Abstract
Purpose:
To improve image quality and CT number accuracy of daily cone-beam computed tomography (CBCT) through a deep-learning methodology with Generative Adversarial Network.
Methods:
150 paired pelvic CT and CBCT scans were used for model training and validation. An unsupervised deep-learning method, 2.5D pixel-to-pixel generative adversarial network (GAN) model with feature mapping was proposed. A total of 12000 slice pairs of CT and CBCT were used for model training, while 10-cross validation was applied to verify model robustness. Paired CT-CBCT scans from an additional 15 pelvic patients and 10 head-and-neck (HN) patients with CBCT images collected at a different machine were used for independent testing purpose. Besides the proposed method above, other network architectures were also tested as: 2D vs. 2.5D; GAN model with vs. without feature mapping; GAN model with vs. without additional perceptual loss; and previously reported models as U-net and cycleGAN with or without identity loss. Image quality of deep-learning generated synthetic CT (sCT) images were quantitatively compared against the reference CT (rCT) image using mean absolute error (MAE) of Hounsfield units (HU) and peak signal-to-noise ratio (PSNR). The dosimetric calculation accuracy was further evaluated with both photon and proton beams.
Results:
The deep-learning generated synthetic CTs (sCT) showed improved image quality with reduced artifact distortion and improved soft tissue contrast. The proposed algorithm of 2.5 Pix2pix GAN with feature matching (FM) was shown to be the best model among all tested methods producing the highest PSNR and the lowest MAE to reference CT (rCT). The dose distribution demonstrated a high accuracy in the scope of photon based planning, yet more work is needed for proton based treatment. Once the model was trained, it took 11–12 ms to process one slice, and could generate a 3D-volume of dCBCT (80 slices) in less than a second using a NVIDIA GeForce GTX Titan X GPU (12GB, Maxwell architecture).
Conclusion:
The proposed deep-learning algorithm is promising to improve CBCT image quality in an efficient way, thus has a potential to support online CBCT-based adaptive radiotherapy.
1. Introduction
Cone-beam CT (CBCT) is widely used in radiotherapy clinics for patient setup and treatment monitoring, and is essential in the context of adaptive radiation therapy. Current work flow with adaptive planning on CBCT includes two major streams, one is to employ CBCT for direct dose calculation and the other is to perform dose evaluation on deformed planning CT with CBCT. Yang et al. is one of the first to evaluate the feasibility and accuracy of both ways1. They concluded it is more beneficial to improve CBCT image quality to CT level for adaptive radiotherapy due to the inherent reduced image quality and inaccurate Hounsfield units (HU) mapping2.
There have been numerous efforts in improving CBCT image quality using scatter correction: such as hardware improvement by adding anti-scatter grid3, x-ray beam blocker with a strip pattern2, or a lattice-shaped lead beam stopper4; or software improvement with iterative filtering5, raytracing6, model-based approach7, or Monte Carlo (MC) modeling 8,9. Especially, raytracing and MC methods have been shown to reproduce HUs to sufficient accuracy for both photon and proton dose calculation. They are, however limited by the time it takes to perform correction, about minutes or hours due to high computational complexity. Alternatively, conventional analytic reconstruction algorithms, such as filtered back projection, remain the mainstream due to fast computation.
Recently, machine-learning based algorithm has been applied to improve image quality and image reconstruction. It has been even shown that synthetic CT could be generated from MRI by using convolutional neural network (CNN) for radiotherapy planning without acquiring the actual CT 10,11. Similar strategy can also be applied to improve image quality of low-dose CT to match high-resolution CT 12. The purpose of this study is to develop unsupervised deep-learning model to improve CBCT image quality to CT level and to further validate the model on different anatomical sites.
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
Data from 30 pelvic patients were included. Each patient had one planning CT and five CBCT scans, a total of 150 pairs of CT-CBCT were used for model training and validation purposes. The CBCTs were from the first week of treatment to ensure the closest anatomy to planning CT. Paired CT-CBCT from an additional 15 pelvic patients and 10 head-and-neck (HN) patients were used for independent testing purpose. The CBCT scans of the validation set were collected at the first day of treatment on a different Varian TrueBeam.
All treatment planning CT images were collected with a GE LightSpeed16 CT scanner (GE Health Systems, Milwaukee, WI) and the CBCT images of the training set were acquired with an on-board-imager (OBI) equipped Varian TrueBeam STx linear accelerator (Varian Medical Systems, Palo Alto, CA). The original CTs had a resolution of 0.91 × 0.91 × 1.99 mm3 and dimensions of 512 × 512 × 210. All CBCTs had a resolution of 1.27 × 1.27 × 1.25 mm3 and dimensions of 512 × 512 × 80. For each patient, the CT images were mapped to each set of CBCT images using Velocity (Varian Medical Systems, Palo Alto, CA) with multi-pass B-spline based free form deformation to create a reference CT (rCT). All the deep-learning generated synthetic CTs (sCT) were compared to this reference.
2.2. Pix2pix GAN Architecture with Feature Matching
A 2.5 dimensional (2.5D) Pix2pix GAN-based deep-learning model with Feature Matching (FM) was proposed and the architecture is shown in Figure 113. The Generator was used to generate synthetic CT (sCT) from the original CBCT, and the Discriminator was used to distinguish the synthetic CT (sCT) from the reference CT (rCT). The Generators and Discriminators competed against each other until they reached an optimum.
Figure 1.
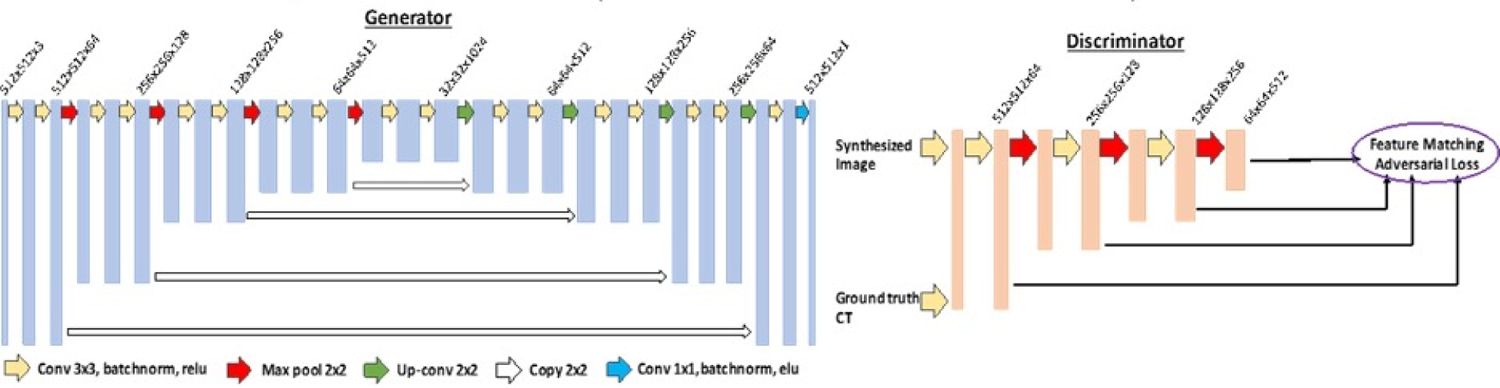
U-Net architecture is used for Generators in GAN. The input data size is 512 × 512 × 3 and the output data size is 512 × 512 × 1; the first two numbers represent resolutions and the third number represents channels.
