Abstract
Leaves provide energy for plants, and consequently for animals, through photosynthesis. Despite their important functions, plant leaf developmental processes and their underlying mechanisms have not been well characterized. Here, we provide a holistic description of leaf developmental processes that is centered on cytokinins and their signaling functions. Cytokinins maintain the growth potential (pluripotency) of shoot apical meristems, which provide stem cells for the generation of leaf primordia during the initial stage of leaf formation; cytokinins and auxins, as well as their interaction, determine the phyllotaxis pattern. The activities of cytokinins in various regions of the leaf, especially at the margins, collectively determine the final leaf morphology (e.g., simple or compound). The area of a leaf is generally determined by the number and size of the cells in the leaf. Cytokinins promote cell division and increase cell expansion during the proliferation and expansion stages of leaf cell development, respectively. During leaf senescence, cytokinins reduce sugar accumulation, increase chlorophyll synthesis, and prolong the leaf photosynthetic period. We also briefly describe the roles of other hormones, including auxin and ethylene, during the whole leaf developmental process. In this study, we review the regulatory roles of cytokinins in various leaf developmental stages, with a focus on cytokinin metabolism and signal transduction processes, in order to shed light on the molecular mechanisms underlying leaf development.
Subject terms: Leaf development, Cytokinin
Introduction
In the late 1950s, a substance that promoted plant cell division was discovered in autoclaved herring sperm DNA and was called kinetin1. A few years later, a class of phytohormones with similar molecular structures, including 6-(γ,γ-dimethylallylamino)-purine, 6-benzyladenines, and zeatin, referred to as cytokinins2, were found to play important regulatory roles in cell division. Since then, the biosynthesis, metabolism, distribution, signaling pathways, and functions of cytokinins have been intensely investigated and characterized.
The main genes currently known to be involved in the cytokinin biosynthesis pathway encode the ISOPENTENYL TRANSFERASE (IPT) and LONELY GUY (LOG) enzymes3,4. The initial step of cytokinin biosynthesis in higher plants is the formation of cytokinin nucleotides, namely, isopentenyladenosine 5′-tri-, di-, or monophosphate (iPRTP, iPRDP, or iPRMP, respectively), from ATP, ADP, or AMP and dimethylallyl pyrophosphate (DMAPP) by IPTs5. LOGs, which encode phosphoribohydrolase-activating enzymes, directly convert a cytokinin nucleotide to an active free-base form of cytokinins in the final step of cytokinin biosynthesis3 (Fig. 1). The levels of active cytokinins can be modulated via irreversible cleavage by CYTOKININ OXIDASE (CKX) enzymes6,7 or through conjugation to glucose by cytokinin glycosyltransferases8,9 (Fig. 1). Plants regulate the concentration of active cytokinins through reversible and irreversible metabolism processes. Therefore, the precise maintenance of the homeostasis of cytokinins through these synthesis and inactivation enzymes is essential for plant development and adaptation to complex and changing environments. Recent studies have indicated that cytokinin biosynthesis varies with tissue and cell type10,11. However, as they are mobile signals, cytokinins rely on PURINE PERMEASES (PUP)12,13, EQUILIBRATIVE NUCLEOSIDE TRANSPORTERS (ENT)12, and G SUBFAMILY ATP-BINDING CASSETTE (ABCG)14 for short- and long-distance transport between roots and shoots (Fig. 1). In Arabidopsis, cytokinin signal transduction begins when cytokinins are received by sensor histidine kinases, HISTIDINE KINASE (HK2, HK3, and HK4), which initiate a phosphorylation signaling cascade in the endoplasmic reticulum15,16. After cytokinin binding, the phosphoryl group is transferred from HKs onto HISTIDINE-CONTAINING PHOSPHOTRANSMITTER (HPT) proteins17. HPTs then translocate from the cytoplasm to the nucleus and activate the transcription of ARABIDOPSIS RESPONSE REGULATORS (ARRs), which are categorized as type A transcriptional repressors18–20 or type B activators20–22, and CYTOKININ RESPONSE FACTOR (CRF)20,23 (Fig. 1). Through this signal transmission, cytokinins influence many aspects of biological processes that affect plant growth and development, such as cell division, apical dominance, shoot initiation and growth, phyllotaxis, vascular bundles, leaf senescence, branching and nodulation, seed germination, nutrient uptake, and biotic and abiotic stress responses20,24,25.
Fig. 1. Schematic model of cytokinin (CK) biosynthesis, metabolism, degradation, and signal transduction.
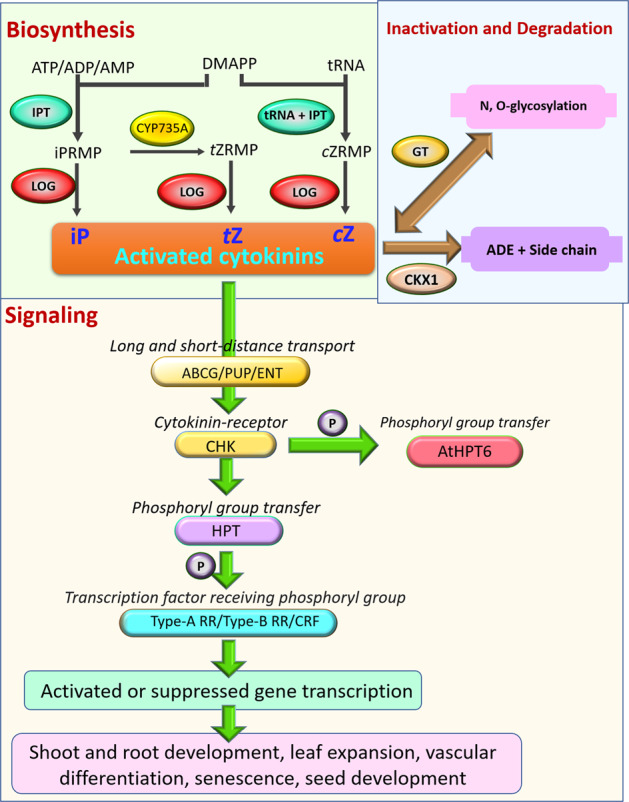
The names of the genes in up panels are shown in ovals, and in low panel are shown in the capsule shapes (see the text for further details). DMAPP: dimethylallyl pyrophosphate; iPRMP: isopentenyladenosine-5-monophosphate; tZRMP, trans-zeatin riboside 5′-monophosphate; cZRMP, cis-zeatin riboside 5′-monophosphate; iP, N6-(Δ2-isopentenyl)adenine; tZ: trans-zeatin; cZ: cis-zeatin; Ade: adenine; IPT, isopentenyltransferases; tRNA-IPT, tRNA-isopentenyltransferase; CYP735A, cytochrome P450 monooxygenase; LOG, LONELY GUY; GT, glycosyltransferase; CKX, cytokinin oxidase/dehydrogenase; ABCG, g subfamily ATP-binding cassette; PUP, purine permeases; ENT, equilibrative nucleoside transporters; HKs, histidine kinase; HPTs, histidine phosphotransfer proteins; ARR, response regulator, CRF, cytokinin response factor. Other abbreviations are as defined in the text
The development of plant leaves, which are the primary organs in plants for capturing light energy and perceiving diverse environmental conditions, is a dynamic process that can be divided into four different phases: the initiation of leaf primordia, the establishment of polarity (EP), the establishment of leaf size and morphology26,27, and leaf senescence28. First, cells at the peripheral zone of the shoot apical meristem (SAM) differentiate into a leaf primordium, whose position is regulated by phyllotactic patterning29. Second, the three growth axes, the adaxial–abaxial, proximal-distal, and mediolateral axes, are determined in the leaf primordium30,31. Even before the EP is completed, leaf primordium cells begin to divide and proliferate, which results in exponential increases in both leaf area and cell number. After the leaf blade and the petiole clearly separate, growth occurs throughout the entire leaf along the mediolateral axis, which results in the formation of the final shape of the leaf. After a growing season, flowering, nutrient deficiency, or unfavorable environmental conditions such as inadequate light or certain abiotic/biotic stresses, leaf senescence is initiated, which constitutes the final stage of the leaf lifespan preceding its death. However, these leaf developmental stages are not completely independent, as they are continuous and interconnected26,27,29.
In plants, cytokinins are essential regulators that are involved in almost every aspect of plant growth and development. During the various stages of leaf development, cytokinins play essential roles by regulating the transcriptional expression of downstream genes. Cytokinin homeostasis is modulated by certain transcription factors or by modulators during leaf development. Therefore, studies of the relationships among cytokinin signal transduction, gene regulation, and cytokinin modulation during various stages of leaf development help to reveal the underlying molecular mechanisms and advance our understanding in order to open novel avenues for improving agricultural and forestry yields. In this review, we focus on cytokinin homeostasis, signal transduction, and gene regulation as well as their regulatory roles in leaf development.
