughs in vegetable production. (5) The study of heterosis can promote the study of ideal plant architecture in vegetable breeding. A breeding strategy that combines heterosis with the ideal plant architecture can achieve substantial gains in vegetable yield and quality. (6) Maintaining heterosis is the core factor of the extensive use of heterosis and has been reflected mainly in F1 hybrid seed production. With the development of gene editing technology, sterile line gene editing systems, MiMe (Cas9) systems and even new biotechnology approaches will have opportunities to be widely applied; this will be of great significance for hybrid seed production. (7) Progressive heterosis caused by the dosage effect in polyploid hybrids is also an important component of the genetic mechanisms of heterosis, and these phenomena have been observed in different plants55,185. Polyploid systems allow experiments to be performed that are impossible in diploid systems; hence, polyploid crossbreeding may lead to different plant performance results than diploid breeding. However, polyploids have highly heterozygous genomes and complex genetic structures, and we may not be able to evaluate their phenotypes and genetic structures using diploid criteria. This topic deserves future investigation.
Supplementary information
Acknowledgements
This work was supported by the National Key Research and Development Program of China [2016YFD0101705], the Earmarked Fund for Modern Agro–industry Technology Research System [CARS–25] and the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, China.
Author contributions
D.Y. and K.B. conducted the literature review and wrote the paper. X.G., S.Z., S.D., H.M. and K.G. helped review the crop heterosis data. K.B. conceived of and supervised the study. All authors reviewed and approved the final submission.
Conflict of interest
The author declares no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00552-9.
References
- 1.Koelreuter, J. In Methods of Plant Breeding (eds. Hayes, H. K. & Immer, F. R. & Smith, B. C.) (Mcgraw Hill Book Co. Inc., 1763).
- 2.Darwin, C. The Effects of Cross and Self Fertilisation in the Vegetable Kingdom (D. Appleton, 1885).
- 3.Beal, W. Re. Michigan Board Agric. P 287-288. Cited in Hallauer, AR and J. B. Miranda (1988). Quantitative genetics in maize breeding. (IOWA State Univ. Press Ames, 1880).
- 4.Shull, G. H. The composition of a field of maize. J. Hered. 4, 296–301 (1908).
- 5.Shull GH. Duplicate genes for capsule-form in Bursa bursa-pastoris. Z. Indukt. Abstamm. Vererbungsl. 1914;12:97–149. [Google Scholar]
- 6.Tian F, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011;43:159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 7.Liedl BE, Anderson NO. Reproductive barriers: identification, uses and circumvention. Plant Breed. Rev. 1993;11:11–154. [Google Scholar]
- 8.Kumar S, Singh P. Mechanisms for hybrid development in vegetables. J. N. Seeds. 2005;6:381–407. doi: 10.1300/J153v06n04_05. [DOI] [Google Scholar]
- 9.Nishi S. F1 seed production in Japan. Proc. XVIII Int. Hort. Cong. 1967;3:231–257. [Google Scholar]
- 10.da Silva Dias JC. Guiding strategies for breeding vegetable cultivars. Agric. Sci. 2014;5:9. [Google Scholar]
- 11.Hayes HK, Jones D. First generation crosses in cucumbers. Conn. Storrs. Agric. Exp. Stn. Res. Rpt. 1917;40:319–322. [Google Scholar]
- 12.Daunay, M. C. Eggplant in Vegetables II 163–220 (Springer, 2008).
- 13.Singh, P. K., Dasgupta, S. K. & Tripathi, S. K. Hybrid Vegetable Development (CRC Press, 2005).
- 14.Colombo N, Galmarini CR. The use of genetic, manual and chemical methods to control pollination in vegetable hybrid seed production: a review. Plant Breed. 2017;136:287–299. doi: 10.1111/pbr.12473. [DOI] [Google Scholar]
- 15.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010;42:459. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- 16.Zhang WL, et al. Cloning and sequence analysis of CsMYB108 gene in cucumber (Cucumis sativa L.) Mol. 2020;18:4555–4561. [Google Scholar]
- 17.Liu T, et al. Enhanced photosynthetic activity in pak choi hybrids is associated with increased grana thylakoids in chloroplasts. Plant J. 2020;103:2211–2224. doi: 10.1111/tpj.14893. [DOI] [PubMed] [Google Scholar]
- 18.Kakizaki Y. Hybrid vigor in egg-plants and its practical utilization. Genetics. 1931;16:1. doi: 10.1093/genetics/16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balwani A, Patel J, Acharya R, Gohil D, Dhruve J. Heterosis for fruit yield and its component traits in brinjal (Solanum melongena L.) J. Pharmacogn. Phytochem. 2017;6:187–190. [Google Scholar]
