Abstract
Group 3 innate lymphoid cells (ILC3s) play critical roles in innate immunity and gut homeostasis. However, how ILC3 homeostasis is regulated remains elusive. Here, we identified a novel circular RNA, circZbtb20, that is highly expressed in ILC3s and required for their maintenance and function. CircZbtb20 deletion causes reduced ILC3 numbers, increasing susceptibility to C. rodentium infection. Mechanistically, circZbtb20 enhances the interaction of Alkbh5 with Nr4a1 mRNA, leading to ablation of the m6A modification of Nr4a1 mRNA to promote its stability. Nr4a1 initiates Notch2 signaling activation, which contributes to the maintenance of ILC3 homeostasis. Deletion of Alkbh5 or Nr4a1 also impairs ILC3 homeostasis and increases susceptibilities to bacterial infection. Thus, our findings reveal an important role of circular RNA in the regulation of innate lymphoid cell homeostasis.
Keywords: circZbtb20, ILC3, Alkbh5, Nr4a1, Homeostasis
Subject terms: Innate immune cells, Cell signalling
Introduction
Innate lymphoid cells (ILCs) play critical roles in sustaining tissue homeostasis, promoting lymphoid organogenesis and orchestrating host defense.1 The development of ILCs is delicately regulated by fate-decision transcription factors (TFs).2 All ILC lineages are derived from common lymphoid progenitors (CLPs), which can differentiate into common innate lymphoid progenitors (CILPs) and then give rise to common helper innate lymphoid progenitors (CHILPs) and NK cell precursors (NKPs). CHILPs can differentiate into lymphoid tissue inducer progenitors and innate lymphoid cell precursors (ILCPs), which finally give rise to all subsets of ILCs. ILCs encompass group 1 ILCs (ILC1s), group 2 ILCs (ILC2s), and group 3 ILCs (ILC3s). ILC1s express T-bet and produce interferon-γ (IFN-γ). ILC2s require Gata3 for maintenance and secrete Th2-like cytokines, such as IL-5 and IL-13. ILC3s express RORγt and mainly produce IL-22 and/or IL-17 after activation.2 ILC3s are classified into tissue inducer (LTi) cells, NKp46+ ILC3s, and CCR6−NKp46− ILC3s.3 ILC3s mainly reside in the gut and play diverse important roles, including promoting lymphoid tissue development, regulating adaptive immunity, and controlling the containment of commensal bacteria4. It has been reported that dysregulation of ILC3s causes some diseases, such as colitis. ILC3 maintenance and function are regulated by a variety of factors. For example, the aryl hydrocarbon receptor (Ahr) is required for the promotion of ILC3 maintenance and function.5 We previously showed that WASH can activate AHR expression to promote the maintenance and effector functions of NKp46+ ILC3s.6 We also demonstrated that the lncRNA lncKdm2b enhances the expansion of ILC3s.7
Circular RNAs (circRNAs) are characterized by a covalent bond linking the 3′ and 5′ ends generated by back-splicing.8 CircRNAs originate from precursor mRNAs (pre-mmRNAs) and may comprise exons, introns, or exon–introns.9 CircRNAs harbor a long half-life and are resistant to exonuclease degradation. CircRNAs are widely expressed in various tissues and cell types. CircRNAs were previously considered byproducts of mRNA splicing.10 Accumulating evidence shows that circRNAs perform critical functions in many biological processes, including tumorigenesis,11 neuropsychiatric disorders12, and immunity.13 For example, the circRNA Cdr1as sponges miR-7 and miR-671 to regulate brain function.12 Fusion circRNAs derived from transcribed exons of some genes contribute to tumorigenesis and resistance to therapy.11 We recently demonstrated that circRNA cia-cGAS is highly expressed in hematopoietic stem cells (HSCs) and maintains HSC homeostasis by blocking cGAS enzymatic activity.10 We also showed that the circRNA circPan3 is required for the self-renewal of intestinal stem cells.14 However, how circRNAs regulate ILC biology remains elusive.
Nr4a orphan nuclear receptors, including Nr4a1 (Nur77), Nr4a2 (Nurr1), and Nr4a3 (Nor1), are members of the TF nuclear receptor family involved in the regulation of tissue responses.15 Increasing evidence demonstrates a close correlation between Nr4a receptors and immune regulation. Deficiency of Nr4a3 and Nr4a1 contributes to the tumorigenesis of acute myeloid leukemia.16 Nr4a1 causes T-cell dysfunction by inhibiting effector T-cell differentiation, while Nr4a1 deletion enhances the T cell-mediated antitumor response.17 Nr4a1 is also involved in the differentiation of Ly6C− monocytes.18 Loss of Nr4a1 arrests the progression of the cell cycle and leads to apoptosis of Ly6C− monocytes. In addition, Nr4a1 promotes Foxp3 transcription and is required for Treg cell development.19 Nr4a1 has been implicated in the activation and differentiation of macrophages in atherosclerosis.20 Moreover, Nr4a1 is also a critical regulator of inflammation, especially neuroinflammation.15 However, how Nr4a receptors modulate ILCs is largely unknown. Here, we identified a novel circRNA, circZbtb20, which originates from the Zbtb20 gene transcript (gene symbol: mmu_circRNA_29739 or mmu_circ_0006298) that is highly expressed in ILC3s. CircZbtb20 promotes ILC3 homeostasis and function via Alkbh5-dependent m6A demethylation of Nr4a1 mRNA.
Results
CircZbtb20 knockout impairs ILC3 maintenance and function
Since the functions of circRNAs in ILC biology are largely unclear, we previously performed a circRNA microarray of ILC3s from Rag1−/− mice to investigate how circRNAs regulate ILC3-mediated innate colitis.21 Based on the microarray analysis, we selected ten circRNAs that were highly expressed in Rag1−/− ILC3s and validated them as circRNAs via PCR (Supplementary Fig. 1a) and RNase R digestion (Supplementary Fig. 1b). To examine whether these circRNAs play critical roles in ILC3s, we knocked them down in isolated lamina propria lymphocytes (LPLs) using a lentivirus (Supplementary Fig. 1c) and analyzed their effects on ILC3 expansion. Of these ten circRNAs, only circZbtb20 depletion inhibited ILC3 proliferation (Supplementary Fig. 1d). CircZbtb20 is formed by back-splicing of Zbtb20 transcripts from exon 8. We then confirmed the back-splicing site by DNA sequencing (Supplementary Fig. 1e). Many circRNAs are conserved across species. The human ortholog circZBTB20 was validated in human LPLs by DNA sequencing (Supplementary Fig. 1f), indicating that circZbtb20 was highly conserved between mice and humans. We observed that circZbtb20 was uniquely expressed in some tissues, especially in LPLs (Fig. 1a). Among the lymphocyte progenitors and lymphocytes we tested, circZbtb20 was most highly expressed in intestinal ILC3s (Fig. 1b and Supplementary Fig. 2a–i), which was further confirmed by a FISH assay (Fig. 1c). However, we noticed that Zbtb20 mRNA was highly expressed in several types of lymphocytes (Supplementary Fig. 2j), suggesting that the back-splicing of circZbtb20 in ILC3s is uniquely active. In addition, circZbtb20 was mainly located in the nucleus of ILC3s (Fig. 1c and Supplementary Fig. 2k).
Fig. 1.
