Abstract
Increasing studies have shown that long non-coding RNAs (lncRNAs) had crucial regulatory roles in many diseases. Nevertheless, the biological relevance and mechanisms of the NNT-AS1 in gliom remain poorly understood. In the present study, NNT-AS1 expression was up-regulated in the glioma cell lines. Functional assays demonstrated that depletion of NNT-AS1 could notably suppress the proliferation, migration, invasion and EMT of U87MG and A172 cells. In addition, miR-582-5p was predicted to be a target of NNT-AS1 and EZH2 was predicted to be a target of miR-582-5p by bioinformatics software, which was further confirmed by luciferase reporter assay. Additionally, results of recue assays proofed that NNT-AS1/miR-582-5p/EZH2 axis aggravated the malignant behaviors of glioma. Ultimately, our findings revealed that NNT-AS1 contributes to progression via targeting miR-582-5p/EZH2 axis, revealing NNT-AS1 as a latent effective target for the diagnosis and treatment of glioma patients.
Keywords: LncRNA-NNT-AS1, MiR-582-5p/EZH2 axis, Proliferation, Metastasis, Glioma
Introduction
Glioma is the most common malignant primary brain cancer in adults, and most aggressive human malignancies (Siegel et al. 2019). Although treatments including maximal safe resection, radiotherapy, and adjuvant temozolomide have evidently improved the outcomes of glioma patients, they still lag behind those of other tumors (Lim et al. 2018; Zhao et al. 2019a). In the clinical practices, glioma is commonly diagnosed at late/advanced stages with extremely poor prognosis and a 5-year survival rate of only 5.5% (Ostrom et al. 2013). The treatment of such brain tumors remains a challenge. Molecularly targeted therapies are essential for better glioma prognosis. Thus, it is urgent to elucidate the underlying lncRNA-based mechanisms in glioma patients.
Long non-coding RNAs (lncRNAs) are a kind of RNA molecules of more than 200nt in length. Growing evidences have pointed out that lncRNAs function as crucial mediators in regulation of gene expression levels (Yuan et al. 2015). Increasing discoveries have found that aberrantly expressed lncRNAs play crucial roles in the development of malignancies, including glioma (Jiang et al. 2018; Xu et al. 2018; Li et al. 2019a). Many evidences reported that lncRNAs are involved in modulation cancer biological activities, such as cell proliferation, differentiation, metastasis, metabolism and apoptosis. For example, LncRNA DICER1-AS1 promotes cell proliferation and autophagy via miR-30b/ATG5 axis in osteosarcoma (Gu et al. 2018). In addition, lncRNA ZEB1-AS1 facilitates cell migration and metastasis of bladder cancer cells by regulating ZEB1 (Zhao et al. 2019b).
Recent reports indicated that lncRNA nicotinamide nucleotide transhydrogenase-antisense RNA1 (NNT-AS1) was proved to play important roles in a variety of cancers. For example, NNT-AS1 elevated cholangiocarcinoma cells proliferation and epithelial-to-mesenchymal transition by regulating miR-203 (Gu et al. 2020). Huang en at showed that NNT-AS1 knockdown attenuated proliferation and enhanced apoptosis of lung cancer cells through miR-3666/E2F2 axis (Huang et al. 2020). Moreover, NNT-AS1 enhances cell growth by targeting miR-1301-3p/PODXL axis through Wnt pathway in bladder cancer (Liu and Wu 2020). However, the NNT-AS1 expression and functional mechanisms of NNT-AS1 in glioma progression remain unknown.
In recent years, growing evidences have reported that lncRNAs could interact with miRNA to affect tumorigenesis and involved in the diagnosis for patients (Xiao et al. 2017; Ding et al. 2018). For example, LncRNA H19 promoted epithelial-mesenchymal transition through miR-29b-3p/PGRN axis in colorectal cancer. LncRNA NORAD contributes to colorectal cancer progression by modulation of miR-202-5p (Zhang et al. 2018). What’s more, LncRNA XIST/miR-34a axis facilitates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling (Liu et al. 2018). Yuan et al. reported that LncRNA HOTTIP functions as an oncogene by regulating miR-637 in papillary thyroid carcinoma (Yuan et al. 2018). Previous studies have identified miR-582-5p was the target of Lnc UCA1 and contributes to the progression of bladder cancer (Wu et al. 2019). Therefore, the aim of this study was to explore the function and the underlying mechanism of NNT-AS1/ miR-582-5p in glioma development.
