Abstract
Surfactin, a quorum sensing signal molecule, is correlated with carbon metabolism in Bacillus amyloliquefaciens. In the present work, we found that mutation of srfA (ΔsrfA) led to an obviously changed carbon metabolism in B. amyloliquefaciens. Firstly, the PTS-glucose system was significantly increased as a feedback to glucose exhaustion. Secondly, the basic carbon metabolism such as glycolysis and TCA cycle was obviously weakened in ΔsrfA. Thirdly, the global regulator of CcpA (carbon catabolite protein A) and P ~ Ser46-HPr (seryl-phosphorylated form of histidine-containing protein) to mediate the CcpA-dependent CCR (carbon catabolite repression) were not increased, but the ability to use extracellular non- and less-preferred carbon sources was down-regulated in ΔsrfA. Fourthly, the carbon overflow metabolism such as biosynthesis of acetate was enhanced while biosynthesis of acetoin/2,3-butanediol and branched-chain amino acids were weakened in ΔsrfA. Finally, ΔsrfA could use most of non- and less-preferred carbon sources except for fatty acids, branched chain amino acids, and some organic acids (e.g. pyruvate, citrate and glutamate) after glucose exhaustion. Collectively, surfactin showed a global influence on carbon metabolism in B. amyloliquefaciens. Our studies highlighted a way to correlate quorum sensing with carbon metabolism via surfactin in Bacillus species.
Keywords: Surfactin, Carbon metabolism, Quorum sensing, CcpA, HPr, Bacillus amyloliquefaciens
Highlights
-
•
The quorum sensing molecule of surfactin globally influenced the carbon metabolism in B. amyloliquefaciens.
-
•
The studies highlighted a way to correlate quorum sensing with carbon metabolism via surfactin in Bacillus species.
-
•
Glycolysis and TCA cycle was obviously weakened after biosynthesis of surfactin was disrupted.
-
•
The CcpA-dependent carbon catabolite repression was not enhanced after disruption of surfactin biosynthesis.
-
•
The ability to use non-preferred carbon sources was down-regulated after disruption of surfactin biosynthesis.
1. Introduction
Generally, the available carbon sources are composed of diverse sugars in the complex environment, and the most favored carbon source (e.g. glucose, fructose or malate) are preferentially utilized by microorganisms for fast growth. The regulatory mechanism to make the sole utilization of preferred carbon sources over other sugars is called carbon catabolite repression (CCR) (Marciniak et al., 2012). Bacillus are generally soil-dwelling Gram-positive bacteria ubiquitously distributed in the natural environment. In B. subtilis, CCR is achieved by the global regulator of CcpA (carbon catabolite protein A), which binds to the operator sites called catabolite responsive elements (cre) in a complex with HPr-Ser46~P (seryl-phosphorylated form of histidine-containing protein, HPr) (Reuβ et al., 2018).
CCR occurs when the repressing sugars are converted to certain glycolytic intermediates, e.g. fructose-1,6-bisphosphate (FBP) (Fujita, 2009). FBP is a signal to enhance the indispensable stage of glycolysis from dihydroxyaceton~P to phosphoenolpyruvate (PEP). PEP stimulates the phosphorylation of HPr at Ser46 under the catalysis of HPr kinase/phosphatase (HPrK/P). HPr is also involved in carbohydrate transport via the PEP: sugar phosphotransferase system. The phosphoryl group of PEP is transferred to PtsI (EI) to form EI ~ P, then transferred from EI ~ P to HPr to form P ~ His15-HPr. Finally, the phosphoryl group is transferred from P ~ His15-HPr to the PTS-sugars. Thereby, HPr is also an energy-coupling protein of the PTS system (Deutscher and Milohanic, 2013).
CcpA in a complex with HPr-Ser46~P can negatively or positively regulate many genes transcription, and this is determined by the location of cre in many cases. Upstream binding of CcpA from the core promoter results in carbon catabolite activation (CCA), and downstream binding of CcpA from the core promoter or binding of CcpA in the core promoter causes CCR (Ishii et al., 2013). For example, CcpA in a complex with HPr-Ser46~P promotes the expression of several genes involved in carbon overflow metabolism to produce acetate and acetoin, and the expression of ilv-leu operon involved in biosynthesis of branched-chain amino acids. However, more genes and operons are repressed by the complex of CcpA and P ~ Ser46-HPr, including the gene citZ for the entrance of TCA cycle, several transporters gene for transportation of the TCA cycle intermediates, some respiration genes, and many catabolic and anabolic genes involved in carbon, nitrogen, and phosphate metabolism (Fujita, 2009).
P ~ His15-HPr also regulates the genes expression involved in utilization of less-preferred carbon source (e.g. glycerol, sucrose, oligo-β-glucoside, aryl-β-glucoside, etc) (Fujita, 2009). P ~ His15-HPr can enhance the activity of several transcriptional regulatory proteins by phosphorylation, including some antiterminators and transcriptional activators, to promote expression of the operons for utilization of less-preferred PTS-sugars. However, P ~ His15-HPr is preferentially used for phosphorylation of preferred PTS-sugars (e.g. glucose); thereby, most of P ~ His15-HPr will be consumed for transporting preferred PTS-sugars. In this case, P ~ His15-HPr is not enough for activating the antiterminators or transcriptional activators involved in utilization of less-preferred PTS-sugars. This is also called the CcpA-independent (P ~ His-HPr-dependent) CCR (Fujita, 2009; Darbon et al., 2002).
Surfactin is a signal molecule to regulate quorum sensing response in B. subtilis. As nutrients is deprived in the environment, the cells will produce surfactin to regulate several adaptive responses to this deprivation including quorum sensing (López et al., 2010). Previously, we reported that the quorum sensing signal molecule of surfactin is correlated with the carbon metabolism in B. amyloliquefaciens, a relative of B. subtilis (Chen et al., 2020). We found that mutation of srfA (ΔsrfA), a gene cluster for biosynthesizing surfactin, resulted in the decreased ability to utilize some carbon sources in B. amyloliquefaciens WH1. After glucose was exhausted, ΔsrfA was lyzed without sporulation although nutrients were still enough for growth during this period, but this could be reversed by addition with surfactin or extra glucose in the medium (Chen et al., 2020). At first, we deduced that surfactin might act as a signal molecule to relieve CCR in this bacterium. Deficiency of surfactin resulted in the disability to relieve CCR for utilizing non-preferred carbon sources after glucose was exhausted. We hypothesized that, besides for quorum sensing, surfactin might function as a signal molecule to regulate utilization of non-preferred carbon sources to counter nutrients limitation (e.g. glucose deprivation) in the environment. In the present work, we compared the difference of carbon metabolic networks between the wild-type strain WH1 and the mutant strain ΔsrfA via multiomics analysis including transcriptome, proteome, phosphoproteome and metabolome. We found that deficiency of surfactin did not result in an increase of CcpA and P ~ Ser46-HPr to enhance the CcpA-dependent CCR in ΔsrfA. In fact, surfactin globally influenced the carbon metabolism, including the PTS-glucose system, the CcpA-activated carbon metabolism, the CcpA-mediated CCR, the CcpA-independent (P ~ His15-HPr-dependent) carbon metabolism, and the carbon metabolic intermediates, in B. amyloliquefaciens.
2. Materials and methods
2.1. Bacterial strains and materials
B. amyloliquefaciens WH1 and ΔsrfA were stored in our lab (Chen et al., 2020; Qi et al., 2010). Chemicals with analytical grade were supplied by Sinopharm Chemical Reagent (China).
2.2. Samples preparation
WH1 and ΔsrfA were cultured in a 250-mL flask with 50 mL LB medium at 28 °C and 180 rpm, respectively. After 18 h (Chen et al., 2020), 1.5 mL culture of 24 test tubes per strain were centrifuged for collecting cell pellets, then used for analysis of transcriptome, proteome, phosphoproteome and metabolome by Novogene Co., Ltd (China).
2.3. RNA-seq analysis
Cell pellets (6 samples per strain) were used for isolating RNA with RNeasy Mini Kit, then mRNA was enriched with RiboZero rRNA Removal Kit (Qiagen, German). Library preparations for paired-end sequencing were performed on the Illumina HiSeq, and the obtained sequences were mapped onto the reference genome of B. amyloliquefaciens (NCBI Accession no: HE617159.1). To quantify gene transcription from the obtained sequence reads, RPKM values (Reads Per Kilobase per Million mapped reads) were calculated using the number of reads mapped to a gene, the total amount of mapped reads in the experiment, and the length in base pairs for a gene. The thresholds for significant changes in gene transcription were fold-changes of greater than 2 or less than 0.5 with p < 0.05 and 0.01, respectively (Kröber et al., 2016).
Some key genes transcription were quantified by Quantitative Real-Time PCR (qRT-PCR). RNA was extracted as above, then cDNA was produced by reverse transcription with 1 μg RNA, iScript Select cDNA Synthesis Kit and random oligonucleotide primers (Bio-Rad). qRT-PCR was performed with cDNA, SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and target-specific primers in CF96 Real-Time System (Bio-Rad). All expression data were normalized to the copy number of 16S rRNA in each sample.
2.4. Analysis of proteome
Pellets (6 samples per strain) were powered in liquid nitrogen for extracting total proteins, then digested by trypsin, and the obtained peptides were desalted, dried, and labeled with TMT10-plex reagents (Thermofisher). The labeled peptides were fractionated, dried, then determined by LC-MS/MS, and the resulting spectra from each fraction were searched separately by PD 2.2 (Thermo). The Reporter Quantification (TMT 10-plex) was used for quantification, and the quantitation results were statistically analyzed by Mann-Whitney Test. The significant ratios, defined as p < 0.05 and p < 0.01, were used to screen differentially expressed proteins (DEP) (Wiśniewski et al., 2009). InterPro (IPR) analysis was conducted using the interproscan-5 program against the non-redundant protein database (Pfam, PRINTS, ProDom, SMART, ProSiteProfiles, PANTHER) (Jones et al., 2014), then enriched with the enrichment pipeline (Huang et al., 2009; Franceschini et al., 2013).
CcpA and HPr were also quantified by Western blot assay. The ccpA and hpr gene were subcloned into the pET28a vector, then the recombinant plasmids were transformed into Escherichia coli BL21, respectively. The recombinant proteins including CcpA and HPr were induced by IPTG for expression, purified by Ni-NAT column, then used for intraperitoneal immunization of mice at 50 μg/mouse for 3 dosages after emulsification with Freund's adjuvant (Bie et al., 2016). The sera were collected, than used for Western blot analysis of CcpA and HPr, respectively. Briefly, the cells of WH1 and ΔsrfA were lyzed by ultrasonification, then the total proteins concentration of each sample was adjusted to 10 μg/μL 10 μL of each protein sample was separated by 15% SDS-PAGE gel, transferred onto the polyvinylidene fluoride (PVDF) membrane (Millipore, USA), then incubated with the above mouse antisera at a dilution of 400 × for 1 h. After extensive wash, the membrane was incubated with goat anti-mouse IgG labeled with Horseradish Peroxidase (HRP) (Boster Biological Technology, Wuhan, China) at a dilution of 5000 × for 30 min. Thereafter, the membrane was washed, reacted with BeyoECL Star (Beyotime Biotechnology, Shanghai, China), then scanned by Monad QuickChemi 5100 (Zhuhai, China).