The complex roles of cytokinins in leaf initiation
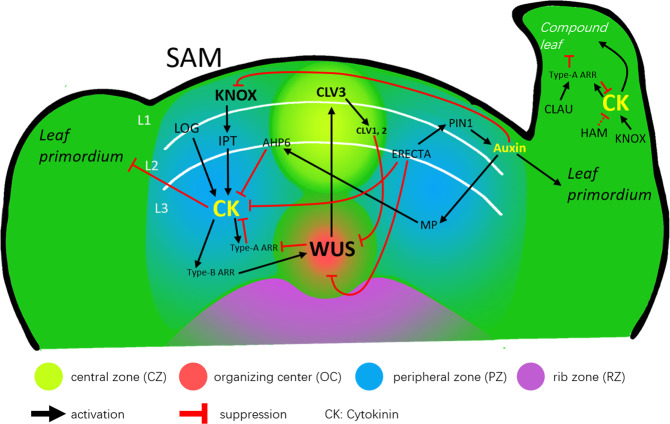
SAMs located at the shoot apexes are highly organized tissues containing pluripotent stem cells that can be divided into different functional zones, including the central zone (CZ), peripheral zone (PZ), and rib zone (RZ). SAMs generate nearly all the aerial organs and tissues of plants during postembryonic growth. Generally, cells in the CZ at the summit of the SAM divide slowly and maintain their pluripotency. The RZ below the CZ is responsible for generating stems. Some of the daughter cells produced by the CZ exhibit an accelerated cell division rate when they emerge in the PZ and eventually lead to the formation of lateral organs such as leaves and flowers (Fig. 2)32,33. Leaf primordia are initiated from the PZ, where cells become responsive to differentiation. Leaf primordia in the PZ are generated in a temporally and spatially controlled manner; this process is referred to as phyllotaxy34. The SAM is anatomically divided into three well-defined cell layers: The epidermal (L1) and subepidermal (L2) layers, known as the tunica, and an inner layer (L3) that is referred to as the corpus32,35. L1 and L2 are single-cell sheets with anticlinal cell division planes that form the epidermis of the plant tissue surface. The cells within L3 divide in all directions and form vascular tissues (Fig. 2)26. In response to plant hormones and external cues, the dynamic balance of cell division and differentiation can be perfectly controlled and maintained in the different subdomains of the SAM by cytokinin and auxin interactions as well as their homeostasis and spatial signaling.
Fig. 2. Schematic diagram of cytokinin (CK) regulation of leaf primordium initiation.
Cells in the shoot apical meristem (SAM) are arranged into the L1, L2, and L3 layers and four distinct zones: the central zone (CZ), peripheral zone (PZ), organizing center (OC), and rib zone (RZ). KNOX is expressed in almost the entire SAM. KNOX positively regulates the synthesis of cytokinins and keeps their levels high. Cytokinins promote the expression of WUS through signal transduction and transcription factors, which maintain a high cell division rate in the OC. ERECTA blocks the effect of cytokinins and promotes the transport of auxin. In areas with higher auxin concentrations, the leaf primordium begins to form. In the early stage of leaf development, KNOX, which is highly expressed in the marginal blastozone, changes the leaf morphology by promoting cytokinin synthesis to form compound leaves. Solid lines indicate direct relationships that have been confirmed; dashed lines represent potential mechanisms. The abbreviations are as defined in the text
Phytohormones, such as auxins and cytokinins, play indispensable but distinct roles during SAM development and maintenance. Auxins are required for leaf formation and organogenesis; in contrast, cytokinins promote meristem maintenance. However, these phytohormones do not exist and function independently; recent studies have shown that auxins and cytokinins function together in multiple cells, tissues, and organs with both antagonistic and synergistic actions36–38. The formation of new leaves in the apical meristem is initiated by the accumulation of auxin36. Unlike those of auxins, the primary functions of cytokinins in maintaining the size and structure of SAMs have been fully demonstrated in multiple experiments25. For instance, a reduction in the concentration of or sensitivity to cytokinins via mutation of IPT39, the overexpression of CKX40, or the modulation of signal transporter genes41 results in a decrease in SAM size and activity. Therefore, cytokinins play a central role in stimulating SAM activity and size through synergistic or antagonistic interactions with auxin. At the same time, many other regulators contribute to modulating cytokinin and auxin concentrations and gradients in different zones of the SAM.
Several regulators have been shown to play important roles in modulating the concentrations and activities of cytokinins. Transcription factor class I KNOTTED-LIKE homeobox (KNOX I) family genes42, including SHOOT-MERISTEMLESS (STM), kn1-like in Arabidopsis thaliana1 (KNAT1), KNAT2, and KNAT6, are essential for establishing and maintaining meristem development by increasing cytokinin levels or sensitivity while simultaneously repressing GA. The expression patterns of KNOX I are primarily limited to the SAM, and KNOX I expression is absent in leaf primordia43,44. In Arabidopsis, STM activates the expression of IPT7 in the SAM to promote cytokinin biosynthesis (Fig. 2). stm mutants, which exhibit shoot meristem loss, can be partially rescued through the application of exogenous cytokinins45,46. In addition, endogenous cytokinin overproduction significantly increases the mRNA levels of the KNOX I genes, which indicates that there may be a positive feedback loop between KNOX I genes and cytokinins in the SAM47. Another enzyme, LOG4, is expressed in the L1 layer and produces active cytokinins that move to the lower cell layer and form a diffusion gradient within the SAM48,49. Thus, KNOX I and LOG4 provide a high level of cytokinin accumulation and activation in the SAM to sustain SAM growth and activity (Fig. 2).
The functions and effects of cytokinins in various cells in different zones are determined not only by their concentrations but also by their spatial signal transduction. Some genes regulate the size of the SAM by regulating the cytokinin signaling pathway to modulate the sensitivity of cytokinins. The main response genes involved in cytokinin signaling, type-B ARRs (ARR1, 10, and 12), directly bind and activate the homeodomain transcription factor WUSCHEL (WUS), which positively governs cell proliferation in the organizing center (OC) under a very high concentration of cytokinins50,51. In the peripheral area where WUS is expressed, multiple feedback systems mediated by hormonal components and transcription factors act in parallel to control the fate of meristems. WUS represses the type-A ARRs ARR7 and 15 to positively strengthen cytokinin response sensitivity in the plant (Fig. 2)52. CLAVATA3 (CLV3), together with CLV1 and CLV2, forms a receptor complex that can also determine the size of the meristem by limiting the expression of WUS. In addition, WUS directly activates the expression of CLV3 in the CZ, forming a negative-regulation feedback loop53. This local WUS–CLV3 feedback loop ensures a constant number of stem cells in the SAM (Fig. 2)54,55. Furthermore, a signaling pathway composed of ERECTA family receptors and epidermal characteristic factor-like ligands can limit the width of the SAM and promote leaf initiation by inhibiting the expression of CLV3 and WUS56,57. Compared with wild-type Arabidopsis seedlings, mutant seedlings lacking all members of the ERECTA family are more sensitive to cytokinin treatment and exhibit increased SAM size and drastic changes in WUS and CLV3 expression; this may occur because the buffer mechanism that maintains stem cell homeostasis against an increase in cytokinins is severely impaired56. Thus, the establishment of cytokinin homeostasis and the cytokinin gradient, as well as spatial signal transduction by cytokinins, play key roles in maintaining the structure of SAMs and their ability to continuously divide and grow.
The spatiotemporal pattern of lateral organ initiation at the SAM is controlled by both auxins and cytokinins58. Changing the auxin/cytokinin ratio requires additional feedback loops that stabilize phyllotaxis. During the initiation of leaf primordia, ERECTA family receptors not only inhibit the effect of cytokinins but also promote the formation of leaf primordia by increasing the expression of PIN-FORMED 1 (PIN1), which increases polar auxin transport59. Auxins acting through MONOPTEROS (MP), an auxin-responsive transcription factor, activate the cytokinin signaling inhibitor ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6) to block the cytokinin signaling pathway (Fig. 2)60. AHP6 is not uniformly distributed in the SAM, resulting in different active regions of cytokinins within the SAM. This small spatial difference causes changes in the ratio of auxins to cytokinins that maintain the orderly production of leaf primordia60,61. However, studies have shown that cytokinins are a prerequisite for leaf initiation. Leaf initiation in tomato shoot apexes ceases in darkness but resumes under light or under cytokinin (zeatin) application to the summit of the meristem62. Cytokinins play two roles in this process: (1) promoting the growth of meristems to provide a source of stem cells as a prerequisite for leaf initiation and (2) affecting the establishment of the auxin gradient by regulating auxin biosynthesis and transport62,63. In monocotyledons, altered phyllotactic patterns are observed in the maize mutant aberrant phyllotaxy1 (abph1)64 and the rice mutant decussate (dec)65. Both ABPH1 and DEC encode proteins that function in cytokinin signaling, and the abph1 and dec mutants have enlarged SAMs. However, mutants with disrupted cytokinin signal transduction pathways do not exhibit a phyllotactic shift22. In the abph1 mutant, maize PIN1 expression and auxins at the incipient leaf primordium are greatly reduced, as cytokinin specifically promotes the expression of maize PIN1 in the incipient leaf primordium66. Collectively, these studies indicate that the phyllotactic shift may be a result of a delay in the initiation of lateral organs stemming from the change in the auxin/cytokinin ratio. In the initial stage of leaf development, the functions of cytokinins are contradictory; they delay the initiation of the leaf development process and suppress the formation of leaf primordia by inhibiting stem cell differentiation. However, cytokinins not only maintain the existence of the SAM but also provide cells for plant apical growth, which is a prerequisite for lateral organ formation and leaf primordium initiation.