The Generator was implemented using U-net architecture, in which each Conv-ReLU-BN block consists of either convolution or de-convolution layers with kernel size of 3×3, a batch normalization layer (BN) and a leaky rectified linear unit (ReLU). Concatenate connections were linked between the corresponding layers of the encoder and decoder. The activation function after the last convolutional layer was Elu. Then the synthesize CT (sCT) slices were used as the input of the Discriminator with the reference CT (rCT) slices as ground truth. The discriminator was a classifier that consisted of 8 stages of Conv-ReLU-BN block same as Generator.
The instability during the training of GAN is a critical issue which affects the output image quality from the generator. To address this issue, we implemented feature matching by changing the adversarial loss function14. This strategy forced the generator to generate images which could match the expected values of the features on the intermediate layers of the discriminator, besides the output of the discriminator. The loss function for the Discriminator was constructed as:
| (1) |
where fl is the output feature map on layer l, and nl represents number of pixels. The sCT and rCT slices were used as input. The corresponding feature maps from the 2nd, 4th, 6th, and 7th layers were obtained with mean absolute error summed together as loss function. fl(D(rCT)) represents the output of the discriminator from the original CT slices. αl is the adaptive weights for the features extracted from each layer. To further preserve the HU values between rCT and sCT, the L1 norm distance was added to the loss function:
| (2) |
where n is the number of pixels on the images, with the final adversarial loss function as:
| (3) |
where α is the weight between two different loss functions.
The 2.5 D architecture used a volume set with adjacent three slices as input of the network. This method stacked neighboring three slices together as different channels of the input to provide the network with 2.5D information, providing more morphology information to reconstruct the high-quality images.
2.3. Other network architectures
Besides feature mapping as mentioned in 2.2, another way to improve the synthesized image quality is to add perceptual loss 15. The architectures as pix2pix GAN model with vs. without additional perceptual loss were tested. VGG16 on ImageNet16 was used to extract the image features for two types of losses: content loss and style loss. The content loss was defined as the Euclidian distance between the feature maps from original and synthesized images of each layer:
| (4) |
where fj(CT) and fj(sCT) stand for the feature maps from the jth layer in the network for the ground-truth image and the synthesized image, respectively, and hj, wj, and cj stands for the size.
The style loss was used to control the similarity of image styles and was defined as the Euclidian distance between the stylistic feature maps from original and synthesized images of each layer:
| (5) |
where Gram matrix was defined as:
| (6) |
where m and n represent different output channels from the same layer. So the loss function becomes
| (7) |
β1 and β2 are the weights.
In addition, we also compared our methods with previously published models as U-net 17,18 and cycleGAN 19. U-net is a popular algorithm in image processing field and some investigators have explored its use in this context 17,18,20. In brief, the basic structure consists of convolution and max-pooling layers at the descending part (the left component of U), and convolution and up-sampling layers at ascending part (the right component of U) 20. In the down-sampling stage, the input image size is divided by the size of the max-pooling kernel size at each max-pooling layer. In the up-sampling stage, the input image size is increased by the operations, which are performed and implemented by convolutions, where kernel weights are learned during training. The arrows between the two components of the U show the incorporation of the information available at the down-sampling stage into the up-sampling stage, by copying the outputs of convolution layers from descending components to the corresponding ascending components. In this way, fine-detailed information captured in descending part of the network is used at the ascending part. The output images share the same size of the input images.
A few works have been done using CycleGAN to obtain synthetic CT from CBCT 19,21. In brief, it consisted of two generators as GA (mapping from CBCT to sCT) and GB (mapping from CT to sCT). It also had two discriminators as DA to distinguish rCT from fake CT, and DB to distinguish real CBCT from fake CBCT. With this bidirectional configurations, cycled CBCT images (cycleCBCT) from sCT and cycled CT images (cycleCT) from sCBCT could be obtained.
Besides adversarial loss from discriminators, cycle loss was added to the final function:
| (3) |
where
| (3) |
| (3) |
and n is the number of pixels on the image and γ is the weight of the cycle loss.
2.4. Model Configuration and Statistical Analysis
Normalized images were used as input, with rescaling HU numbers to the mean values of 0 and the standard deviation to 1. All models were trained with Adam optimization with a mini-batch size of 2 and epoch number of 100. All weights were initialized from He normal initializer. Batch normalization was used after each convolutional layer. The learning rate was set to 0.0001 with momentum term 0.5 to stabilize training. The generator was trained twice while the discriminator was trained once to keep the balance between the two components. To control the overfitting, three methods were utilized. First, before training, all images were augmented by horizontally flipping, a small angle rotation, as well as adding some background noise. Then L2 regularization term was added to the final loss function. Lastly, during the training process, early stop was applied based on the lowest validation loss to obtain the optimized model.
10-fold cross validation was used to evaluate the performance of the model. Each slice was used as an independent case. The training and validation sets included 150 CBCT-CT pairs, and 90% of cases were used for training while remaining 10% were used for validation purpose. The results from the validation sets were calculated. A separate dataset with additional 15 pelvic patients and 10 head-and-neck patients with paired CT and first-day CBCT, with CBCTs collected at a different linac machine, was used as an independent testing set to evaluate the robustness of proposed algorithm.
Synthetic CT slices (sCT) were firstly generated using the proposed model then rendered into 3D volumes to compare to the reference CT (rCT) images. Two metrics as peak signal-to-noise ratio (PSNR), and mean average error (MAE) were calculated by comparing synthetic CT and reference CT. PSNR measured the maximum possible power of a signal, with higher value indicating better image quality. MAE measured absolute HU differences of every single pixel between target and reconstructed image, with lower value indicating closer similarity to target. A total of 8 models were tested and compared: (1) 2.5D Pix2pix GAN with feature matching (FM) – as proposed in this study; (2) 2D Pix2pix GAN without feature matching, using single slice as network input; (3) 2D Pix2pix GAN with feature matching; (4) 2.5D Pix2pix GAN without feature matching; (5) 2.5D Pix2pix GAN with feature matching and perceptual loss; (6) U-net; and (7) cycleGAN and (8) cycleGAN with identity loss21,22. To further prove that the sCT can carry the dose calculation with comparable accuracy as CT, both photon and proton treatment plans were transferred to the corresponding sCT. The differences were also compared to that was directly generated from the rCT.
3. Results
Figure 2 shows the intermediate results of training and testing curves when using different network architectures. Due to the large number of training iteration, only the first 9000 training iterations were recorded for assessment. Figure 2(a) compares the pix2pix GAN with or without feature matching (FM), and the one without FM showed obvious instability during the training process. The performance of adding FM to various or different layers was further evaluated. As shown in Figure 2(b), adding FM to all layers could lead to overfitting as the testing MAE increased when training iterations increased. While if adding FM to limited number of layers as layers of 6 and 7, the stability cannot be obtained as shown in Figure 2(c). Experiments have been conducted by applying FM to various combination of intermediated layers, the pix2pix GAN with feature matching added to layer 2,4,6,7 was determined as the final architecture to obtain a balance between instability and overfitting. For illustration purpose, Figure 2(d) shows the training and testing process comparing pix2pix GAN and cycleGAN. The stability was not well maintained in training dataset and the MAE increased with iterations for testing dataset indicating potential overfitting for cycleGAN compared to proposed method on our dataset.
Figure 2.