- 20.Makani A, Patel A, Bhatt M, Patel P. Heterosis for yield and its contributing attributes in brinjal (Solanum melongena L.) bioscan. 2013;8:1369–1371. [Google Scholar]
- 21.Tamta S, Singh J. Heterosis in tomato for growth and yield traits. Int. J. Veg. Sci. 2018;24:169–179. doi: 10.1080/19315260.2017.1407857. [DOI] [Google Scholar]
- 22.Spaldon S, Hussain S, Jabeen N, Lay P. Heterosis studies for earliness, fruit yield and yield attributing traits in chilli (Capsicum Annum L.) Bioscan. 2015;10:813–818. [Google Scholar]
- 23.Kong X, et al. Transcriptome analysis of biological pathways associated with heterosis in Chinese cabbage. Genomics. 2020;112:4732–4741. doi: 10.1016/j.ygeno.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Li H, et al. Transcriptome and DNA methylome reveal insights into yield heterosis in the curds of broccoli (Brassica oleracea L. var. italic) BMC Plant Biol. 2018;18:1–17. doi: 10.1186/s12870-017-1213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma M, Singh Y, Singh SK, Dhangrah V. Exploitation of gynoecious lines in cucumber (Cucumis Sativus L.) for heterosis breeding. Int. J. Bio-Resour. Stress Manag. 2016;7:184–190. doi: 10.23910/IJBSM/2016.7.2.1557. [DOI] [Google Scholar]
- 26.El-Adl A, Abd El-Hadi A, Fathy HM, Abdein M. Heterosis, Heritability and Combining Abilities for some Earliness Traits in Squash (Cucurbita pepo, L.) Alex. Sci. Exch. J. 2014;35:203–214. [Google Scholar]
- 27.Liu J, Li M, Zhang Q, Wei X, Huang X. Exploring the molecular basis of heterosis for plant breeding. J. Integr. Plant Biol. 2020;62:287–298. doi: 10.1111/jipb.12804. [DOI] [PubMed] [Google Scholar]
- 28.Qin J, et al. Identification and characterization of a repertoire of genes differentially expressed in developing top ear shoots between a superior hybrid and its parental inbreds in Zea mays L. Mol. Genet. Genom. 2013;288:691–705. doi: 10.1007/s00438-013-0781-5. [DOI] [PubMed] [Google Scholar]
- 29.Kumar R, Kumar S, Kumar D, Kansal S. Heterosis, combining ability and gene action studies for insect-pest and disease resistance in cucumber. Electron. J. Plant Breed. 2018;9:815–828. doi: 10.5958/0975-928X.2018.00102.3. [DOI] [Google Scholar]
- 30.Kumar A, Sharma V, Jain BT, Kaushik P. Heterosis breeding in eggplant (Solanum melongena L.): Gains and provocations. Plants. 2020;9:403. doi: 10.3390/plants9030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujimoto, R. et al. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed. Sci. 68, 145–158 (2018). [DOI] [PMC free article] [PubMed]
- 32.Shang LG, Gao ZY, Qian Q. Progress in Understanding the Genetic Basis of Heterosis in Crops. Chin. Bull. Bot. 2017;52:10–18. [Google Scholar]
- 33.Herath, H. N., Rafii, M., Ismail, S., Nakasha, J. J. & Ramlee, S. Improvement of important economic traits in chilli through heterosis breeding: a review. J. Hortic. Sci. Biotechnol.96, 1–10 (2020).
- 34.Mallikarjunarao K, Badu M, Bandi HRK, Tripathy B. Heterosis breeding in Bitter Gourd (Momordica charantia L.): a review. J. Pharmacogn. Phytochem. 2020;9:308–313. [Google Scholar]
- 35.Davenport CB. Degeneration, albinism and inbreeding. Science. 1908;28:454–455. doi: 10.1126/science.28.718.454-b. [DOI] [PubMed] [Google Scholar]
- 36.Bruce A. The Mendelian theory of heredity and the augmentation of vigor. Science. 1910;32:627–628. doi: 10.1126/science.32.827.627-a. [DOI] [PubMed] [Google Scholar]
- 37.Jones DF. Dominance of linked factors as a means of accounting for heterosis. Genetics. 1917;2:466. doi: 10.1093/genetics/2.5.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.East EM. Heterosis. Genetics. 1936;21:375. doi: 10.1093/genetics/21.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dranginis A. Binding of yeast al and α2 as a heterodimer to the operator DNA of a haploid-specific gene. Nature. 1990;347:682–685. doi: 10.1038/347682a0. [DOI] [PubMed] [Google Scholar]
- 40.Hollick JB, Chandler VL. Epigenetic allelic states of a maize transcriptional regulatory locus exhibit overdominant gene action. Genetics. 1998;150:891–897. doi: 10.1093/genetics/150.2.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stuber CW, Lincoln SE, Wolff D, Helentjaris T, Lander E. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics. 1992;132:823–839. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnable, P. S. & Springer, N. M. Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 64, 71–88 (2013). [DOI] [PubMed]
- 43.Wright, S. Evolution and the Genetics of Populations, volume 1: Genetic and Biometric Foundations (University of Chicago press, 1984).
- 44.Sheridan, A. Cross breeding and heterosis Animal Breeding. Anim. Breed. Abst. 49, 131–144 (1981).