2.5. Analysis of phosphoproteome
Pellets (6 samples per strain) were powered in liquid nitrogen for extracting total proteins, then digested by trypsin as above. The obtained peptides were desalted, dried, and enriched using phos-select iron affinity gel, then used for shotgun proteomics analyses via EASY-nLCTM 1200 UHPLC system coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Resulting spectra from each fraction were searched separately by PD 2.2. The precursor quantification based on intensity was used for Label-free quantification. Analysis of IPR was done as the methods mentioned above, then the motif-x algorithm was used to identify motifs enriched within a set of phosphosites (Wagih et al., 2016).
The phoshorylated protein P ~ Ser46-HPr was also quantified by Western blot assay. The rabbit antiserum against P ~ Ser46-HPr was prepared by DIA.AN Biotech (Wuhan, China). Briefly, the peptide containing P ~ Ser (GKTVNLKSIMG) was cross-linked with keyhole limpet hemocyanin (KLH), then used for intramuscular injection of rabbits at 100 μg/rabbit for 3 dosages after emulsification with Freund's adjuvant. The serum was collected, than used for Western blot analysis of P ~ Ser46-HPr as the methods mentioned above.
2.6. Untargeted metabolomics analysis
Pellets (6 samples per strain) were grounded with liquid nitrogen, then the homogenate was resuspended with prechilled 80% methanol and 0.1% formic acid. The samples were incubated on ice for 5 min, then centrifuged at 15,000 g, 4 °C for 20 min. The supernatant was diluted to a final concentration containing 60% methanol by LC-MS grade water, filtrated with 0.22 μm filter, then injected into the LC-MS/MS system coupled with Hyperil Gold column (100 × 2.1 mm, 1.9 μm) and analyzed using Vanquish UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Eluents for the positive polarity mode were eluent A1 (0.1% formic acid in water) and eluent B (methanol). Eluents for the negative polarity mode were eluent A2 (5 mM ammonium acetate, pH 9.0) and eluent B (methanol). Solvent gradient was set as follows: 2% B, 1.5 min; 2–100% B, 12.0 min; 100% B, 14.0 min; 100 - 2% B, 14.1 min; 2% B, 16 min. Q Exactive HF-X mass spectrometer was operated in positive/negative polarity mode with spray voltage of 3.2 kV, capillary temperature of 320 °C, sheath gas flow rate of 35 arb and aux gas flow rate of 10 arb. The raw data generated by UHPLC-MS/MS were processed using the Compound Discoverer 3.0 (Thermo Fisher) to perform peak alignment, peak picking, and quantitation for each metabolite. Peak intensities were normalized to the total spectral intensity. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks and fragment ions, and then peaks were matched with the mzCloud and ChemSpider databases to obtain the accurate qualitative and relative quantitative results (Want et al., 2010; Kieffer et al., 2016).
2.7. Ability to utilize carbon sources
WH1 and ΔsrfA were cultured in LB medium added with different carbon sources including glucose, sucrose, starch, glycerol, lactose, mannose, mannitol, xylose, glucan, pyruvate, citrate, fumarate, malate, leucine, isoleucine, valine, glutamate, fatty acids (vegetable oil, VO), acetoin and 2,3-butanediol at a final concentration of 4 g/L, at 28 °C and 180 rpm, respectively. After 24 h, the cells were stained with crystal violet for observation of spores and viable cells in the broth (Chen et al., 2020).
3. Results
3.1. Neither CcpA nor HPr-Ser46~P was increased in ΔsrfA
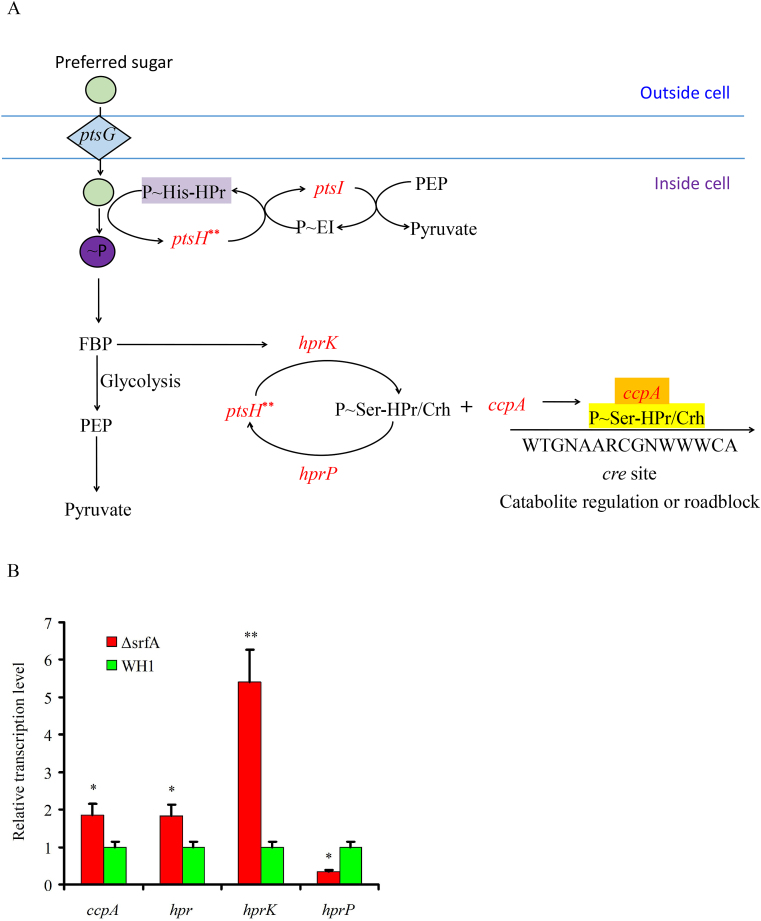
RNA-seq analysis showed that the transcription of ptsH encoding HPr (PtsH), was significantly increased in ΔsrfA compared to WH1 (Fig. 1A). Also, the transcription of ptsI encoding EI (PtsI), and the transcription of hprK, crh, and ccpA were all increased in ΔsrfA (Fig. 1A). The transcription of some key genes such as ccpA, hpr, hprK and hprP were also further verified by qRT-PCR. Consistently, these genes transcription were significantly increased in the strain ΔsrfA when compared to the wild-type strain WH1 (Fig. 1B).
Fig. 1.
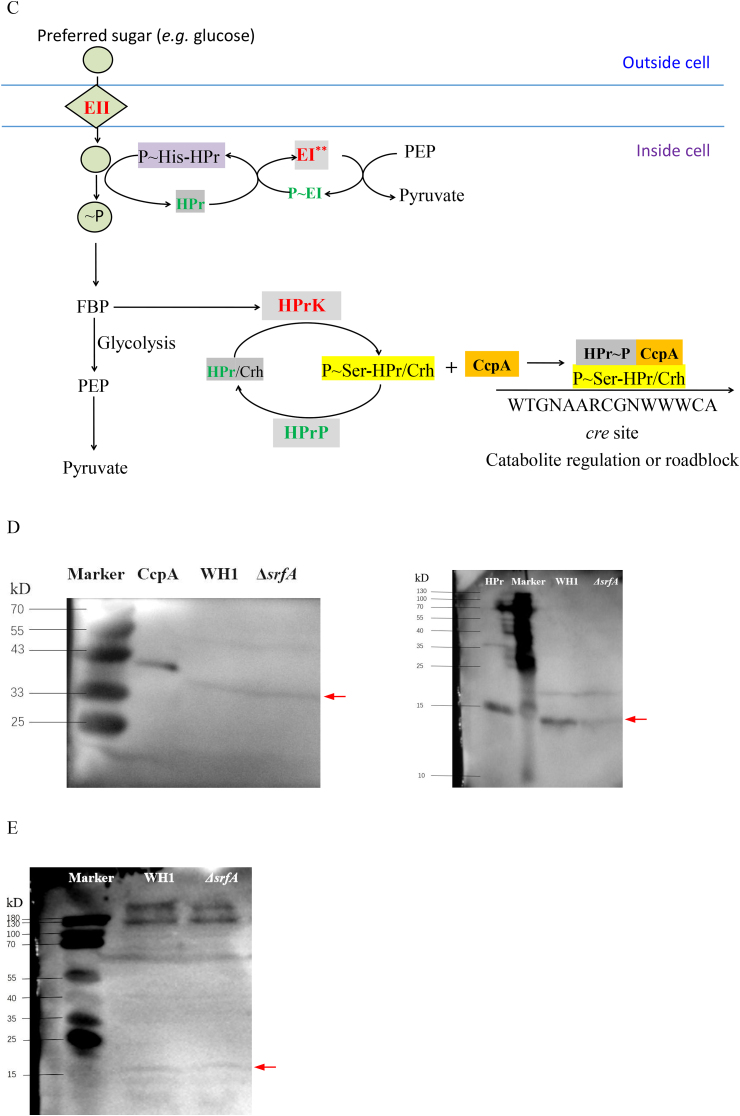
Comparison of CcpA, HPr and PtsG between ΔsrfA and WH1. A and B: Difference of CcpA, HPr and PtsG between ΔsrfA and WH1 at the level of transcription (A) and protein (C), respectively. Red: up-regulation; Green: down-regulation. B: Verification of the transcription of ccpA, hpr, hprK and hprP by qRT-PCR. D: Verification of CcpA and HPr by Western blot. The purified recombinant CcpA and HPr was used as control. Due to introduction of an extra sequence from the pET28a vector, the recombinant CcpA and HPr showed a higher molecular weight than the proteins in WH1 and ΔsrfA. E: Verification of P ~ Ser46-HPr by Western blot. Red arrows direct the objective proteins. Single and double stars represent significant (p < 0.05) and very significant (p < 0.01) difference between two groups, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
At the protein level, proteome analysis showed that PtsG (EII), PtsI (EI) and HPrK were all increased in ΔsrfA (Fig. 1C), consistent with the results of transcription. However, phosphoproteomic analysis showed EI ~ P (PtsI ~ P) was decreased in ΔsrfA (Fig. 1C). CcpA was also decreased in ΔsrfA, but without significant difference from WH1 (Fig. 1C), and this result was also confirmed by Western blot assay (Fig. 1D). HPr (PtsH) was decreased in the strain ΔsrfA, and this result was further verified by Western blot assay (Fig. 1D). Not only HPr, the content of HPr ~ P (PtsH ~ P) was also determined by Western blot assay, and found it was similar between ΔsrfA and WH1 (Fig. 1E). All of above results suggested that the complex of CcpA and HPr-Ser46~P was not increased in ΔsrfA.
