Aicardi–Goutières syndrome (AGS) is a rare genetic disease caused by mutations in nine genes that are all involved in nucleic acid metabolism or sensing.1,2 The three RNASEH2 subunits represent the most frequently mutated genes in AGS patients,1,3 and mutations in RNASEH2 subunits lead to the accumulation of endogenous RNA:DNA hybrids that may trigger an interferon-α-mediated immune response4 through the activation of pattern recognition receptors (PRRs).5 PRRs perform surveillance on extracellular, endosomal, and cytosolic compartments to identify signs of infection: endogenous nucleic acids that are inappropriately cleared may enter and accumulate in the cytoplasm, driving inflammation and autoimmune diseases.6 This accumulation may be the cause of the clinical autoimmune phenotype of AGS patients carrying RNASEH2 mutations. A proven effective cure for AGS has not been discovered, and targeting these two pathways may lead to a treatment that improves patients’ immunological symptoms. Hydroxychloroquine (HCQ) interferes with normal antigen processing and presentation and is widely used in the clinical treatment of autoimmune diseases such as systemic lupus erythematosus and rheumatoid arthritis.7 HCQ is also a well-known inhibitor of autophagy that prevents the degradation of autolysosomes. This drug inhibits acidification and maturation of endosomes and increases the pH in lysosomes, resulting in the inhibition of their main functions, such as downstream cell signaling through TLRs.7 Therefore, the aim of our work was to investigate whether HCQ is able to modulate the abnormal inflammatory response driven by RNA:DNA hybrids.
Considering the noteworthy difficulties in obtaining primary cells from children affected by this rare disease, we used immortalized lymphoblastoid cell lines (LCLs): one from a patient carrying the RNASEH2A p.R108W + p.F230L mutations, one from a patient carrying the RNASEH2B p.A177T mutation and one from a healthy control. We investigated the presence of RNA:DNA hybrids using the S9.6 antibody and flow cytometry. Quantification of the mean S9.6 fluorescence revealed a significant increase in RNA:DNA hybrids only in the RNASEH2B-mutant LCL (Fig. S1A). RNA:DNA hybrids were primarily localized in the cytoplasm in all three cell lines, while in the control and RNASEH2A-mutant LCLs, RNA:DNA hybrids were distributed homogeneously; in the RNASEH2B-mutant LCL, they accumulated in a specific region of the cytoplasm (Fig. S1B, white arrow). These results were confirmed by immunogold staining of the RNA:DNA hybrids (Fig. S1C).
The S9.6 antibody may also bind double-stranded RNA but with a lower affinity than with RNA:DNA hybrids, and therefore, to verify the cytoplasmic accumulation of the hybrids in these cell lines, we treated the cells with RNase H.8 After RNase H treatment, the RNA:DNA hybrid fluorescence signal considerably decreased in the RNASEH2B-mutant LCL (Fig. S1D), the only cell line that presented significant accumulation of RNA:DNA hybrid. We also decided to investigate the subcellular localization of RNA:DNA hybrids by staining cells with monoclonal S9.6 antibody and specific vital dyes. While we did not find any colocalization between RNA:DNA hybrids and mitochondria or the endoplasmic reticulum in either mutant cell line, we observed colocalization of hybrids in the lysosomes in the RNASEH2B-mutant LCL (Fig. S1E).