Cytokinins control leaf shape
The leaves of most plants have a flat and broad structure to support photosynthesis and gas exchange. Given their adaptations to different natural habitats, leaf forms can be broadly grouped into two categories: simple leaves, which consist of one entire unit with a single lamina, and compound leaves, which consist of multiple subunits called leaflets, each resembling a simple leaf67. Generally, the final shape of leaves has been shown to be determined by two biological processes: primary morphogenesis (PM), which determines the basic leaf form and structures, such as leaflets, lobes, and leaf margins, and secondary morphogenesis (SM), which includes most leaf expansion and differentiation and involves the production of cell types that are typical in mature leaves33. The development of a compound leaf requires the maintenance of temporospatial morphogenetic activity in the early stage of leaf development. For example, at the leaf margin, a region called the marginal blastozone (MB) is responsible for the organogenesis of structures such as lobes in simple leaves or leaflets in compound leaves68. In this case, the meristematic or stem cell identity in the MB or the marginal regions of leaflets needs to be maintained longer than that in the sinus regions to support compound leaf formation.
For prolonged activity in the MB and leaflet formation, cytokinins are involved in the maintenance of extended morphogenetic activity69. Increasing or decreasing endogenous cytokinin levels or readjusting cytokinin sensitivity in the developing leaf marginal meristem alters leaf complexity70. This change has been linked to the timely maintenance of morphogenetic capacity and regulation of cell proliferation by cytokinins along the margins of developing leaves69,71. During the formation of compound leaves, cytokinins also interact with auxins. The discrete distribution of the auxin response in the leaf margin is the key factor in the formation of compound leaves70. Both the local application of auxin in the developing leaf primordium and the increase in auxin sensitivity inhibit the supercompound leaf phenotype, which develops owing to an increased cytokinin concentration69.
In monocotyledons and dicotyledons, various mechanisms of compound leaf formation have been identified; some of them are common in both types of plants, whereas others have not been observed in dicotyledons. Previous studies have shown that the mechanisms regulating compound leaf development in dicotyledonous plant species, such as tomato, pea, Cardamine, and Medicago, are not entirely consistent. During simple leaf development, the downregulation of the KNOX I gene in leaf primordia is permanent. Unlike in the development process for simple leaves, KNOX I expression during compound leaf development is upregulated in the leaf primordium after leaf initiation, which leads to leaflet development; KNOX I expression eventually ceases, which leads to the acceleration of leaf maturation72,73. Overexpression or silencing KNOX I results in increased or decreased leaflet numbers in alfalfa74 and tomato plants19,75, respectively. During PM, KNOX I promotes cytokinin biosynthesis, similar to its effect in SAMs69,76. The reductions in cytokinin levels suppress the effect of KNOX I on leaf shape, and cytokinins can substitute for KNOX I activity at the leaf margin. Thus, cytokinins act downstream of KNOX I in the leaf margin (Fig. 2)69. Similar regulatory mechanisms that involve elevated cytokinins have also been shown to give rise to dissected or deeply lobed leaf morphogenesis in Araceae, a monocotyledonous family.
Increasing sensitivity to cytokinins has a similar effect on leaf morphology in dicots as elevating the level of cytokinins. One study has indicated that a change in the sensitivity of cytokinin signal transduction can affect leaf shape in tomato77. The CIN-TCP transcription factor family affects leaf shape by promoting differentiation. Overexpression of the CIN-TCP family gene LANCEOLATE (LA) in tomato leads to premature leaf differentiation and the production of smaller leaflets77. CIN-TCP in Arabidopsis inhibits cytokinin signal transduction and advances leaf cells to the expansion stage78. The decrease in MB activity in tomato may be mediated by the same mechanism of CIN-TCP regulation as in Arabidopsis. This mechanism has not been reported in Cardamine, pea, or Medicago. However, UNIFOLIATA (UNI)79 and SINGLE LEAFLET1 (SGL1)80, which are homologs of LEAFY (LFY) in Arabidopsis that promote the cytokinin effect by inhibiting type-A ARR7 expression81, have been reported to affect leaf shapes in pea and Medicago, respectively. UNI is expressed in the leaf blastozone and plays an active role in maintaining the blastozone. Both pea uni79 and Medicago sgl180 mutants exhibited reduced leaf complexity because of the previously initiated differentiation of leaf cells. Therefore, UNI and SGL1 may regulate compound leaf development by altering cytokinin signaling. In tomato, other genes have been found to alter leaf morphology through cytokinin signaling. For example, the CLAUSA (CLAU) gene encodes an MYB transcription factor that regulates various aspects of tomato plant development. The tomato clau mutant is characterized by ectopic meristematic activity in leaves, which are highly complex and have many more secondary leaflets than wild-type tomato leaves82. Recent research has shown that CLAU attenuates cytokinin signaling by upregulating the expression of type-A ARRs, which negatively regulate the cytokinin response71. CLAU is of particular spatiotemporal relevance in compound leaves with regard to the cytokinin signaling pathway (Fig. 2). The HAIRY MERISTEM (HAM) gene encodes a GRAS family transcription factor that functions in meristem maintenance in compound leaf primordia83,84. In tomato, ham mutants exhibit overproliferation of meristematic cells in the compound leaf rachis; this phenomenon resembles the response to the elevation of cytokinin levels or sensitivity, leading to the proliferation of the ectopic shoot on the adaxial side of the compound leaf rachis83. The reduction in cytokinin levels in the ham mutant leaves completely suppresses the overproliferation phenotype. Thus, HAM, along with cytokinins, is required for the proper morphogenesis of the compound leaf (Fig. 2)83. In short, the final morphological development of leaves is likely accomplished through multiple concurrent regulatory mechanisms, and the temporospatial elevation of cytokinins and the altered sensitivities of their signal pathways in different leaf areas likely contribute greatly to compound leaf formation.
There are also other regulatory mechanisms related to compound leaf formation in monocotyledons. For instance, it has been reported that in the transition from cell division to cell expansion, the palm leaf primordium undergoes the second stage of leaflet separation, in which the number of cells on the folds of the ridge decreases. As a result, when the leaflets expand, mechanical force eventually separates them, resulting in the development of pinnate compound leaves85. In the other case, programmed cell death (PCD) has been suggested to play a role in the dissection of Monstera leaves; specifically, PCD causes perforations between two adjacent lateral veins during the early period of leaf expansion, and these perforations become enlarged as the leaf grows86. In brief, simple leaves subdivide into compound leaves through three mechanisms: (1) controlling marginal growth, which occurs in both dicotyledons and monocotyledons; (2) tissue abscission, which occurs in monocotyledons; and (3) PCD, which also occurs in monocotyledons. At present, cytokinins are known to be involved in controlling leaf marginal growth but not in leaf dissection through tissue abscission or PCD.
Recent studies have shown that cytokinins can affect the morphological development of simple leaves in monocotyledons. The leaf of a monocotyledon is composed of a leaf blade, ligule, auricle, and leaf sheath. In maize, the semidominant HAIRY SHEATH FRAYED1 (Hsf1) mutation displays a mutant leaf phenotype that resembles the leaf pattern at the sheath-blade boundary, with outgrowths consisting of an organized auricle/ligule and a sheath emanating from the distal blade margin. Analysis of three independent Hsf1 alleles revealed gain-of-function missense mutations in the maize cytokinin receptor ZEA MAYS HISTIDINE KINASE1 (ZmHK1); the mutated residues near the cytokinin binding pocket enhance the cytokinin binding affinity and thus cause changes in the leaf pattern. Treating wild-type seedlings with exogenous cytokinins gives rise to the leaf phenotype of the Hsf1 mutant87. Thus, cytokinins can influence the specifications of leaf patterning and alter leaf developmental programs in monocotyledons.
Among the various regulatory mechanisms that have been found to change leaf morphology, most increase the cytokinin concentration or cytokinin sensitivity to promote cell division activity in some leaf areas and ultimately result in dissected or altered leaf morphology in leaves. However, how these mechanisms function coordinately in through space and time and adapt to different environmental cues needs to be studied in greater detail.