3.2. Difference of CcpA-mediated carbon metabolism between ΔsrfA and WH1
3.2.1. CcpA-activated carbon metabolism
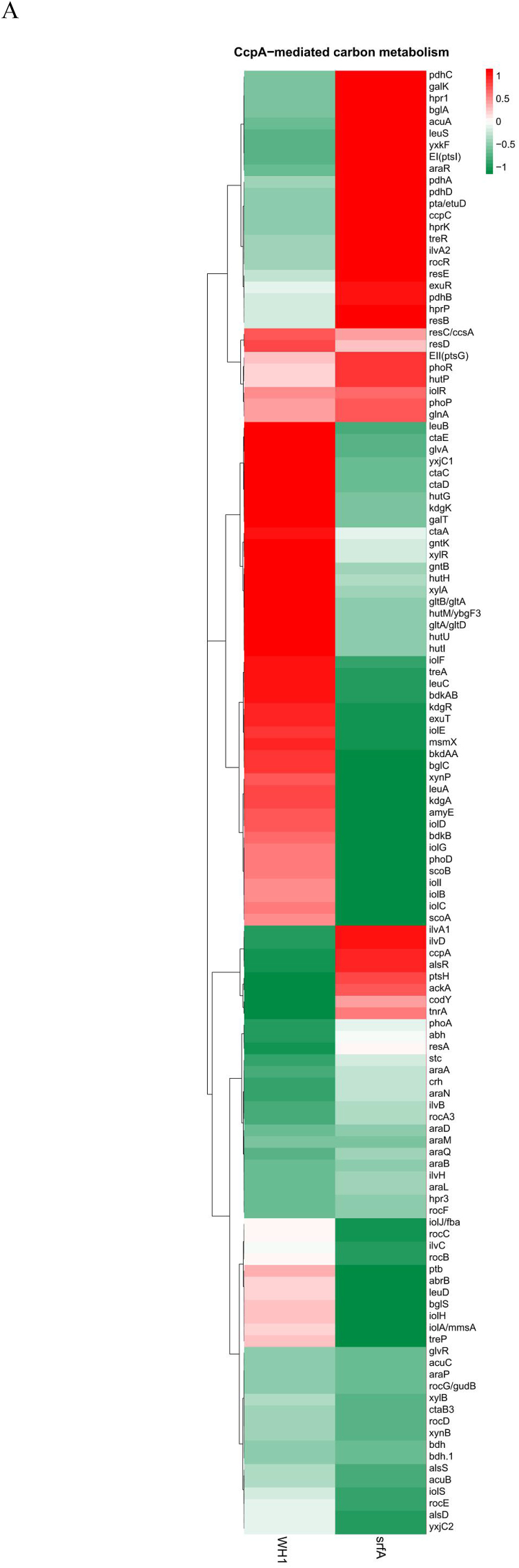
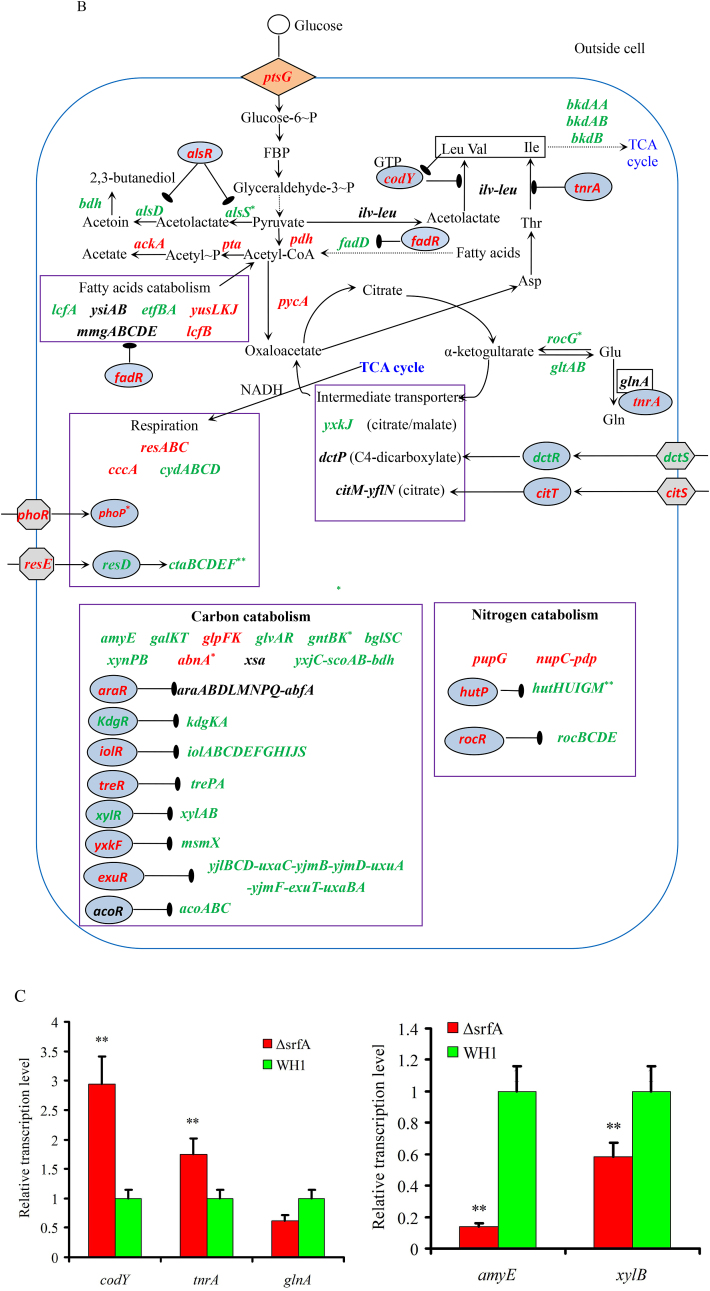
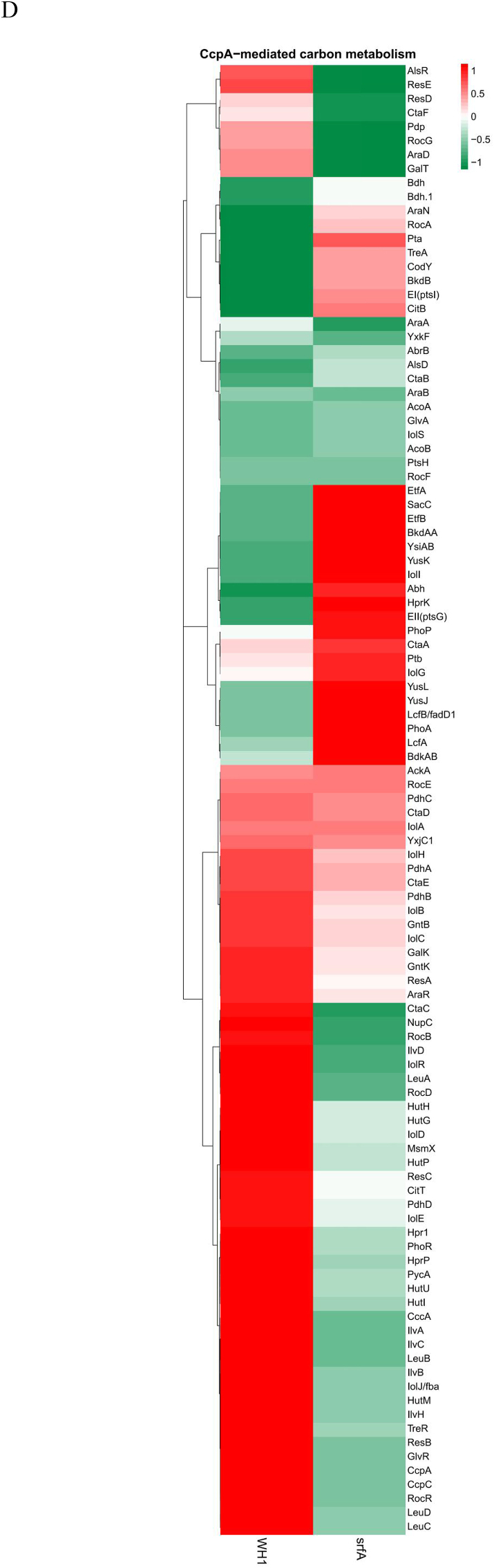
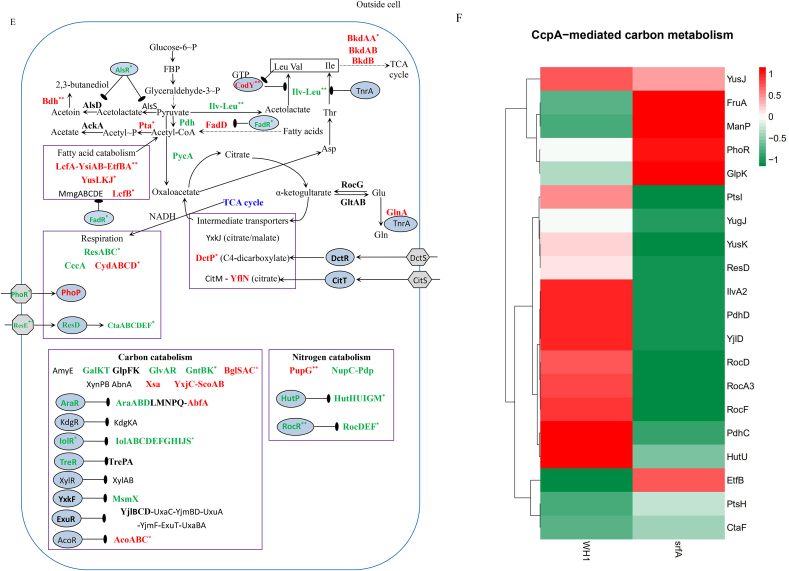
Several genes transcription (Fig. 2A) and proteins expression (Fig. 2D) involved in the CcpA-mediated carbon metabolism showed significant difference between ΔsrfA and WH1. Compared to WH1, the transcription of alsS, alsD and bdh for biosynthesis of acetoin and 2,3-butanediol were decreased, while the transcription of alsR (repressor) was increased in ΔsrfA, indicating biosynthesis of acetoin and 2,3-butanediol was weakened in this strain. Differently, the transcription of pdh, pta and ackA for biosynthesis of acetate were enhanced in ΔsrfA (Fig. 2B). At the protein level, Pta was significantly increased in ΔsrfA, indicating the carbon overflow metabolism towards biosynthesis of acetate was enhanced in this strain. As a feedback, the carbon overflow metabolism towards biosynthesis of 2,3-butanediol was consequently enhanced to counter the stress of acetate, accompanying with a significant decrease of AlsR, and a significant increase of Bdh in ΔsrfA (Fig. 2E). The key enzymes to catalyze pyruvate to acetyl-CoA, including PdhC ~ P and PdhD ~ P, were also decreased in ΔsrfA, suggested that the metabolic pathway from EMP to TCA was weakened in this strain (Fig. 2F). The transcription of repressors involved in biosynthesis of branched-chain amino acids, e.g. codY and tnrA (Wünsche et al., 2012), were increased in ΔsrfA (Fig. 2B&C). At the protein level, CodY was significantly increased, and the key enzymes of Ilv-Leu were significantly decreased consequently in ΔsrfA (Fig. 2E). These results indicated that biosynthesis of branched-chain amino acids was repressed in this mutant strain.