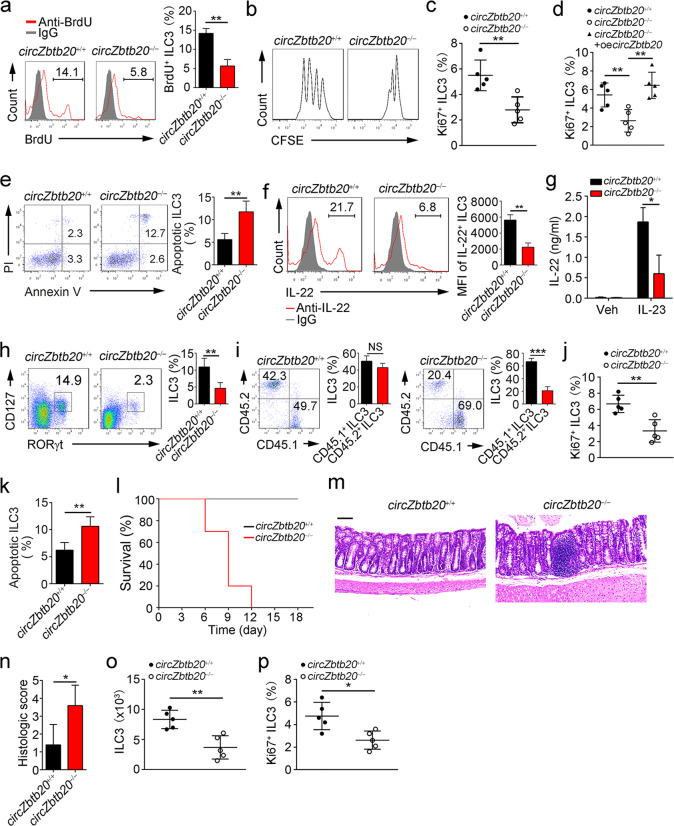
CircZbtb20 knockout impairs ILC3 maintenance and function. a CircZbtb20 expression was analyzed in the indicated tissues using Northern blotting. Probes were biotin labeled, and 18S was used as a loading control. b Relative expression of circZbtb20 was measured in the indicated hematopoietic cells using qRT-PCR. Fold changes were normalized to endogenous Actb. HSCs (Lin−c-Kit+Sca-1+CD150+CD48−), MPPs (Lin−c-Kit+Sca-1+CD150−CD48+), CLPs (Lin−CD127+c-KitintSca-1intFlt3+α4β7−), CHILPs (Lin−CD25−CD127+Flt3−α4β7+Id2GFP), and ILCPs (Lin−CD127+Flt3−c-Kit+α4β7+PLZFGFP) were isolated from BM cells. ILC1s (CD3−CD19−CD127+NK1.1+NKp46+), ILC2s (Lin−CD127+Thy1.2+KLRG1+), and ILC3s (CD3−CD19−CD45+RORγtGFP) were isolated from small intestines. NK cells (NK1.1+) and B cells (CD19+) were isolated from mouse spleens. T cells (CD3+) were isolated from mouse thymuses. c CircZbtb20 expression was measured using biotin-labeled probes by FISH. ILC3s (Lin−CD45lowCD90high) were isolated from small intestines by FACS. Scale bar, 5 μm. d CLPs, CHILPs, and ILCPs were analyzed in circZbtb20+/+ and circZbtb20−/− mice by FACS. CLPs were gated on Lin−CD127+α4β7−Flt3+. CHILPs were gated on Lin−CD25−Flt3−CD127+. ILCPs were gated on Lin−Flt3−PLZF+CD127+. e ILC3s were analyzed in the small intestines of circZbtb20+/+ and circZbtb20−/− mice by FACS. ILC3s were gated on Lin−CD45+. f ILC3s in small intestines from circZbtb20+/+ and circZbtb20−/− mice were visualized by immunofluorescence staining in situ. Yellow arrows indicate ILC3s. Scale bars, 20 μm. g Bacterial counts (CFUs) in spleens, livers, and feces were measured. CircZbtb20+/+ and circZbtb20−/− mice were infected with C. rodentium for 6 days. n = 5 for each group. h Colon lengths were analyzed after infection with C. rodentium. i Colon tissues from circZbtb20+/+ and circZbtb20−/− mice were analyzed by H&E staining. Scale bars, 100 μm. j Histological scores of colons from circZbtb20+/+ and circZbtb20−/− mice in (i). k Body weight changes of circZbtb20+/+ and circZbtb20−/− mice were analyzed after infection with C. rodentium. n = 5 for each group. l Survival rates of circZbtb20+/+ and circZbtb20−/− mice were measured after infection with C. rodentium. n = 10 for each group. m Analysis of IL-22+ ILC3s in circZbtb20+/+ and circZbtb20−/− mice after C. rodentium infection. n The amount of IL-22 protein secreted by LPLs from C. rodentium-infected mice was measured via ELISA after stimulation with IL-23 for 24 h. *P < 0.05, **P < 0.01, and ***P < 0.001. Data are shown as the mean ± SD. Data are representative of at least three independent experiments.
RNA-binding proteins (RBPs) are required for the biogenesis of circRNAs and can bind to flanking introns of back-splicing junctions.22–24 Using Zbtb20 pre-mRNA biotin probes against the upstream intron, we performed RNA pulldown of LPL cell lysates, followed by mass spectrometry. Rbpms2, an RBP, was identified as a potential interaction candidate with Zbtb20 mRNA (Supplementary Fig. 3a, b). The interaction of Rbpms2 with Zbtb20 mRNA was validated by a RIP assay (Supplementary Fig. 3c). Notably, Rbpms2 was specifically expressed in ILC3s among the lymphocytes we tested (Supplementary Fig. 3d). We then depleted Rbpms2 expression in ILC3s (Supplementary Fig. 3e). We found that Rbpms2 depletion dramatically suppressed circZbtb20 expression (Supplementary Fig. 3f). In contrast, Zbtb20 expression was not affected by Rbpms2 knockdown (Supplementary Fig. 3g).
To further determine the physiological role of circZbtb20 in the regulation of ILC3 function, we sought to generate circZbtb20-deficient mice. The complementary elements flanking circRNA sequences are essential for their generation.9 We screened the complementary sequences in the introns flanking circZbtb20 and constructed plasmids for minigene assays. We defined the complementary sequence for circZbtb20 formation (Supplementary Fig. 3h). We thus generated circZbtb20 knockout mice (circZbtb20−/−) by deleting the complementary element in the genome with CRISPR/Cas9 technology (Supplementary Fig. 3i). CircZbtb20 deletion was validated in circZbtb20−/− mice through PCR, Northern blot, and qPCR analyses (Supplementary Fig. 3j–l). Notably, deletion of circZbtb20 did not affect the expression of its maternal gene Zbtb20 (Supplementary Fig. 3l, m). Additionally, circZbtb20−/− mice displayed normal reproductive capacity, with Mendelian ratios but lost body weight compared to their littermate controls. We next overexpressed Rbpms2 in ILC3s (Supplementary Fig. 3n), and Rbpms2 overexpression promoted circZbtb20 expression in ILC3s (Supplementary Fig. 3o). In contrast, Rbpms2 overexpression failed to enhance circZbtb20 expression in circZbtb20-deficient ILC3s (Supplementary Fig. 3n, o). We conclude that Rbpms2 is uniquely expressed in ILC3s and is required for the biogenesis of circZbtb20 in ILC3s.
We found that circZbtb20−/− mice and circZbtb20+/+ littermate WT control mice displayed comparable numbers of ILC progenitors, including CLPs, CHILPs, and ILCPs in the bone marrow (BM) (Fig. 1d). Moreover, circZbtb20 deficiency did not affect the number of intestinal ILC1s or ILC2s (Supplementary Fig. 4a–c). Notably, circZbtb20 knockout did not impact the number of intestinal T cells (Supplementary Fig. 4d). However, circZbtb20 deletion decreased intestinal ILC3 percentages and numbers (Fig. 1e and Supplementary Fig. 4c), and these changes were further verified by immunofluorescence staining in situ (Fig. 1f). ILC3s are quite heterogeneous, including CCR6+ LTi-like, T-bet+, and CCR6−T-bet− ILC3 subsets.14 We noticed that circZbtb20 was expressed in these three subgroups of ILC3s and most highly expressed in the CCR6−T-bet− ILC3 subset (Supplementary Fig. 4e). In addition, circZbtb20 deletion decreased the abundance of all three subgroups of ILC3s (Supplementary Fig. 4f). ILC3s are essential for gut homeostasis and bacterial defense due to secretion of IL-22.1,25 As expected, circZbtb20−/− mice were more susceptible to C. rodentium infection (Fig. 1g), which was accompanied by colon shortening and intestinal damage (Fig. 1h–j). Moreover, circZbtb20−/− mice lost more body weight and exhibited a lower survival rate than did their circZbtb20+/+ littermates after C. rodentium infection (Fig. 1k, l). IL-22 plays a critical role in protection against C. rodentium.26 In parallel, circZbtb20−/− ILC3s showed an attenuated ability to produce IL-22 after infection (Fig. 1m, n). Notably, Il22 mRNA was dramatically reduced in circZbtb20−/− ILC3s after C. rodentium infection (Supplementary Fig. 4g). Moreover, the numbers of IL-22+ ILC3s were much lower in circZbtb20−/− mice (Supplementary Fig. 4h). In addition, we noticed that C. rodentium infection did not affect circZbtb20 expression (Supplementary Fig. 4i).
To exclude the effect of Zbtb20 on the circZbtb20-mediated function of ILC3s, we also established Zbtb20flox/flox (Zbtb20+/+) mice. To conditionally delete Zbtb20 in ILC3s, we generated Zbtb20flox/floxRorc-Cre (Zbtb20−/−) mice by crossing Zbtb20flox/flox mice with Rorc-Cre mice. Zbtb20 was completely deleted in ILC3s of Zbtb20−/− mice (Supplementary Fig. 5a). Of note, we deleted the exon 7 locus of the Zbtb20 gene for the Zbtb20 deletion model (Supplementary Fig. 5a), which did not affect circZbtb20 expression (Supplementary Fig. 5b), suggesting that exon 7 deletion of the Zbtb20 gene does not impact circZbtb20 formation. We found that Zbtb20 deficiency did not affect the number of intestinal ILC3s (Supplementary Fig. 5c). In addition, Zbtb20−/− mice and Zbtb20+/+ littermate control mice showed comparable resistance to C. rodentium infection (Supplementary Fig. 5d, e), suggesting that Zbtb20 has no influence on ILC3 development and function. Taken together, these findings show that circZbtb20 is highly expressed in ILC3s and required for ILC3 maintenance and function.
CircZbtb20 is required for the proliferation and survival of ILC3s
To determine the turnover rate of ILC3s in vivo, 1 mg of BrdU was intraperitoneally injected into mice for 18 h. Then, BrdU uptake was analyzed by FACS. We found that ILC3s derived from circZbtb20−/− mice took up much less BrdU than those from littermate control mice (Fig. 2a). We then isolated ILC3s from circZbtb20+/+ and circZbtb20−/− mice, labeled them using CFSE, and transferred them into Rag1−/−Il2rg−/− mice for the cell proliferation assay. We noticed that circZbtb20−/− ILC3 proliferation was reduced compared to that of littermate WT ILC3s (Fig. 2b). Consistently, there were far fewer Ki67+ ILC3s in circZbtb20−/− mice than in circZbtb20+/+ mice (Fig. 2c). Notably, circZbtb20 overexpression in circZbtb20−/− ILC3s restored the number of Ki67+ circZbtb20−/− ILC3s (Fig. 2d and Supplementary Fig. 5f). In addition, circZbtb20 deficiency promoted the apoptosis of ILC3s (Fig. 2e). Interestingly, circZbtb20−/− ILC3s showed decreased potential for IL-22 or IL-17A production (Fig. 2f, g and Supplementary Fig. 5g). Overall, circZbtb20 is required for the proliferation and survival of ILC3s.
Fig. 2.