- 45.Sao A, Mehta N. Heterosis in relation to combining ability for yield and quality attributes in brinjal (Solanum melongena L.) Electron. J. Plant Breed. 2010;1:783–788. [Google Scholar]
- 46.Yu S, et al. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. P. Natl Acad. Sci. USA. 1997;94:9226–9231. doi: 10.1073/pnas.94.17.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong JC. Active gene effect hypothesis. J. Southwest Nationalities Nat. Sci. Ed. 1994;20:2. [Google Scholar]
- 48.Kermicle J. Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics. 1970;66:69. doi: 10.1093/genetics/66.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang M, et al. Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 2014;24:167–176. doi: 10.1101/gr.155879.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bao WK. Opportunity and risk–considerations for 40 years of breeding research. Plant J. 1990;4:4–5. [Google Scholar]
- 51.Goff SA. A unifying theory for general multigenic heterosis: energy efficiency, protein metabolism, and implications for molecular breeding. N. Phytol. 2011;189:923–937. doi: 10.1111/j.1469-8137.2010.03574.x. [DOI] [PubMed] [Google Scholar]
- 52.Yao H, Gray AD, Auger DL, Birchler JA. Genomic dosage effects on heterosis in triploid maize. Proc. Natl Acad. Sci. USA. 2013;110:2665–2669. doi: 10.1073/pnas.1221966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu D, et al. Utilization of crop heterosis: a review. Euphytica. 2014;197:161–173. doi: 10.1007/s10681-014-1103-7. [DOI] [Google Scholar]
- 54.Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010;15:57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. Heterosis. Plant Cell. 2010;22:2105–2112. doi: 10.1105/tpc.110.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C, Wang M, Wang L, Guo Q, Liang G. Extensive genetic and DNA methylation variation contribute to heterosis in triploid loquat hybrids. Genome. 2018;61:437–447. doi: 10.1139/gen-2017-0232. [DOI] [PubMed] [Google Scholar]
- 57.Zhao X, Chai Y, Liu B. Epigenetic inheritance and variation of DNA methylation level and pattern in maize intra-specific hybrids. Plant Sci. (Amst., Neth.) 2007;172:930–938. [Google Scholar]
- 58.He G, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen H, et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell. 2012;24:875–892. doi: 10.1105/tpc.111.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauss K, et al. Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol. 2018;176:1627–1645. doi: 10.1104/pp.17.01054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC. Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 2012;31:257–266. doi: 10.1038/emboj.2011.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cerruti, E. et al. Epigenetic bases of grafting-induced vigour in eggplant. bioRxiv10.1101/831719 (2019).
- 63.Kenchanmane, R. S. K. & Niederhuth, C. E. Epigenetic diversity and application to breeding. Adv. Bot. Res. 88, 49–86 (2018).
- 64.Gouil Q, Novák O, Baulcombe DC. SLTAB2 is the paramutated SULFUREA locus in tomato. J. Exp. Bot. 2016;67:2655–2664. doi: 10.1093/jxb/erw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hagemann RSomatische. Konversion (Paramutation) am sulfurea Locus von Lycopersicon esculentum Mill. Theor. Appl. Genet. 1969;39:295–305. doi: 10.1007/BF00281910. [DOI] [PubMed] [Google Scholar]
- 66.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kundariya H, et al. MSH1-induced heritable enhanced growth vigor through grafting is associated with the RdDM pathway in plants. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-020-19140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barber WT, et al. Repeat associated small RNAs vary among parents and following hybridization in maize. Proc. Natl Acad. Sci. USA. 2012;109:10444–10449. doi: 10.1073/pnas.1202073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nogueira, F. T. Tomato epigenetics: deciphering the “beyond” genetic information in a vegetable fleshy-fruited crop in Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications 247–265 (Springer, 2019).
- 70.Molnar A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 71.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 72.Vance V, Vaucheret H. RNA silencing in plants-defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 73.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 74.Huang X, Han B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014;65:531–551. doi: 10.1146/annurev-arplant-050213-035715. [DOI] [PubMed] [Google Scholar]
- 75.Huang X, et al. Genomic architecture of heterosis for yield traits in rice. Nature. 2016;537:629–633. doi: 10.1038/nature19760. [DOI] [PubMed] [Google Scholar]
- 76.Lippman ZB, Zamir D. Heterosis: revisiting the magic. Trends Genet. 2007;23:60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 77.Xiao J, Li J, Yuan L, Tanksley SD. Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics. 1995;140:745–754. doi: 10.1093/genetics/140.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Groszmann M, et al. Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc. Natl Acad. Sci. USA. 2015;112:E6397–E6406. doi: 10.1073/pnas.1519926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song GS, et al. Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol. Plant. 2010;3:1012–1025. doi: 10.1093/mp/ssq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen ZJ. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013;14:471–482. doi: 10.1038/nrg3503. [DOI] [PubMed] [Google Scholar]
- 81.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baranwal VK, Mikkilineni V, Zehr UB, Tyagi AK, Kapoor S. Heterosis: emerging ideas about hybrid vigour. J. Exp. Bot. 2012;63:6309–6314. doi: 10.1093/jxb/ers291. [DOI] [PubMed] [Google Scholar]
- 83.Chen, S. R. Vegetable Breeding (Agricultural Press, 1980).