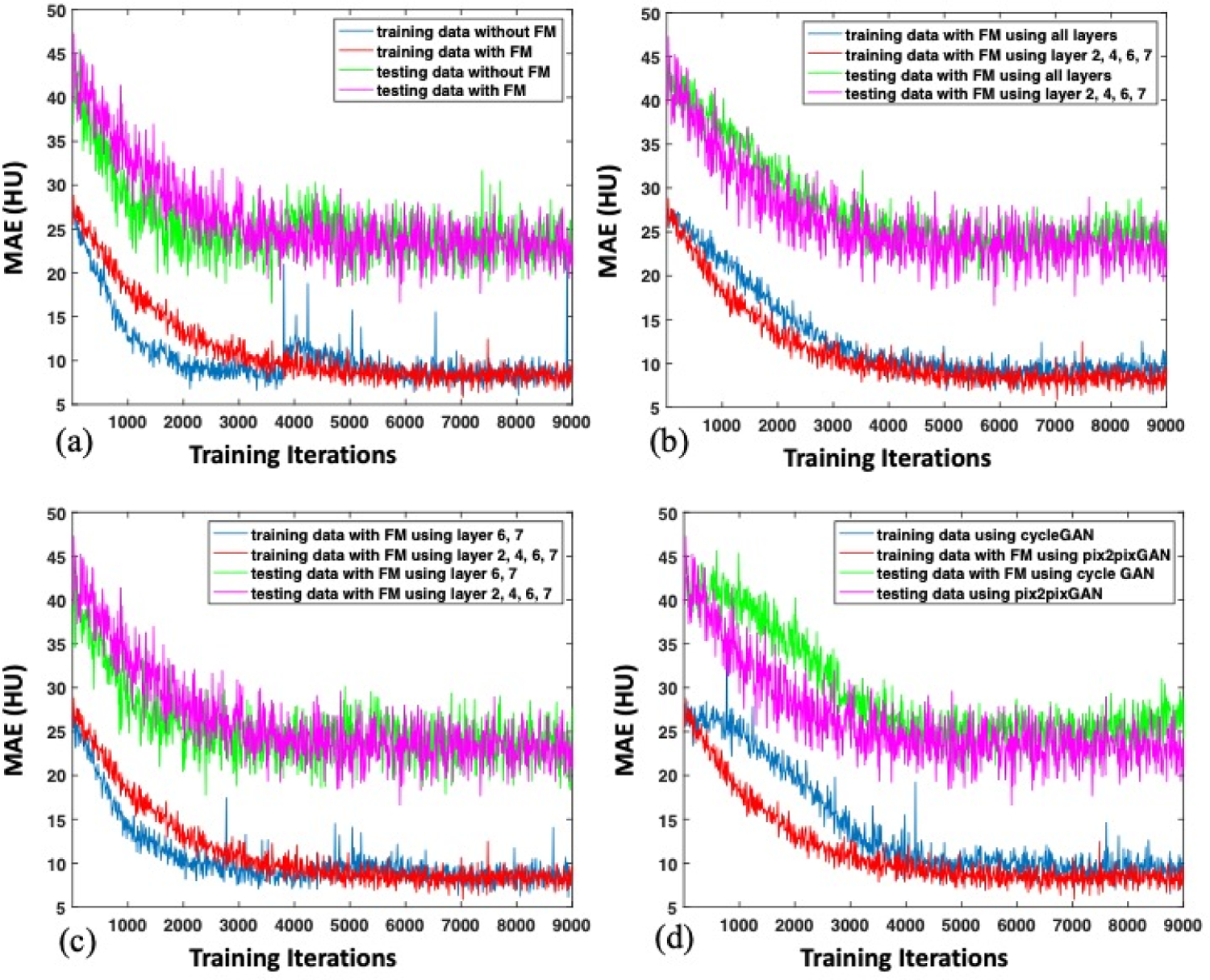
The intermediate results of training and testing curves to compare (1) the pix2pix GAN with or without FM, (2) the pix2pix GAN with FM at all layers vs. at layers of 2,4,6,7; (3) the pix2pix GAN with FM at layers of 6,7 vs. at layers of 2,4,6,7; (4) the pix2pix GAN vs. cycleGAN. Due to the different training and testing datasets, training and testing MAEs were different.
Figure 3 shows two case examples with reference CT (rCT) images, raw CBCT images and deep-learning generated synthetic CT (sCT). The intensity differences in Hounsfield Unit (HU) are also displayed. It can be clearly seen that the synthetic CT had much closer HU level to the reference CT compared to the raw CBCT.
Figure 3.
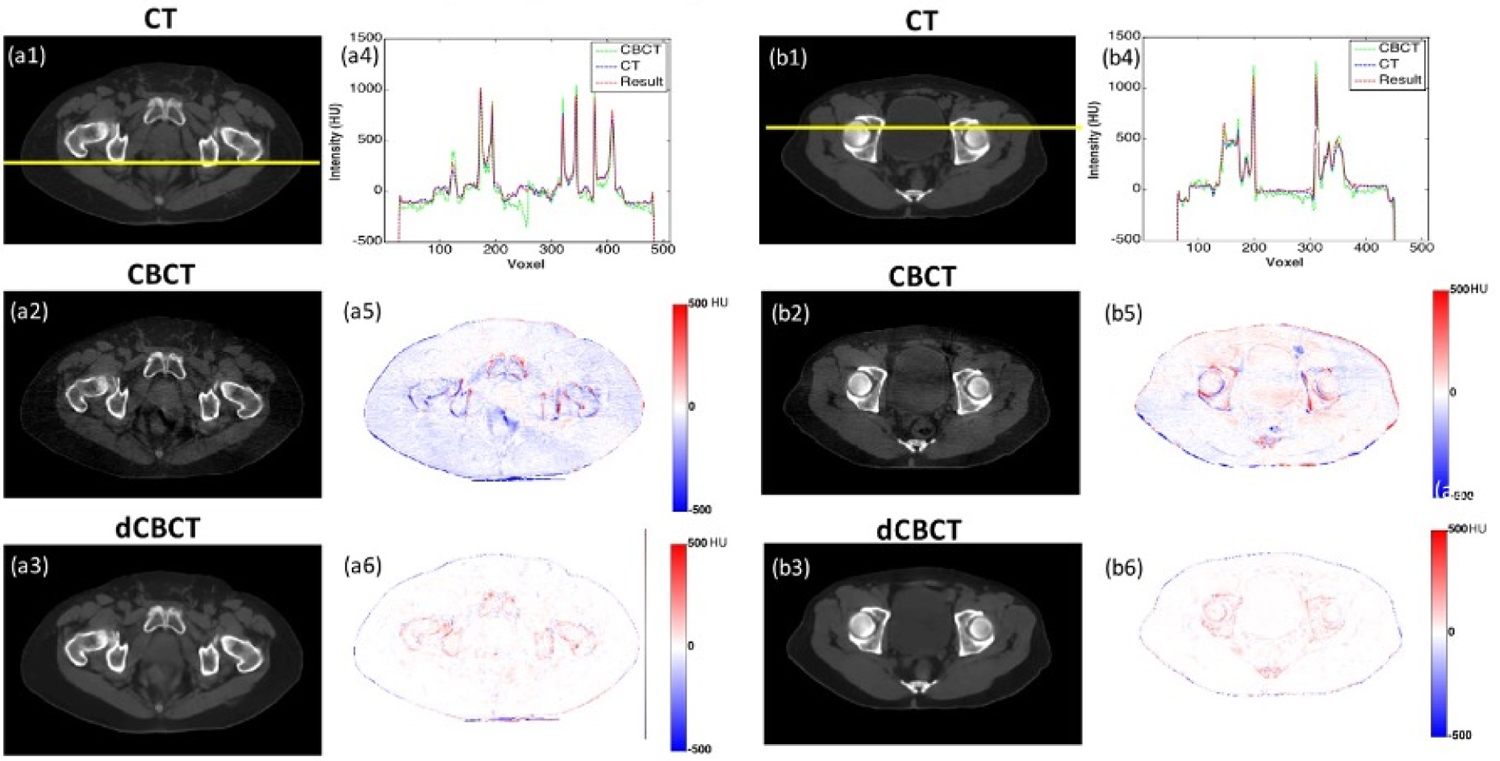
Two case examples: (1) CT image, (2) CBCT image, (3) deep-learning based CBCT (dCBCT) predicted using 2.5D GAN with feature matching, (4) line plot showing intensity profile of CT (blue), CBCT (green) and dCBCT (red) in range of [−500, 1500] HU, (5) HU differences between CBCT to CT in range of [−500, 500] HU, (6) HU differences between dCBCT to CT in range of [−500, 500] HU.