Material and methods
Cell culture and transfection
Human glioma cell lines (U87MG, A172, LN229, SHG-44, U251) and epithelial cell line (HA1800) were obtained from the Chinese Science institute (Shanghai, China). Cells were maintained in RPMI-1640 medium (Invitrogen) supplemented with 10% FBS at 37 °C under 5% CO2.
Sh-NNT-AS1, miR-582-5p inhibitor, sh-EZH2 and negative controls were purchased from GenePharma (Shanghai, China). For transfection, cells were seeded in 6-well plates overnight and cell transfection was carried out using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Transfection efficiency was determined by RT-qPCR analysis. Cells were harvested for futher analysis after 48 h transfection.
RNA isolation and real-time quantitative PCR (RT-qPCR) assay
The total RNA of the cells was extracted using TRIzol (Invitrogen, USA) according to the manufacturer’s instructions and then reverse-transcribed to cDNA with a Reverse Transcription Kit (Beyotime, Haimen, China). RT-qPCR was performed on SYBR Green qPCR Master Mixes (Thermo Fisher Scientific). Quantification of the miRNA was performed with SYBR-Green PrimeScript miRNA RT-PCR kit (Takara, Japan). The levels of total RNA were normalized to GAPDH and miRNA RNA were normalized to U6 snRNA (small nuclear RNA). RNA expression was assessed using 2−ΔΔCt method. The specific primers were as follows: NNT-AS1 forward (F): 5′-ACGTGCAGACAACATCTACCT-3′ and reverse (R): 5′-TACAACACCTTCCCGCAT-3′; miR-582-5p forward (F): 5′-GCGGTTACAGTTGTTCAACC-3′ and reverse (R): 5′-CTCAACTGGTGTCGTGGA-3′; U6 forward (F): 5′-CTCGCTTCGGCAGCACA-3′ and reverse (R): 5′-AACGCTTCACGAATTTGCGT-3′; EZH2 U6 forward (F): 5′-TTGTTGGCGGAAGCGTGTAAAATC-3′ and reverse (R): 5′- TCCCTAGTCCCGCGCAATGAGC-3′; E-cadherin forward (F): 5′- GACAACAAGCCCGAATT-3′ and reverse (R): 5′-GGAAACTCTCTCGGTCCA-3′; N-cadherin forward (F): 5′-CGGGTAATCCTCCCAAATCA-3′ and reverse (R): 5′-CTTTATCCCGGCGTTTCATC-3′; GAPDH forward (F): 5′-ATGACCCCTTCATTGACCTCA-3′ and reverse (R): 5′-GAGATGATGACCCTTTTGGCT-3'.
Cell counting Kit-8 (CCK-8) assay
Cell visibility was evaluated with the Cell Counting Kit 8 (Dojindo, Japan) and optical density (OD) was measured at wavelength of 450 nm with the BioTek Gen5 system (BioTek, USA).
5-ethynyl-2′-deoxyuridine (EdU) analysis
Cell proliferation was measured by a 5-ethynyl-20-deoxyuridine (EdU) assay kit (Thermo Fisher Scientific) according with the instructions. Cells (1 × 105) were maintained in 6-well plate. After 48 h, 100 μL of EdU was added for 2 h. Afterwards, cells were treated with 4% paraformaldehyde and then added 0.5% Triton X-100. Finally, cells were stained with anti-EdU solution. The proportion of cells that incorporated EdU was determined with a fluorescence microscope (Nikon C2, Tokyo, Japan) at × 200 magnification.
Western blot assay
Protein was got through RIPA lysis buffer (Beyotime, China), and protein concentration was examined by BCA kit (Beyotime, Haimen, China). After isolating protein samples through 10% SDS-PAGE and transferred to PVDF membrane, following by blocked with 5% non-fat milk for 1 h at 37 °C. Subsequently, the membranes were probed with primary antibodies specific against E-cadherin (ab15148), N-cadherin (ab95440) at 4 °C overnight. After washing with TBS, the membrane was probed with HRP-conjugated secondary antibodies (ab6728, 1:2000) for 2 h at 37 °C. Detection was performed using an enhanced chemiluminescence reagent (Beckman Coulter, USA) and bands were got through Image J Software (Bio-Rad). GAPDH was used as an internal reference.