- 84.Preuss SB, et al. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol. Cell. 2008;32:673–684. doi: 10.1016/j.molcel.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang L, et al. Hybrid mimics and hybrid vigor in Arabidopsis. Proc. Natl Acad. Sci. USA. 2015;112:E4959–E4967. doi: 10.1073/pnas.1514190112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paschold A, et al. Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents. Genome Res. 2012;22:2445–2454. doi: 10.1101/gr.138461.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li D, et al. Integrated analysis of phenome, genome, and transcriptome of hybrid rice uncovered multiple heterosis-related loci for yield increase. Proc. Natl Acad. Sci. USA. 2016;113:E6026–E6035. doi: 10.1073/pnas.1610115113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Springer NM, Stupar RM. Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res. 2007;17:264–275. doi: 10.1101/gr.5347007. [DOI] [PubMed] [Google Scholar]
- 89.Barboza, G. E. et al. Solanaceae in Flowering plants. Eudicots 295–357 (Springer, 2016).
- 90.Khush GS. Rice germplasm enhancement at IRRI. Phillipp. J. Crop Sci. 2000;25:45–51. [Google Scholar]
- 91.Donald CT. The breeding of crop ideotypes. Euphytica. 1968;17:385–403. doi: 10.1007/BF00056241. [DOI] [Google Scholar]
- 92.Yeager A. Determinate growth in the tomato. J. Hered. 1927;18:263–265. doi: 10.1093/oxfordjournals.jhered.a102869. [DOI] [Google Scholar]
- 93.Jiang K, Liberatore KL, Park SJ, Alvarez JP, Lippman ZB. Tomato yield heterosis is triggered by a dosage sensitivity of the florigen pathway that fine-tunes shoot architecture. PLoS Genet. 2013;9:e1004043. doi: 10.1371/journal.pgen.1004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khan A, Jindal S. Exploiting yield potential in tomato (Solanum lycopersicum L.) through heterosis breeding. Plant Gene Trait. 2016;7:1–7. doi: 10.1016/j.plgene.2016.07.001. [DOI] [Google Scholar]
- 95.Wellington, R. Influence of Crossing in Increasing the Yield of the Tomato (New York Agricultural Experiment Station, 1912).
- 96.Tschermak, E. V. Steigerung der Ertragsfähigkeit der Tomaten durch Bastardierung in der F1 Generation. Nachr. Dtseh. Landwirtsch. Ges. Öst. 51 (1918).
- 97.Laterrot H. Résistance de la tomate au virus de la mosaïque du tabac. Difficultés rencontrées pour la sélection de variétés résistantes. Ann. Am.élior Plantes. 1973;23:287–316. [Google Scholar]
- 98.Soost R. Tobacco mosaic resistance. TGC Rep. 1959;9:46. [Google Scholar]
- 99.Lapushner, D. & Frankel, R. Rationale and practice of tomato F1 hybrid breeding and seed production. Monographic di Genetica Agraria IV. Roma 259–273 (1979).
- 100.Philouze, J. Les hybrides de la tomate: leur intérêt, les techniques d’hybridation, l’utilisation de la stérilité mâle. PHM Revue Hortieole.164, 11–18 (1976).
- 101.Naresh BV, Dubey A, Tiwari P, Dabbas M. Line x Tester analysis for yield components and cercospora leaf spot resistance in brinjal (Solanum melongena L.) Electron. J. Plant Breed. 2014;5:230–235. [Google Scholar]
- 102.Pandey S, Singh B, Singh M, Rai M. Heterosis in cucumber (Cucumis sativus L.) Veg. Sci. 2005;32:143–145. [Google Scholar]
- 103.Xu Y, et al. Whole-genome strategies for marker-assisted plant breeding. Mol. Breed. 2012;29:833–854. doi: 10.1007/s11032-012-9699-6. [DOI] [Google Scholar]
- 104.Xing L, et al. Heterotic group classification of 63 inbred lines and hybrid purity identification by using SSR markers in winter cabbage (Brassica oleracea L. var. capitata) Hortic. Plant J. 2018;4:158–164. doi: 10.1016/j.hpj.2018.03.010. [DOI] [Google Scholar]
- 105.Muthuselvi R, Praneetha S. Molecular basis of self-incompatibility in vegetable crops. Int. J. Chem. Stud. 2019;7:506–510. [Google Scholar]
- 106.Earle, E., Stephenson, C., Walters, T. & Dickson, M. Cold-tolerant Ogura CMS brassica vegetables for horticultural use. Cruciferae Newsletter16, 80–81 (1994).
- 107.Pelletier G, et al. Intergeneric cytoplasmic hybridization in Cruciferae by protoplast fusion. Mol. Gen. Genet. MGG. 1983;191:244–250. doi: 10.1007/BF00334821. [DOI] [Google Scholar]
- 108.Xiao Z, et al. Overcoming cabbage crossing incompatibility by the development and application of self-compatibility-QTL-specific markers and genome-wide background analysis. Front. Plant Sci. 2019;10:189. doi: 10.3389/fpls.2019.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Atanassova, B. & Georgiev, H. Using genic male sterility in improving hybrid seed production in tomato (Lycopersicon esculentum Mill.). Acta Hortic579, 185–188 (2000).