The group result in the validation dataset is summarized in Table 1. All deep-learning generated synthetic CTs showed improved image quality with less discrepancies (smaller MAE) to reference CT. The proposed algorithm as 2.5 Pix2pix GAN with feature matching was shown to be the best model among all tested methods with the highest PSNR and the lowest MAE. The mean MAE improved from 26.1±9.9 HU (CBCT vs. rCT) to 8.0±1.3 HU (sCT vs. rCT). The PSNR also increased significantly from 16.7±10.2 (CBCT vs. rCT) to 24.0±7.5 (sCT vs. rCT) in the validation set. The results showed that changing from 2D to 2.5D input had slight improvement for the PSNR and MAE but not statistically significant, due to only 3 slices information added into the model.
Table 1:
The Mean Average Error (MAE) and Peak Signal-to-Noise Ratio (PSNR) of the original CBCT and the synthetic CT generated by using 8 deep learning architectures compared to the referece CT: (1) 2.5D Pix2pix GAN with feature matching – as proposed in this study; (2) 2D Pix2pix GAN without feature matching, using single slice as network input; (3) 2D Pix2pix GAN with feature matching; (4) 2.5D Pix2pix GAN without feature matching; (5) 2.5D Pix2pix GAN with feature matching and perceptual loss; (6) U-net; and (7) cycleGAN and (8) cycleGAN with identity loss.
| Network | Mean Average Error (MAE) | Peak Signal-to-Noise Ratio (PSNR) | |
|---|---|---|---|
| 0 | Original CBCT | 26.1±9.9* | 16.7±10.2 |
| 1 | 2.5D GAN with FM | 8.1±1.3 | 24±7.5 |
| 2 | 2D GAN without FM | 9.4±1.2 | 22.4±3.8 |
| 3 | 2D GAN with FM | 8.1±1.4 | 23.8±1.8 |
| 4 | 2.5D GAN without FM | 9.3±2.1 | 22.7±2.9 |
| 5 | 2.5D GAN with FM and Perceptual Loss | 9.2±1.5 | 23.2±7.8 |
| 6 | U-net | 19.2±6.4** | 18.9±6.7 |
| 7 | CycleGAN | 9.2±1.5 | 23.2±7.8 |
| 8 | CycleGAN with Identity Loss | 8.9±3.1 | 22.1±5.5 |
MAE between the original CBCT and CT is significantly higher compared to other methods
MAE between the U-net generated synthetic CT and original CT is significantly higher compared to other methods
U-net was under-performed than any of GAN networks. As shown in Figure 4, the U-net generated blurred images and lost detailed information especially at the tissue boundaries. Overall, the deep-learning based CBCT generated through the pix2pix GAN methods had greatly reduced artifacts compared to the corresponding raw CBCT.
Figure 4.
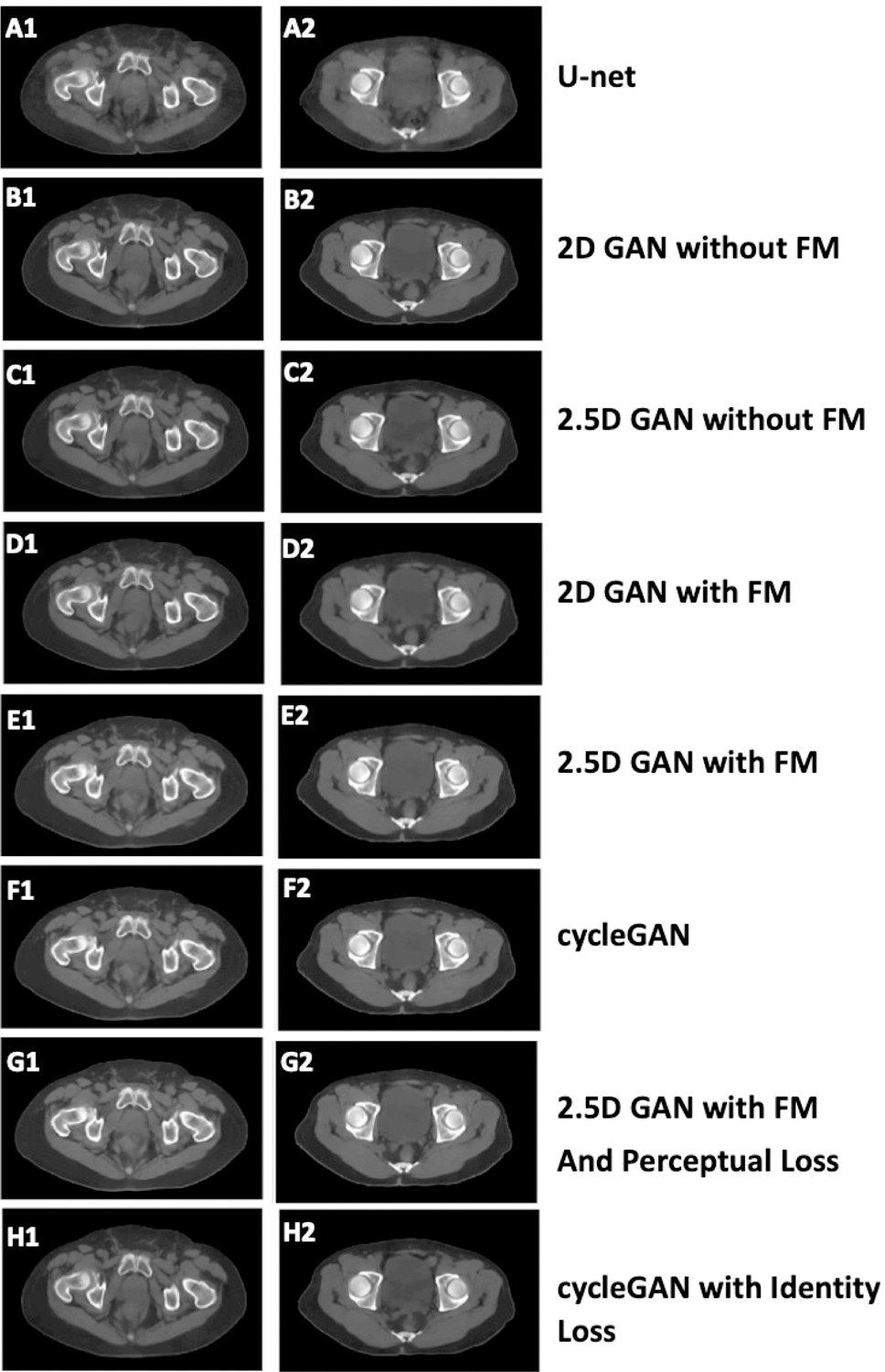
Comparison among the presented algorithm and other algorithms as prediction results using: (1) U-net; (2) 2D GAN without feature matching (FM); (3) 2.5D GAN without FM; (4) 2D GAN with FM; (5) 2.5D GAN with feature matching; (6) CycleGAN; (7) 2.5D GAN with FM and perceptual loss; and (8) cycleGAN with identify loss.