CircZbtb20 is required for the proliferation and survival of ILC3s. a BrdU+ ILC3s in circZbtb20+/+ and circZbtb20−/− mice were analyzed by FACS. 1 mg of BrdU was intraperitoneally injected into mice. Eighteen hours later, ILC3s in the small intestine were detected. n = 5 for each group. b ILC3s (Lin−CD45lowCD90high) were isolated from circZbtb20+/+ and circZbtb20−/− mice, labeled with CFSE and transferred into Rag1−/−Il2rg−/− mice for cell proliferation assays. Seven days later, CFSE-labeled ILC3s were analyzed through FACS. c Ki67+ ILC3s in circZbtb20+/+ and circZbtb20−/− mice were measured by FACS. n = 5 for each group. d Analysis of Ki67+ ILC3s isolated from circZbtb20+/+ and circZbtb20−/− mice. LPLs were infected with control or circZbtb20-overexpressing (OE) virus. Infected cells were cultured at 37 °C for 24 h. e Detection of ILC3 apoptosis by Annexin-V/PI staining. n = 5 for each group. f LPLs were isolated from circZbtb20+/+ and circZbtb20−/− mice and stimulated with IL-23 for 4 h. Then, IL-22-producing ILC3s were analyzed by FACS. The MFI of IL-22+ ILC3s is presented on the right. n = 5 for each group. g LPLs were isolated from circZbtb20+/+ and circZbtb20−/− mice and stimulated with IL-23. Then, IL-22 protein levels were measured by ELISA. h A total of 5 × 106 CD45.2+ BM cells from circZbtb20+/+ or circZbtb20−/− mice were transferred into lethally irradiated CD45.1+ recipients. Eight weeks later, ILC3s (Lin−CD45.2+CD127+RORγt+) were analyzed. n = 5 for each group. i A 1:1 mixture of CD45.1+ wild-type and CD45.2+ circZbtb20+/+ or circZbtb20−/− BM cells was transferred into lethally irradiated CD45.1+ recipient mice for 8 weeks. Then, the ratios of CD45.1+ to CD45.2+ ILC3s in chimeras were analyzed by gating on CD45.2+Lin−RORγt+ (circZbtb20+/+ or circZbtb20−/−) and CD45.1+Lin−RORγt+ (WT) populations. n = 5 for each group. j Analysis of Ki67 ILC3s in reconstituted mice of (h). k Analysis of ILC3 apoptosis in reconstituted mice of (h). l Reconstituted mice were infected with C. rodentium, and survival rates were measured. n = 10 for each group. m HE staining of colons isolated from reconstituted mice infected with C. rodentium. n Histological scores of colons from circZbtb20+/+ and circZbtb20−/− reconstituted mice in (m). o, p CHILPs (Lin−Flt3−CD25−CD127+α4β7+CD244+) were adoptively transferred into Rag1−/−Il2rg−/− mice. Six weeks later, ILC3 numbers and Ki67+ ILC3s were analyzed. n = 5 for each group. *P < 0.05, **P < 0.01, and ***P < 0.001. Data are shown as the mean ± SD. Data are representative of at least three independent experiments.
To further determine whether the role of circZbtb20 in ILC3s is intrinsic or extrinsic, we transplanted 5 × 106 CD45.2+ circZbtb20−/− or circZbtb20+/+ mouse BM cells into lethally irradiated CD45.1+ recipients. After 8 weeks, we found that recipients of circZbtb20−/− BM cells had decreased ILC3 numbers (Fig. 2h). We also conducted competitive BM transplantation assays. We transferred a 1:1 mixture of CD45.1+ WT and CD45.2+ circZbtb20+/+ or circZbtb20−/− BM cells into lethally irradiated recipient mice for 8 weeks. Recipients reconstituted with circZbtb20−/− BM cells showed reduced ILC3 numbers compared to those of recipients reconstituted with circZbtb20+/+ BM cells (Fig. 2i). Furthermore, the proliferation and survival of ILC3s derived from circZbtb20−/− BM cells were impaired (Fig. 2j, k). Consequently, circZbtb20−/− BM cell-derived recipients were also susceptible to C. rodentium infection (Fig. 2l–n). To further validate the role of circZbtb20 in the regulation of ILC3s, we transferred circZbtb20−/− and circZbtb20+/+ CHILPs into Rag1−/−Il2rg−/− mice, followed by examination of ILC3 numbers and proliferation at 6 weeks. We observed that circZbtb20−/− CHILPs generated fewer ILC3s and that ILC3s displayed reduced proliferation rates (Fig. 2o, p). These data indicate that circZbtb20 regulates ILC3 proliferation and survival in an intrinsic manner.
CircZbtb20 interacts with Nr4a1 mRNA to enhance its stability
To explore the downstream signaling of circZbtb20-mediated ILC3 regulation, we isolated ILC3s from circZbtb20+/+ and circZbtb20−/− mice to conduct microarray analysis. We focused on the top 10 downregulated TFs in circZbtb20−/− ILC3s (Fig. 3a and Supplementary Table 1). Among them, Nr4a1 was the most downregulated TF (Fig. 3b). Nr4a1 downregulation was confirmed by western blotting (Fig. 3c), which was further validated by immunofluorescence staining (Fig. 3d). Notably, circZbtb20 was not enriched on the promoter of Nr4a1 (Fig. 3e). Moreover, circZbtb20 deletion did not affect the luciferase activity of the Nr4a1 promoter (Fig. 3f), suggesting that circZbtb20 has no effect on Nr4a1 gene transcription. Intriguingly, circZbtb20 deficiency caused rapid degradation of Nr4a1 mRNA in ILC3s (Fig. 3g), suggesting that circZbtb20 may regulate the stability of Nr4a1 mRNA.
Fig. 3.
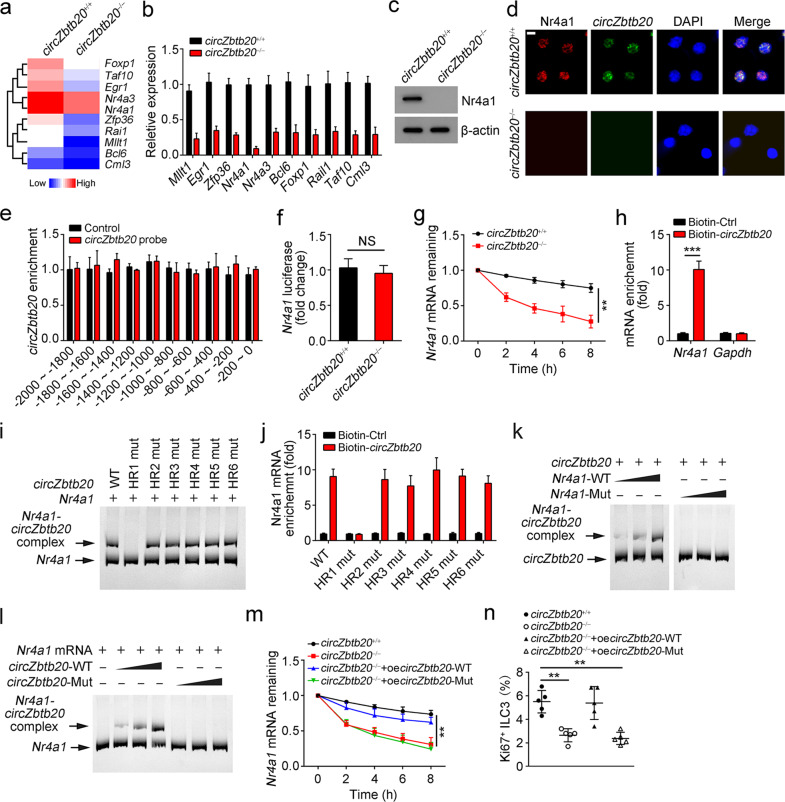
CircZbtb20 interacts with Nr4a1 mRNA to enhance its stability. a Heatmap of the ten most downregulated TFs in circZbtb20−/− ILC3s versus circZbtb20+/+ ILC3s by microarray analysis. b Relative expression levels of the top 10 TFs were analyzed in circZbtb20+/+ and circZbtb20−/− ILC3s by qRT-PCR. Fold changes were normalized to endogenous Actb. c Protein levels of Nr4a1 were analyzed in circZbtb20+/+ and circZbtb20−/− LPLs by western blotting. d Immunofluorescence staining of Nr4a1 in circZbtb20+/+ and circZbtb20−/− ILC3s. Scale bar, 5 μm. e Enrichment of circZbtb20 on the Nr4a1 gene promoter was analyzed by a CHIRP assay. The CircZbtb20 probe was biotin labeled. f A luciferase reporter assay was performed using circZbtb20+/+ and circZbtb20−/− ILC3s ater transfection with an Nr4a1 promoter-containing reporter vector. NS no significance. g Nr4a1 mRNA degradation in ILC3s treated with 2 μg/ml actinomycin-D for the indicated times was analyzed by qRT-PCR. Residual RNAs were normalized to 0 h. h Enrichment of Nr4a1 mRNA was analyzed by qPCR. Biotin-labeled linear circZbtb20 was incubated with LPL lysates for the RNA pulldown assay. Gapdh mRNA was used as a negative control. i RNA mobility-shift assay of Nr4a1 mRNA with WT circZbtb20 and mutated circZbtb20 transcript (Mut). CircZbtb20-Mut, loop-hairpin (HR) mutation of HR1–HR6. j RNA pulldown using biotin-labeled WT circZbtb20 or mutant circZbtb20 to test the enrichment of Nr4a1 mRNA in LPL lysates. k RNA mobility-shift assay of circZbtb20 with WT Nr4a1 mRNA and the indicated mutant (Mut). CircZbtb20 was biotin labeled. l RNA mobility-shift assay of Nr4a1 mRNA with WT circZbtb20 and the indicated mutant (Mut). Nr4a1 mRNA was biotin labeled. m Nr4a1 mRNA degradation in ILC3s treated with 2 μg/ml actinomycin-D for the indicated times was analyzed by qRT-PCR. Residual RNAs were normalized to 0 h. circZbtb20−/− ILC3s were infected with WT or Mut circZbtb20. n Ki67+ ILC3s were analyzed after infection with lentivirus overexpressing WT or Mut circZbtb20. **P < 0.01 and ***P < 0.001. Data are shown as the mean ± SD. Data are representative of at least three independent experiments.