Fig. 2.
Difference of the CcpA-mediated carbon metabolism between ΔsrfA and WH1. A, D and F: Heatmaps at the level of transcription (A), protein (D) and phosphorylated protein (F), respectively. The colour from red to green indicates the richness from high to low. B and E: Difference of CcpA-mediated carbon metabolism between ΔsrfA and WH1 at the level of transcription (B) and protein (E), respectively. Red: up-regulation; Green: down-regulation. C: Verification of genes transcription by qRT-PCR. Single and double stars represent significant (p < 0.05) and very significant (p < 0.01) difference between two groups, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2.2. Utilization of non-preferred carbon sources
Several proteins (e.g. LcfA, LcfB, YsiAB, EtfBA and YusLKJ) involved in degradation of fatty acids, were significantly increased in ΔsrfA (Fig. 2D). Genes transcription involved in histidine metabolism (e.g. the hut operon) was decreased in ΔsrfA, consistent with an increase of hutP (repressor) transcription in this strain (Fig. 2B). Consistently, the proteins such as HutHUIGM (Fig. 2E) and the phosphorylated proteins such as HutU ~ P (Fig. 2F) were significantly decreased in ΔsrfA, indicating the ability to utilize histidine was weakened in this strain. The transcription of rocBCDE operon for arginine catabolism was also decreased in ΔsrfA, consistent with an increase of rocR (repressor) transcription in this strain (Fig. 2B). Consistently, the proteins such as RocD (Fig. 2E) and the phosphorylated proteins such as RocA~P, RocD ~ P and RocF ~ P (Fig. 2F) were all significantly decreased in ΔsrfA. These results all suggested that the catabolism of arginine was weakened in this mutant strain. The transcription of bdk operon involved in degradation of leucine and valine was down-regulated (Fig. 2B), and consistently BkdAA, BkdAB, and BkdB were significantly decreased in ΔsrfA (Fig. 2E), indicated the ability to utilize these amino acids was weakened in this strain.
Most of the genes transcription involved in utilization of extracellular non-preferred carbon sources, including the kdg operon for utilization of hexuronate, the iol operon for inositol, the tre operon for trehalose, the xyl operon for xylose, the yxkF-msmX operon for transporting unknown sugars, and the acu operon for utilization of acetoin, were all decreased in ΔsrfA. Consistently, the transcription of repressors such as iolR, treR, yxkF, stc, exuR, araR, were all increased in this mutant strain, respectively. Besides, the genes transcription including the amyE for digestion of starch, the galKT for galactose metabolism, the glvAR for 6-P-α-glucoside metabolism, the gntBK for gluconate metabolism, the bglSC for β-glucoside metabolism, the xynPB for β-xyloside, the yxjC-scoAB-bdh for β-hydroxybutyrate metabolism, were all down-regulated in ΔsrfA (Fig. 2B). qRT-PCR also showed the genes transcription for utilizing extracellular non-preferred carbon sources such as amyE and xylB were significantly decreased in ΔsrfA (Fig. 2C). At the protein level, AraA, AraB and AraD for utilizing arabinose, and IolD, IolH and IolJ for utilization of inositol, were all significantly decreased in ΔsrfA. Besides, MsmX, GalK, GlvR and GntBK, were also decreased in this strain. However, AcoAB for utilization of acetoin was significantly increased in ΔsrfA (Fig. 2E).
3.2.3. Respiration
Transcription of the two component system phoR and phoP was significantly increased in ΔsrfA (Fig. 2 A&B). At the protein level, the sensor PhoR was significantly decreased (Fig. 2E), and the phosphorylated PhoR (PhoR ~ P) was significantly increased in ΔsrfA (Fig. 2F), indicating more PhoR was converted to PhoR ~ P in this strain. Transcription of the resDE operon and the ctaABCDE operon involved in respiration, were both significantly decreased in ΔsrfA (Fig. 2B). Consistently, the proteins complex like ResDE and CtaABCDE were both decreased in this strain (Fig. 2 D&E). All of these results suggested that the respiration was weakened in ΔsrfA.
3.3. Comparison of CcpA-independent carbon metabolism between ΔsrfA and WH1
3.3.1. Utilization of less-preferred carbon sources
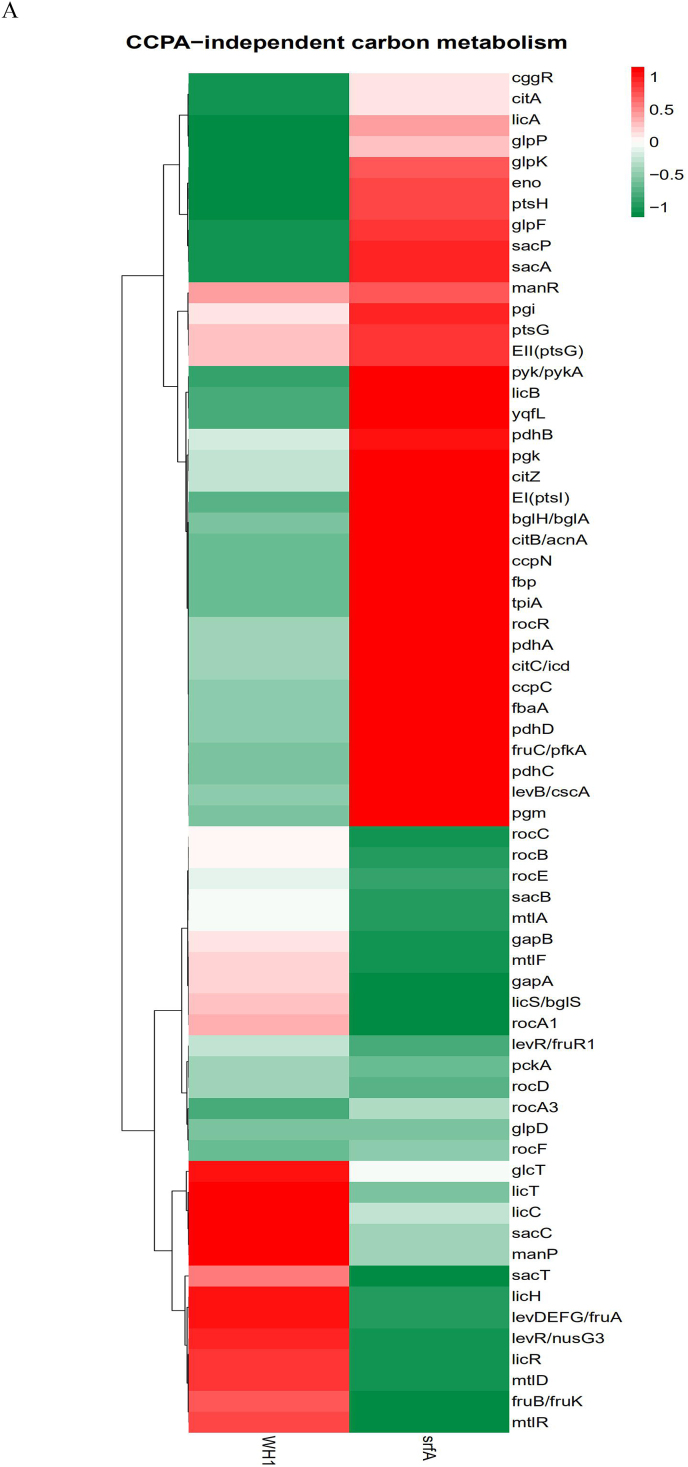
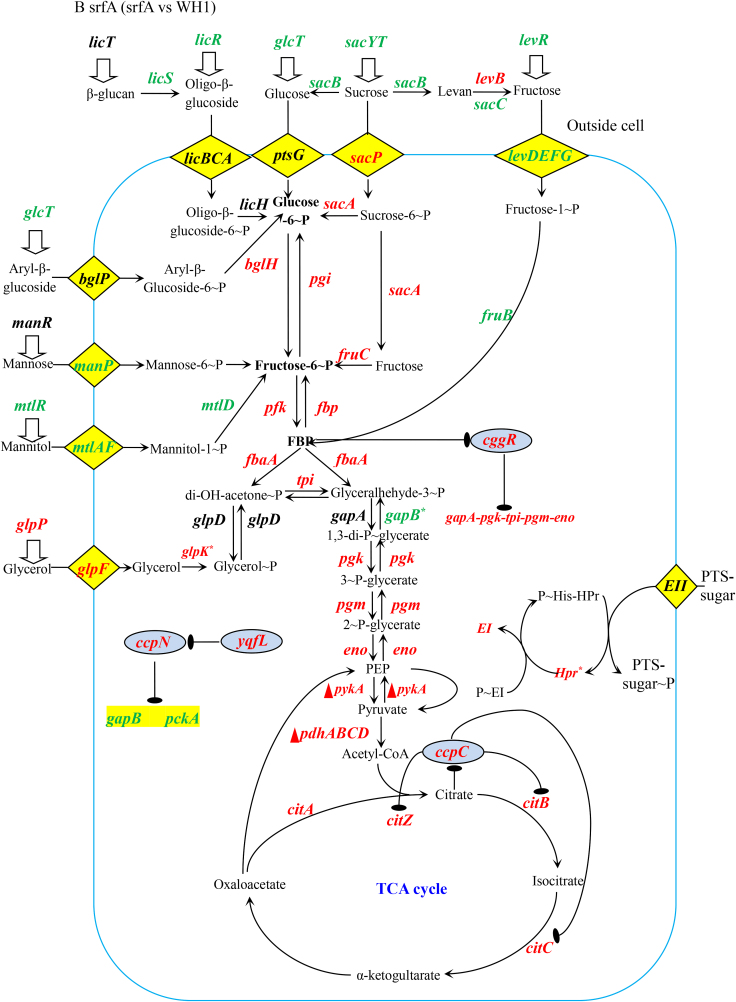
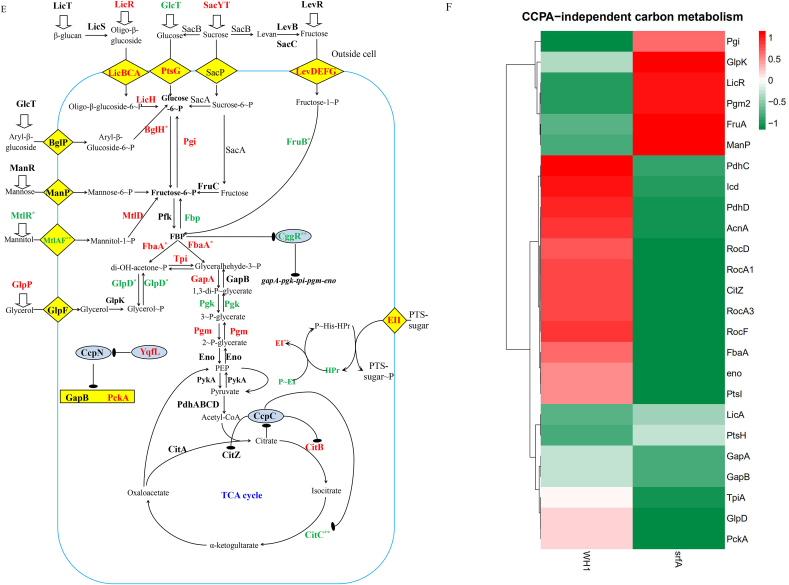
In the CcpA-independent carbon metabolism, several genes transcription (Fig. 3A) and proteins expression (Fig. 3D) showed significant differences between ΔsrfA and WH1. Compared to WH1, the transcription of mtlAF (EII) and mtlR (transcriptional activator) (Fig. 3B) were both decreased, and consistently, MtlR and MtlAF (Fig. 3E) were also significantly decreased in ΔsrfA, indicating the ability to utilize mannitol was weakened in this mutant strain. Besides, GlpD for catalyzing Glycerol ~ P to di-OH-acetone ~ P was significantly decreased (Fig. 3E), indicating the ability to utilize glycerol was also weakened in ΔsrfA.