The proposed algorithm was further applied to the independent testing dataset. Due to different linac machine setting, the image discrepancies from raw CBCT to CT was larger compared to the training/validation set. The average MAE was 43.8±6.9 HU for pelvic cases originally, but was improved to 23.6±4.5 with deep-learning. The pSNR was improved from 14.53±6.7 to 20.09±3.4. When extended to head-and-neck regions, the model still produced less MAE discrepancies to 24.1±3.8 from original 32.3±5.7 HU. The pSNR was improved from 20.34±1.6 to 22.79±3.4. This indicated that the GAN model pre-trained with pelvic region might be able transferred to other region. The testing performance showed improvement, with yet less extent, also indicating task-specific performance may be needed for further improvement. An example of the head-and-neck cases is shown in Figure 5. It shows improved image quality with much closer HU to reference CT.
Figure 5.
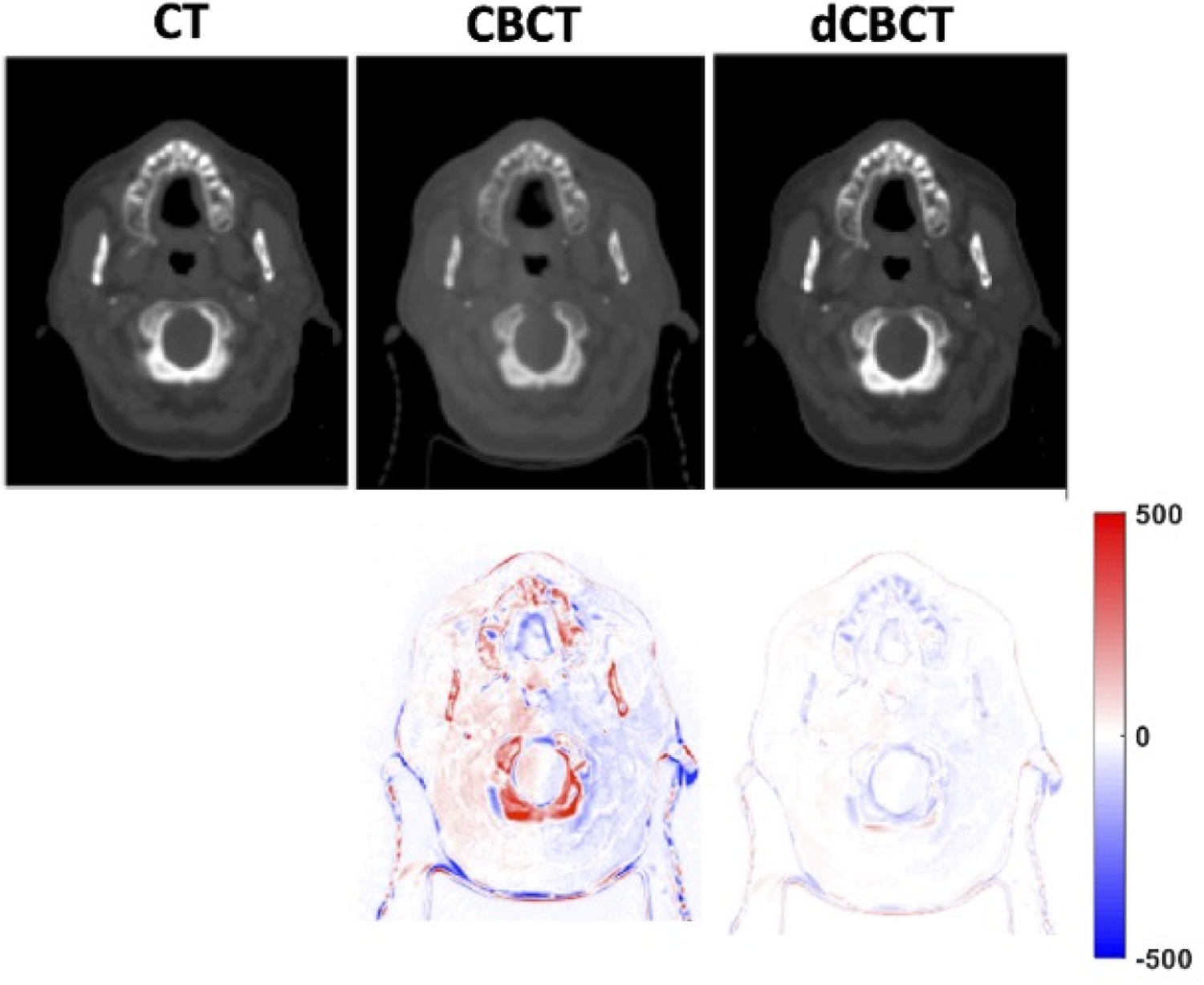
One head-and-neck case example from an independent testing dataset. The deep-learning based CBCT showed much closer HU to reference CT.
Figure 6 and 7 shows the dose difference map of a representative patient with both photon and proton dose calculation, respectively. Photon plan was delivered with VMAT using 6X beam, originally prescribed at 5760 cGy in 32 fractions. Proton plan was designed with double scattering technique using two lateral beams under the same prescription. The plan was designed on reference CT and recalculated on synthetic CT. For the VMAT plan, dose differences were confined close to the patient surface and minimal differences (< 1%) inside the patient, showing high accuracy for photon based dose calculation. Yet for proton plans, which the dose distribution is more sensitive HU differences and water equivalent depth (WED), an over/under-estimation of the proton range was observed. A median range difference of 2 mm was observed for the representative case, in line with a MAE of a 9.8 HU.
Figure 6.
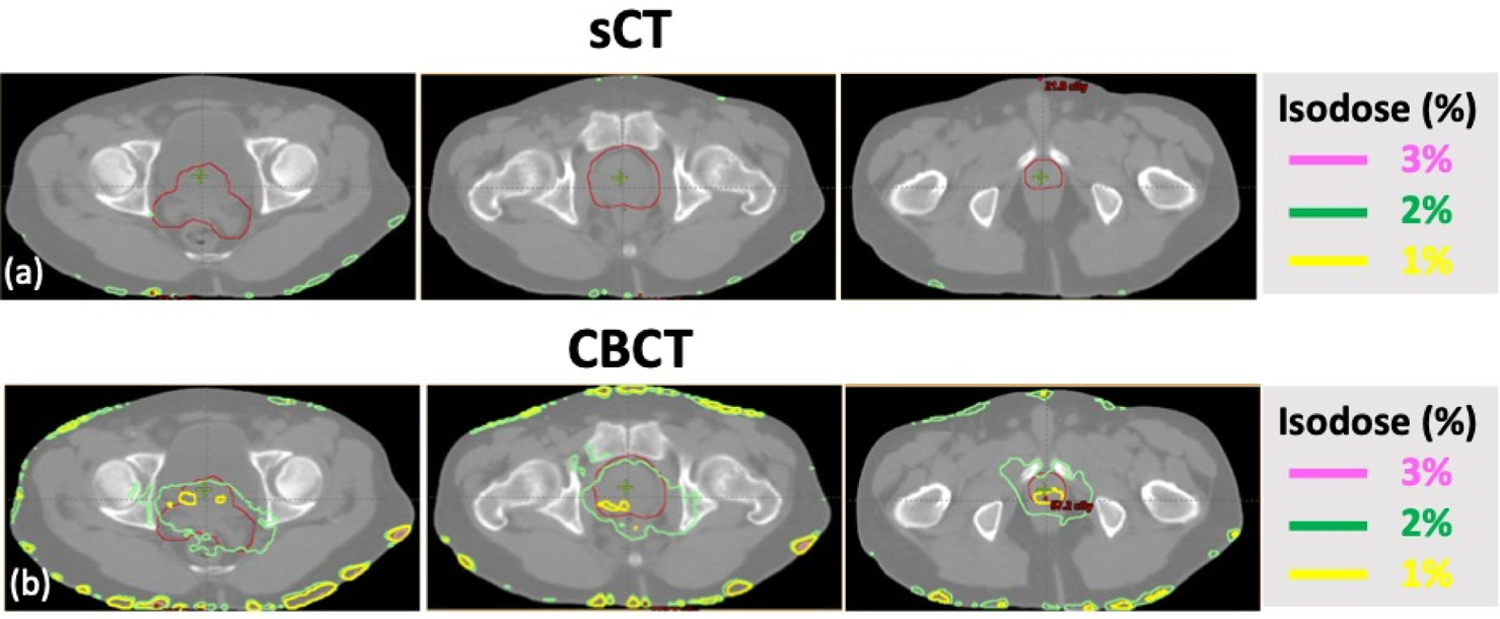
VMAT based photon plans showing dose differences calculated on (a) synthetic CT (sCT) and (b) CBCT with relative to the reference CT (rCT), dose calculation accuracy can be obtained with generated sCT. The planning target is shown in red.