Through an RNA pulldown assay, we found that circZbtb20 interacted with Nr4a1 mRNA (Fig. 3h). CircRNA loops play critical roles in the RNA interactome.27 Thus, we predicted the loop structure of circZbtb20 with a bioinformatics tool, and six hairpin regions (HR1–6) were predicted (Supplementary Fig. 5h). We then mutated each loop of the circZbtb20 transcript to determine which loop was a potential loop mediating their interaction. Through an RNA mobility-shift assay, we found that only HR1 mutation abrogated the direct interaction of circZbtb20 with Nr4a1 mRNA (Fig. 3i), which was further verified by an RNA pulldown assay (Fig. 3j). Through sequence alignment, we noticed that pairing of complementary bases occurred between the HR1 sequence of circZbtb20 (nt: 238–248) and Nr4a1 mRNA (nt: 1144–1154) (Supplementary Fig. 5i). To test whether these pairing bases are required for their interaction, we then mutated the pairing bases of circZbtb20 or Nr4a1 mRNA, followed by an RNA mobility-shift assay. We noticed that mutation of the pairing bases from either circZbtb20 or Nr4a1 mRNA abrogated their interaction (Fig. 3k, l). Importantly, only overexpression of WT circZbtb20 in circZbtb20−/− ILC3s maintained the stability of Nr4a1 mRNA (Fig. 3m and Supplementary Fig. 5j). Consequently, only WT circZbtb20 overexpression rescued the proliferation of ILC3s (Fig. 3n). Collectively, these finding show that circZbtb20 enhances the stability of Nr4a1 mRNA via direct interaction.
CircZbtb20 recruits Alkbh5 to ablate the m6A modification of Nr4a1 mRNA to promote its stability
To further reveal how circZbtb20 regulated the stability of Nr4a1 mRNA, we performed an RNA pulldown assay using biotin-labeled linearized circZbtb20 as bait, followed by silver staining and mass spectrometry. We identified Alkbh5 as a potential candidate interacting with circZbtb20 in LPL lysates (Fig. 4a and Supplementary Fig. 5k). The interaction of circZbtb20 with Alkbh5 was validated by RNA pulldown assay (Fig. 4b, c). Through domain mapping, we found that the region (1200–1605) of the circZbtb20 transcript was needed for its interaction with Alkbh5 (Fig. 4d, e). Moreover, the circZbtb20 region (1200–1605) directly interacted with Alkbh5 in an RNA-EMSA (Fig. 4f). Notably, circZbtb20 enhanced the interaction of Alkbh5 with Nr4a1 mRNA (Fig. 4g). As expected, circZbtb20 deletion impaired the interaction of Nr4a1 mRNA with Alkbh5 in circZbtb20-deficient LPLs, whereas circZbtb20 overexpression in circZbtb20-deficient LPLs restored this interaction (Fig. 4h). Notably, the interaction between Alkbh5 and Nr4a1 mRNA in circZbtb20+/− LPLs was attenuated, whereas the addition of exogenous circZbtb20 enhanced their interaction (Supplementary Fig. 5l). Importantly, we found that Alkbh5 also interacted with circZbtb20 and Nr4a1 mRNA in LPL lysates by RIP assay (Supplementary Fig. 5m). As expected, circZbtb20 deficiency dramatically decreased the interaction of Alkbh5 with Nr4a1 mRNA (Supplementary Fig. 5n).
Fig. 4.
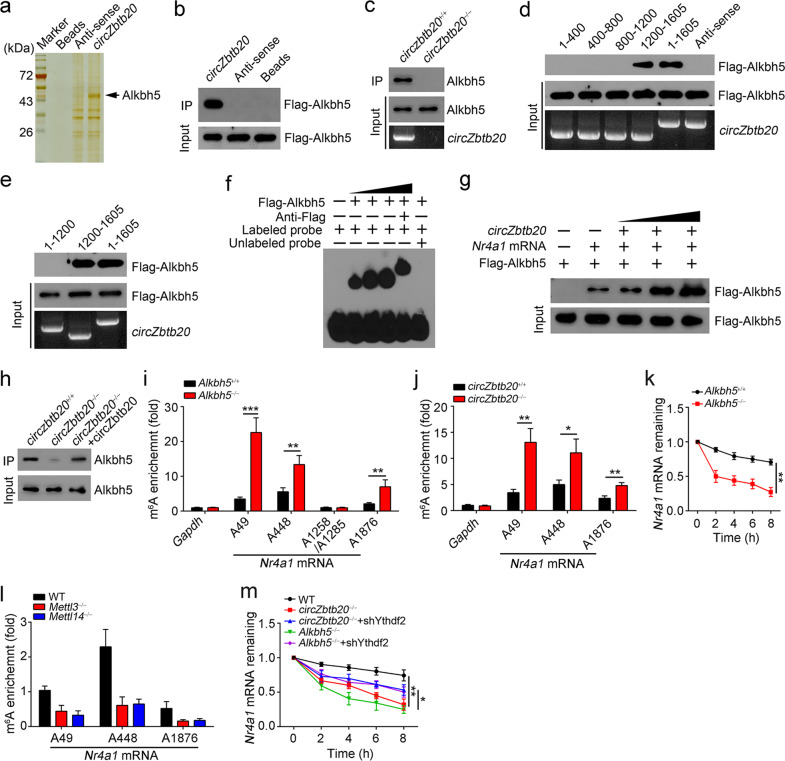
CircZbtb20 recruits Alkbh5 to ablate the m6A modification of Nr4a1 mRNA to promote its stability. a Biotin-labeled circZbtb20 and antisense antibodies were used for the RNA pulldown assay. Eluted fractions were resolved by SDS-PAGE followed by silver staining and mass spectrometry. b Alkbh5-overexpressing 293T cells were lysed and incubated with biotin-labeled circZbtb20, antisense, or bead control, followed by western blotting. c RNA pulldown assay using biotin-labeled circZbtb20 probes with circZbtb20+/+ and circZbtb20−/− LPL lysates. d, e RNA pulldown assay using biotin-labeled circZbtb20 truncations and Alkbh5-overexpressing 293T lysates. f RNA-EMSA using biotin-labeled circZbtb20 (1200–1605) and Flag-Alkbh5 protein. g RNA pulldown was performed using Nr4a1 mRNA with or without circZbtb20. Nr4a1 mRNA was precipitated by biotin probes and purified. h RNA pulldown was carried out using a biotin-labeled Nr4a1 mRNA probe and circZbtb20+/+ or circZbtb20−/− LPL lysates. i m6A enrichment on the A49, A448, A1258/A1285, or A1876 site of Nr4a1 mRNA in Alkbh5+/+ and Alkbh5−/− LPL (Lin−CD45+) lysates was analyzed by m6A-RIP-qPCR. The fold changes were 6.5-fold at A49, 2.4-fold at A448 and 3.3-fold at A1876 (Alkbh5−/− group versus Alkbh5+/+ group). IgG was used as a normalization controls. Gapdh: m6A negative control. j m6A enrichment on the A49, A448, or A1876 site of Nr4a1 mRNA in circZbtb20+/+ and circZbtb20−/− LPL (Lin−CD45+) lysates was measured by m6A-RIP-qPCR. Fold changes were 3.8-fold at A49, 2.2-fold at A448 and 2.0-fold at A1876 (circZbtb20−/− group versus circZbtb20+/+ group). k Nr4a1 mRNA degradation in Alkbh5+/+ or Alkbh5−/− ILC3s treated with 2 μg/ml actinomycin-D for the indicated times was analyzed by qRT-PCR. Residual RNAs were normalized to 0 h. l m6A enrichment on the A49, A448, or A1876 site of Nr4a1 mRNA in Mettl3−/−, Mettl14−/−, and WT LPL (Lin−CD45+) lysates was determined by m6A-RIP-qPCR. m Nr4a1 mRNA degradation in the indicated ILC3s treated with 2 μg/ml actinomycin-D for the indicated times was analyzed by qRT-PCR. Residual RNAs were normalized to 0 h. *P < 0.05, **P < 0.01, and ***P < 0.001. Data are shown as the mean ± SD. Data are representative of at least three independent experiments.