Fig. 3.
Comparison of the CcpA-independent carbon metabolism between ΔsrfA and WH1. A, D and F: Heatmaps at the level of transcription (A), protein (D) and phosphorylated protein (F), respectively. The colour from red to green indicates the richness from high to low. B and E: Difference of CcpA-independent carbon metabolism between ΔsrfA and WH1 at the level of transcription (B) and protein (E), respectively. Red: up-regulation; Green: down-regulation. C: Verification of genes transcription by qRT-PCR. Single and double stars represent significant (p < 0.05) and very significant (p < 0.01) difference between two groups, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3.2. Glycolysis, TCA and gluconeogenesis
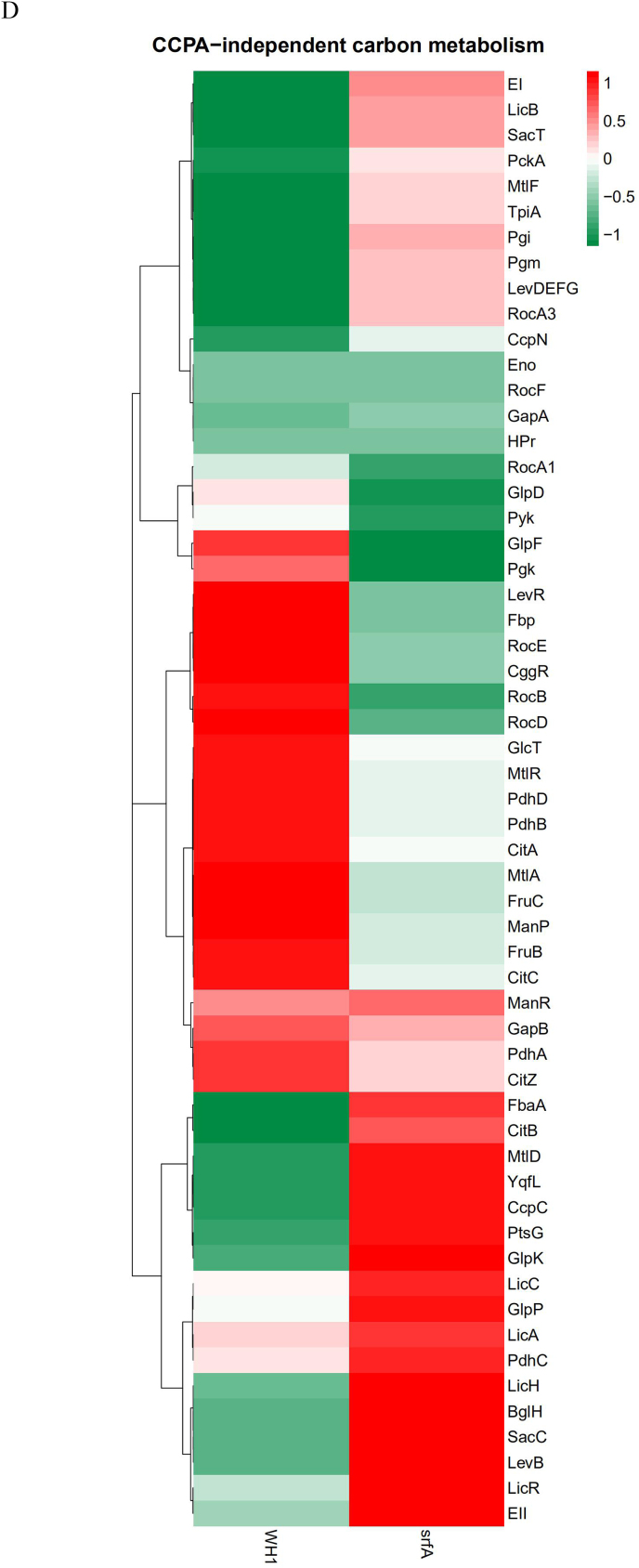
Transcription of the genes involved in glycolysis, including pfk, fbaA, tpi, pgk, pgm, eno, pykA, and pdhABCD, were all increased in ΔsrfA (Fig. 3 A & B). The operon of gapA-pgk-tpi-pgm-eno is repressed by CggR (Rud et al., 2011), and CggR is inhibited by FBP, an intermediate of glycolysis. Here, the transcription of cggR was increased in ΔsrfA (Fig. 3B), and this result was further verified by qRT-PCR (Fig. 3C). Nevertheless, the protein CggR was significantly decreased in ΔsrfA (Fig. 3E), and the transcription of gapA-pgk-tpi-pgm-eno was consequently increased in this strain. Consistently, several proteins involved in the glycolytic pathway, including Pgi, FbaA, Tpi, GapA, Pgm, were all increased in ΔsrfA (Fig. 3E). At the phosphorylated protein level, except for Pgi ~ P, most of the phosphorylated proteins such as FbaA~P, Tpi ~ P, GapA~P, GapB ~ P, Pgm ~ P, Eno~P, were all decreased in ΔsrfA (Fig. 3F).
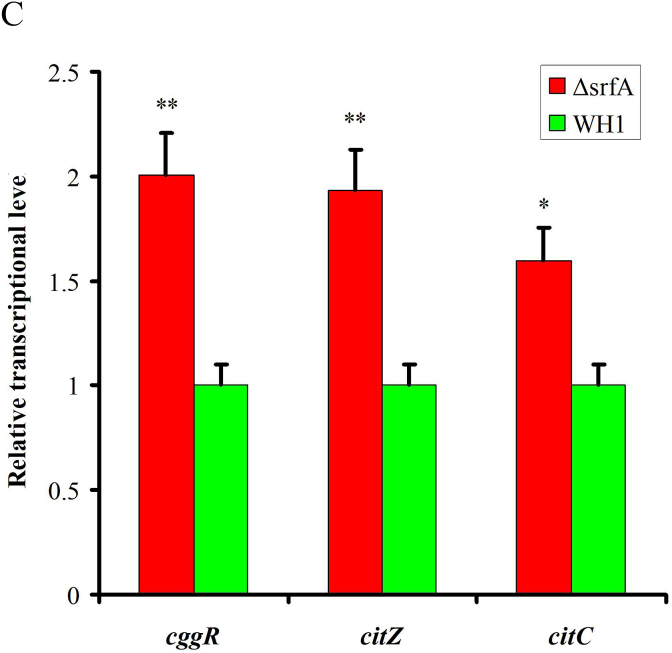
In the metabolic pathway from pyruvate to acetyl-CoA, the transcription of pdhABCD was increased in ΔsrfA (Fig. 3B). Transcription of the genes involved in the TCA cycle, including citA, citB, citC and citZ, were all increased in ΔsrfA (Fig. 3B), consistent with the results of qRT-PCR (Fig. 3C). However, at the protein level, CitC that catalyzes isocitrate to α-ketogultarate, was significantly decreased (Fig. 3E), indicating the TCA cycle was weakened in this mutant strain. Transcription of the genes involved in gluconeogenesis, such as gapB and pckA, is repressed by CcpN (Fujita, 2009). Here, the transcription of ccpN was increased, and the transcription of gapB and pckA were both decreased in ΔsrfA (Fig. 3B), indicating that the gluconeogenesis was weakened in this mutant strain.