Figure 7.
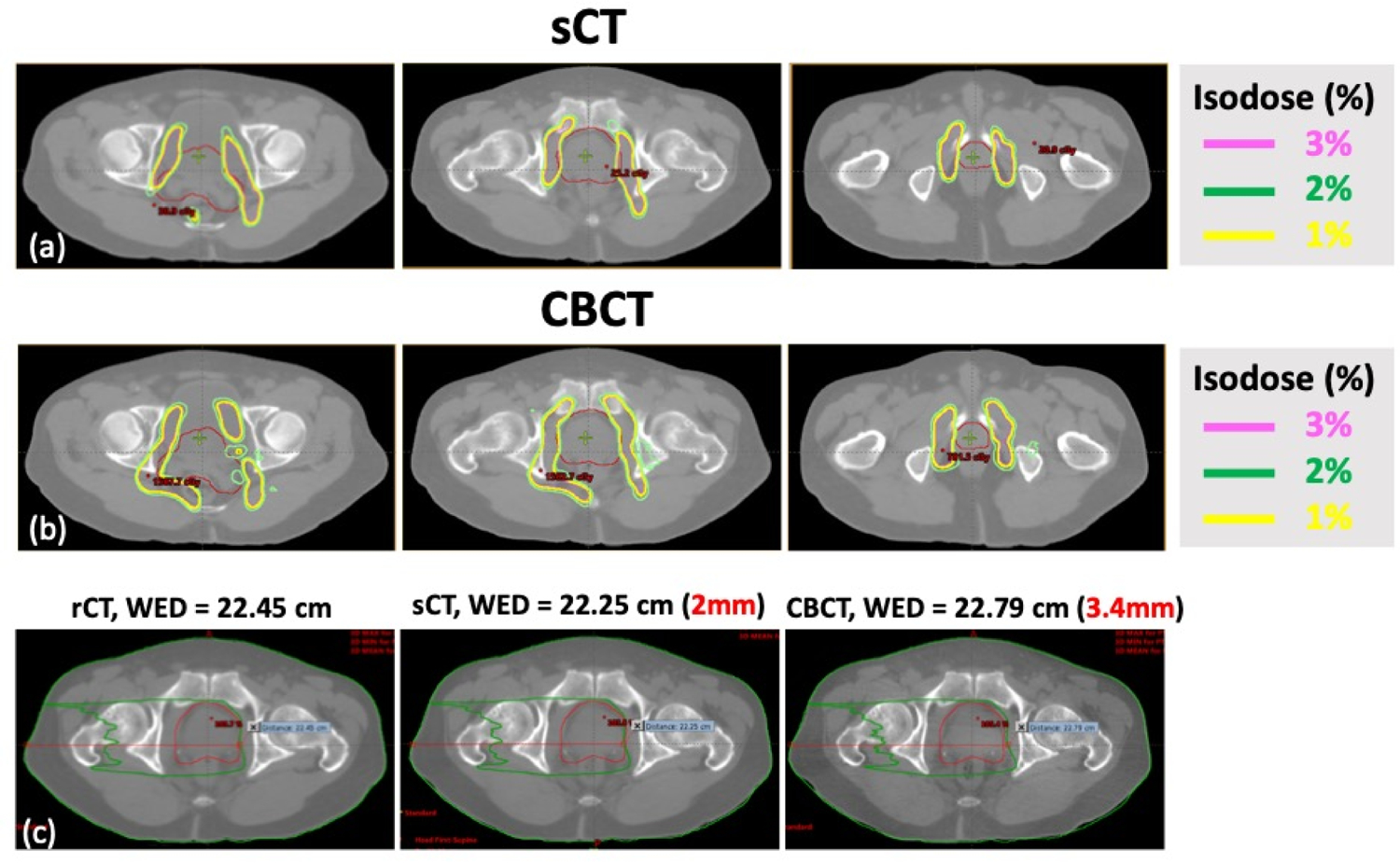
Double scattering based proton plans showing dose differences calculated on (a) synthetic CT (sCT), (b) CBCT with relative to the reference CT (rCT), with two lateral beams; and (c) the field water equivalent depth (WED) of single beam was displayed for rCT, sCT and CBCT. The sCT showed less proton range differences. The planning target is shown in red.
The network code was written in Python 3.6 and TensorFlow 2.0 and experiments were performed on a GPU-optimized workstation with a single NVIDIA GeForce GTX Titan X (12GB, Maxwell architecture). Once the model was trained, it took 11–12 ms to process one slice and generate a 3D volume of synthetic CT in less than a second.
4. Discussions and Conclusions
We have developed a deep-learning based model to generate synthetic CT from routine CBCT images based on pixel-to-pixel (Pix2pix) GAN. The model was built and validated on 30 pelvic patients with 150 paired CT-CBCT images, and further tested with an independent cohort with 15 additional pelvic cases and 10 head-and-neck cases collected at another linac machine. The image quality of the deep-learning based synthetic CT had been overall improved with much less MAE discrepancies to reference CT in both validation and testing datasets. The dose distribution also demonstrated a high accuracy for photon based calculation. This proof-of-concept technique provides substantial improvement in terms of speed, which can be directly generated within a second and thus be implemented real time. More investigations are needed for direct clinical adaption as well as for proton related applications.
The online CBCT has been widely used for daily positioning and target alignment. It may also allow early assessment of treatment response and be a prognostic factor of treatment outcomes. However, its use in adaptive radiotherapy is limited due to large scattering and inaccurate mapping of HU. Numerous mathematical algorithms have been proposed for past decades to improve CBCT image quality, including model-based approach7, Monte Carlo (MC) modeling 8,9 and iterative reconstruction (IR) 5 and raytracing6 with literatures cited in Table 2. The high demand on computational complexity was the major concern. Jia et al. developed an advanced MC algorithm with ray-tracing 6. With GPU, the computational time was greatly reduced from hours to minutes. Xu et al. extended the work using planning CT as prior information and was able to further shorted the computational time in 30s9. Yet, most of the work was tested on phantom or limited number of patient data and has not commonly implemented for clinical use. Alternatively, conventional analytic reconstruction algorithms, such as filtered back projection, remain the mainstream due to its fast computation.
Table 2.