The relationship between cytokinins and leaf size
The size of a mature leaf is largely determined by the leaf cell numbers and cell sizes. After a leaf primordium emerges from the PZ as a rod-shaped protrusion at the flanking region of the SAM, all leaf cells undergo two biological processes, cell division and cell expansion, which determine cell numbers and cell sizes, respectively88,89. In addition, the timing of the transition between cell division and cell expansion indirectly affects leaf size90. In dicotyledonous plants, such as the model plant Arabidopsis thaliana, leaf growth is most often described as being influenced temporally by cell division. When the leaf primordium attains a certain size and position, leaf cell division begins to arrest at the distal tip of the leaf, which is termed the arrest front88. The arrest front boundary then moves downwards; when it reaches a certain point in the midleaf area, it stops for some time (generally a few days) before moving to the bottom of the leaf base91,92. After that, all leaf blade cells rapidly become committed to cell expansion92 (Fig. 3). In contrast, leaf development in monocotyledons is often seen as spatially regulated; that is, cell division occurs primarily at the base of a leaf, cell expansion in the middle of the leaf, and cell maturation at the tip of the leaf93. Therefore, the temporospatial distribution of cytokinins and their functional loci during leaf development may be quite different and determine the final leaf size.
Fig. 3. Schematic diagram of cytokinin functions in different stages of leaf development.
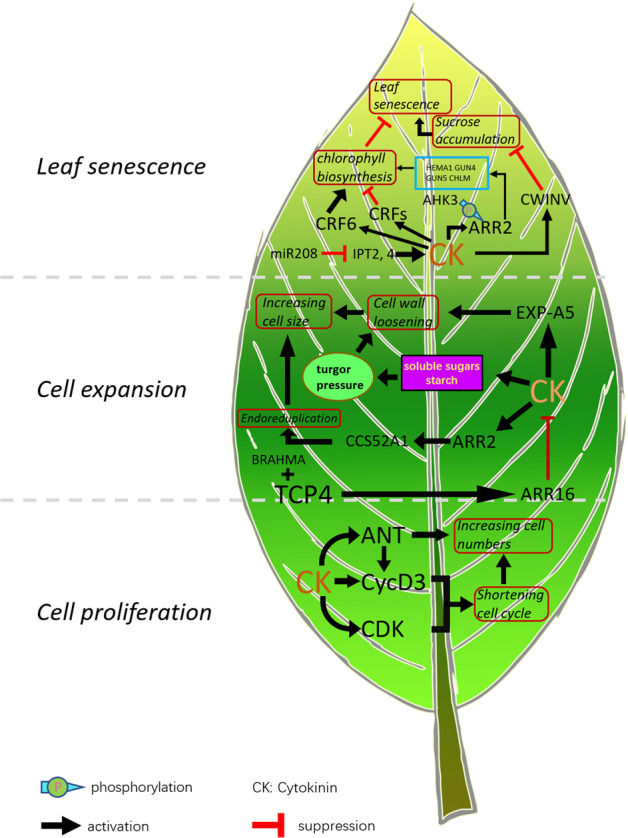
The gray dashed lines divide the three stages of leaf development; the bright green area represents cells in the proliferation stage; the green area represents cells in the expansion stage; and the yellow-green area represents the senescence stage. The terms inside the red rounded rectangles are biological functions; the genes in the blue box are involved in chlorophyll synthesis. The solid arrows and blocked bars indicate activation and suppression, respectively
Cytokinins control leaf size by regulating both leaf cell division and expansion; this has been known for many years94–97. During the leaf cell division phase, cytokinins, together with auxin, activate cell proliferation (Fig. 3)98,99. For instance, a cell culture study with cells in suspension showed that cell division arrests without auxin and that the addition of auxin to the arrested cell culture restores cell division activity99, suggesting that auxin provides the necessary signal that allows cells to enter the cell cycle100–102. However, cytokinins shorten the transition between any two adjacent cell cycle phases103 and extend the period of cell proliferation by delaying the onset of cell differentiation. Conversely, the onset of cell differentiation is brought about by cytokinin degradation caused by the upregulation of CKX3, which slows cell proliferation but expedites the onset of cell expansion104,105. After leaf development enters the cell expansion period, an excess of cytokinins stimulates cell expansion, resulting in plants with larger leaves that consist of larger cells, which leads to higher shoot biomass104,106,107. During leaf cell expansion, cytokinins are responsible for cell wall elongation108, turgor pressure increases104, and endoreduplication109. Endoreduplication, also known as endoreplication, gives rise to cells with extra copies of genomic DNA110 and contributes to the sizes of some specific types of cells in certain plant species during leaf expansion. Furthermore, auxin is also involved in cell expansion. Auxins are known to promote cell wall loosening and endoreduplication; this topic has been reviewed by Tsukaya111 and Perrot-Rechenmann99. In other tissues, such as roots, cell expansion is probably and mainly determined by the crosstalk between cytokinins and auxins112. However, how the temporospatial distribution and interaction of cytokinins and auxins change during the expansion phase of leaf cells remains largely unknown. In shaded environments, which significantly decrease the cytokinin contents in young, fully developed, and mature leaves, leaf size growth is impeded by a lack of cell expansion. Shaded environments reduce leaf expansion; however, when shaded leaves are treated with exogenous cytokinins, they achieve the same size as leaves grown under normal light conditions113. Defects in cytokinin signaling also result in reduced cell expansion. The sizes of the cotyledons of the triple crf1,2,5 mutant are much smaller than those of the wild type, by nearly 96%; this is due primarily to the decline in cell expansion114. In conclusion, multifaceted cytokinin activities at distinct phases of leaf development shape the ultimate sizes of leaves. Recent studies have begun to unveil the underlying mechanisms by which cytokinins regulate cell division and expansion.
In the cell division phase, one of the mechanisms by which cytokinins control cell mitosis in leaf development is by modulating the expression of D3-TYPE CYCLINS (CycD3)115, CYCLIN-DEPENDENT KINASES (CDKs), and AINTEGUMENTA (ANT); CycD3, CDKs, and ANT encode a cell cycle regulatory protein116, serine/threonine kinases103, and a transcription factor117, respectively. During the cell proliferation stage of leaf development, cytokinins control cell division, which activates the G1/S and G2/M transitions in the cell cycle by promoting the expression of CycD398 and CDKs103, respectively (Fig. 3). The more rapid transition from the G1 phase of the cell cycle to the S phase is crucial for the upregulation of eukaryotic cell proliferation118. CycD3 is a cell cycle regulatory protein that binds and activates CDK. The overexpression of CycD3 is sufficient to induce cytokinin-independent shoot formation in the calli98, and the loss of CycD3 activity reduces the ability of exogenous cytokinins to induce shoot formation115. CycD3 promotes mitotic cell division and inhibits endoreduplication and cell differentiation. Therefore, CycD3 is considered the main means by which cytokinins interact with cell cycle control mechanisms. ANT is required for normal cell proliferation but not cell growth119. In situ hybridization experiments have shown that ANT mRNA accumulates in the primordia of all lateral bud organs. Shortly after the appearance of the primordium, ANT mRNA is localized in the growing regions of immature organs and functions120. The overexpression of ANT results in an increase in leaf and flower size, while loss-of-function ant mutants produce smaller leaves119. In response to exogenous cytokinins, ANT transcript levels increased relative to those in untreated plants (Fig. 3)117. Cytokinin deficiency in CKX-overexpression lines or in ipt mutants leads to a reduction in ANT transcript levels during root secondary growth or early leaf proliferation104,117. Some genes control leaf size by regulating cytokinin accumulation or signaling pathways to change the number of leaf cells and ultimately change the size of the leaves. GROWTH-REGULATING FACTORs (GRFs) are a gene family of transcription factors that regulate various aspects of plant growth and development. In a variety of plant species, the overexpression of most GRF genes leads to the enlargement of lateral organs. In A. thaliana, the number of leaf cells in transgenic plants with GRF5 overexpression increased, and the leaves became larger. The expression of CRF2, which is a gene downstream of GRF5, is significantly upregulated, and the corresponding sensitivity of cytokinins is increased121. In poplar, GRF5-1 can bind to the promoter of CKX1 and inhibit its expression. The cytokinin concentration in the apical buds and immature leaves of GRF5-1-overexpressing transgenic plants increased, which increased the number of mesophyll cells and leaf area122. In brief, during the leaf cell proliferation stage, cytokinins increase the number of leaf cells by promoting the mitotic replication cycle and accelerating cell division in different facets, thereby regulating leaf size.