To further examine the physiological role of Alkbh5 in the regulation of Nr4a1 mRNA stability, we generated Alkbh5-deficient mice via CRISPR/Cas9 technology (Supplementary Fig. 5o). Alkbh5 is a demethylase for m6A modification of mRNA and is involved in the regulation of mRNA stability.28 Previous reports suggested that GGACU is the motif with the greatest potential for m6A modification.29,30 We thus used SELECT (a single-base elongation- and ligation-based qPCR amplification method) assays to determine whether the GGACU motifs (A49, A448, A1258, A1285, and A1876) in Nr4a1 mRNA underwent m6A modification. We found that the m6A modification existed on the A49, A448, and A1876 sites in Nr4a1 mRNA (Supplementary Fig. 6a, b). Then, we performed an m6A-RIP-qPCR assay and found that Alkbh5 deletion resulted in an increase in the m6A modification on the A49, A448, or A1876 site of Nr4a1 mRNA in ILC3s (Fig. 4i), similar to that of circZbtb20-deficient ILC3s (Fig. 4j). Consequently, Alkbh5−/− ILC3s accelerated the degradation of Nr4a1 mRNA (Fig. 4k). Mettl3 and Mettl14 are two major methyltransferases for m6A modification.31 We observed that Mettl3 or Mettl14 deficiency suppressed the m6A modification of Nr4a1 mRNA (Fig. 4l). We observed that circZbtb20 did not undergo m6A modification (Supplementary Fig. 6c). Of note, in the absence of circZbtb20, a weak interaction of Alkbh5 with m6A-modified Nr4a1 mRNA existed (Supplementary Fig. 6d), suggesting that m6A-modified Nr4a1 mRNA is a substrate of Alkbh5. It has been reported that Ythdf2 can recognize m6A-modified mRNA and facilitate its degradation.32 We noticed that Ythdf2 interacted with Nr4a1 mRNA in LPL lysates (Supplementary Fig. 6e). Ythdf2 was then silenced in circZbtb20- and Alkbh5-deficient ILC3s (Supplementary Fig. 6f). We found that Ythdf2 depletion rescued the stability of Nr4a1 mRNA in circZbtb20- or Alkbh5-deficient ILC3s (Fig. 4m). Taken together, these findings show that circZbtb20 recruits Alkbh5 to remove the m6A modification of Nr4a1 mRNA, promoting Nr4a1 mRNA stability.
Nr4a1 initiates Notch2 signaling to maintain ILC3 homeostasis
Whether Nr4a1 has a role in ILC3s remains unclear. To define this hypothesis, we generated Nr4a1−/− mice via a CRISPR/Cas9 approach (Supplementary Fig. 6g). We found that Nr4a1 deficiency remarkably reduced ILC3 numbers compared to those of littermate WT control mice (Fig. 5a, b). Consistently, Nr4a1 deficiency also decreased ILC3 proliferation (Fig. 5c). Moreover, Nr4a1 deletion also suppressed the secretion of IL-22 by ILC3s (Fig. 5d). Importantly, DKO (circZbtb20−/−Nr4a1−/−) ILC3s displayed a much lower proliferation rate than Nr4a1−/− ILC3s (Fig. 5e). Notably, Nr4a1 overexpression in Nr4a1−/− and DKO ILC3s rescued ILC3 proliferation (Fig. 5e and Supplementary Fig. 6h). In addition, Nr4a1 overexpression in circZbtb20-deficient CHILPs rescued ILC3-producing ability in in vitro differentiation assays (Supplementary Fig. 6i). Notably, DKO mice showed far fewer total or IL-22-producing ILC3s than Nr4a1−/− mice (Fig. 5f–h). Furthermore, DKO mice were much more susceptible to bacterial infection than littermate WT control mice (Supplementary Fig. 6j–m), suggesting that Nr4a1 is involved in the regulation of circZbtb20-mediated ILC3 homeostasis.
Fig. 5.
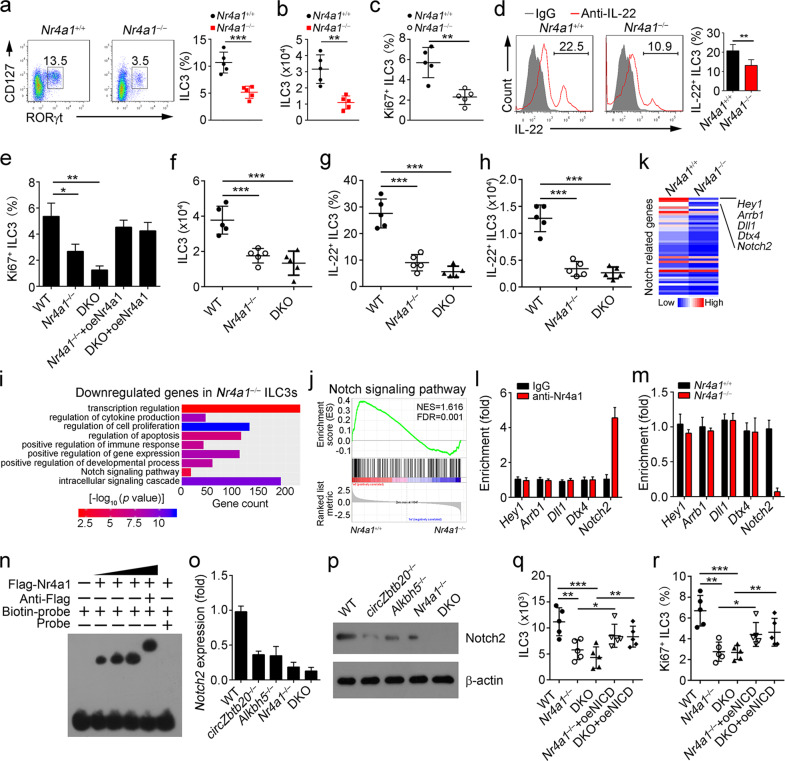
Nr4a1 initiates Notch2 signaling to maintain ILC3 homeostasis. a ILC3 percentages were analyzed in Nr4a1+/+ and Nr4a1−/− mice by FACS. ILC3s were gated on Lin−CD45+. n = 5 for each group. b ILC3 numbers were counted in Nr4a1+/+ and Nr4a1−/− mice. n = 5 for each group. c Ki67+ ILC3s were analyzed via FACS in Nr4a1+/+ and Nr4a1−/− mice. n = 5 for each group. d IL-22+ ILC3s were measured in Nr4a1+/+ and Nr4a1−/− mice. LPLs were isolated and stimulated with IL-23. e Ki67+ ILC3s were analyzed via FACS after infection with the indicated plasmids. f Analysis of ILC3 numbers in Nr4a1−/−, circZbtb20−/−Nr4a1−/− (DKO), or WT mice. g, h IL-22+ ILC3s were measured in Nr4a1−/−, DKO, or WT mice after infection with C. rodentium for 6 days. n = 5 for each group. i GO analysis using downregulated genes in Nr4a1−/− ILC3s compared to Nr4a1+/+ ILC3s. j The gene set of the Notch signaling pathway was analyzed by GSEA. k Heatmap using the expression values of Notch-related genes according to microarray data in Nr4a1+/+ and Nr4a1−/− ILC3s. l WT ILC3s were isolated, and a ChIP assay was performed to analyze the enrichment of Nr4a1 in the indicated gene promoters. m Nr4a1+/+ and Nr4a1−/− ILC3s were isolated, and a ChIP assay was conducted to measure the enrichment of Nr4a1. n EMSA was performed to analyze the interaction of Nr4a1 with the Notch2 promoter. o Relative expression levels of Notch2 in WT, circZbtb20−/−, Alkbh5−/−, Nr4a1−/−, or DKO ILC3s were analyzed by qPCR. Fold changes were normalized to endogenous Actb. p Notch2 expression levels in ILC3s were tested by western blotting. q, r The indicated CHILPs (Lin−Flt3−CD25−CD127+α4β7+CD244+) were adoptively transferred into Rag1−/−Il2rg−/− mice. Six weeks later, ILC3 numbers and Ki67+ ILC3s were analyzed. n = 5 for each group. *P < 0.05, **P < 0.01, and ***P < 0.001. Data are shown as the mean ± SD. Data are representative of at least three independent experiments.
To further explore the mechanism by which Nr4a1 regulated ILC3s, we performed microarray analysis of Nr4a1+/+ and Nr4a1−/− ILC3s. Downregulated genes in Nr4a1−/− ILC3s were used for GO analysis. These downregulated genes were correlated with several biological processes, especially the Notch signaling pathway, which was most strongly downregulated (Fig. 5i). In GSEA, Nr4a1 deletion was negatively correlated with the Notch signaling pathway (Fig. 5j). As expected, many Notch-related genes were downregulated in Nr4a1−/− ILC3s (Fig. 5k). We then focused on the top 5 most downregulated genes: Hey1, Arrb1, Dll1, Dtx4, and Notch2. In a chromatin immunoprecipitation (ChIP) assay, we found that Nr4a1 was enriched only at the promoter of Notch2 (Fig. 5l). Moreover, Nr4a1 deficiency abrogated its enrichment on the Notch2 promoter (Fig. 5m). The direct interaction of Nr4a1 with the Notch2 promoter was further validated by an ESMA assay (Fig. 5n), suggesting that Nr4a1 may regulate Notch2 gene transcription. Notably, Nr4a1 deletion suppressed Notch2 expression in ILC3s (Fig. 5o, p). Consistently, deletion of circZbtb20 or Alkbh5 inhibited Notch2 expression in ILC3s (Fig. 5o, p). These data suggest that Nr4a1 promotes Notch2 expression by initiating Notch2 transcription.
To further test whether Nr4a1 regulated ILC3 homeostasis through Notch2 signaling, we overexpressed the Notch2 intracellular domain (NICD) in CHILPs (Supplementary Fig. 6n) and performed an adoptive transfer assay. We noticed that Nr4a1−/− or circZbtb20−/−Nr4a1−/− (DKO) CHILPs generated fewer ILC3s, while NICD overexpression in Nr4a1−/− or DKO CHILPs restored the number of ILC3s (Fig. 5q). Consequently, NICD overexpression in Nr4a1−/− or DKO CHILPs dramatically rescued the proliferation of ILC3s (Fig. 5r). Taken together, these findings show that Nr4a1-mediated Notch2 signaling is required for the maintenance of ILC3 expansion and function.