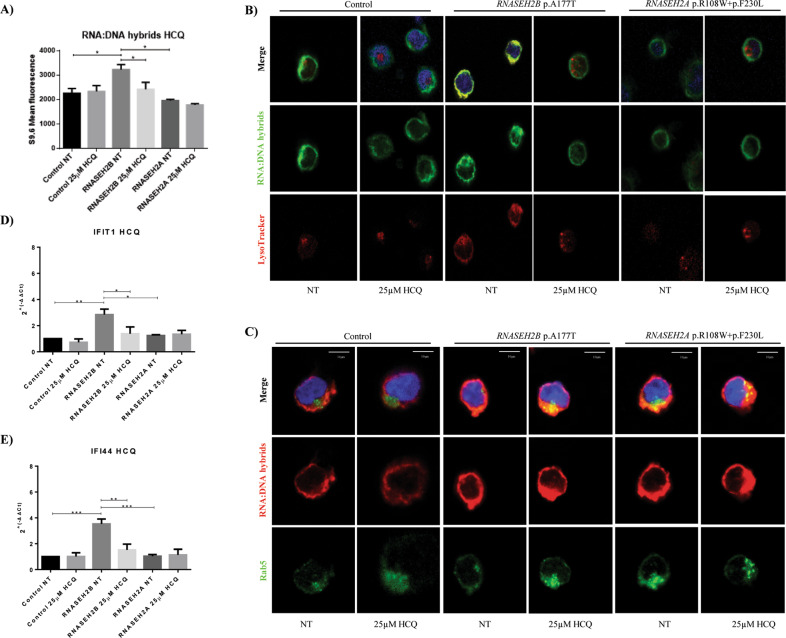
We then evaluated how the RNA:DNA hybrid level changed in the healthy control LCL and the RNASEH2B- and RNASEH2A-mutant LCLs after treatment with 25 µM HCQ for 24 h. We found that in the treated RNASEH2B-mutant LCL, the RNA:DNA hybrid level decreased (Fig. 1A), and a loss of colocalization between lysosomes and RNA:DNA hybrids was observed (Fig. 1B). Since LysoTracker may stain both lysosomes and endosomes, we stained cells with Rab5, an early endosome marker, to clarify the RNA:DNA hybrid localization. The RNASEH2B-mutant LCL showed no colocalization of S9.6 and Rab5, confirming that, at the basal level, hybrids were internalized by lysosomes, whereas we discovered weak colocalization in the untreated and treated RNASEH2A-mutant LCL. Interestingly, Rigby et al.9 described the accumulation of RNA:DNA hybrids in endosomes in the presence of retroviral infection in B3T3 fibroblasts. After treatment with HCQ, we observed colocalization between Rab5 and RNA:DNA hybrids in the RNASEH2B-mutant LCL, highlighting a possible role of endosomes in RNA:DNA hybrid elimination. No colocalization between Rab5 and RNA:DNA hybrids was observed in the control LCL (Fig. 1C). AGS mimics congenital viral brain infections; therefore, endosomal localization, which was not evident in the control LCL, might suggest a physiological way to eliminate dangerous nucleic acids, which was not observed in the RNASEH2B-mutant LCL.
Fig. 1.
HCQ reduces and changes the localization of RNA:DNA hybrids, inhibiting the activation of interferon-stimulated genes (ISGs). A Accumulation of cytosolic RNA:DNA hybrids in the RNASEH2B-mutant LCL compared to the healthy control and RNASEH2A-mutant LCLs was evaluated by flow cytometry with 25 μM HCQ and without (NT) HCQ treatment. The data are presented as the means ± SEM, and significance was determined by paired t-test. *P < 0.05. B Immunofluorescence of the LCL derived from the healthy control and RNASEH2A- and RNASEH2B-mutant LCLs before (NT) and 24 h after HCQ treatment (25 μM). RNA:DNA hybrids stained with S9.6-specific mouse monoclonal antibody (green), lysosomes were stained with the endolysosomal marker LysoTrackerTM (red), and nuclei were stained with DAPI (blue). C Immunofluorescence of the LCLs derived from healthy controls and AGS patients with mutations in the RNASEH2B and RNASEH2A genes before (NT) and 24 h after HCQ treatment (25 μM). RNA:DNA hybrids were stained with S9.6-specific mouse monoclonal antibody (red), endosomes were stained with Rab5 (green), and nuclei were stained with DAPI (blue). D, E mRNA expression levels of IFIT1 and IFI44, two ISGs, in healthy controls and AGS patients before and after HCQ treatment. The data are presented as the means ± SEM, and significance was determined by ANOVA and Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001
Since HCQ also interferes with autophagy,7 we investigated whether RNA:DNA hybrid is depleted upon HCQ treatment via the autophagy pathway. After treating the cells with HCQ, we observed an increase in the LC3-II/LC3-I ratio for each cell line, especially in the RNASEH2B-mutant LCL, with a significant P value < 0.001 (Fig. S2A). Moreover, LC3 colocalized with RNA:DNA hybrids in RNASEH2B-mutant LCL after HCQ treatment (Fig. S2C). In accordance with these results, we observed increased p62 protein levels after HCQ treatment and p62 colocalization with RNA:DNA hybrids in the RNASEH2B-mutant LCL, which became more evident after HCQ treatment (Fig. S2B, D), strengthening the possible role of autophagy in RNA:DNA hybrid discards.