Cytokinins are also involved in the cellular state transition from cell division to cell expansion. The TEOSINTE BRANCHED1/CYCLOIDEA/PCF transcription factor family (TCP), which targets growth-related genes, is composed of two classes that antagonistically control leaf growth in a spatially restricted manner78,123. TCP4, belonged to class II proteins (also known as CIN-TCPs), promotes the change from leaf cell division to leaf expansion by activating cell differentiation and accelerating the progression of the cell cycle arrest front78,107. CIN-TCPs recruit BRAHMA, a component of the SWI/SNF chromatin remodeling complex, to bind the promoter of ARABIDOPSIS RESPONSE REGULATOR16 (ARR16), which is a type-A negative regulator of cytokinin response, and activate the expression of ARR16107. Thus, the reduction in sensitivity to cytokinins due to the expression of ARR16 is thought to be associated with differentiation in leaf growth (Fig. 3). CIN-TCP genes control cell division arrest in the early stage of leaf development and thereby ensure that the leaves remain flat. A reduction in CIN-TCPs expression results in delayed basal node progression and increased cell proliferation before ultimately blocking mitosis78. In short, the blockage of cytokinin signaling is critical to the formation of the arrest front boundary.
In the cell expansion phase, cytokinins are shown to be involved in at least three mechanisms that contribute to the final cell size. During this phase, cell walls undergo loosening, remodeling, and biosynthesis124. Growing plant cells characteristically exhibit faster cell elongation under acidic conditions, which are induced by auxin through the stimulation of plasma membrane H+-ATPase proton pump activity125,126. Expansins are cell wall proteins that induce pH-dependent wall extension and stress relaxation and comprise a large superfamily with at least two major branches (identified as α-expansins (EXPA) and β-expansins (EXPB))127. The interplay between cytokinins and expansins in cell growth has been reported in a few plant species, such as Arabidopsis128, Melilotus108, soybean129, Rosa130, and poplar131. In poplar trees, the highest levels of EXPA3 mRNA are observed in young leaves that will expand in size. Furthermore, the expression of EXPA3 is inducible by exogenous cytokinins131. In addition, a recent study showed that CKX2 activation induces the expression of EXPA5 within 3 h (Fig. 3)104. In Arabidopsis, constitutive overexpression of CKX1 leads to a decrease in the number of leaf cells but an increase in leaf cell size40; these responses might be a compensatory mechanism related to the enhancement of postmitotic cell expansion in response to a decrease in cell number during lateral organ development132. In the root growth zone of Arabidopsis, both cytokinins and auxins can induce the expression of certain expansin genes133. Presumably, this is true in leaves as well, and the underlying activation mechanisms are worth investigating. Proteome profiling of leaves with excessive cytokinin during the cell expansion phase revealed that carbohydrate metabolism and energy-associated processes were stimulated. These processes result in significantly increased contents of major soluble sugars and starch in response to an excess of cytokinins104, which increases turgor pressure and is required for the biochemical loosening of the cell wall (Fig. 3)127. Endoreduplication, the increase in ploidy by chromosome replication without subsequent cell division, is also often involved in the process of increasing cell sizes134. In Arabidopsis, type-B ARR2 binds to and activates the CCS52A1 gene (null function alleles of which reduce endoreduplication expansion in leaf cells109) and promotes the onset of the endocycle135. In cytokinin receptor mutants, CCS52A1 expression was reduced136; consequently, reduced cell endoreduplication was observed135 (Fig. 3). In summary, cytokinins increase cell size in plants by promoting cell wall elongation, increasing turgor pressure, and enhancing endoreduplication.
In conclusion, cytokinins regulate the rate of cell division, the time of transition, and the extent of cell expansion, thereby affecting the numbers and sizes of cells and eventually the leaf size. However, the modes of action of cytokinins during the above processes remain largely unclear or unknown, and further research should focus on the molecular and biochemical mechanisms underlying these processes.
Cytokinins delay leaf senescence
The final stage of leaf development is senescence, which can significantly affect the survival, health, and productivity of plants during the growing season. Senescence is characterized by color changes in both perennial and annual plants in the late summer and throughout autumn. In this phase, the most perceptible phenotypic change that embodies senescence is the appearance of variegated leaves, which develop due to the disassembly of chloroplasts and the degradation of proteins, lipids, nucleic acids, and pigments111,137. The nutrients that are generated from the degradation of senescing leaves are transported to developing seeds and fruits in annual plants or to new leaves or flowers in perennial trees, resulting in the death of the senescing leaves137,138. Therefore, although leaf senescence is an adverse process for leaf organs, it represents an altruistic death that plays a vital role in maintaining plant adaptability by ensuring the production of fit offspring and improving plant survival with a given spatiotemporal ecological niche139. Leaf senescence is influenced by various endogenous signals (plant hormones and age) and environmental signals (darkness, shading by other plants, UV-B or ozone exposure, nutrient limitation, extreme temperatures, drought, high salinity, and pathogen attacks)137,140,141. Under most environmental conditions, leaf senescence is initiated and develops due primarily to leaf age. Various plants shed their old leaves at different times during the growing season. Abiotic and biotic stresses can also enhance this process and may reduce plant biomass accumulation.
Leaf senescence is not a passive, unregulated degeneration process. Phytohormones, especially cytokinins142 and ethylene143, have been reported to affect leaf senescence; specifically, cytokinins are thought to delay leaf senescence, whereas ethylene is thought to induce it. Cytokinins are believed to serve as negative regulators of leaf senescence in a variety of monocotyledonous144–147 and dicotyledonous148–151 species. A reduction in cytokinin levels before the onset of senescence is believed to be a key signal for the initiation of senescence142. The exogenous application of cytokinins or the transgenic expression of cytokinin biosynthesis genes prevents the degradation of chlorophyll, photosynthetic proteins, and RNA, resulting in delayed senescence (Fig. 3)142,147,152,153. For example, transgenic tobacco with a senescence-specific gene (SAG12) promoter fused with the IPT gene significantly impedes leaf shedding and other symptoms of senescence142. Moreover, because of the prolongation of photosynthetic activity, the biomass of the transgenic plants and their seed productivity were greatly augmented. In tomato, overexpression of the IPT gene (pSAG12::IPT and pSAG13::IPT) inhibits leaf senescence, promotes earlier flowering, and increases fruit weight and total soluble solids154. The increase in biomass accumulation is to a large extent due to the extended photosynthetic period and nutrient transport in senescent leaves. In contrast, cytokinin biosynthesis mutants have a shorter leaf life span148,155. miRNAs are important posttranscriptional regulators of plant growth and development that participate in the process of leaf senescence by modulating cytokinins. Recent studies have shown that a newly identified microRNA (miR208) in tomato that reduces cytokinin biosynthesis by regulating IPT2 and IPT4 post-transcriptionally promotes leaf senescence (Fig. 3)148. Cytokinins can postpone leaf aging processes caused by unfavorable or adverse environmental conditions (such as drought or darkness) and delay the occurrence of leaf senescence146,156. Various experiments on different genes in many plant species have consistently shown that the levels of cytokinins play a major role in both single-leaf and whole-plant senescence.
Although the senescence-delaying effects of cytokinins have been well established, the mechanisms behind this phenomenon remain largely unknown. However, recent research on cytokinin signal transduction has shed new light on the underlying mechanisms. Cytokinin signal transduction genes such as AHK3, type-B ARR2, and CRFs are closely associated with the development of leaf senescence157,158. In Arabidopsis, the overexpression of either AHK3 or ARR2 leads to a delay in leaf senescence. However, the overexpression of ARR2 without the AHK3 phosphorylation site does not lead to phenotypic changes. This result suggests that AHK3 plays a major role in controlling cytokinin-mediated leaf longevity through the specific phosphorylation of the response regulator ARR2 (Fig. 3)157. Additional research has shown that in the akh2,3 double mutant, the expression levels of chlorophyll synthesis genes HEMA1, GUN4, GUN5, and CHLM were reduced, which suggests that cytokinins can increase chlorophyll content and delay leaf senescence (Fig. 3)159. CRFs, which encode cytokinin signal transduction transcription factor family genes, are induced by cytokinins and exhibit different biological functions during the process of leaf senescence regulation. In dark-induced excised-leaf assays, leaves overexpressing CRF6 retain more chlorophyll than the wild type leaves without exogenous cytokinins, indicating that CRF6 negatively regulates leaf senescence158. However, the growth phenotype of the crf6 mutant line did not differ from that of the wild type, and no premature senescence was observed. In contrast, leaves of CRF1-, CRF3-, and CRF5-overexpressing transgenic lines developed senescence earlier than the wild type leaves, while the crf1,3,5,6 and crf1/CRF1,2,5,6 mutants exhibited delayed leaf senescence160. These results may be caused by differences in the regulation of plant development by cytokinins in different environments. Sucrose accumulation is an important signal for the induction of leaf senescence137. During plant development, young leaves (a substantial sink) need the hexoses provided by old leaves (a source) as energy until the young leaves are mature enough to supply themselves with energy through photosynthesis. After the demand from other organs decreases, sucrose then gradually accumulates in the old leaves and induces senescence140. Cytokinins have long been shown to accelerate nutrient mobilization during the establishment of new source-sink relationships161. Cytokinins are known to increase the activity of cell wall invertase (CWINV), which plays a key role in regulating the source-sink relationship and is considered one of the main modulators of sink activity162–165. The CWINV enzyme catalyzes the cleavage of sucrose into hexose monomers on the cell wall and transports them to sink organs. The increase in CWINV activity reduces the accumulation of sucrose in old leaves, which delays leaf senescence. Restricting the activity of CWINV has been shown to block the function of cytokinins in inhibiting leaf senescence162. However, the interrelationship between cytokinins and CWINV with regard to leaf senescence can be complicated by other factors; thus, more experiments are needed to characterize their effects on leaf senescence. In conclusion, cytokinin signal transduction genes delay the biological process of leaf senescence by increasing the chlorophyll content and reducing sucrose accumulation in leaves.