Cell migration and invasion assays
Cell migration and invasion assays were performed using a transwell chamber (BD Biosciences, Bedford, MA, USA). For migration assay, 1 × 105 cells were seeded into the upper chamber with serum-free medium, while the lower compartments were filled with complete medium supplemented with 10% FBS. After 48 h, on the upper surfaces the non-migrated cells were removed. For invasion assay, the upper chamber was pre-coated with Matrigel and other operations are the same as above. Cells were fixed and stained with 1% crystal violet. The numbers of cells were counted under a light microscope.
Luciferase reporter assay
Cells were cultured in 96-well plate. WT/Mut 3′UTR of NNT-AS1 or EZH2 were cloned into psiCHECK-2 plasmids (Invitrogen). The cells were cotransfected with miR-582-5p mimic (scramble as control) and plasmids using Lipofectamine 2000 Kit (Invitrogen). The luciferase activity was determined by using the dual-luciferase reporter analysis system and Renilla luciferase activity was normalization.
Statistical analysis
Experimental results were shown as the mean ± SD and data analysis was conducted using SPSS 19.0. Student’s t-test was performed to evaluate significant differences between two groups and one-way analysis of variance (ANOVA) followed Tukey’s poc host was used to test differences among multiple groups. P < 0.05 was considered as statistical significance.
Results
NNT-AS1 was up-regulated in glioma
To investigate the potential involvement of NNT-AS1 in glioma, we analyzed the level of NNT-AS1 in Cancer Genome Atlas (TCGA) and the OncomiR database showed that high NNT-AS1 expression represents a more malignant tumor types. In addition, we found that the level of NNT-AS1 was up-regulated in glioma patients (Fig. 1a, b). Furthermore, we used RT-qPCR assay to measure the expression NNT-AS1 in glioma cells and results showed that NNT-AS1 expression in glioma cell lines (U87MG, A172, LN229, SHG-44, U251) were notably raised contrasted with epithelial cell line (HA1800) (Fig. 1c). Our data revealed that NNT-AS1 was overexpressed in glioma.
Fig. 1.
NNT-AS1 was up-regulated in glioma. a The Cancer Genome Atlas (TCGA) database shows the level of NNT-AS1 in various types of cancers. b The expression of NNT-AS1 in glioma patients and in control groups. c The expression NNT-AS1 in glioma cells. *P < 0.05, **P < 0.01 vs. HA1800 cells
NNT-AS1 silencing inhibits human glioma cell proliferation in vitro
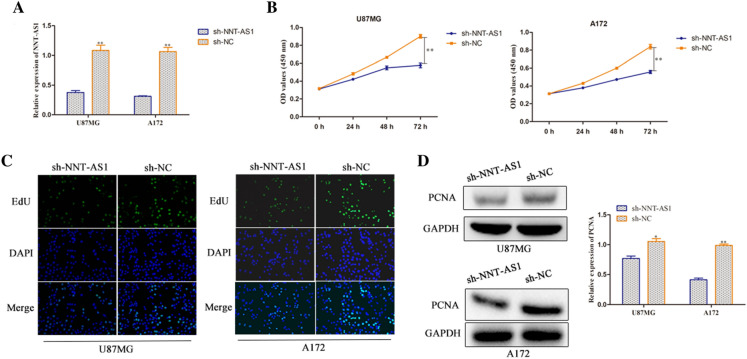
To explore the function of NNT-AS1 on glioma, we knocked down NNT-AS1 expression through transfection with sh-NNT-AS1 in U87MG and A172 cells. The successful transfection was confirmed by RT-qPCR expression. Cells transfected with sh-NNT-AS1 exhibited a decreased level compared to the NC group (Fig. 2a). The CCK-8 and EdU assays were used to investigate the impact of NNT-AS1 on glioma cell proliferation. As hypothesized, we found that inhibition of NNT-AS1 expression led to cell growth inhibition at 48 and 72 h in U87MG and A172 cells (Fig. 2b). Further study of cell proliferation using EdU assay also showed obvious attenuation of cell proliferation in U87MG and A172 cells transfected with sh-NNT-AS1 (Fig. 2c). In addition, we detected the protein expression level of PCNA by western blot assay. As expected, silencing of NNT-AS1 expression suppressed the expression level of PCNA accompanied with NC group (Fig. 2d). Collectively, these data demonstrated that knockdown of NNT-AS1 inhibited cell proliferation of glioma cells.
Fig. 2.