Summary of previous publications on conventional algorithms in improving CBCT image quality.
| Paper | Method | Time | Dataset |
|---|---|---|---|
| Zbijewski et al. 20068 | Monte Carlo | ~ hours | a digital rat abdomen phantom |
| Wang et al. 20095 | Interative reconstruction | ~5 hours | a CT quality assurance phantom and an anthropomorphic head phantom |
| Sun et al. 20107 | Scatter Kernel | ~15 mins (8-core thread) | Pelvis phantom |
| Tian et al. 201123 | Iterative reconstruction | 6s per slices (GPU) | Thorax phantom, chest phantom and Catphan phantom |
| Jia et al. 2011 24 | Iterative reconstruction | 5 mins (GPU) | thorax phantom and Catphan phantom |
| Jia et al. 20126 | Ray-Tracing | ~ mins (GPU) | Catphan phantom and 1 H&N patient |
| Xu et al. 20159 | Modified Monte Carlo with planning CT as prior information | 30s (GPU) | Full-fan headneck case and the half-fan prostate case |
| Park et al. 201525 | Modified Monte Carlo with planning CT as prior information | 6 mins(GPU) | Anthropomorphic phantoms and a prostate patient |
Recently, deep learning based approaches have emerged as a potential solution to overcome computational complexity of prior mathematical algorithms in improving CBCT image quality. Some efforts have been done at 2D projection level 26–28. Nomura et al. used U-net convolutional neural network (CNN) based algorithm to perform scatter correction with lung phantom27. Jiang et al. performed scatter correction of CBCT using a deep residual CNN and also claimed computational efficient 28. Another route is directly applying deep-learning technique on reconstructed 3D volume with recent publications summarized in Table 3. Kida et al. used a U-net CNN for the pelvic CBCT-to-sCT generation, and reported improvement of MAE from 92 to 31HU with 20 patient cases 29. Similarly, Li et al.30 used an improved U-net architecture with residual block and trained the architecture on 50 H&N patients. Improved MAE was also reported. Yuan et al. also applied similar technique for head-and-neck patients, but with CBCT collected at fast-scan low-dose acquisition18. Recently, cycleGAN has been proposed to deal with the unpaired training data in multiple applications in medical imaging such as MRI-based sCT generation 10, organ segmentation 31, and CBCT-based sCT generation 19,32,33. CycleGAN incorporates an inverse transformation to better constrain the training model toward one-to-one mapping. In the application of CBCT-to-sCT generation, Liang et al. applied cycleGAN to train the CBCT-planning CT dataset without performing deformable registration 19. The cycleCBCT generated from CT was used to restrain the network. The algorithm was tested on 4 H&N patients and the MAE was improved from 70 to 30 HU. Kruz et al. used a similar algorithm to process the pelvic images34. The resulted MAE was improved to 87 HU, compared to the original 103 HU. Harms et al. published a CBCT-to-sCT generation method using cycleGAN with the incorporation of residual blocks and a novel compound loss in the cycle consistency loss function with improved results 33. The authors mentioned that although cycleGAN was initially designed for unpaired mapping, rigid registration should still be recommended to preserve the qualitative values. Liu et al. were the first to extend the deep-learning algorithm to abdomen regions with large motion artifacts32. Improved MAE was reported from 81HU to 57HU. We have compared our proposed deep-learning model with some previous reported methods. It was found the U-net CNN underperformed than any GAN based methods on our datasets. This might be due to the fact that the algorithm started with multi-layer image smoothing which would in-turn resulted in large signal discrepancies at boundaries. Another tested algorithm as CycleGAN has been widely applied to match unpaired images. Yet, with the co-registration done in the preprocessing step, the input CBCT and the reference CT were matched with similar morphologies. Since the purpose of this study is to generate synthetic CT from CBCT and further to match with reference CT, with this to-match purpose, the cycle loss as used in CycleGAN was not deemed necessary. In addition, we tried to add perceptual loss into the model. The initial weights merely captured the features of natural images, and it actually disturbed the training process. By comparing all deep-learning algorithms, 2.5D pix2pix GAN with feature matching was identified as the best model. The model was built on a large pelvic datasets with 150 pairs of CBCT-CT. The pelvic dataset contained enough variation of the anatomy structures, which helped to improve the robustness of the GAN model. The co-registration results contributed to the good correspondence between CT slices and CBCT slices, thus the conversion difficulty was reduced. Notably, the current model not only showed improved results in the validation set, it was further extended to an independent image set with two disease sets collected on a different machine. The improvement was again confirmed by a significant reduction of MAE discrepancies. All these demonstrated its robustness in clinical image sets and potential clinical use.
Table 3.
Summary of recent publications on deep-learning based algorithms on reconstructed volume to further improve CBCT image quality.
| Paper | Algorithm | Original-> Result mean MAE | Dataset | Slice matching |
|---|---|---|---|---|
| Kida et al. 201829 | Unet | 92*->16 | Pelvis: 20 | Yes |
| Li et al. 201930 | Unet | (60, 120)**-> (6,27) | H&N: 50(training)+10(validation) +10(testing) | Yes |
| Harms et al. 201933 | cycleGAN | Brain:24->13 Pelvic:53->16 |
Brain:24 Pelvic:20 |
Yes |
| Liang et al. 201919 | cycleGAN | 72->28 | H&N: 13(training)+4(testing) | No |
| Chen et al. 202035 | Unet | 44->19 | H&N: 37(training)+7(testing) | Yes*** |
| Kruz et al. 202034 | cycleGAN | 103->87 | Pelvic: 25(training)+8(testing) | No |
| Liu et al. 202032 | Deep-Attention cycleGAN | 81->57 | Abdomen: 30 | Yes |
| Yuan et al. 202018 | Unet | 172–49 | H&N: 37(training)+15(testing) | Yes*** |
the analysis is the was evaluated in ROIs on selected slices in terms of spatial nonuniformity
this paper only gave the range of MAE
CBCT and CT images were acquired on the same day.
Despite the promising results, we acknowledge several limitations. Due to technical limitation of the GPU capacity, only three adjacent slices as 2.5D information were used as input. The performance did not show significant improvement compared to 2D single-slice method. We also performed patch-based approach to incorporate more slices 32. Experiments were done with 1/4 sized patches cropped from original images with 16 slices (4×4), and 1/8 sized patches with 64 slices (8×8). However, the MAEs were not better than the presented method. The 3D patch-based method involves more parameters to fit the loss function33, which requires significant larger training samples to avoid overfitting before comparing with current model. The future direction is to include a true 3D information with larger dataset and computer power. The second limitation is that signals between tissue boundaries, as body-to-air or bone-to-soft tissue, were not preserved. This can be seen from photon based planning as dose differences retain at the body contour. These differences may not be clinically significant for photon based planning but can result range over/underestimation for proton based planning. This may be due to the signal loss during pre-processing as volumetric resizing and image interpolation. To overcome this issue, high-resolution images with original details need to be retained during the pre-processing for which again high computational power is needed. In addition, proton beam is sensitivity to HU change, with a 5HU difference resulting ~1mm range shift. The adaption of current and similar techniques to proton based planning warrants more investigation. Thirdly, lack of the same day paired CT and CBCT at the same position prevented us to precisely evaluate the exact HU mapping. Identifying matched CT/CBCT pair taken at the same position is extremely challenging in a retrospective setting. None of previous published literatures except Yuan et al. used 10 paired same day CT/CBCT 18 and all in proof-of-concept stage. To truly adapt the technique in clinic, rigorous verification with precise ground truth is needed. The data collection of the same day paired CT/CBCTs and with various disease types are undergoing and will be included in our future study.