Concluding remarks
Given the urgent demand for renewable energy and the increased interest in bioenergy, understanding the biochemical and molecular mechanisms underlying leaf development has become increasingly important; such studies can help scientists find a way to increase plant growth and biomass in various agricultural crops and woody plant species. In this review, we summarized the roles of cytokinins in various leaf developmental stages. At the initial stage of leaf development, cytokinins sustain the growth of SAMs and provide sufficient stem cells for the leaf primordium to protrude. Cytokinins then regulate the synthesis and transport of auxin to promote the emergence of leaf primordia. At later stages, cytokinins promote cell proliferation, which increases the number of leaf cells in a short period and represses the transition of leaf cells into the expansion stage. The development of leaves from simple to compound morphology is a complicated process that is still not fully understood. Nevertheless, it is clear that cytokinins play a role in this process by maintaining the activity of leaf margin meristems. After leaf cells develop and grow into the expansion stage, cytokinins promote cell expansion by promoting cell wall elongation, increasing turgor pressure, and enhancing endoreduplication. In the final stage of leaf development, cytokinins maintain chlorophyll synthesis while slowing the process of leaf senescence. Thus, cytokinins play an important role during the entire process of leaf development. However, as most of the underlying molecular mechanisms remain unclear, there is a need for more work to be done to advance our understanding of these mechanisms. Cytokinins shorten the cell replication cycle by activating the transcription of CycD3, but little is known about the factors that mediate between cytokinins and CycD3. ARR and CRF genes, which can activate or inhibit the transcription of downstream genes, belong to large gene families, and which of these genes are regulated by these transcription factors at different stages of leaf development remains to be determined. Excessive cytokinins can change the leaf morphology of both monocotyledons and dicotyledons, but whether the leaves of these two types of plants are regulated by the same molecular mechanism requires further study. Plant hormones such as cytokinin, auxin, ethylene, brassinosteroid, gibberellins, abscisic acid, jasmonic acid, and others interact with each other and affect all stages of leaf development. Understanding the relationships between the ratio of these hormones and leaf development will benefit plant tissue culture and provide new insights into plant development. Therefore, more focused studies on the mechanisms of action of cytokinins and their synergistic interactions with other hormones are needed to advance our understanding of leaf development.
Author contributions
W.W. and H.W. planned the outline of the review. W.W. and K.D. collected the available literature and completed the first draft of the paper. H.W. and X.K. carefully compiled and revised the paper. All authors approved the final paper.
Conflict of interest
The authors declare no competing interests.
Contributor Information
Xiangyang Kang, Email: kangxy@bjfu.edu.cn.
Hairong Wei, Email: hairong@mtu.edu.
References
- 1.Miller CO, Skoog F, Von Saltza MH, Strong FM. Kinetin, a cell division factor from deoxyribonucleic acid. J. Am. Chem. Soc. 1955;77:1392–1392. doi: 10.1021/ja01610a105. [DOI] [Google Scholar]
- 2.Skoog F, Strong FM, Miller CO. Cytokinins. Science. 1965;148:532–533. doi: 10.1126/science.148.3669.532-a. [DOI] [PubMed] [Google Scholar]
- 3.Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 4.Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- 5.Taya Y, Tanaka Y, Nishimura S. 5'-AMP is a direct precursor of cytokinin in Dictyostelium discoideum. Nature. 1978;271:545–547. doi: 10.1038/271545a0. [DOI] [PubMed] [Google Scholar]
- 6.Houba-Herin N, Pethe C, d'Alayer J, Laloue M. Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J. 1999;17:615–626. doi: 10.1046/j.1365-313X.1999.00408.x. [DOI] [PubMed] [Google Scholar]
- 7.Schmulling T, Werner T, Riefler M, Krupkova E, Bartrina y Manns I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003;116:241–252. doi: 10.1007/s10265-003-0096-4. [DOI] [PubMed] [Google Scholar]
- 8.Brzobohaty B, et al. Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science. 1993;262:1051–1054. doi: 10.1126/science.8235622. [DOI] [PubMed] [Google Scholar]
- 9.Hou B, Lim E-K, Higgins GS, Bowles DJ. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 2004;279:47822–47832. doi: 10.1074/jbc.M409569200. [DOI] [PubMed] [Google Scholar]
- 10.Kamada-Nobusada T, Sakakibara H. Molecular basis for cytokinin biosynthesis. Phytochemistry. 2009;70:444–449. doi: 10.1016/j.phytochem.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Hirose N, et al. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008;59:75–83. doi: 10.1093/jxb/erm157. [DOI] [PubMed] [Google Scholar]
- 12.Kudo T, Kiba T, Sakakibara H. Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 2010;52:53–60. doi: 10.1111/j.1744-7909.2010.00898.x. [DOI] [PubMed] [Google Scholar]
- 13.Girke C, Daumann M, Niopek-Witz S, Mohlmann T. Nucleobase and nucleoside transport and integration into plant metabolism. Front Plant Sci. 2014;5:443. doi: 10.3389/fpls.2014.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borghi L, Kang J, Ko D, Lee Y, Martinoia E. The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 2015;43:924–930. doi: 10.1042/BST20150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wulfetange K, et al. The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 2011;156:1808–1818. doi: 10.1104/pp.111.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. Ligand-binding properties and subcellular localization of maize cytokinin receptors. J. Exp. Bot. 2011;62:5149–5159. doi: 10.1093/jxb/err220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki T, Imamura A, Ueguchi C, Mizuno T. Histidine-containing phosphotransfer (HPt) signal transducers implicated in His-to-Asp phosphorelay in Arabidopsis. Plant Cell Physiol. 1998;39:1258–1268. doi: 10.1093/oxfordjournals.pcp.a029329. [DOI] [PubMed] [Google Scholar]
- 18.To JPC, et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shani E, et al. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell. 2009;21:3078–3092. doi: 10.1105/tpc.109.068148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieber, J. J. & Schaller, G. E. Cytokinin signaling in plant development. Development145, dev149344 (2018). [DOI] [PubMed]
- 21.Mason MG, et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Z, et al. Hormonal control of the shoot stem-cell niche. Nature. 2010;465:1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]
- 23.Hwang I, Sheen J, Muller B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- 24.Zurcher E, Muller B. Cytokinin synthesis, signaling, and function–advances and new insights. Int. Rev. Cell Mol. Biol. 2016;324:1–38. doi: 10.1016/bs.ircmb.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Werner T, Schmulling T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009;12:527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Kalve S, De Vos D, Beemster GTS. Leaf development: a cellular perspective. Front Plant Sci. 2014;5:362. doi: 10.3389/fpls.2014.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukaya H. Leaf development. Arabidopsis Book. 2013;11:e0163. doi: 10.1199/tab.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wojciechowska N, Sobieszczuk-Nowicka E, Bagniewska-Zadworna A. Plant organ senescence - regulation by manifold pathways. Plant Biol. (Stuttg.) 2018;20:167–181. doi: 10.1111/plb.12672. [DOI] [PubMed] [Google Scholar]
- 29.Du F, Guan C, Jiao Y. Molecular mechanisms of leaf morphogenesis. Mol. Plant. 2018;11:1117–1134. doi: 10.1016/j.molp.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Nakata M, Okada K. The leaf adaxial-abaxial boundary and lamina growth. Plants (Basel) 2013;2:174–202. doi: 10.3390/plants2020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sluis A, Hake S. Organogenesis in plants: initiation and elaboration of leaves. Trends Genet. 2015;31:300–306. doi: 10.1016/j.tig.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Shi B, Vernoux T. Patterning at the shoot apical meristem and phyllotaxis. Curr. Top. Dev. Biol. 2019;131:81–107. doi: 10.1016/bs.ctdb.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Bar M, Ori N. Leaf development and morphogenesis. Development. 2014;141:4219–4230. doi: 10.1242/dev.106195. [DOI] [PubMed] [Google Scholar]
- 34.Reinhardt D. Regulation of phyllotaxis. Int. J. Dev. Biol. 2005;49:539–546. doi: 10.1387/ijdb.041922dr. [DOI] [PubMed] [Google Scholar]
- 35.Lee ZH, Hirakawa T, Yamaguchi N, Ito T. The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int. J. Mol. Sci. 2019;20:4065. doi: 10.3390/ijms20164065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaller GE, Bishopp A, Kieber JJ. The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell. 2015;27:44–63. doi: 10.1105/tpc.114.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Showk S, Ruonala R, Helariutta Y. Crossing paths: cytokinin signalling and crosstalk. Development. 2013;140:1373–1383. doi: 10.1242/dev.086371. [DOI] [PubMed] [Google Scholar]