NNT-AS1 silencing inhibits human glioma cell proliferation in vitro. a RT-qPCR analysis of the expression of NNT-AS1 in the U87MG and A172 cells (transfected with sh-NNT-AS1). b, c Effect of NNT-AS1 on U87MG and A172 cellular viabilities was determined by CCK-8 and EdU assays (magnification × 200). d Expression of PCNA was examined via western blot in both U87MG and A172 cells. *P < 0.05, **P < 0.01 vs. sh-NC group
NNT-AS1 promotes metastasis of glioma cells in vitro
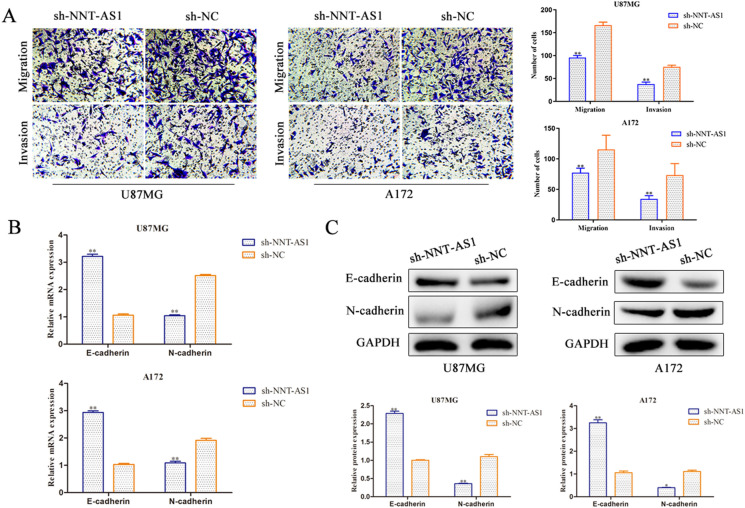
Then we attempted to investigate the effect of NNT-AS1 on migration and invasion in glioma cells. As shown in Fig. 3a, down-regulation of NNT-AS1 inhibited the migration capacity of U87MG and A172 cells through transwell assay. Moreover, transwell chamber assay indicated that depletion of NNT-AS1 weakened the invasion ability of U87MG and A172 cells compared with NC group. EMT is very important to tumor progression (Kalluri and Weinberg 2009) and accompanies by the loss of expression of E-cadherin, induction of N-cadherin and vimentin (Davalos et al. 2012; Xing et al. 2014). To further explore the effect of NNT-AS1 in glioma cell metatasis, western blot analysis was adapted to determine the expression levels of E-cadherin and N-cadherin. The results of RT-qPCR and western blot analysis revealed that the mRNA and protein expression levels of N-cadherin was lessened by suppression of NNT-AS1, while level of E-cadherin was notably raised both in U87MG and A172 cells (Fig. 3b, c). Overall, these findings demonstrated that NNT-AS1 knockdown exerted suppressive impacts on glioma cell metastasis.
Fig. 3.
NNT-AS1 promotes metastasis of glioma cells in vitro. a Transwell migration and invasion assays show that NNT-AS1 knockdown suppressed cell migration and invasion ability (magnification × 200). b, c Expression of E-cadherin and N-cadherin were examined via RT-qPCR and western blot in both U87MG and A172 cells. *P < 0.05, **P < 0.01 vs. sh-NC group
MiR-582-5p was a direct target of NNT-AS1
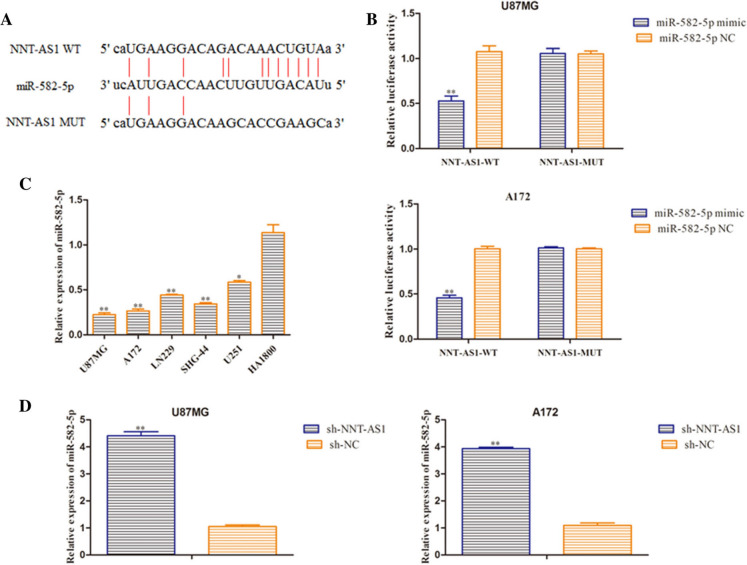
Bioinformatics prediction program (Database) was employed to provide NNT-AS1 contained binding sequences with the miR-582-5p at the 3′-untranslated region (3′-UTR) (Fig. 4a). The dual luciferase reporter experiment showed that luciferase activities of the vectors carrying NNT-AS1-WT notably reduced in U87MG and A172 cells transfected with miR-582-5p mimic (Fig. 4b). Furthermore, the expression miR-582-5p in glioma cells were assessed and results showed that miR-582-5p expression was significantly down-regulated in the glioma cells (Fig. 4c). The expression level of miR-582-5p was detected using RT-qPCR analysis and results revealed that miR-582-5p expression was increased in sh-NNT-AS1 group compared with NC group (Fig. 4d). These data indicated that miR-582-5p is negatively regulated by NNT-AS1 in glioma cells, which is the novel target of NNT-AS1.