- 110.Dhall, R. Status of male sterility in vegetables for hybrid development. A review. Adv. Hortic. Sci.24, 263–279 (2010).
- 111.Georgiev, H. Heterosis in tomato breeding in Genetic improvement of tomato 83–98 (Springer, 1991).
- 112.Mizanur, M., Khan, R. & Isshiki, S. Cytoplasmic male sterility in eggplant. Hortic. J. 85, 1–7 (2016).
- 113.Krommydas KS, et al. Development and fertility restoration of CMS eggplant lines carrying the cytoplasm of Solanum violaceum. J. Agric. Sci. 2016;8:10–26. [Google Scholar]
- 114.Lin SW, et al. Restorer breeding in sweet pepper: introgressing Rf allele from hot pepper through marker-assisted backcrossing. Sci. Hortic. 2015;197:170–175. doi: 10.1016/j.scienta.2015.09.036. [DOI] [Google Scholar]
- 115.Liu C, Ma N, Wang PY, Fu N, Shen HL. Transcriptome sequencing and de novo analysis of a cytoplasmic male sterile line and its near-isogenic restorer line in chili pepper (Capsicum annuum L.) PLoS ONE. 2013;8:e65209. doi: 10.1371/journal.pone.0065209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kumar S. Male sterility in vegetables. Olericulture Fundam. Veg. Prod. 2013;1:431–439. [Google Scholar]
- 117.Du, M. et al. A biotechnology-based male-sterility system for hybrid seed production in tomato. Plant J. 102, 1090–1100 (2020). [DOI] [PMC free article] [PubMed]
- 118.Papadopoulou E, Grumet R. Brassinosteriod-induced femaleness in cucumber and relationship to ethylene production. HortScience. 2005;40:1763–1767. doi: 10.21273/HORTSCI.40.6.1763. [DOI] [Google Scholar]
- 119.Shannon S, Guardia deLa. M. D. Sex expression and the production of ethylene induced by auxin in the cucumber (Cucumis sativum L.) Nature. 1969;223:186–186. doi: 10.1038/223186a0. [DOI] [Google Scholar]
- 120.Melchinger AE, Gumber RK. Overview of heterosis and heterotic groups in agronomic crops. Concepts Breed. Heterosis Crop Plants. 1998;25:29–44. [Google Scholar]
- 121.Chen J, et al. Genome‐wide association analyses reveal the genetic basis of combining ability in rice. Plant Biotechnol. J. 2019;17:2211–2222. doi: 10.1111/pbi.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mather K. The balance of polygenic combinations. J. Genet. 1942;43:309–336. doi: 10.1007/BF02982906. [DOI] [Google Scholar]
- 123.Griffing B. Concept of general and specific combining ability in relation to diallel crossing systems. Aust. J. Biol. Sci. 1956;9:463–493. doi: 10.1071/BI9560463. [DOI] [Google Scholar]
- 124.Toker, C. et al. Association between heterosis and genetic distance based on morphological traits and SSR markers in Cicer species. In Pre Breeding–fishing in the Gene Pool». Abstract of Oral Presentation And Posters of the European Plant Genetic Resources Conference 85 (2013).
- 125.Jagosz B. The relationship between heterosis and genetic distances based on RAPD and AFLP markers in carrot. Plant Breed. 2011;130:574–579. doi: 10.1111/j.1439-0523.2011.01877.x. [DOI] [Google Scholar]
- 126.Sharma BB, Sharma VK, Dhakar MK, Punetha S. Combining ability and gene action studies for horticultural traits in garden pea: A review. Afr. J. Agric. Res. 2013;8:4718–4725. [Google Scholar]
- 127.Fasahat P, Rajabi A, Rad J, Derera J. Principles and utilization of combining ability in plant breeding. Biom. Biostat. Int J. 2016;4:1–24. [Google Scholar]
- 128.Rajendrakumar, P., Hariprasanna, K. & Seetharama, N. Prediction of heterosis in crop plants—status and prospects. J. Exp. Agric. Int.9, 1–16 (2015).
- 129.Singh A, Pan R, Bhavana P. Heterosis and combining ability analysis in bittergourd (Momordica charantia L.) Bioscan. 2013;8:1533–1536. [Google Scholar]
- 130.Kenga R, Alabi S, Gupta S. Combining ability studies in tropical sorghum (Sorghum bicolor (L.) Moench) Field Crops Res. 2004;88:251–260. doi: 10.1016/j.fcr.2004.01.002. [DOI] [Google Scholar]
- 131.Franco MC, et al. Combining ability for nodulation in common bean (Phaseolus vulgaris L.) genotypes from Andean and Middle American gene pools. Euphytica. 2001;118:265–270. doi: 10.1023/A:1017560118666. [DOI] [Google Scholar]
- 132.Gerdes JT, Tracy WF. Pedigree diversity within the Lancaster surecrop heterotic group of maize. Crop Sci. 1993;33:334–337. doi: 10.2135/cropsci1993.0011183X003300020025x. [DOI] [Google Scholar]
- 133.Arunachalam, V. & Bandyopadhyay, A. Limits to genetic divergence for occurrence of heterosis Experimental evidence from crop plants. Indian J. Genet. Plant Breed. (India)44, 548–554 (1984).