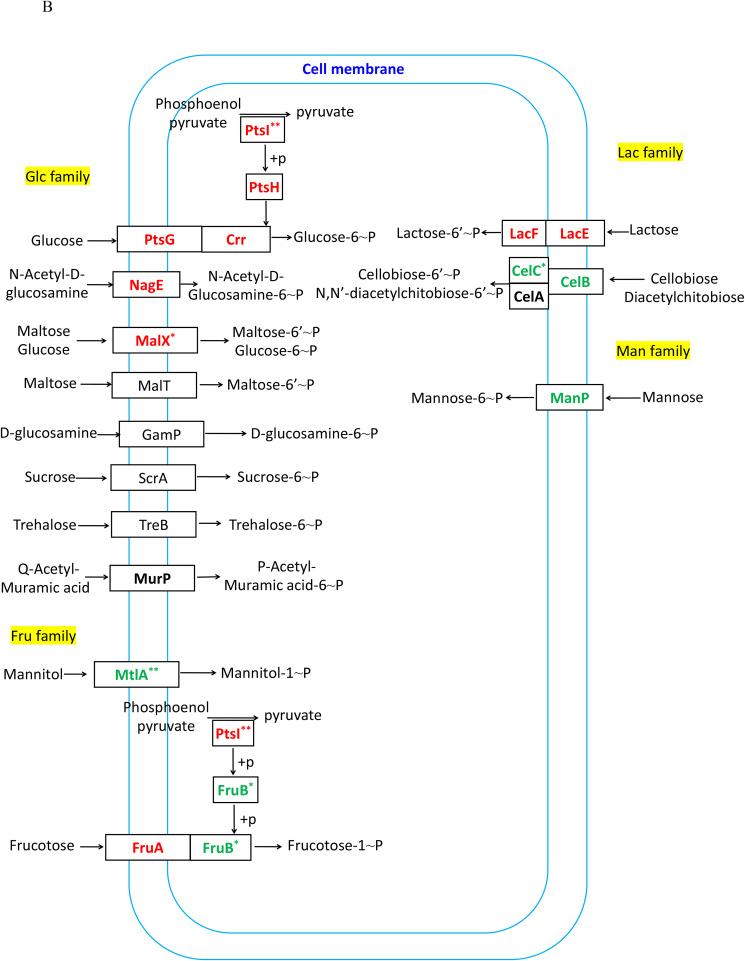
3.4. Difference of PTS system between ΔsrfA and WH1
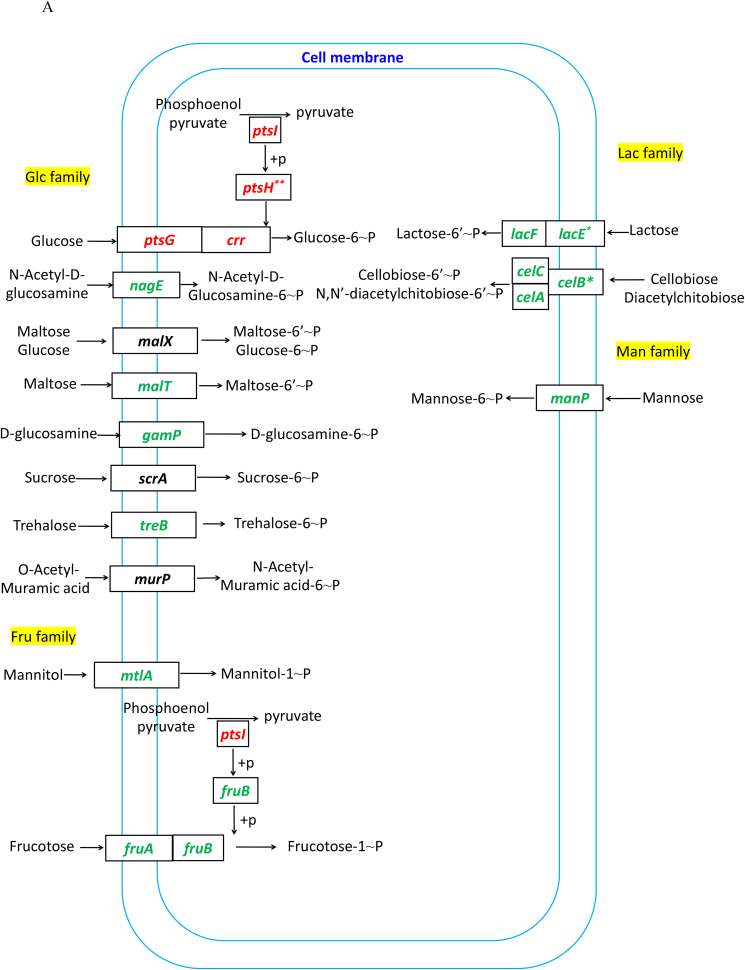
The PTS system is responsible for transporting and phosphorylating sugars. Transcription of the genes in several PTS systems, including the nagE for utilizing N-acetyl-D-glucosamine, the malT for maltose, the gamP for D-glucosamine, the treB for trehalose, the mtlA for mannitol, the fruAB for fructose, the lacEF for lactose, the celABC for cellobiose diacetylchitobiose, and the manP for mannose, were all decreased in ΔsrfA (Fig. 4A). At the protein level, MtlA, FruB and CelC, were all significantly decreased, and CelB and ManP were also decreased in this mutant strain (Fig. 4B). These data suggested that the ability to transport and phosphorylate less - preferred sugars was decreased in ΔsrfA.
Fig. 4.
Difference of the PTS system between ΔsrfA and WH1. A: The level of transcription; B: The level of protein. Red: up-regulation; Green: down-regulation. Single and double stars represent significant (p < 0.05) and very significant (p < 0.01) difference between two groups, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
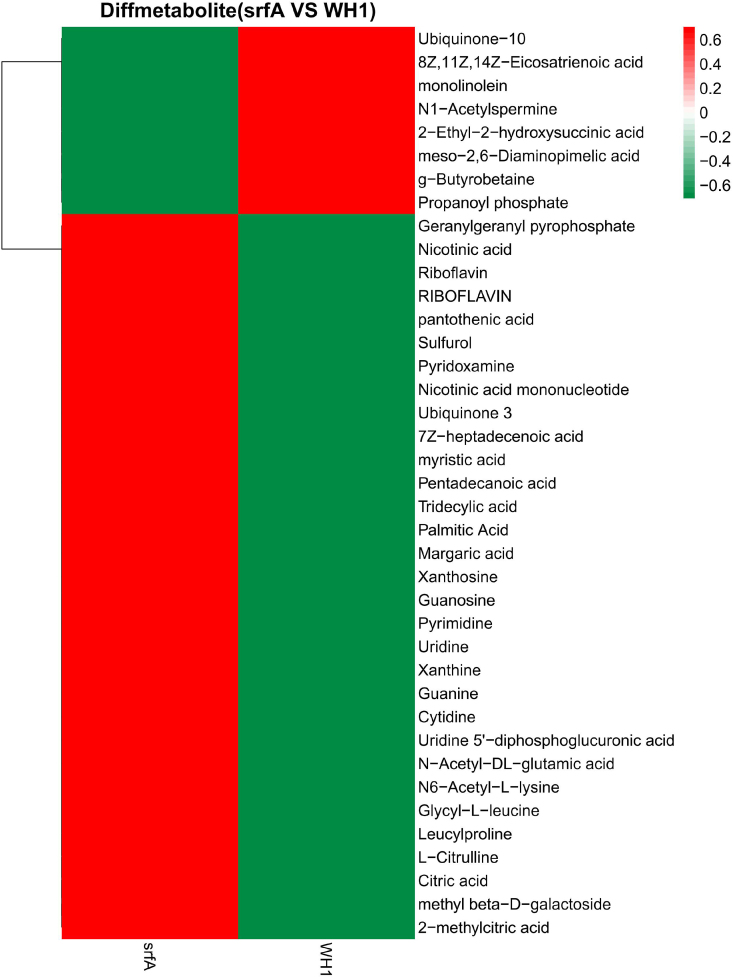
3.5. Comparison of carbon metabolic intermediates between ΔsrfA and WH1
Many carbon metabolic intermediates showed significant differences between ΔsrfA and WH1. Compared to WH1, butyrobetaine, propanoyl phosphate, 2-Ethyl-2-hydroxysuccinic acid, and meso-2,6-diaminopimelic acid were all significantly decreased in ΔsrfA (Fig. 5). However, citric acid, 2-methylcitric acid, leucylproline and glycyl-L-leucine were all significantly increased in ΔsrfA. Most of fatty acids, such as margaric acid, palmitic acid, tridecylic acid, pentadecanoic acid, myristic acid and 7Z-heptadecenoic acid, were also significantly increased in this mutant strain. Most of coenzymes such as ubiquinone 3, nicotinic acid mononucleotide, pyridoxamine, sulfuro, pantothenic acid, riboflavin, nicotinic acid, were increased in ΔsrfA.
Fig. 5.
Comparison of the carbon metabolic intermediates between ΔsrfA and WH1. The colour from red to green means the richness from high to low. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
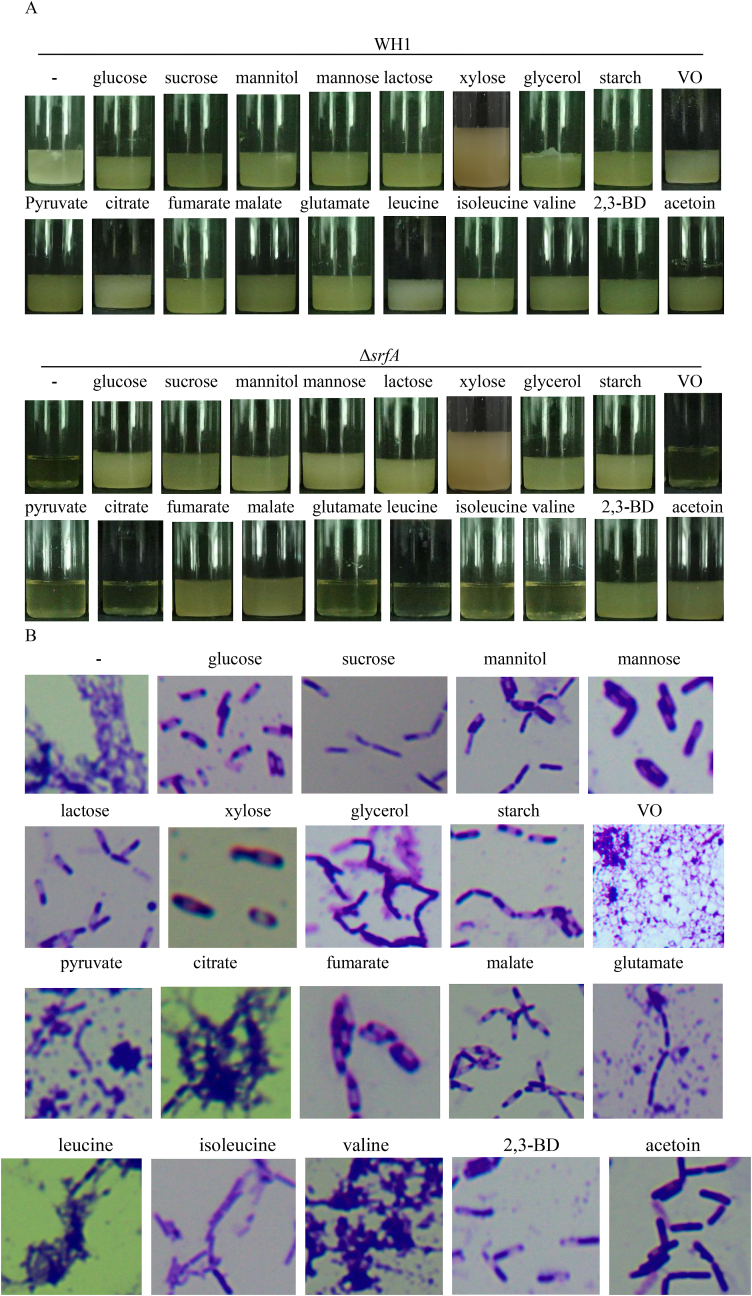
3.6. Effects of addition with carbon sources on the cell survival of ΔsrfA
We further determined whether ΔsrfA could use different carbon sources such as sucrose, starch, glycerol, lactose, mannose, mannitol, xylose, glucan, pyruvate, citrate, fumarate, malate, leucine, isoleucine, valine, glutamate, fatty acids (vegetable oil, VO), acetoin and 2,3-butanediol, for survival after glucose was exhausted in the medium. WH1 could use all of these carbon sources for survival. Compared to WH1, ΔsrfA was died after glucose was exhausted in the LB medium, and addition with glucose could alleviate the cell death as described previously (Chen et al., 2020). Besides for glucose, other carbon sources such as sucrose, mannitol, mannose, lactose, xylose, glycerol, starch, fumarate, malate, 2,3-BD and acetoin (Thanh et al., 2010), could also alleviate the cell death (Fig. 6A) and sporulation (Fig. 6B) of ΔsrfA. However, VO, pyruvate, citrate, glutamate, leucine, isoleucine and valine could not alleviate the cell death and sporulation of ΔsrfA.