Fig. 4.
MiR-582-5p was a direct target of NNT-AS1. a The target matching sequence of NNT-AS1 and miR-582-5p. b Luciferase activity was measured after co-transfection with NNT-AS1-WT or NNT-AS1-MUT and miR-582-5p mimic or NC mimic. c Expression levels of miR-582-5p in U87MG, A172, LN229, SHG-44, U251 and HA1800 cells were examined by RT-qPCR assay. *P < 0.05, **P < 0.01 vs. miR-582-5p NC group. d The mRNA level of miR-582-5p was tested through qRT-PCR after transfection with sh-NNT-AS1 and the relative control. *P < 0.05, **P < 0.01 vs. sh-NC group
MiR-582-5p targets EZH2 in glioma
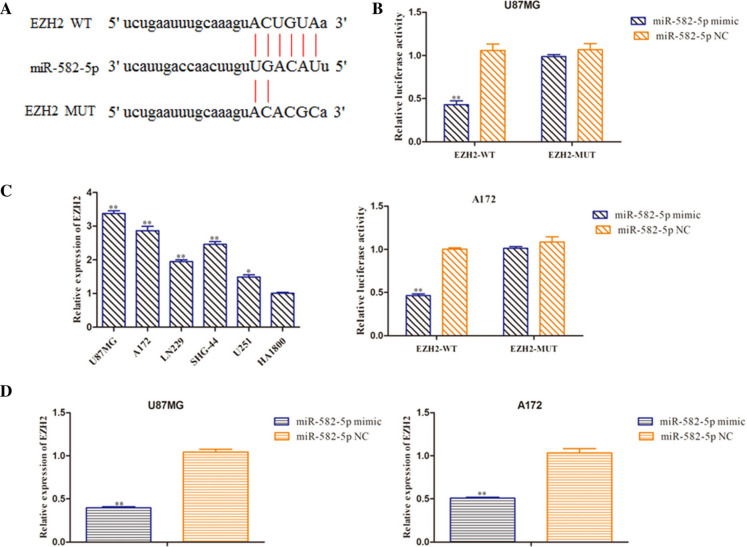
Bioinformatics databases were employed to predict the target gene of miR-582-5p. The data revealed that EZH2 had a complementary sequence to miR-582-5p, and the predicted binding sites of miR-582-5p seed sequence and 3′-UTR of EZH2 were shown in Fig. 5a. Dual-luciferase reporter analysis indicated that miR-582-5p mimic inhibited luciferase activity of wild-type 3′-UTR of EZH2, leaving the mutant one unaltered (Fig. 5b). Next, we detected the level of EZH2 using RT-qPCR assay and EZH2 mRNA level was discovered to be up-regulated in glioma cell lines (Fig. 5c). Moreover, RT-qPCR analysis was performed to assess the EZH2 expression and results revealed that EZH2 expression was decreased in miR-582-5p mimic group compared with NC group (Fig. 5d). These data revealed that miR-582-5p negatively regulated EZH2 expression by directly targeting the 3′-UTR of EZH2.
Fig. 5.