- 134.Zhong-hu H. An investigation of the relationship between the F 1 potential and the measures of genetic distance among wheat lines. Euphytica. 1991;58:165–170. doi: 10.1007/BF00022817. [DOI] [Google Scholar]
- 135.Kaeppler, S. Heterosis: many genes, many mechanisms—end the search for an undiscovered unifying theory. Int. Sch. Res. Not.2012, 1–12 (2012).
- 136.Poehlman, J. & Sleper, D. Breeding Field Crops (Iowa State University Press, 1995).
- 137.Tomkowiak A, Bocianowski J, Kwiatek M, Kowalczewski P. Ł. Dependence of the heterosis effect on genetic distance, determined using various molecular markers. Open Life Sci. 2020;15:1–11. doi: 10.1515/biol-2020-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xiao J, Li J, Yuan L, McCouch S, Tanksley S. Genetic diversity and its relationship to hybrid performance and heterosis in rice as revealed by PCR-based markers. Theor. Appl. Genet. 1996;92:637–643. doi: 10.1007/BF00226083. [DOI] [PubMed] [Google Scholar]
- 139.Geleta L, Labuschagne M, Viljoen C. Relationship between heterosis and genetic distance based on morphological traits and AFLP markers in pepper. Plant Breed. 2004;123:467–473. doi: 10.1111/j.1439-0523.2004.01017.x. [DOI] [Google Scholar]
- 140.José MA, Iban E, Silvia A, Pere A. Inheritance mode of fruit traits in melon: Heterosis for fruit shape and its correlation with genetic distance. Euphytica. 2005;144:31–38. doi: 10.1007/s10681-005-0201-y. [DOI] [Google Scholar]
- 141.Xu S, Xu Y, Gong L, Zhang Q. Metabolomic prediction of yield in hybrid rice. Plant J. 2016;88:219–227. doi: 10.1111/tpj.13242. [DOI] [PubMed] [Google Scholar]
- 142.Bo K, et al. QTL mapping and genome-wide association study reveal two novel loci associated with green flesh color in cucumber. BMC Plant Biol. 2019;19:1–13. doi: 10.1186/s12870-019-1835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.WEI, S. et al. Evaluation and Genome-wide Association Study (GWAS) of seedling thermotolerance in cucumber core germplasm. J. Plant Genet. Resour.23, 1223–1231 (2019).
- 144.Huang, X. et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun.6, 1–9 (2015). [DOI] [PMC free article] [PubMed]
- 145.Frascaroli, E. et al. Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics176, 625–644 (2007). [DOI] [PMC free article] [PubMed]
- 146.Guo T, et al. Genetic basis of grain yield heterosis in an “immortalized F 2” maize population. Theor. Appl. Genet. 2014;127:2149–2158. doi: 10.1007/s00122-014-2368-x. [DOI] [PubMed] [Google Scholar]
- 147.Alpert K, Grandillo S, Tanksley S. fw 2.2: a major QTL controlling fruit weight is common to both red-and green-fruited tomato species. Theor. Appl. Genet. 1995;91:994–1000. doi: 10.1007/BF00223911. [DOI] [PubMed] [Google Scholar]
- 148.Zhuang J, Fan Y, Wu J, Xia Y, Zheng K. Identification of over-dominance QTL in hybrid rice combinations. Hereditas. 2000;22:205–208. [Google Scholar]
- 149.Mei H, et al. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations. Theor. Appl. Genet. 2005;110:649–659. doi: 10.1007/s00122-004-1890-7. [DOI] [PubMed] [Google Scholar]
- 150.Li L, et al. Dominance, overdominance and epistasis condition the heterosis in two heterotic rice hybrids. Genetics. 2008;180:1725–1742. doi: 10.1534/genetics.108.091942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jiang G, Zeng J, He Y. Analysis of quantitative trait loci affecting chlorophyll content of rice leaves in a double haploid population and two backcross populations. Gene. 2014;536:287–295. doi: 10.1016/j.gene.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 152.Wei X, et al. Heterotic loci for various morphological traits of maize detected using a single segment substitution lines test-cross population. Mol. Breed. 2015;35:94. doi: 10.1007/s11032-015-0287-4. [DOI] [Google Scholar]
- 153.Li, H. et al. Quantitative trait locus analysis of heterosis for plant height and ear height in an elite maize hybrid zhengdan 958 by design III. BMC Genom.18, 36 (2017). [DOI] [PMC free article] [PubMed]
- 154.Guo X, et al. Mapping heterotic loci for yield and agronomic traits using chromosome segment introgression lines in cotton. J. Integr. Plant Biol. 2013;55:759–774. doi: 10.1111/jipb.12054. [DOI] [PubMed] [Google Scholar]
- 155.Tian S, et al. Overdominance is the major genetic basis of lint yield heterosis in interspecific hybrids between G. hirsutum and G. barbadense. Heredity. 2019;123:384–394. doi: 10.1038/s41437-019-0211-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang M, et al. Genomic architecture of biomass heterosis in Arabidopsis. Proc. Natl Acad. Sci. USA. 2017;114:8101–8106. doi: 10.1073/pnas.1705423114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.DeVicente M, Tanksley S. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics. 1993;134:585–596. doi: 10.1093/genetics/134.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou G, et al. Genetic composition of yield heterosis in an elite rice hybrid. Proc. Natl Acad. Sci. USA. 2012;109:15847–15852. doi: 10.1073/pnas.1214141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Radoev, M., Becker, H. C. & Ecke, W. J. G. Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics179, 1547–1558 (2008). [DOI] [PMC free article] [PubMed]
- 160.Li C, et al. Genetic basis of heterosis for yield and yield components explored by QTL mapping across four genetic populations in upland cotton. BMC Genom. 2018;19:910. doi: 10.1186/s12864-018-5289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Luo X, et al. Additive and over‐dominant effects resulting from epistatic loci are the primary genetic basis of heterosis in rice. J. Integr. Plant Biol. 2009;51:393–408. doi: 10.1111/j.1744-7909.2008.00807.x. [DOI] [PubMed] [Google Scholar]