- 38.Wolters H, Jurgens G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009;10:305–317. doi: 10.1038/nrg2558. [DOI] [PubMed] [Google Scholar]
- 39.Miyawaki K, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl Acad. Sci. USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner T, et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higuchi M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl Acad. Sci. USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng L, et al. Functional divergence and adaptive selection of KNOX gene family in plants. Open Life Sci. 2020;15:346–363. doi: 10.1515/biol-2020-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 44.Tsuda K, Hake S. Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 2015;27:91–96. doi: 10.1016/j.pbi.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Scofield S, Dewitte W, Nieuwland J, Murray JAH. The Arabidopsis homeobox gene SHOOT MERISTEMLESS has cellular and meristem-organisational roles with differential requirements for cytokinin and CYCD3 activity. Plant J. 2013;75:53–66. doi: 10.1111/tpj.12198. [DOI] [PubMed] [Google Scholar]
- 46.Yanai O, et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005;15:1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 47.Rupp HM, Frank M, Werner T, Strnad M, Schmulling T. Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J. 1999;18:557–563. doi: 10.1046/j.1365-313X.1999.00472.x. [DOI] [PubMed] [Google Scholar]
- 48.Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM. Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl Acad. Sci. USA. 2012;109:4002–4007. doi: 10.1073/pnas.1200636109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuroha T, et al. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21:3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang T-Q, et al. A two-step model for de Novo activation of WUSCHEL during plant shoot regeneration. Plant Cell. 2017;29:1073–1087. doi: 10.1105/tpc.16.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindsay DL, Sawhney VK, Bonham-Smith PC. Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in Arabidopsis. Plant Sci. (Amst., Neth.) 2006;170:1111–1117. [Google Scholar]
- 52.Leibfried A, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 53.Yadav RK, et al. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 55.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 56.Uchida N, Shimada M, Tasaka M. ERECTA-family receptor kinases regulate stem cell homeostasis via buffering its cytokinin responsiveness in the shoot apical meristem. Plant Cell Physiol. 2013;54:343–351. doi: 10.1093/pcp/pcs109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, L., DeGennaro, D., Lin, G., Chai, J. & Shpak, E. D. ERECTA family signaling constrains CLAVATA3 and WUSCHEL to the center of the shoot apical meristem. bioRxiv 2020.02.24.962787 10.1101/2020.02.24.962787 (2020). [DOI] [PubMed]
- 58.Bartlett ME, Thompson B. Meristem identity and phyllotaxis in inflorescence development. Front Plant Sci. 2014;5:508. doi: 10.3389/fpls.2014.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen M-K, Wilson RL, Palme K, Ditengou FA, Shpak ED. ERECTA family genes regulate auxin transport in the shoot apical meristem and forming leaf primordia. Plant Physiol. 2013;162:1978–1991. doi: 10.1104/pp.113.218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Besnard F, et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature. 2014;505:417–421. doi: 10.1038/nature12791. [DOI] [PubMed] [Google Scholar]
- 61.Besnard, F., Rozier, F. & Vernoux, T. The AHP6 cytokinin signaling inhibitor mediates an auxin-cytokinin crosstalk that regulates the timing of organ initiation at the shoot apical meristem. Plant Signal. Behav.9, https://www.ncbi.nlm.nih.gov/pubmed/24732036. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4091322/pdf/psb-9-e28788.pdf (2014). [DOI] [PMC free article] [PubMed]
- 62.Yoshida S, Mandel T, Kuhlemeier C. Stem cell activation by light guides plant organogenesis. Genes Dev. 2011;25:1439–1450. doi: 10.1101/gad.631211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohammed B, et al. Converging light, energy and hormonal signaling control meristem activity, leaf initiation, and growth. Plant Physiol. 2018;176:1365–1381. doi: 10.1104/pp.17.01730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jackson, D. & Hake, S. Control of phyllotaxy in maize by the abphyl1 gene. Development 126, 315–323, https://www.ncbi.nlm.nih.gov/pubmed/9847245, https://dev.biologists.org/content/develop/126/2/315.full.pdf (1999). [DOI] [PubMed]
- 65.Itoh J-i, Hibara K-i, Kojima M, Sakakibara H, Nagato Y. Rice DECUSSATE controls phyllotaxy by affecting the cytokinin signaling pathway. Plant J. 2012;72:869–881. doi: 10.1111/j.1365-313x.2012.05123.x. [DOI] [PubMed] [Google Scholar]
- 66.Lee B-h, et al. Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 2009;150:205–216. doi: 10.1104/pp.109.137034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogden MS, Lacroix CR. Comparative development of simple and compound leaves in the genus Cecropia. Botany. 2017;95:185–193. doi: 10.1139/cjb-2016-0148. [DOI] [Google Scholar]
- 68.Hagemann W, Gleissberg S. Organogenetic capacity of leaves: the significance of marginal blastozones in angiosperms. Plant Syst. Evol. 1996;199:121–152. doi: 10.1007/BF00984901. [DOI] [Google Scholar]
- 69.Shani E, et al. Cytokinin regulates compound leaf development in tomato. Plant Cell. 2010;22:3206–3217. doi: 10.1105/tpc.110.078253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shwartz I, Levy M, Ori N, Bar M. Hormones in tomato leaf development. Dev. Biol. 2016;419:132–142. doi: 10.1016/j.ydbio.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 71.Bar M, et al. CLAUSA is a MYB transcription factor that promotes leaf differentiation by attenuating cytokinin signaling. Plant Cell. 2016;28:1602–1615. doi: 10.1105/tpc.16.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bharathan G, et al. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 73.Hay A, Tsiantis M. KNOX genes: versatile regulators of plant development and diversity. Development. 2010;137:3153–3165. doi: 10.1242/dev.030049. [DOI] [PubMed] [Google Scholar]
- 74.Champagne CEM, et al. Compound leaf development and evolution in the legumes. Plant Cell. 2007;19:3369–3378. doi: 10.1105/tpc.107.052886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim M, et al. Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development. 2003;130:4405–4415. doi: 10.1242/dev.00655. [DOI] [PubMed] [Google Scholar]
- 76.Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 2006;38:942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- 77.Ori N, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 2007;39:787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- 78.Efroni I, Blum E, Goldshmidt A, Eshed Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell. 2008;20:2293–2306. doi: 10.1105/tpc.107.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gourlay CW, Hofer JM, Ellis TH. Pea compound leaf architecture is regulated by interactions among the genes UNIFOLIATA, cochleata, afila, and tendril-lessn. Plant Cell. 2000;12:1279–1294. doi: 10.1105/tpc.12.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang H, et al. Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 2008;146:1759–1772. doi: 10.1104/pp.108.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chahtane H, et al. A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 2013;74:678–689. doi: 10.1111/tpj.12156. [DOI] [PubMed] [Google Scholar]
- 82.Avivi Y, et al. Clausa, a tomato mutant with a wide range of phenotypic perturbations, displays a cell type-dependent expression of the homeobox gene LeT6/TKn2. Plant Physiol. 2000;124:541–552. doi: 10.1104/pp.124.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hendelman A, Kravchik M, Stav R, Frank W, Arazi T. Tomato HAIRY MERISTEM genes are involved in meristem maintenance and compound leaf morphogenesis. J. Exp. Bot. 2016;67:6187–6200. doi: 10.1093/jxb/erw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Engstrom EM, et al. Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 2011;155:735–750. doi: 10.1104/pp.110.168757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nowak JS, Bolduc N, Dengler NG, Posluszny U. Compound leaf development in the palm Chamaedorea elegans is KNOX-independent. Am. J. Bot. 2011;98:1575–1582. doi: 10.3732/ajb.1100101. [DOI] [PubMed] [Google Scholar]
- 86.Gunawardena AH, Sault K, Donnelly P, Greenwood JS, Dengler NG. Programmed cell death and leaf morphogenesis in Monstera obliqua (Araceae) Planta. 2005;221:607–618. doi: 10.1007/s00425-005-1545-1. [DOI] [PubMed] [Google Scholar]
- 87.Muszynski MG, et al. The Maize Hairy Sheath Frayed1 (Hsf1) mutation alters leaf patterning through increased cytokinin signaling. Plant Cell. 2020;32:1501–1518. doi: 10.1105/tpc.19.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- 89.Tsukaya H. Leaf shape diversity with an emphasis on leaf contour variation, developmental background, and adaptation. Semin Cell Dev. Biol. 2018;79:48–57. doi: 10.1016/j.semcdb.2017.11.035. [DOI] [PubMed] [Google Scholar]