Fig. 6.
Effects of addition with carbon sources on the survive and sporulation of ΔsrfA. A: Survive of ΔsrfA; B: Sporulation of ΔsrfA.
4. Discussion
Bacillus species are classic Gram-positive microorganisms. The carbon metabolism in Bacillus has been studied for several decades, but it is remained unknown in many aspects. For example, the relationship between carbon metabolism and quorum sensing is rarely reported until now. Previously, we found that surfactin, a quorum sensing signal molecule, is also involved in the carbon metabolism in B. amyloliquefaciens (Chen et al., 2020). Mutation of srfA to disrupt surfactin production led to the cell death in B. amyloliquefaciens after glucose was exhausted, but this could be reversed by compensation with surfactin or glucose in the medium. We deduced that surfactin might act as a link between quorum sensing and carbon metabolism (Chen et al., 2020). Quorum sensing system has been used as a good tool for dynamically regulating fermentation (Gupta et al., 2017), while carbon metabolism is a kind of basal metabolism that determines the balance between biomass and products (Cui et al., 2019; Cao et al., 2018). Thereby, clarification of the link between quorum sensing and carbon metabolism is very important for using the signal molecule surfactin and quorum sensing system to dynamically regulate carbon distribution and effectively balance biomass and products in the broth. However, the effects of surfactin on carbon metabolism are poorly understood. Here, we investigated the global influence of surfactin on the carbon metabolism in B. amyloliquefaciens via multiomics analysis.
CcpA is a key regulator to control the carbon metabolism in B. subtilis (Fujita, 2009). In the environment with glucose, CcpA in a complex with HPr-Ser46~P plays a key role in the carbon catabolite repression to utilize non-preferred carbon sources (Fujita, 2009). Unexpectedly, CcpA was not increased in this srfA-mutant strain. Moreover, P ~ Ser46-HPr was also similar between WH1 and ΔsrfA. Thereby, it seemed that mutation of srfA to disrupt surfactin production did not enhance the CcpA mediated CCR in B. amyloliquefaciens. This was further confirmed by the fact that addition with non-preferred carbon sources could also alleviate the death of ΔsrfA like addition with glucose.
Biosynthesis of acetoin/2,3-butanediol and acetate is belonged to carbon overflow metabolism (Fujita, 2009; Qi et al., 2014). Here, biosynthesis of acetate was enhanced while biosynthesis of acetoin/2,3-butanediol was weakened in ΔsrfA. Enhanced biosynthesis of acetate will lead to a decrease of broth pH that is deleterious for the bacterial survival and growth. Conversely, acetoin and 2,3-butanediol is favorable for protecting cells from the detriment of low pH stress (Fan et al., 2018). In addition to biosynthesis of acetate and acetoin/2,3-butanediol, biosynthesis of branched-chain amino acids is also activated by CcpA (Marciniak et al., 2012). Here, CodY (Wünsche et al., 2012) was significantly increased, and the key enzymes of Ilv-Leu were significantly decreased, indicating biosynthesis of these amino acids were weakened in ΔsrfA. Vigorous biosynthesis of branched-chain amino acids is considered as a marker of enough nutrients for cell propagation in the environment (Fujita, 2009). Thereby, the weakened biosynthesis of branched-chain amino acids means a decreased growth in ΔsrfA (Chen et al., 2020).
Utilization of some carbon sources (e.g. fatty acids and branched-chain amino acids) is repressed by CcpA (Fujita, 2009). Here, several key proteins involved in degradation of fatty acids and branched-chain amino acids were all significantly increased in ΔsrfA as a response to glucose exhaustion in the medium (Chen et al., 2020). But in fact, this mutant strain could not use fatty acids and branched-chain amino acids (e.g. Val, Leu and Ile) to alleviate the cell death after glucose exhaustion. Possibly, this can be explained that the cells of ΔsrfA could not use these carbon sources, so the enzymes were increased as a feedback to this disability. Differently, the CcpA-controlled enzymes for utilization of non-preferred carbon sources such as inositol, hexuronate, xylose, arabinose, starch, galactose, 6-P-α-glucoside, gluconate, β-xyloside, β-hydroxybutyrate, acetoin, etc, were all down-regulated, indicated the ability to utilize these extracellular non-preferred carbon sources was weakened in this mutant strain. However, in fact this strain could use many carbon sources such as lactose, xylose, starch, 2,3-BD and acetoin after glucose was exhausted in the medium. This could be explained that although the ability to utilize these carbon sources was weakened, ΔsrfA could still use them after glucose exhaustion. Thereby, addition with these carbon sources could alleviate the cell death of ΔsrfA.
The resABCDE operon encodes a protein complex for cytochrome c biogenesis, and a two-component system ResED to control the electron transfer in response to oxygen availability. This operon is indispensable for respiration, and is also subject to the CcpA-mediated CCR in B. subtilis (Fujita, 2009). Here, ResABCDE and CtaABCDE were both decreased, suggested the respiration was obviously down-regulated in ΔsrfA. This was further verified by the result that ResD ~ P was significantly decreased in this strain. Thereby, the bacterial growth was weakened in ΔsrfA after glucose was exhausted in the medium (Chen et al., 2020).
After mutation of srfA, the proteins of PtsG (EII), PtsH (HPr) and PtsI (EI) that are responsible for uptake of glucose, were all significantly increased as a feedback to glucose exhaustion. In addition to glucose, ΔsrfA could use many less-preferred carbon sources such as mannose, mannitol, glycerol, glucoside and sucrose for growth after glucose exhaustion. P ~ His15-HPr is essential for utilization of these less-preferred carbon sources by phosphorylation of the PTS-sugars (Fujita, 2009; Darbon et al., 2002). In the CcpA-independent carbon metabolism, a low amount of P ~ His15-HPr as to HPr, which is generated through active transfer of preferred PTS-sugars (e.g. glucose), results in a dephosphorylation of antiterminators or transcriptional activators for utilization of less-preferred PTS-sugars. Here, EI ~ P (PtsI ~ P) was decreased in ΔsrfA, suggested that EI ~ P was insufficient for phosphorylation of HPr to activate antiterminators or transcriptional activators in this strain. Thereby, the genes expression under the control of antiterminators or transcriptional activators were decreased in ΔsrfA. For example, the PTS for transferring mannitol including the transcriptional activator of MtlR and the EII of MtlAF were both significantly decreased in this mutant strain.
It is very interesting that the genes transcription involved in the glycolytic pathway, including pfk, fbaA, tpi, pgk, pgm, eno, pykA, and pdhABCD, were all increased in ΔsrfA. Consistently, the proteins like Pgi, FbaA, Tpi, GapA, Pgm, were all increased, corresponding to a decrease of FbaA~P, Tpi ~ P, GapA~P, GapB ~ P, Pgm ~ P, Eno~P in this strain. Although these proteins were increased, the glycosis was weakened due to a decrease of phosphorylated proteins in this mutant strain. As a feedback, these proteins were increased as a response to the weakened glycolysis in ΔsrfA. Similarly, CitC involved in the TCA cycle was significantly decreased, indicating the TCA cycle was down-regulated in ΔsrfA. Thereby, citric acid was deposited, and addition with citrate could not alleviate the death of ΔsrfA. CitB is responsible for producing 2-methylcitric acid in B. subtilis (Reddick et al., 2017). Here, CitB was increased, possibly as a response to the weakened TCA cycle, so more 2-methylcitric acid was produced in ΔsrfA when compared to WH1.
Several carbon metabolic intermediates showed significant differences between ΔsrfA and WH1. Butyrobetaine was significantly decreased, indicating the ability to counter stress was weakened in ΔsrfA. Leucylproline and glycyl-L-leucine were both increased in ΔsrfA. Leucylproline is considered as an inhibitor of proteases to suppress growth (Hebert et al., 2008), and glycyl-L-leucine can inhibit biosynthesis of isoleucine in bacteria (Vonder Haar and Umbarger, 1972). Thereby, increase of these dipeptides resulted in an impaired growth in ΔsrfA. Most of fatty acids were significantly increased in ΔsrfA, implied this strain was disable to use these fatty acids, consistent with the result that addition with fatty acids could not alleviate the cell death after glucose was exhausted in the medium. As a feedback, the expression of ysiA regulon that is responsible for degradation of fatty acids (Matsuoka et al., 2007), was significantly increased in ΔsrfA compared to WH1. In addition, some fatty acids like pentadecanoic acid and myristic acid are substrates for biosynthesis of lipopeptides such as surfactin (Ding et al., 2019). Here, they were both accumulated in ΔsrfA because biosynthesis of surfactin was disrupted in this strain (Chen et al., 2020).
Most of the coenzymes such as nicotinic acid, nicotinic acid mononucleotide, pyridoxamine, sulfuro, pantothenic acid, riboflavin, were all significantly increased in ΔsrfA. Nicotinic acid and nicotinic acid mononucleotide are substrates for biosynthesis of NAD that is very important for producing energy. Pyridoxamine is a coenzyme of transaminase that catalyzes 2-oxoglutarate to glutamate. Sulfurol is a component of thiamine that is the coenzyme of pyruvate dehydrogenase complex and α-ketoglutarate dehydrogenase complex. These enzymes play very important roles in the TCA cycle. Pantothenic acid can be catalyzed to CoA. Riboflavin plays a role in transferring hydrogen in the oxidation-reduction reactions. These coenzymes were induced as a feedback to energy crisis in the cells after glucose exhaustion, consistent with a significant decrease of proteins involved in respiration such as ResE, CtaABCDEF and ResABC in ΔsrfA.