Alkbh5-mediated m6A demethylation is required for ILC3 maintenance and gut immunity
We found that Alkbh5 deletion also downregulated Nr4a1 expression (Fig. 6a, b). H205A mutation causes catalytic inactivation of Alkbh5.33 To test whether Alkbh5-modulated Nr4a1 mRNA stability was dependent on its enzymatic activity, we overexpressed WT or H205A-mutated Alkbh5 in Alkbh5−/− ILC3s. We found that only overexpression of WT Alkbh5 could rescue Nr4a1 mRNA expression in Alkbh5−/− ILC3s (Fig. 6c). Consistently, overexpression of WT Alkbh5 in Alkbh5−/− ILC3s was able to sustain the stability of Nr4a1 mRNA (Supplementary Fig. 6o), suggesting that Alkbh5 sustains Nr4a1 mRNA stability via its m6A modification. We then examined the effect of Alkbh5 on ILC3 maintenance. We observed that Alkbh5 deletion decreased the number of ILC3s and suppressed ILC3 proliferation (Fig. 6d–g). Moreover, Alkbh5 deficiency caused apoptosis of ILC3s, while Nr4a1 overexpression in Alkbh5−/− ILC3s suppressed cell death of ILC3s (Fig. 6h). We next isolated CHILPs from Alkbh5+/+ and Alkbh5−/− mice and overexpressed Nr4a1 in Alkbh5−/− CHILPs via lentivirus infection (Supplementary Fig. 6p), followed by adoptive transfer into Rag1−/−Il2rg−/− mice. Six weeks later, we observed that Alkbh5−/− CHILPs generated fewer ILC3s and had lower proliferation rates, whereas Nr4a1 overexpression in Alkbh5−/− CHILPs restored ILC3 production and proliferation ability (Fig. 6i, j). In parallel, Alkbh5 deletion impaired IL-22 production by ILC3s (Fig. 6k), and Alkbh5−/− mice were much more susceptible to C. rodentium infection (Fig. 6l–o). Collectively, Alkbh5-mediated m6A demethylation is required for ILC3 maintenance and gut immunity.
Fig. 6.
Alkbh5-mediated m6A demethylation is required for ILC3 maintenance and gut immunity. a mRNA levels of Nr4a1 in Alkbh5+/+ or Alkbh5−/− ILC3s were analyzed by qPCR. Fold changes were normalized to endogenous Actb. b Protein levels of Nr4a1 were measured in Alkbh5+/+ or Alkbh5−/− ILC3s by western blotting. c mRNA levels of Nr4a1 in the indicated ILC3s were analyzed by qPCR. Fold changes were normalized to endogenous Actb. d ILC3s were analyzed in the small intestines of Alkbh5+/+ and Alkbh5−/− mice by FACS. ILC3s were gated on Lin−CD45+. n = 5 for each group. e BrdU+ ILC3s in Alkbh5+/+ and Alkbh5−/− mice were analyzed by FACS. n = 5 for each group. f ILC3s (Lin−CD45lowCD90high) were isolated from Alkbh5+/+ and Alkbh5−/− mice, labeled with CFSE and transferred into Rag1−/−Il2rg−/− mice for the cell proliferation assay. Seven days later, CFSE-labeled ILC3s were analyzed through FACS. g Ki67+ ILC3s in Alkbh5+/+ and Alkbh5−/− mice were measured by FACS. n = 5 for each group. h ILC3 apoptosis was analyzed by Annexin-V/PI staining. n = 5 for each group. i, j The indicated CHILPs (Lin−Flt3−CD25−CD127+α4β7+CD244+) were adoptively transferred into Rag1−/−Il2rg−/− mice. Six weeks later, ILC3 numbers and Ki67+ ILC3s were analyzed. n = 5 for each group. k LPLs were isolated from Alkbh5+/+ and Alkbh5−/− mice and stimulated with IL-23. IL-22 protein levels were measured by ELISA. l Bacterial counts (CFUs) in the feces were measured. Alkbh5+/+ and Alkbh5−/− mice were infected with C. rodentium for 6 days. n = 5 for each group. m Colon lengths were analyzed after infection with C. rodentium. n Colon tissues from Alkbh5+/+ and Alkbh5−/− mice were analyzed by H&E staining. Scale bars, 100 μm. o Histological scores of colons from Alkbh5+/+ and Alkbh5−/− mice in (n). *P < 0.05, **P < 0.01, and ***P < 0.001. Data are shown as the mean ± SD. Data are representative of at least three independent experiments.
Discussion
ILC3s mainly reside on mucosal surfaces that are involved in defense against pathogens at the early stage as innate effectors. ILC3s play critical roles in innate immunity and gut homeostasis. Dysregulation of ILC3 homeostasis may cause severe intestinal diseases, such as inflammation. Here, we found that a novel circular RNA, circZbtb20, is highly expressed in ILC3s and required for their maintenance and function. CircZbtb20 deletion causes reduced ILC3 numbers, increasing susceptibility to C. rodentium infection. Mechanistically, circZbtb20 enhances the interaction between Alkbh5 and Nr4a1 mRNA, leading to ablation of the Nr4a1 m6A modification to promote its stability. Nr4a1 initiates Notch2 signaling activation, which contributes to the maintenance of ILC3 homeostasis.
Many key ILC progenitors have been identified to date, such as CILPs, CHILPs, and ILCPs.34–36 Several TFs, including Tcf7, Id2, PLZF, and Gata3,34–36 play critical roles in the process of ILC maturation. Dysregulation of ILC3 homeostasis leads to apparent pathologic disorders.37 Many extrinsic factors have been reported to regulate ILC3 functions. For instance, ILC3s can be directly activated by IL-23 to maintain intestinal barrier integrity and regulate mucosal healing.38 It has been reported that the microbiota induces CX3CR1+ mononuclear phagocytes to produce TL1A to enhance ILC3 activation and IL-22 secretion.39 On the other hand, ILC3 activation could be restrained by Treg cells via suppression of the generation of IL-23 from CX3CR1+ macrophages.40 Maintenance of the ILC3 population is important for its function and gut immunity. However, how ILC3 homeostasis is regulated remains poorly understood. AHR is acknowledged as an essential regulator of ILC3 maintenance.5,41 We previously demonstrated that the lncRNA lncKdm2b is required for ILC3 expansion.7 Here, we identified a novel circRNA, circZbtb20, that is highly expressed in ILC3s and is required for the maintenance and function of ILC3s. In addition, we showed that circZbtb20 expression is not affected by C. rodentium infection. CircZbtb20 deficiency has no effect on Ahr expression. These data suggest that circZbtb20-mediated ILC3 homeostasis is independent of other known pathways, including Ahr- and microbiota-mediated ILC3 activation.
Accumulating evidence shows that circRNAs play important roles in various biological processes, especially in tumorigenesis.11 CircRNAs may be derived from introns, exons, or intron–exon sequences depending on the presence of complementary elements. Most circRNAs contain miRNA-binding elements and are considered to act as miRNA sponges to perform their functions.42 By contrast, circRNAs without obvious miRNA-binding sites also function through alternative mechanisms. For example, several circRNAs accumulate in the nucleus to regulate transcription.43 CircACC1 regulates the assembly and activation of the AMPK complex to participate in the regulation of metabolism.44 We recently reported that cia-cGAS blocks the activity of cGAS to protect HSCs from cGAS-mediated exhaustion.10 In this study, we found that highly expressed circZbtb20 is critical for ILC3 homeostasis because it regulates the stability of Nr4a1 mRNA via modulation of the m6A modification. Zbtb20, a member of the subfamily of zinc finger proteins containing C2H2 Krüppel-type zinc fingers and BTB/POZ domains, is involved in glucose and lipid homeostasis.45 We noticed that circZbtb20 knockout had no effect on Zbtb20 expression. Furthermore, Zbtb20 deficiency does not affect ILC3 homeostasis, indicating that circZbtb20 has a function independent from that of its parental gene.
Unlike classical ligand-activated nuclear receptors, Nr4a1 is not activated by small-molecule ligands, while its activation is dependent on posttranslational modifications, such as phosphorylation.46 Nr4a1 gradually came to be considered a critical regulator of immune cells and the immune response. Herein, we showed that Nr4a1 mRNA stability is modulated by m6A modification mediated by circZbtb20 in ILC3s. Nr4a1 can promote Notch2 expression by initiating Notch2 transcription, which triggers Notch2 signaling activation that maintains ILC3 homeostasis. Notch signaling is required for the generation of adult ILC progenitors.47 Moreover, Notch2 is essential for the development of Nkp46+ ILC3s.48,49 However, how Nr4a1 regulates the Notch signaling pathway remains elusive. In this study, we revealed that the circZbtb20-Alkbh5-Nr4a1-Notch2 axis is required for the regulation of ILC3 homeostasis and function. A previous report showed that Nr4a1-deficient T cells have increased expression of IL-17 and IFN-γ.50 Here, we demonstrate that Nr4a1 deletion increases IL-17 production in ILC3s during C. rodentium infection. This may be caused by Nr4a1-mediated downstream signaling pathways in T cells and ILC3s.
In eukaryotes, m6A methylation is the most common chemical modification in mRNA.51 The methyltransferases METTL3, METTL14, and WTAP are responsible for m6A methylation, which is recognized by a “reader,” YTHDF1, YTHDF2, or YTHDF3,33,52 and erased by Alkbh5 or FTO. Increasing evidence indicates that the m6A modification has important effects on various cellular processes, such as regulating mRNA stability, translation efficiency, and splicing.32,53,54 Consequently, m6A modification is correlated with a broad set of biological functions, while its dysregulation causes homeostasis disorder.55 For instance, deficiency of METTL3 or YTHDF2 protects mice from virus infection by increasing the induction of interferon-stimulated genes.56 Alkbh5 sustains FOXM1 expression by erasing m6A modification and promotes tumorigenesis of glioblastoma stem-like cells.28 In addition, m6A-mediated decay of mRNAs is also involved in the regulation of T-cell homeostasis.57 Herein, we showed that circZbtb20 enhances the interaction of Nr4a1 mRNA with Alkbh5, which removes m6A modification in Nr4a1 mRNA to promote its stability. Alkbh5 deficiency impairs ILC3 maintenance and function, suggesting that Alkbh5-mediated m6A removal is involved in the regulation of ILC3 biology. In conclusion, circZbtb20 promotes ILC3 maintenance and function via Alkbh5-dependent m6A demethylation of Nr4a1 mRNA.