EZH2 was a direct target of miR-582-5p. a The target matching sequence of NNT-AS1 and miR-582-5p. b Luciferase activity was measured after co-transfection with EZH2-WT or EZH2-MUT and miR-582-5p mimic or NC mimic. c Expression levels of EZH2 in glioma cells were tested by RT-qPCR assay. d The mRNA level of EZH2 was tested through qRT-PCR after transfection with miR-582-5p mimic and the relative control. **P < 0.01 vs. miR-582-5p NC group
Knockdown of NNT-AS1 expression inhibits glioma progression via miR-582-5p/EZH2 axis
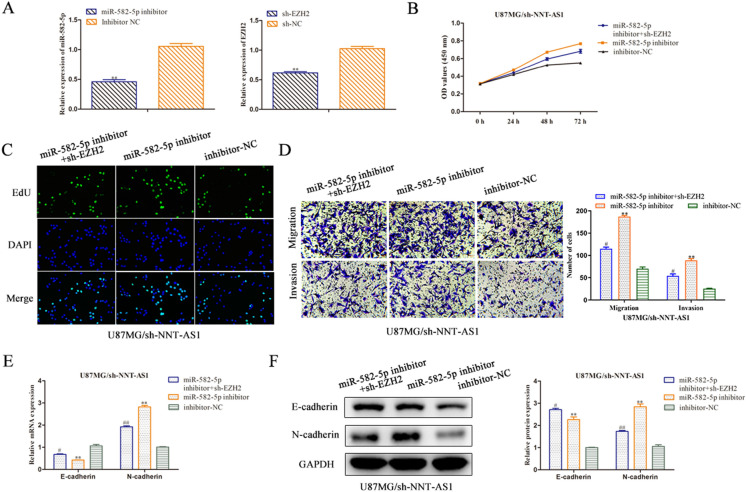
Based on the above findings, rescue assays were conducted to certify whether NNT-AS1 elicited its performance through miR-582-5p/EZH2 axis in glioma. We knocked the miR-582-5p and EZH2 expression in U87MG cells transfected with miR-582-5p inhibitor or sh-EZH2. The knockdown efficiency were detected using RT-qPCR analysis and results revealed that the expression level of miR-582-5p was decreased after transfection miR-582-5p inhibitor and EZH2 expression was repressed in sh-EZH2 group compared with NC group in U87MG cells (Fig. 6a).
Fig. 6.
Knockdown of NNT-AS1 expression inhibits glioma progression via miR-582-5p/EZH2 axis. a The RT-qPCR assay was implemented to certify transfection efficiency for miR-582-5p inhibitor and sh-EZH2. b–d CCK-8, EdU, transwell assays were employed to estimate the role of NNT-AS1/miR-582-5p/EZH2 axis in U87MG cell proliferation, migration and invasion (magnification × 200). e, f RT-qPCR and western blot analyses were performed to detected mRNA and protein expressions of E-cadherin and N-cadherin. **P < 0.01 vs. inhibitor-NC, #P < 0.05, ##P < 0.01 vs. smiR-582-5p inhibitor
CCK-8 assay depicted that NNT-AS1 knockdown-induced reduction in cell proliferative ability of U87MG cells was promoted by the miR-582-5p inhibitor, while the effect of miR-582-5p inhibitor was subsequently recovered by EZH2 depletion (Fig. 6b). Consistently, the EdU assay also indicated the similar results (Fig. 6c). Additionally, the transwell assays manifested that the migration and invasion capacity of U87MG cells was restrained by sh-NNT-AS1 and retrieved by miR-582-5p inhibitor. However, co-transfection of sh-EZH2 reversed the cell migration and invasion caused by miR-582-5p inhibitor (Fig. 6d).
As expected, RT-qPCR and western blot analyses demonstrated that knockdown of NNT-AS1 enhanced the expression level of E-cadherin, while repressed the expression level of N-cadherin, and abolished by miR-582-5p inhibitor, while co-transfected with sh-EZH2 antagonized the impact of miR-582-5p inhibitor in the sh-NNT-AS1 cells (Fig. 6e, f). These results provided strong evidence that silencing NNT-AS1 expression inhibits glioma progression via targeting miR-582-5p/EZH2 axis.
Discussion
Various studies have uncovered the crucial roles of lncRNAs in regulating various kinds of cancer initiation and development. Accumulative evidence has shown that deregulated lncRNAs can affect some biological processes of cancers. For instance, lncRNA SNHG4 contributes cervical cancer progression through miR-148a-3p/c-Met (Li et al. 2019b). Previous studies have reported that DIO3OS interacts with miR-122 to facilitate pancreatic cancer cell proliferation by regulating ALDOA (Cui et al. 2019). Hao et al. reported that Long noncoding RNA MAGI1-IT1 affects ovarian cancer cell invasion and metastasis by miR-200a/ZEB axis (Hao et al. 2019). However, the involvement of lncRNAs in glioma progression is not widely studied. Specifically, the roles and potential mechanism of NNT-AS1 in glioma remain obscure.