ΔsrfA could utilize most of the carbon sources used in this study, including sucrose, mannitol, mannose, lactose, xylose, glycerol, starch, fumarate, malate, 2,3-BD and acetoin after glucose was exhausted in the medium. However, some carbon sources could not be used by ΔsrfA after glucose was exhausted, including fatty acids, organic acids (pyruvate, citrate, and glutamate), and branched-chain amino acids (leucine, isoleucine, valine). It was very interesting that ΔsrfA could use fumarate and malate, rather than pyruvate, citrate and glutamate after glucose was exhausted. Fumarate and malate are both of the preferred carbon sources (Meyer et al., 2011), thereby they could be used to relieve the death of ΔsrfA after glucose was exhausted. Pdh that catalyzes pyruvate to acetyl-CoA, and PycA that catalyzes acetyl-CoA to oxaloacetate, were both decreased in ΔsrfA. This was unfavorable for utilization of pyruvate, so addition with pyruvate could not alleviate the death of ΔsrfA. The transcription of rogG that is responsible for catalyzing glutamate to α-ketogultarate, was significantly decreased in ΔsrfA. This result could partially explain the reason why addition with glutamate could not alleviate the death of ΔsrfA after glucose was exhausted. CitC that catalyzes isocitrate to α-ketogultarate was significantly decreased, so addition with citrate could not alleviate the death of ΔsrfA.
Collectively, mutation of srfA to disrupt surfactin production led to an obvious change in the carbon metabolism in B. amyloliquefaciens. Firstly, the PTS-glucose system was significantly enhanced as a feedback to glucose exhaustion. Secondly, the basic carbon metabolism such as glycolysis and TCA cycle was obviously weakened. Thirdly, CcpA and P ~ Ser46-HPr was not significantly increased, but the ability to use non-preferred carbon sources was down-regulated. Fourthly, biosynthesis of acetate was enhanced while biosynthesis of acetoin/2,3-butanediol and branched-chain amino acids was weakened. Finally, the mutant strain could use most of the non- or less-preferred carbon sources except for fatty acids, branched chain amino acids (e.g. Ile, Leu, Val) and some organic acids (e.g. pyruvate, citrate and glutamate), to alleviate the cell death after glucose exhaustion.
Author statement
Gaofu Qi: Conceptualization, Writing, Supervision. Jiahong Wen: Methodology, Investigation, Data curation. Xiuyun Zhao: Writing, Reviewing and Editing. Fengmei Si: Methodology, Investigation.
Funding
Project 31870030 is supported by National Natural Science Foundation of China.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Bie N., Zhao X., Li Z., Qi G. Constructing tumor vaccines targeting for vascular endothelial growth factor (VEGF) by DNA shuffling. J. Immunother. 2016;39(7):260–268. doi: 10.1097/CJI.0000000000000129. [DOI] [PubMed] [Google Scholar]
- Cao H., Villatoro-Hernandez J., Weme R.D.O., Frenzel E., Kuipers O.P. Boosting heterologous protein production yield by adjusting global nitrogen and carbon metabolic regulatory networks in Bacillus subtilis. Metab. Eng. 2018;49:143–152. doi: 10.1016/j.ymben.2018.08.001. [DOI] [PubMed] [Google Scholar]
- Chen B., Wen J., Zhao X., Ding J., Qi G. Surfactin: a quorum-sensing signal molecule to relieve CCR in Bacillus amyloliquefaciens. Front. Microbiol. 2020;11:631. doi: 10.3389/fmicb.2020.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S., Lv X., Wu Y., Li J., Du G., Ledesma-Amaro R., Liu L. Engineering a bifunctional Phr60-Rap60-Spo0A quorum-sensing molecular switch for dynamic fine-tuning of menaquinone-7 synthesis in Bacillus subtilis. ACS Synth. Biol. 2019;8:1826–1837. doi: 10.1021/acssynbio.9b00140. [DOI] [PubMed] [Google Scholar]
- Darbon E., Servant P., Poncet S., Deutscher J. Antitermination by GlpP, catabolite repression via CcpA and inducer exclusion triggered by P-GlpK dephosphorylation control Bacillus subtilis glpFK expression. Mol. Microbiol. 2002;43:1039–1052. doi: 10.1046/j.1365-2958.2002.02800.x. [DOI] [PubMed] [Google Scholar]
- Deutscher J., Milohanic E. Transcription regulators controlled by interaction with enzyme IIB components of the phosphoenolpyruvate: sugar phosphotransferase system. Biochim. Biophys. Acta. 2013;1834:1415–1424. doi: 10.1016/j.bbapap.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Ding L., Guo W., Chen X. Exogenous addition of alkanoic acids enhanced production of antifungal lipopeptides in Bacillus amyloliquefaciens Pc3. Appl. Microbiol. Biotechnol. 2019;103:5367–5377. doi: 10.1007/s00253-019-09792-1. [DOI] [PubMed] [Google Scholar]
- Fan X., Wu H., Jia Z., Li G., Li Q., Chen N., Xie X. Metabolic engineering of Bacillus subtilis for the co-production of uridine and acetoin. Appl. Microbiol. Biotechnol. 2018;102:8753–8762. doi: 10.1007/s00253-018-9316-7. [DOI] [PubMed] [Google Scholar]
- Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Jensen L.J. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 2009;73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- Gupta A., Reizman I.M., Reisch C.R., Prather K.L. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 2017;35:273–279. doi: 10.1038/nbt.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert E.M., Mamone G., Picariello G., Raya R.R., Savoy G., Ferranti P., Addeo F. Characterization of the pattern of alphas1- and beta-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581. Appl. Environ. Microbiol. 2008;74:3682–3689. doi: 10.1128/AEM.00247-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H., Tanaka T., Ogura M. The Bacillus subtilis response regulator gene degU is positively regulated by CcpA and by catabolite-repressed synthesis of ClpC. J. Bacteriol. 2013;195:193–201. doi: 10.1128/JB.01881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H., Fraser M., Li W., Mcanulla C., Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer D.A., Piccolo B.D., Vaziri N.D., Liu S., Lau W.L., Khazaeli M., Nazertehrani S., Moore M.E., Marco M.L., Martin R.J., Adams S.H. Resistant starch alters gut microbiome and metabolomics profiles concurrent with amelioration of chronic kidney disease in rats. Am. J. Physiol. Ren. Physiol. 2016;310:F857–F871. doi: 10.1152/ajprenal.00513.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröber M., Verwaaijen B., Wibberg D., Winkler A., Pühler A., Schlüter A. Comparative transcriptome analysis of the biocontrol strain Bacillus amyloliquefaciens FZB42 as response to biofilm formation analyzed by RNA sequencing. J. Biotechnol. 2016;231:212–223. doi: 10.1016/j.jbiotec.2016.06.013. [DOI] [PubMed] [Google Scholar]
- López D., Vlamakis H., Kolter R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak B.C., Pabijaniak M., de Jong A., Dűhring R., Seidel G., Hillen W., Kuipers O.P. High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis. BMC Genom. 2012;13:401. doi: 10.1186/1471-2164-13-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka H., Hirooka K., Fujita Y. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J. Biol. Chem. 2007;282:5180–5194. doi: 10.1074/jbc.M606831200. [DOI] [PubMed] [Google Scholar]
- Meyer F.M., Jules M., Mehne F.M., Le Coq D., Landmann J.J., Görke B., Aymerich S., Stülke J. Malate-mediated carbon catabolite repression in Bacillus subtilis involves the HPrK/CcpA pathway. J. Bacteriol. 2011;193:6939–6949. doi: 10.1128/JB.06197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G., Zhu F., Du P., Yang X., Qiu D., Yu Z., Chen J., Zhao X. Lipopeptide induces apoptosis in fungal cells by a mitochondria-dependent pathway. Peptides. 2010;31:1978–1986. doi: 10.1016/j.peptides.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Qi G., Kang Y., Li L., Xiao A., Zhang S., Wen Z., Xu D., Chen S. Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced D-2,3-butanediol production in Bacillus licheniformis. Biotechnol. Biofuels. 2014;7(1):16. doi: 10.1186/1754-6834-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddick J.J., Sirkisoon S., Dahal R.A., Hardesty G., Hage N.E., Booth W.T., Quattlebaum A.L., Mills S.N., Meadows V.G., Adams S.L.H., Doyle J.S., Kiel B.E. First biochemical characterization of a methylcitric acid cycle from Bacillus subtilis strain 168. Biochemistry. 2017;56:5698–5711. doi: 10.1021/acs.biochem.7b00778. [DOI] [PubMed] [Google Scholar]
- Reuß D.R., Rath H., Thürmer A., Benda M., Daniel R., Völker U., Mäder U., Commichau F.M., Stülke J. Changes of DNA topology affect the global transcription landscape and allow rapid growth of a Bacillus subtilis mutant lacking carbon catabolite repression. Metab. Eng. 2018;45:171–179. doi: 10.1016/j.ymben.2017.12.004. [DOI] [PubMed] [Google Scholar]
- Rud I., Naterstad K., Bongers R.S., Molenaar D., Kleerebezem M., Axelsson L. Functional analysis of the role of CggR (central glycolytic gene regulator) in Lactobacillus plantarum by transcriptome analysis. Microb. Biotechnol. 2011;4:345–356. doi: 10.1111/j.1751-7915.2010.00223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh T.N., Jürgen B., Bauch M., Liebeke M., Lalk M., Ehrenreich A., Evers S., Maurer K.H., Antelmann H., Ernst F., Homuth G., Hecker M., Schweder T. Regulation of acetoin and 2,3-butanediol utilization in Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2010;87:2227–2235. doi: 10.1007/s00253-010-2681-5. [DOI] [PubMed] [Google Scholar]
- Vonder Haar R.A., Umbarger H.E. Isoleucine and valine metabolism in Escherichia coli. XIX. Inhibition of isoleucine biosynthesis by glycyl-leucine. J. Bacteriol. 1972;112:142–147. doi: 10.1128/JB.112.1.142-147.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O., Sugiyama N., Ishihama Y., Beltrao P. Uncovering phosphorylation-based specificities through functional interaction networks. Mol. Cell. Proteomics. 2016;15:236–245. doi: 10.1074/mcp.M115.052357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want E.J., Wilson I.D., Gika H., Theodoridis G., Plumb R.S., Shockcor J., Holmes E., Nicholson J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010;5:1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Wünsche A., Hammer E., Bartholomae M., Völker U., Burkovski A., Seidel G., Hillen W. CcpA forms complexes with CodY and RpoA in Bacillus subtilis. FEBS J. 2012;279:2201–2214. doi: 10.1111/j.1742-4658.2012.08604.x. [DOI] [PubMed] [Google Scholar]