- 162.Tang J, et al. Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor. Appl. Genet. 2010;120:333–340. doi: 10.1007/s00122-009-1213-0. [DOI] [PubMed] [Google Scholar]
- 163.Liu, H. et al. Genome‐wide identification and analysis of heterotic loci in three maize hybrids. Plant Biotechnol. J.18, 185–194 (2020). [DOI] [PMC free article] [PubMed]
- 164.Ma L, Wang Y, Ijaz B, Hua J. Cumulative and different genetic effects contributed to yield heterosis using maternal and paternal backcross populations in Upland cotton. Sci. Rep. UK. 2019;9:1–11. doi: 10.1038/s41598-019-40611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li Z-K, et al. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass. Grain Yield. 2001;158:1737–1753. doi: 10.1093/genetics/158.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Luo L, et al. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain Yield Compon. 2001;158:1755–1771. doi: 10.1093/genetics/158.4.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhu D, Zhou G, Xu C, Zhang Q. Genetic components of heterosis for seedling traits in an elite rice hybrid analyzed using an immortalized F2 population. J. Genet. Genom. 2016;43:87–97. doi: 10.1016/j.jgg.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 168.Song F, et al. Heterosis for plant height and ear position in maize revealed by quantitative trait loci analysis with triple testcross design. Acta Agronomica Sin. 2011;37:1186–1195. doi: 10.3724/SP.J.1006.2011.01186. [DOI] [Google Scholar]
- 169.Liang, Q., Shang, L., Wang, Y. & Hua, J. Partial dominance, overdominance and epistasis as the genetic basis of heterosis in upland cotton (Gossypium hirsutum L.). PLoS ONE10 (2015). [DOI] [PMC free article] [PubMed]
- 170.Liu R, Wang B, Guo W, Wang L, Zhang T. Differential gene expression and associated QTL mapping for cotton yield based on a cDNA-AFLP transcriptome map in an immortalized F 2. Theor. Appl. Genet. 2011;123:439–454. doi: 10.1007/s00122-011-1597-5. [DOI] [PubMed] [Google Scholar]
- 171.Semel Y, et al. Overdominant quantitative trait loci for yield and fitness in tomato. P. Natl Acad. Sci. USA. 2006;103:12981–12986. doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Larièpe A, et al. The genetic basis of heterosis: multiparental quantitative trait loci mapping reveals contrasted levels of apparent overdominance among traits of agronomical interest in maize (Zea mays L.) Genetics. 2012;190:795–811. doi: 10.1534/genetics.111.133447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li, Q. et al. The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot.66, 2515–2526 (2015). [DOI] [PubMed]
- 174.Hua, J. et al. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. P. Natl Acad. Sci. USA.100, 2574–2579 (2003). [DOI] [PMC free article] [PubMed]
- 175.Zhang L, et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 2017;8:1–10. doi: 10.1038/s41467-016-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yu Y, et al. Genome sequence and QTL analyses using backcross recombinant inbred lines (BILs) and BILF1 lines uncover multiple heterosis-related loci. Int. J. Mol. Sci. 2020;21:780. doi: 10.3390/ijms21030780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu Y, et al. Identification of quantitative trait loci for kernel-related traits and the heterosis for these traits in maize (Zea mays L.) Mol. Genet. Genom. 2020;295:121–133. doi: 10.1007/s00438-019-01608-1. [DOI] [PubMed] [Google Scholar]
- 178.Xu M, et al. Heterotic loci analysis for root traits of maize seedlings using an SSSL test population under different nitrogen conditions. Mol. Breed. 2020;40:1–12. doi: 10.1007/s11032-019-1080-6. [DOI] [Google Scholar]
- 179.Li X, et al. Maize GOLDEN2-LIKE genes enhance biomass and grain yields in rice by improving photosynthesis and reducing photoinhibition. Commun. Biol. 2020;3:1–12. doi: 10.1038/s42003-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mohapatra PP, Batra V. Tissue culture of potato (Solanum tuberosum L.): A review. Int. J. Curr. Microbiol. Appl. Sci. 2017;6:489–495. doi: 10.20546/ijcmas.2017.604.058. [DOI] [Google Scholar]
- 181.Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature. 2019;565:91–95. doi: 10.1038/s41586-018-0785-8. [DOI] [PubMed] [Google Scholar]
- 182.Wang C, et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 2019;37:283–286. doi: 10.1038/s41587-018-0003-0. [DOI] [PubMed] [Google Scholar]
- 183.Williams W. The isolation of ‘pure lines’ from F1 hybrids of tomato, and the problem of heterosis in inbreeding crop species. J. Agric. Sci. 1959;53:347–353. doi: 10.1017/S002185960002075X. [DOI] [Google Scholar]
- 184.Sarawat P, Stoddard F, Marshall D. Derivation of superior F5 lines from heterotic hybrids in pea. Euphytica. 1993;73:265–272. doi: 10.1007/BF00036705. [DOI] [Google Scholar]
- 185.Manrique-Carpintero NC, et al. Genome reduction in tetraploid potato reveals genetic load, haplotype variation, and loci associated with agronomic traits. Front. plant Sci. 2018;9:944. doi: 10.3389/fpls.2018.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 187.Li, X. et al. Establishment of Agrobacterium-mediated genetic transformation and application of CRISPR/Cas9 gene-editing system to Chinese cabbage (Brassica rapa L. ssp. pekinensis). (2020).