Overall, CBCT plays a very important role in image-guided radiation therapy (IGRT). Enhancement of its quality can contribute to daily patient setup and adaptive dose delivery, thus enabling higher confidence in patient treatment accuracy. The results of this study demonstrate that the artificial intelligence (AI) technique can improve CBCT image quality without hardware improvement. Once the model is trained, it takes less than a second to process a deep-learning based volumetric CBCT set. The results also show that the improved CBCT can achieve high image quality to be close to the level of conventional CT, thus have the potential to be used for adaptive planning. Overall, the method presented in this study may provide a time-efficient and economic-efficient solution for machines that are coupled with CBCT capability. The output may improve the soft-tissue definition that is necessary for accurate visualization, contouring, deformable image registration, and may enable new applications, such as CBCT-based online adaptive radiotherapy.
Acknowledgement
Funding support: This study was supported in part by NIH R01CA127927, National Natural Science Foundation of China (11704108).
Footnotes
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record.
References:
- 1.Yang Y, Schreibmann E, Li T, Wang C, Xing L. Evaluation of on-board kV cone beam CT (CBCT)-based dose calculation. Physics in Medicine & Biology. 2007;52(3):685. [DOI] [PubMed] [Google Scholar]
- 2.Zhu L, Xie Y, Wang J, Xing L. Scatter correction for cone-beam CT in radiation therapy. Medical physics. 2009;36(6Part1):2258–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siewerdsen JH, Moseley D, Bakhtiar B, Richard S, Jaffray DA. The influence of antiscatter grids on soft-tissue detectability in cone-beam computed tomography with flat-panel detectors: Antiscatter grids in cone-beam CT. Medical physics. 2004;31(12):3506–3520. [DOI] [PubMed] [Google Scholar]
- 4.Cai W, Ning R, Conover D. Scatter correction using beam stop array algorithm for cone-beam CT breast imaging. Vol 6142: SPIE; 2006. [Google Scholar]
- 5.Wang J, Li T, Xing L. Iterative image reconstruction for CBCT using edge-preserving prior. Medical Physics. 2009;36(1):252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia X, Yan H Fau Cervino L, Cervino L Fau Folkerts M, Folkerts M Fau Jiang SB, Jiang SB. A GPU tool for efficient, accurate, and realistic simulation of cone beam CT projections. Med Phys. 2012;39(12)(0094–2405 (Print)):7368–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun M, Star-Lack J. Improved scatter correction using adaptive scatter kernel superposition. Physics in Medicine & Biology. 2010;55(22):6695. [DOI] [PubMed] [Google Scholar]
- 8.Zbijewski W, Beekman FJ. Efficient Monte Carlo based scatter artifact reduction in cone-beam micro-CT. IEEE transactions on medical imaging. 2006;25(7):817–827. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Bai T, Yan H, et al. A practical cone-beam CT scatter correction method with optimized Monte Carlo simulations for image-guided radiation therapy. Physics in Medicine & Biology. 2015;60(9):3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maspero M, Savenije MHF, Dinkla AM, et al. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy [published online ahead of print 2018/08/16]. Phys Med Biol. 2018;63(18):185001. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Lei Y, Wang T, et al. MRI-based treatment planning for liver stereotactic body radiotherapy: validation of a deep learning-based synthetic CT generation method. Br J Radiol. 2019;92(1100):20190067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Paper presented at: Proceedings of the IEEE conference on computer vision and pattern recognition2017. [Google Scholar]
- 13.Mroueh Y, Sercu T, Goel V. Mcgan: Mean and covariance feature matching gan. arXiv preprint arXiv:170208398. 2017.
- 14.Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X. Improved techniques for training gans. Paper presented at: Advances in neural information processing systems2016. [Google Scholar]
- 15.Johnson J, Alahi A, Fei-Fei L. Perceptual losses for real-time style transfer and super-resolution. Paper presented at: European conference on computer vision2016. [Google Scholar]
- 16.Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. CoRR. 2015;abs/1409.1556.
- 17.Hansen DC, Landry G, Kamp F, et al. ScatterNet: A convolutional neural network for cone-beam CT intensity correction [published online ahead of print 2018/09/11]. Med Phys. 2018;45(11):4916–4926. [DOI] [PubMed] [Google Scholar]
- 18.Yuan N, Dyer B, Rao S, et al. Convolutional neural network enhancement of fast-scan low-dose cone-beam CT images for head and neck radiotherapy [published online ahead of print 2019/12/17]. Phys Med Biol. 2020;65(3):035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang X, Chen L, Nguyen D, et al. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy [published online ahead of print 2019/05/21]. Phys Med Biol. 2019;64(12):125002. [DOI] [PubMed] [Google Scholar]
- 20.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Paper presented at: International Conference on Medical image computing and computer-assisted intervention2015. [Google Scholar]
- 21.Zhu J-Y, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. Paper presented at: Proceedings of the IEEE international conference on computer vision2017. [Google Scholar]
- 22.Taigman Y, Polyak A, Wolf L. Unsupervised cross-domain image generation. arXiv preprint arXiv:161102200. 2016.
- 23.Tian Z, Jia X, Yuan K, Pan T, Jiang SB. Low-dose CT reconstruction via edge-preserving total variation regularization. Physics in Medicine & Biology. 2011;56(18):5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia X, Dong B, Lou Y, Jiang SB. GPU-based iterative cone-beam CT reconstruction using tight frame regularization. Physics in Medicine & Biology. 2011;56(13):3787. [DOI] [PubMed] [Google Scholar]
- 25.Park YK, Sharp GC, Phillips J, Winey BA. Proton dose calculation on scatter-corrected CBCT image: Feasibility study for adaptive proton therapy. Medical physics. 2015;42(8):4449–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen L, Zhao W, Xing L. Patient-specific reconstruction of volumetric computed tomography images from a single projection view via deep learning. Nature biomedical engineering. 2019;3(11):880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomura Y, Xu Q, Shirato H, Shimizu S, Xing L. Projection-domain scatter correction for cone beam computed tomography using a residual convolutional neural network. Medical Physics. 2019;46(7):3142–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y, Yang C, Yang P, et al. Scatter correction of cone-beam CT using a deep residual convolution neural network (DRCNN). Physics in Medicine & Biology. 2019;64(14):145003. [DOI] [PubMed] [Google Scholar]
- 29.Kida S, Nakamoto T, Nakano M, et al. Cone Beam Computed Tomography Image Quality Improvement Using a Deep Convolutional Neural Network [published online ahead of print 2018/07/03]. Cureus. 2018;10(4):e2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Zhu J, Liu Z, et al. A preliminary study of using a deep convolution neural network to generate synthesized CT images based on CBCT for adaptive radiotherapy of nasopharyngeal carcinoma. Physics in Medicine & Biology. 2019;64(14):145010. [DOI] [PubMed] [Google Scholar]
- 31.Lei Y, Wang T, Tian S, et al. Male pelvic multi-organ segmentation aided by CBCT-based synthetic MRI [published online ahead of print 2019/12/19]. Phys Med Biol. 2020;65(3):035013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Lei Y, Wang T, et al. CBCT-based synthetic CT generation using deep-attention cycleGAN for pancreatic adaptive radiotherapy [published online ahead of print 2020/03/07]. Med Phys. 2020. [DOI] [PMC free article] [PubMed]
- 33.Harms J, Lei Y, Wang T, et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography [published online ahead of print 2019/06/18]. Med Phys. 2019;46(9):3998–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurz C, Maspero M, Savenije MH, et al. CBCT correction using a cycle-consistent generative adversarial network and unpaired training to enable photon and proton dose calculation. Physics in Medicine & Biology. 2019;64(22):225004. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Liang X, Shen C, Jiang S, Wang J. Synthetic CT generation from CBCT images via deep learning. Medical physics. 2020;47(3):1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]