Methods
Antibodies and reagents
Anti-Nr4a1 (M-210) and anti-Zbtb20 (E-11) were purchased from Santa Cruz Biotechnology. Anti-m6A (202003) was from Synaptic System. Anti-β-actin (Cat# 3700) was purchased from Cell Signaling Technology (Danvers, USA). Anti-CD127 (A7R34), anti-c-Kit (2B8), anti-CD3 (17A2), anti-CD4 (GK1.5), anti-CD19 (1D3), anti-NK1.1 (PK136), anti-CD150 (mShad150), anti-CD34 (RAM34), anti-CD45 (30-F11), anti-CD90 (HIS51), anti-Sca-1 (D7), anti-CD25 (PC61.5), anti-Flt3 (A2F10), anti-α4β7 (DATK32), anti-RORγt (AFKJS-9), anti-NKp46 (29A1.4), anti-Gata3 (TWAJ), anti-KLRG1 (2F1), anti-PLZF (Mags.21F7), Lineage cocktail (88-7772-72), anti-CD48 (HM48-1), anti-Ki67 (SolA15), anti-IL-22 (IL22JOP), anti-BrdU (BU20A), anti-CD45.2 (104), anti-CD45.1 (A20), and anti-CD16/32 (93) were purchased from eBiosciences (San Diego, USA). Anti-Alkbh5 (PA5-100873) was purchased from Invitrogen. Paraformaldehyde (PFA) and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Sigma. The IL-22 ELISA kit was purchased from eBiosciences.
Generation of knockout mice with CRISPR/Cas9 technology
For the generation of circZbtb20−/−, Alkbh5−/−, Nr4a1−/−, and Zbtb20flox/flox mice, CRISPR-mediated single-stranded oligodeoxynucleotide donors were synthesized as previously described.58 Approximately 250 zygotes from C57BL/6 mice were injected with sgRNAs and subsequently transferred to the uterus of pseudopregnant ICR females from which viable founder mice were obtained. Genomic DNA mutations were identified by PCR screening and DNA sequencing, followed by western blotting or Northern blotting. sgRNA sequences are listed in Supplementary Table 2. Rorc-Cre, PLZFGFPcre, Rorc(γt)+/GFP, and Id2+/GFP mice were purchased from the Jackson Laboratory. Mettl3flox/flox and Mettl14flox/flox mice were provided by Dr. Minghan Tong (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences). Zbtb20flox/floxRorc-Cre mice were obtained by crossing Zbtb20flox/flox mice with Rorc-Cre mice. Rag1−/−Il2rg−/− mice were generated by crossing Rag1+/− mice (from Model Animal Research Center of Nanjing University, China) with Il2rg+/− mice. For animal studies, 8–12-week-old knockout and wild-type littermate control mice were used for experimental groups, irrespective of sex. We were not blinded to the phenotype prior to experiments. The results from five mice were randomly chosen if over five mice from one group were analyzed. All mouse strains were on the C57BL/6 background and maintained under specific pathogen-free conditions with approval from the Institutional Committee of Institute of Biophysics, Chinese Academy of Sciences. The study was compliant with all relevant ethical regulations regarding animal research.
Intestinal lymphocyte separation
Intestinal lymphocytes were isolated as described previously.7 Briefly, the intestines were cut open longitudinally and washed using phosphate-buffered saline (PBS) five times. Then, the intestines were cut into pieces, washed with solution I buffer (10 mM HEPES and 5 mM EDTA in HBSS) five times, and digested with solution II buffer (DNaseI, 5% FBS, and 0.5 mg/ml collagenase II and collagenase III) three times. Finally, the digested LPLs were sifted through 70 μm strainers and used for experiments.
Flow cytometry
For analysis of BM cells, cells were flushed out from femurs using PBS containing 5% FBS and sifted through 70 μm strainers. LPL isolation was described above. For flow cytometric analysis, HSC (Lin−Sca-1+c-Kit+CD150+CD48−), MPP (Lin−Sca-1+c-Kit+CD150−CD48+), CLP (Lin−CD127+c-KitintSca-1intFlt3+α4β7−), siILC1 (CD3−CD19−CD127+NK1.1+NKp46+), siILC3 (Lin−CD127+RORγt+CD45+), NK1.1+ NK, CD19+ B, and CD3+ T populations were analyzed or sorted with a FACSAria III instrument (BD Biosciences). PLZFGFPcre mice were used for ILCP (Lin−CD127+α4β7+PLZFGFP) isolation, and Id2+/GFP mice were used for CHILP (Lin−CD25−CD127+Flt3−α4β7+Id2GFP) isolation by FACS.
Immunofluorescence staining
ILC3s (Lin−CD90highCD45low) were fixed using 4% PFA (Sigma-Aldrich) for 20 min at room temperature, permeabilized with PBS containing 1% Triton X-100 for 20 min, blocked with 5% donkey and 5% rat serum for 1 h at room temperature, incubated with appropriate primary antibodies at 4 °C overnight, and then incubated with fluorescence-conjugated secondary antibodies. DAPI was used for nuclear staining. Cells were visualized with an Olympus FV1200 laser scanning confocal microscope (Olympus, Japan).
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated using an RNA Miniprep Kit (Tiangen, Beijing, China) following the manufacturer’s instructions as described previously.59 cDNA was synthesized using M-MLV reverse transcriptase (Promega, Madison, USA) and analyzed on an ABI 7300 qPCR system using the specific primer pairs listed in Supplementary Table 3. Relative expression was calculated and normalized to endogenous 18S or Gapdh.
m6A-RIP-qPCR
Total RNA was isolated from Lin−CD45+ LPLs using TRIzol reagent (Invitrogen). RNA was fragmented into 300–400 nt fragments and incubated with anti-m6A antibody or rabbit IgG for 2 h at 4 °C, followed by incubation with Protein A beads (Thermo Fisher Scientific) for another 2 h at 4 °C. Then, the captured RNA was eluted, purified, and analyzed by RT-qPCR.
Chromatin immunoprecipitation (ChIP) assay
ILC3s were cross-linked with 1% formaldehyde at 37 °C for 10 min. Then, the cells were washed twice with PBS, lysed with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris) and sonicated to generate 200–500-bp DNA fragments. Lysates were precleared with Protein A Agarose/Salmon Sperm DNA (50% Slurry) and then incubated with 4 μg antibody overnight at 4 °C. Then, Protein A Agarose/Salmon Sperm DNA (50% Slurry) beads were added and incubated for 4 h. After washing, DNA was eluted from the beads and purified. DNA fragments were analyzed using the primer pairs listed in Supplementary Table 4.
C. rodentium infection
Mice were fasted for 8 h prior to infection. Then, the indicated mice were orally infected with 5 × 109 C. rodentium as described previously.60 C. rodentium was a gift from B. Ge (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences). After infection for 6 days, the mice were sacrificed. The colons were collected and analyzed for pathology. The spleen, liver, and feces were analyzed for bacterial loads. LPLs were isolated for analyses of ILC3s and IL-22 production. To test weight changes, mice infected with C. rodentium were weighed every 3 days.
Histologic analysis
Mouse colons were cut open longitudinally and fixed in 4% PFA, followed by paraffin sectioning and H&E staining. The severity of colitis was assigned a score and analyzed as described previously.41
Adoptive transfer assay
A total of 1 × 104 CHILPs (Lin−CD25−CD127+Flt3−α4β7+) were adoptively transferred into Rag1−/−Il2rg−/− mice. Six weeks later, ILC3s from the transferred mice were analyzed by FACS.
Bone marrow transplantation
A total of 5 × 106 CD45.2+ BM cells from the indicated mice were transplanted into lethally irradiated CD45.1+ recipient mice. Eight weeks after transplantation, ILC3s derived from donor cells were analyzed by FACS. For competitive transplantation, 1 × 106 CD45.2+ BM cells from the indicated mice and 1 × 106 wild-type CD45.1+ BM cells were injected into lethally irradiated CD45.1+ recipient mice. Eight weeks after transplantation, the ratio of CD45.2+ ILC3s to CD45.1+ ILC3s was examined.
Lentivirus preparation and infection
RNA interference was performed as previously described.10 The target sequences are listed in Supplementary Table 5. For circZbtb20 overexpression, genomic exon regions of circZbtb20 were ligated into split GFP sites on the pSIN-EF2-GFP vector flanked by upstream and downstream complementary elements. Lentiviral vector (pSIN-EF2-GFP) was cotransfected with the packaging plasmids pVSVg and psPAX2 into HEK293T cells for 48 h, followed by culture medium collection and ultracentrifugation at 25,000 × g for 1.5 h. The pellets were resuspended in IMDM medium, and viral titers were determined by infecting HEK293T cells with diluted viruses. Cells were incubated with lentiviruses (MOI = 10) and centrifuged at 500 × g for 2 h in the presence of 8 μg/ml polybrene. The cells were cultured for 24 h to allow GFP expression, followed by sorting of GFP-positive cells through a flow cytometer. HEK293T cells were obtained from ATCC (CRL-11268) and tested negative for mycoplasma contamination.