In the present study, the upregulation of NNT-AS1 was identified by TCGA database and was further verified by RT-qPCR in glioma cells, indicating that the NNT-AS1 may functional as a critical oncogene in glioma. Functionally, loss-of-function assay disclosed that depletion of NNT-AS1 contributed to the suppression of glioma cell proliferation and metastasis.
Increasing numbers of studies suggest that a widespread interaction network of ceRNA is involved in cancer carcinogenesis, in which lncRNAs functioned as a miRNA sponge to regulate cancer development, including glioma (Ye and Jiao 2019; Zhang and Ju 2018; Cesana 2011; Zong et al. 2019). Chen et al. revealed that HOXD-AS1/miR-130a sponge modulates glioma development by targeting E2F8 (Chen et al. 2018). Silencing of lncRNA LUCAT1 inhibits glioma cell invasion via targeting miR-375 (Gao et al. 2018).
In the current study, accordance with bioinformatics prediction, we revealed that miR-582-5p physically binds with NNT-AS1 and EZH2 is a potential target of miR-582-5p. In addition, luciferase reporter assay and RNA pull-down assay miR-582-5p sponged NNT-AS1 and EZH2 sponged miR-582-5p to depress glioma cell proliferation and metastasis. Mechanistically, rescue experiments revealed that the function of NNT-AS1 in glioma tumorigenicity was mediated by miR-582-5p/EZH2 axis.
To summarize, our findings unveiled that NNT-AS1 might serves as an oncogene in glioma. NNT-AS1 knockdown inhibits the cell proliferation, migration and invasion in glioma. Deleption of NNT-AS1 exerted its tumor inhibitory effects on glioma progression by modulation of miR-582-5p/EZH2 axis, which offered a potential new biomarker or therapeutic target for glioma treatment.
Acknowledgements
We deeply appreciate the supports by all participants.
Author contributions
MX conceived and designed the study and drafted the manuscript. TP collected and analyzed the data. All authors read and approved the final manuscript.
Funding
This study was supported by National Youth Natural Science Foundation (Grant No. 81500914) and Chinese Postdoctoral Science Foundation (Grant No. 2018M632381).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors state that there is no conflict of interest.
Footnotes
The original online version of this article was revised: In Figure 3 and Figure 6 of this article, the results of transwell experiment were not very ideal and the data organization problem caused some confusion in the inter group data. The Figure 3 and Figure 6 should have appeared as shown below.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/22/2024
A Correction to this paper has been published: 10.1007/s10616-024-00636-z
References
- Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhao F, Cui D. HOXD-AS1/miR-130a sponge regulates glioma development by targeting E2F8. Int J Cancer. 2018;142:2313. doi: 10.1002/ijc.31262. [DOI] [PubMed] [Google Scholar]
- Cui K, Jin S, Du Y. Long noncoding RNA DIO3OS interacts with miR-122 to promote proliferation and invasion of pancreatic cancer cells through upregulating ALDOA. Cancer Cell Int. 2019;19:1. doi: 10.1186/s12935-019-0922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V, Moutinho C, Villanueva A, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Li C, Zhao T. LncRNA H19/miR-29b-3p/PGRN axis promoted epithelial-mesenchymal transition of colorectal cancer cells by acting on Wnt signaling. Mol Cells. 2018;41:423–435. doi: 10.14348/molcells.2018.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y-S, Liu X-Z, Zhang Y-G. Knockdown of Long Noncoding RNA LUCAT1 Inhibits Cell Viability and Invasion by Regulating miR-375 in Glioma. Oncol Res. 2018;26:307. doi: 10.3727/096504017X15088061795756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Hou Z, Zheng L. LncRNA DICER1-AS1 promotes the proliferation, invasion and autophagy of osteosarcoma cells via miR-30b/ATG5. Biomed Pharmacother. 2018;104:110–118. doi: 10.1016/j.biopha.2018.04.193. [DOI] [PubMed] [Google Scholar]