- 188.Liu S, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ma C, et al. Efficient BoPDS gene editing in cabbage by the CRISPR/Cas9 system. Hortic. Plant J. 2019;5:164–169. doi: 10.1016/j.hpj.2019.04.001. [DOI] [Google Scholar]
- 190.Kitashiba H, et al. Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res. 2014;21:481–490. doi: 10.1093/dnares/dsu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kim K, et al. Quantitative Trait Loci (QTLs) Associated with Microspore Culture in Raphanus sativus L.(Radish) Genes. 2020;11:337. doi: 10.3390/genes11030337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Consortium TG. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lee MH, et al. Temporal and spatial expression analysis of shoot-regeneration regulatory genes during the adventitious shoot formation in hypocotyl and cotyledon explants of tomato (cv. Micro-Tom) Int. J. Mol. Sci. 2020;21:5309. doi: 10.3390/ijms21155309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Consortium PGS. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 195.Zhang, C., Wang, D., Yang, Y. & Chen, Q. Construction of Potato DM1-3-516-R44 (DM) transgenic system based on agrobacterium transformation. (2020).
- 196.Kim S, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014;46:270–278. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- 197.Qin C, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl Acad. Sci. USA. 2014;111:5135–5140. doi: 10.1073/pnas.1400975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chee MJY, Lycett GW, Chin CF. Development of a direct transformation method by GFP screening and in vitro whole plant regeneration of Capsicum frutescens L. Electron. J. Biotechnol. 2018;34:51–58. doi: 10.1016/j.ejbt.2018.05.005. [DOI] [Google Scholar]
- 199.Hirakawa H, et al. Draft genome sequence of eggplant (Solanum melongena L.): the representative solanum species indigenous to the old world. DNA Res. 2014;21:649–660. doi: 10.1093/dnares/dsu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Khan, H. An Efficient Plant Regeneration System Via Leaf Derived Callus of Solanum melongena L in Propagation and Genetic Manipulation of Plants 93-100 (Springer, 2021).
- 201.Huang S, et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 202.Du, C., Fan, H., Liu, C. & Si, Y. Improved the Agrobacterium tumefaciens-mediated transformation of cucumber by a modified the using of antibiotics and acetosyringone. (2020).
- 203.Garcia-Mas J, et al. The genome of melon (Cucumis melo L.) Proc. Natl Acad. Sci. USA. 2012;109:11872–11877. doi: 10.1073/pnas.1205415109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chenarani Z, Shokouhifar F, Mamarabadi M. Study the Effects of Explants and Hormonal Levels onDirect Regeneration in Melon (Cucumis melo L., cv. Khatooni) J. Hortic. Sci. 2018;32:13–22. [Google Scholar]
- 205.Guo B, et al. Comparative proteomic analysis of embryos between a maize hybrid and its parental lines during early stages of seed germination. PLoS ONE. 2013;8:e65867. doi: 10.1371/journal.pone.0065867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vasudevan V, Siva R, Krishnan V, Manickavasagam M. Polyamines, sonication and vacuum infiltration enhances the Agrobacterium-mediated transformation in watermelon (Citrullus lanatus Thunb.) S. Afr. J. Bot. 2020;128:333–338. doi: 10.1016/j.sajb.2019.11.031. [DOI] [Google Scholar]
- 207.Montero‐Pau J, et al. De novo assembly of the zucchini genome reveals a whole‐genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2018;16:1161–1171. doi: 10.1111/pbi.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sun H, et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant. 2017;10:1293–1306. doi: 10.1016/j.molp.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 209.Guo J, Li Y, He C, Yan Y, Yu X. Establishing a High-efficiency Regeneration System in Pumpkin (Cucurbita moschata) Chin. Bull. Bot. 2019;54:539. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.