RNA pulldown assay
Biotin-labeled Zbtb20 pre-mRNA probes targeting the upstream intron (targeting site: −450 to −50 nt from exon 8 of the Zbtb20 transcript) and control RNAs were obtained with Biotin RNA labeling Mix (Roche) in vitro, followed by incubation with LPL cell lysates. Pulldown components were separated by SDS-PAGE for silver staining. Differential bands enriched by Zbtb20 pre-mRNA probes were analyzed by LTQ Orbitrap XL mass spectrometry. To identify interacting proteins of circZbtb20, LPL cells were lysed, and supernatants were incubated with biotin-labeled circZbtb20 or antisense. Similarly, precipitated components were separated by SDS-PAGE for silver staining. Differential bands enriched by circZbtb20 were analyzed by LTQ Orbitrap XL mass spectrometry or immunoblotting.
Microarray assay
For the microarray, ILC3s (Lin−CD90highCD45low) were isolated from circZbtb20+/+, circZbtb20−/−, Nr4a1+/+, or Nr4a1−/− mice. Then, RNAs were isolated using TRIzol reagent (Invitrogen) and used for Affymetrix mRNA microarray assays by Beijing Cnkingbio Biotechnology (GSE148456).
RNA mobility-shift assays
For analysis of the interaction between circZbtb20 and Alkbh5, biotin-labeled linearized circZbtb20 was synthesized by T7 transcription in vitro with Biotin RNA Labeling Mix (Roche). CircZbtb20 and Alkbh5 proteins were incubated in binding buffer, and a mobility-shift assay was performed using a LightShift Chemiluminescent RNA-EMSA Kit (Thermo Scientific) according to the manufacturer’s protocol. For analysis of the interaction between circZbtb20 and Nr4a1, biotin-labeled RNAs were obtained using an in vitro transcription assay. After incubation of circZbtb20 and Nr4a1 for 30 min according to the LightShift Chemiluminescent RNA-EMSA Kit (Thermo Scientific), components were separated by native PAGE and then transferred onto positively charged nitrocellulose film (Beyotime Biotechnology). After UV cross-linking, biotin signals were detected with HRP-conjugated streptavidin according to the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific).
ELISA
LPLs or ILC3s were isolated and cultured for 24 h with the indicated cytokines. Then, supernatants were collected, and cytokines were detected using an ELISA kit (eBioscience) according to the manufacturer’s instructions.
SELECT assay
m6A modification was analyzed using the SELECT method according to a previous report.30 In brief, total RNA treated with Alkbh5 or vehicle was recovered by phenol–chloroform extraction and ethanol precipitation. Then, RNA was mixed with up primers, down primers, and dNTPs in 1× CutSmart buffer (50 mM KAc, 20 mM Tris-HAc, 10 mM MgAc2, and 100 μg/ml BSA, pH 7.9). After annealing, a mixture containing Bst 2.0 DNA polymerase, 0.5 U SplintR ligase, and 10 nmol ATP was added for the reaction, followed by qPCR analysis. Primer sequences are listed in Supplementary Table 6.
TBE-PAGE assay
SELECT products were analyzed using TBE-PAGE as described previously.30 Briefly, SELECT products were amplified using 2× Taq Plus Master Mix (Vazyme) and run on a 12% native TBE-PAGE gel with 0.5× TBE buffer. Then, PAGE gels were stained and photographed.
Statistical analysis
For statistical evaluation, an unpaired Student’s t test was applied to calculate statistical probabilities in this study. For all panels, at least three independent experiments were performed with similar results, and representative experiments are shown. Data were analyzed by using Microsoft Excel or SPSS 22. P values ≤ 0.05 were considered significant.
Supplementary information
Acknowledgements
We thank Shu Meng, Dongdong Fan, Yan Teng, Junying Jia, and Xiang Shi for technical support. We also thank Jing Li (Cnkingbio Company, Ltd., Beijing, China) for technical support. This work was supported by the Ministry of Science and Technology of China (2020YFA0803501 and 2019YFA0508501), the National Natural Science Foundation of China (31930036, 81921003, 92042302, 31870883, 91940305, 31728006, 81772646, and 31871494), the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDB19030203), the Beijing Natural Science Foundation (5192018), the Biological Resource Program of the Chinese Academy of Science (KFJ-BRP-017-04), and the Young Elite Scientist Sponsorship Program of CAST (2018QNRC001).
Author contributions
B.L. and N.L. performed experiments; B.L. designed the project, analyzed the data, and wrote the paper; and X.Z. constructed genetic mouse strains. L.Y., B.Y., H.L., P.Z., and T.L. analyzed data; Y.T. initiated the study and analyzed data; and Z.F. initiated the study and organized, designed, and wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Benyu Liu, Nian Liu, Xiaoxiao Zhu
Contributor Information
Benyu Liu, Email: benyuliu@zzu.edu.cn.
Yong Tian, Email: ytian@ibp.ac.cn.
Zusen Fan, Email: fanz@moon.ibp.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00680-1.
References
- 1.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivier E, et al. Innate lymphoid cells : 10 years on. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 4.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 5.Qiu J, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia P, et al. WASH maintains NKp46(+) ILC3 cells by promoting AHR expression. Nat. Commun. 2017;8:15685. doi: 10.1038/ncomms15685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat. Immunol. 2017;18:499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 8.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XO, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Xia P, et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48:688–701.e687. doi: 10.1016/j.immuni.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Guarnerio J, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Piwecka M, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 13.Liu CX, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880.e821. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 14.Gury-BenAri M, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 2016;166:1231–1246.e1213. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 15.Shaked I, et al. Transcription factor Nr4a1 couples sympathetic and inflammatory cues in CNS-recruited macrophages to limit neuroinflammation. Nat. Immunol. 2015;16:1228–1234. doi: 10.1038/ni.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullican SE, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat. Med. 2007;13:730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 17.Liu, X. D. et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature567, 525–529 (2019). [DOI] [PMC free article] [PubMed]
- 18.Hanna RN, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C-monocytes. Nat. Immunol. 2011;12:778–785. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekiya T, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- 20.Hanna RN, et al. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ. Res. 2012;110:416–427. doi: 10.1161/CIRCRESAHA.111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, et al. An inducible circular RNA circKcnt2 inhibits ILC3 activation to facilitate colitis resolution. Nat. Commun. 2020;11:4076. doi: 10.1038/s41467-020-17944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell. 2017;67:214–227.e217. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 23.Conn SJ, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Errichelli L, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 27.Zhu P, et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat. Immunol. 2019;20:183–194. doi: 10.1038/s41590-018-0297-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 30.Xiao Y, et al. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N(6)-methyladenosine modification. Angew. Chem. 2018;57:15995–16000. doi: 10.1002/anie.201807942. [DOI] [PubMed] [Google Scholar]
- 31.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng GQ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Q, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 2015;16:1044–1050. doi: 10.1038/ni.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klose CSN, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 36.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo X, et al. Innate lymphoid cells control early colonization resistance against intestinal pathogens through ID2-dependent regulation of the microbiota. Immunity. 2015;42:731–743. doi: 10.1016/j.immuni.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longman RS, et al. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castellanos JG, et al. Microbiota-induced TNF-like ligand 1A drives group 3 innate lymphoid cell-mediated barrier protection and intestinal T cell activation during colitis. Immunity. 2018;49:1077–1089.e1075. doi: 10.1016/j.immuni.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauche D, et al. LAG3(+) regulatory T cells restrain interleukin-23-producing CX3CR1(+) gut-resident macrophages during group 3 innate lymphoid cell-driven colitis. Immunity. 2018;49:342–352.e345. doi: 10.1016/j.immuni.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 43.Li ZY, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 44.Li Q, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30:157–173.e157. doi: 10.1016/j.cmet.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Liu G, et al. Regulation of hepatic lipogenesis by the zinc finger protein Zbtb20. Nat. Commun. 2017;8:14824. doi: 10.1038/ncomms14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fahrner TJ, Carroll SL, Milbrandt J. The Ngfi-B protein, an inducible member of the thyroid steroid-receptor family, is rapidly modified posttranslationally. Mol. Cell. Biol. 1990;10:6454–6459. doi: 10.1128/mcb.10.12.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seehus CR, et al. The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat. Immunol. 2015;16:599–608. doi: 10.1038/ni.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rankin LC, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mielke LA, et al. TCF-1 controls ILC2 and NKp46+RORgammat+ innate lymphocyte differentiation and protection in intestinal inflammation. J. Immunol. 2013;191:4383–4391. doi: 10.4049/jimmunol.1301228. [DOI] [PubMed] [Google Scholar]
- 50.Wang LM, et al. Nr4a1 plays a crucial modulatory role in Th1/Th17 cell responses and CNS autoimmunity. Brain. Behav. Immun. 2018;68:44–55. doi: 10.1016/j.bbi.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 51.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 52.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molinie B, et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods. 2016;13:692–698. doi: 10.1038/nmeth.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 2017;18:1094–1103. doi: 10.1038/ni.3830. [DOI] [PubMed] [Google Scholar]
- 56.Winkler R, et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2019;20:173–182. doi: 10.1038/s41590-018-0275-z. [DOI] [PubMed] [Google Scholar]
- 57.Li HB, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu X, et al. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep. 2014;4:6420. doi: 10.1038/srep06420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu B, et al. IL-7Ralpha glutamylation and activation of transcription factor Sall3 promote group 3 ILC development. Nat. Commun. 2017;8:231. doi: 10.1038/s41467-017-00235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu B, et al. Yeats4 drives ILC lineage commitment via activation of Lmo4 transcription. J. Exp. Med. 2019;216:2653–2668. doi: 10.1084/jem.20182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.