- Gu Y, Zhu Z, Pei H, et al. Long non-coding RNA NNT-AS1 promotes cholangiocarcinoma cells proliferation and epithelial-to-mesenchymal transition through down-regulating miR-203. Aging (Albany NY) 2020;12:2333. doi: 10.18632/aging.102747. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hao G, Xiaofeng L, Guangxi Z. Long noncoding RNA MAGI1-IT1 promoted invasion and metastasis of epithelial ovarian cancer via the miR-200a/ZEB axis. Cell cycle (Georgetown, TX) 2019;18:1393. doi: 10.1080/15384101.2019.1618121. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huang JW, Luo XY, Li ZH, et al. LncRNA NNT-AS1 regulates the progression of lung cancer through the NNT-AS1/miR-3666/E2F2 axis. Eur Rev Med Pharmacol Sci. 2020;24:238. doi: 10.26355/eurrev_202001_19916. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem Biophys Res Commun. 2018;495:238. doi: 10.1016/j.bbrc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Investig. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Cai Y, Wang C. LncRNA GAS5 regulates the proliferation, migration, invasion and apoptosis of brain glioma cells through targeting GSTM3 expression. The effect of LncRNA GAS5 on glioma cells. J Neuro-Oncol. 2019;143:1–12. doi: 10.1007/s11060-019-03185-0. [DOI] [PubMed] [Google Scholar]
- Li H, Hong J, Wijayakulathilaka WSMA. Long non-coding RNA SNHG4 promotes cervical cancer progression through regulating c-Met via targeting miR-148a-3p. Cell Cycle. 2019;18:3313. doi: 10.1080/15384101.2019.1674071. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lim M, Xia Y, Bettegowda C, et al. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15:422. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wu G. NNT-AS1 enhances bladder cancer cell growth by targeting miR-1301-3p/PODXL axis and activating Wnt pathway. Neurourol Urodyn. 2020;39:547. doi: 10.1002/nau.24238. [DOI] [PubMed] [Google Scholar]
- Liu H, Deng H, Zhao Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res. 2018;37:1. doi: 10.1186/s13046-018-0950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15:ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Wu J, Li W, Ning J. Long noncoding RNA UCA1 targets miR-582–5p and contributes to the progression and drug resistance of bladder cancer cells through ATG7-mediated autophagy inhibition. OncoTargets Ther. 2019;12:495–508. doi: 10.2147/OTT.S183940. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xiao JN, Yan TH, Yu RM, Gao Y, Zeng WL, Lu SW, Que HX, Liu ZP, Jiang JH. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143:981–990. doi: 10.1007/s00432-017-2370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang J, Li R, et al. TGF-beta-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci Signal. 2014;7:91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- Xu R, Zhu X, Chen F. LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 2018;18:41. doi: 10.1186/s12935-018-0540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Jiao Y. LncRNA FAL1 promotes the development of oral squamous cell carcinoma through regulating the microRNA-761/CRKL pathway. Eur Rev Med Pharmacol Sci. 2019;23:5779–5786. doi: 10.26355/eurrev_201907_18316. [DOI] [PubMed] [Google Scholar]
- Yuan S, Wang J, Yang F. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2015;63:499. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Liu Y, Fan Y. LncRNA HOTTIP promotes papillary thyroid carcinoma cell proliferation, invasion and migration by regulating miR-637. Int J Biochem Cell Biol. 2018;98:1. doi: 10.1016/j.biocel.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Zhang LM, Ju HY. Long non-coding RNA ANRIL promotes tumorgenesis through regulation of FGFR1 expression by sponging miR-125a-3p in head and neck squamous cell carcinoma. Am J Cancer Res. 2018;8:2296–2310. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li X-Y, Hu P. lncRNA NORAD contributes to colorectal cancer progression by inhibition of miR-202-5p. Oncol Res Featur Preclin Clin Cancer Ther. 2018;26:1411. doi: 10.3727/096504018X15190844870055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Chen AX, Gartrell RD, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25:462. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wang D, Ding Y. lncRNA ZEB1-AS1 promotes migration and metastasis of bladder cancer cells by post-transcriptional activation of ZEB1. Int J Mol Med. 2019;44:196. doi: 10.3892/ijmm.2019.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Z, Song Y, Xue Y. Knockdown of LncRNA SCAMP1 suppressed malignant biological behaviours of glioma cells via modulating miR-499a-5p/LMX1A/NLRC5 pathway. J Cell Mol Med. 2019;23:5048. doi: 10.1111/jcmm.14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.