After HCQ treatment, cGAS protein levels slightly decreased in the RNASEH2B-mutant LCL (Fig. S3A), and subsequently, the same behavior was described for IRF3 transcript levels. We also found decreased MYD88 and IRF7 transcript levels, whereas no difference in NF-kB transcripts was evident in the AGS LCLs (Fig. S3C). To confirm these results, we used flow cytometry to evaluate the phosphorylation of TBK1, which links the recognition of nucleic acids to the development of a type I IFN response through cGAS activation.10 No significant differences in the phosphorylation profile were identified before or after treatment with HCQ (Fig. S3B). We also evaluated how the expression of two ISGs, IFI44 and IFIT1, changed in RNASEH2A- and RNASEH2B-mutant LCLs after treatment. IFI44 and IFIT1 expression levels decreased after HCQ treatment in the RNASEH2B-mutant LCL, with a greater decrease in IFIT1, whereas no differences in the RNASEH2A-mutant LCL were observed (Fig. 1D, E).
In conclusion, we found that RNA:DNA hybrids accumulate mainly in the cytoplasm and colocalize with lysosomes in the LCL derived from an AGS patient carrying a RNASEH2B mutation, which may indicate an impairment in the RNA:DNA hybrid degradation process. On the other hand, the RNASEH2A-mutant LCL carries RNA:DNA hybrids in endosomes, possibly explaining the lack of impaired RNA:DNA hybrid degradation in this cell line. Moreover, we found that HCQ is a drug that can induce decreased activation of the IFN-α immune cascade only in the RNASEH2B-mutant LCL, the only cell line presenting high ISGs. Therefore, we hypothesized that HCQ, a European Medicines Agency-approved drug, may be an effective treatment for AGS patients who present abnormal activation of the innate immune response.
Supplementary information
Acknowledgements
We thank Dr. Chiara Baldo of the Laboratorio di Genetica Umana, IRCCS Istituto Giannina Gaslini, Genoa, for the establishment of the LCLs derived from the healthy control and AGS patient B cells. We also thank Dr. Serena Mazzucchelli and Dr. Raffaele Allevi of the Department of Biomedical and Clinical Sciences “L. Sacco,” Milan for confocal images and analysis and for TEM images and analysis, respectively. We also thank the International AGS Association (IAGSA) for its commitment and support to our project. We extend a special thank you to the few patients with AGS and their families. This study was supported by grants from the Italian Ministry of Health RC 2018–2019 to IRCCS Mondino Foundation, Pavia, Italy.
Author contributions
Conceptualization: J.G., D.S., and C.C.; formal analysis: J.G., D.S., F.D., O.P., and C.C.; funding acquisition: C.C. and S.O.; investigation: J.G., D.S., F.D., C.S., E.P., S.C., and A. Tesser; methodology: J.G., D.S., O.P., and C.C.; resources: D.S., D.T., E.P., A. Tommasini, S.O., O.P., and C.C.; supervision: C.C.; visualization: J.G., D.S., F.D., and O.P.; writing—original draft: J.G., D.S., O.P., and C.C.; writing—review and editing: D.T., S.C., G.V.Z., A. Tommasini, O.P., C.C.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jessica Garau, Daisy Sproviero
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00657-0.
References
- 1.Crow YJ, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A. 2015;167A:296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uggenti C, et al. cGAS-mediated induction of type I interferon due to inborn errors of histone pre-mRNA processing. Nat. Genet. 2020;52:1364–1372. doi: 10.1038/s41588-020-00737-3. [DOI] [PubMed] [Google Scholar]
- 3.Garau J, et al. Molecular genetics and interferon signature in the Italian Aicardi Goutières Syndrome Cohort: report of 12 new cases and literature review. J. Clin. Med. 2019;8:750. doi: 10.3390/jcm8050750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum. Mol. Genet. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roers A, Hiller B, Hornung V. Recognition of endogenous nucleic acids by the innate immune system. Immunity. 2016;44:739–754. doi: 10.1016/j.immuni.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Amarante-Mendes GP, et al. Pattern recognition receptors and the host cell death molecular machinery. Front. Immunol. 2018;9:2379. doi: 10.3389/fimmu.2018.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schrezenmeier E, Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 8.Vanoosthuyse V. Strengths and weaknesses of the current strategies to map and characterize R-loops. Noncoding RNA. 2018;4:9. doi: 10.3390/ncrna4020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigby RE, et al. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J. 2014;33:542–558. doi: 10.1002/embj.201386117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.