- 90.Gonzalez N, Vanhaeren H, Inze D. Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 2012;17:332–340. doi: 10.1016/j.tplants.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 91.Kazama T, Ichihashi Y, Murata S, Tsukaya H. The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1046–1054. doi: 10.1093/pcp/pcq051. [DOI] [PubMed] [Google Scholar]
- 92.Andriankaja M, et al. Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell. 2012;22:64–78. doi: 10.1016/j.devcel.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 93.Nelissen H, Gonzalez N, Inze D. Leaf growth in dicots and monocots: so different yet so alike. Curr. Opin. Plant Biol. 2016;33:72–76. doi: 10.1016/j.pbi.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 94.Volkenburgh EV. Leaf expansion – an integrating plant behaviour. Plant, Cell Environ. 1999;22:1463–1473. doi: 10.1046/j.1365-3040.1999.00514.x. [DOI] [Google Scholar]
- 95.Engelke AL, Hamzi HQ, Skoog F. Cytokinin-gibberellin regulation of shoot development and leaf form in tobacco plantlets. Am. J. Bot. 1973;60:491–495. doi: 10.1002/j.1537-2197.1973.tb05949.x. [DOI] [Google Scholar]
- 96.Brock TG. Combined effects of hormones and light during growth promotion in primary leaves of Phaseolus vulgaris. Can. J. Bot. 1993;71:501–505. doi: 10.1139/b93-054. [DOI] [Google Scholar]
- 97.Leopold AC, Kawase M. Benzyladenine effects on bean leaf growth and senescence. Am. J. Bot. 1964;51:294–298. doi: 10.1002/j.1537-2197.1964.tb06633.x. [DOI] [Google Scholar]
- 98.Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- 99.Perrot-Rechenmann C. Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2010;2:a001446. doi: 10.1101/cshperspect.a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stals H, Inze D. When plant cells decide to divide. Trends Plant Sci. 2001;6:359–364. doi: 10.1016/S1360-1385(01)02016-7. [DOI] [PubMed] [Google Scholar]
- 101.Trehin C, et al. Cell cycle regulation by plant growth regulators: involvement of auxin and cytokinin in the re-entry of Petunia protoplasts into the cell cycle. Planta. 1998;206:215–224. doi: 10.1007/s004250050393. [DOI] [PubMed] [Google Scholar]
- 102.Inze D, De Veylder L. Cell cycle regulation in plant development. Annu. Rev. Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 103.Zhang K, Diederich L, John PCL. The cytokinin requirement for cell division in cultured Nicotiana plumbaginifolia cells can be satisfied by yeast Cdc25 protein tyrosine phosphatase: implications for mechanisms of cytokinin response and plant development. Plant Physiol. 2005;137:308–316. doi: 10.1104/pp.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Skalak J, et al. Multifaceted activity of cytokinin in leaf development shapes its size and structure in Arabidopsis. Plant J. 2019;97:805–824. doi: 10.1111/tpj.14285. [DOI] [PubMed] [Google Scholar]
- 105.Holst K, Schmulling T, Werner T. Enhanced cytokinin degradation in leaf primordia of transgenic Arabidopsis plants reduces leaf size and shoot organ primordia formation. J. Plant Physiol. 2011;168:1328–1334. doi: 10.1016/j.jplph.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 106.Carabelli M, et al. A novel regulatory circuit underlying plant response to canopy shade. Plant Signal Behav. 2008;3:137–139. doi: 10.4161/psb.3.2.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Efroni I, et al. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell. 2013;24:438–445. doi: 10.1016/j.devcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee A, Giordano W, Hirsch AM. Cytokinin induces expansin gene expression in Melilotus alba Desr. wild-type and the non-nodulating, non-mycorrhizal (NodMyc) mutant Masym3. Plant Signal Behav. 2008;3:218–223. doi: 10.4161/psb.3.4.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Larson-Rabin Z, Li Z, Masson PH, Day CD. FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 2009;149:874–884. doi: 10.1104/pp.108.132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/S0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 111.Tsukaya H. Re-examination of the role of endoreduplication on cell-size control in leaves. J. Plant Res. 2019;132:571–580. doi: 10.1007/s10265-019-01125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moubayidin L, Di Mambro R, Sabatini S. Cytokinin-auxin crosstalk. Trends Plant Sci. 2009;14:557–562. doi: 10.1016/j.tplants.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 113.Pons TL, Jordi W, Kuiper D. Acclimation of plants to light gradients in leaf canopies: evidence for a possible role for cytokinins transported in the transpiration stream. J. Exp. Bot. 2001;52:1563–1574. doi: 10.1093/jexbot/52.360.1563. [DOI] [PubMed] [Google Scholar]
- 114.Rashotte AM, et al. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl Acad. Sci. USA. 2006;103:11081–11085. doi: 10.1073/pnas.0602038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dewitte W, et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl Acad. Sci. USA. 2007;104:14537–14542. doi: 10.1073/pnas.0704166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schaller GE, Street IH, Kieber JJ. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014;21:7–15. doi: 10.1016/j.pbi.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 117.Randall RS, et al. AINTEGUMENTA and the D-type cyclin CYCD3;1 regulate root secondary growth and respond to cytokinins. Biol. Open. 2015;4:1229–1236. doi: 10.1242/bio.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bertoli C, Skotheim JM, de Bruin RAM. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl Acad. Sci. USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elliott RC, et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vercruyssen L, et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell. 2014;26:210–229. doi: 10.1105/tpc.113.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu W, et al. Growth-Regulating Factor 5 (GRF5)-mediated gene regulatory network promotes leaf growth and expansion in poplar. New Phyto. 2021;230:612–628. doi: 10.1111/nph.17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011;68:147–158. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bashline L, Lei L, Li S, Gu Y. Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant. 2014;7:586–600. doi: 10.1093/mp/ssu018. [DOI] [PubMed] [Google Scholar]
- 125.Cosgrove DJ. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- 126.Takahashi K, Hayashi K, Kinoshita T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012;159:632–641. doi: 10.1104/pp.112.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- 128.Pacifici, E., Di Mambro, R., Dello Ioio, R., Costantino, P. & Sabatini, S. Acidic cell elongation drives cell differentiation in the Arabidopsis root. EMBO J. 37, (2018). [DOI] [PMC free article] [PubMed]
- 129.Downes BP, Crowell DN. Cytokinin regulates the expression of a soybean beta-expansin gene by a post-transcriptional mechanism. Plant Mol. Biol. 1998;37:437–444. doi: 10.1023/A:1005920732211. [DOI] [PubMed] [Google Scholar]
- 130.Roman H, Girault T, Le Gourrierec J, Leduc N. In silico analysis of 3 expansin gene promoters reveals 2 hubs controlling light and cytokinins response during bud outgrowth. Plant Signal Behav. 2017;12:e1284725. doi: 10.1080/15592324.2017.1284725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kuluev BR, Knyazev AV, Mikhaylova EV, Chemeris AV. The role of expansin genes PtrEXPA3 and PnEXPA3 in the regulation of leaf growth in poplar. Russ. J. Genet. 2017;53:651–660. doi: 10.1134/S1022795417060084. [DOI] [Google Scholar]
- 132.Hisanaga T, Kawade K, Tsukaya H. Compensation: a key to clarifying the organ-level regulation of lateral organ size in plants. J. Exp. Bot. 2015;66:1055–1063. doi: 10.1093/jxb/erv028. [DOI] [PubMed] [Google Scholar]
- 133.Samalova, M. et al. Expansin-controlled cell wall stiffness regulates root growth in Arabidopsis. bioRxiv 2020.06.25.170969 10.1101/2020.06.25.170969 (2020).
- 134.Czesnick H, Lenhard M. Size control in plants-lessons from leaves and flowers. Cold Spring Harb. Perspect. Biol. 2015;7:a019190. doi: 10.1101/cshperspect.a019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lammens T, et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl Acad. Sci. USA. 2008;105:14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Takahashi N, et al. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr. Biol. 2013;23:1812–1817. doi: 10.1016/j.cub.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 137.Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 138.Bleecker AB, Patterson SE. Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell. 1997;9:1169–1179. doi: 10.1105/tpc.9.7.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Woo HR, Kim HJ, Nam HG, Lim PO. Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 2013;126:4823–4833. doi: 10.1242/jcs.109116. [DOI] [PubMed] [Google Scholar]
- 140.Lim PO, Woo HR, Nam HG. Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 2003;8:272–278. doi: 10.1016/S1360-1385(03)00103-1. [DOI] [PubMed] [Google Scholar]
- 141.Zhang, Y., Wang, H.-L., Li, Z. & Guo, H. Genetic Network between Leaf Senescence and Plant Immunity: Crucial Regulatory Nodes and New Insights. Plants (Basel)9, 10.3390/plants9040495 (2020). [DOI] [PMC free article] [PubMed]
- 142.Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- 143.Hunter DA, Yoo SD, Butcher SM, McManus MT. Expression of 1-aminocyclopropane-1-carboxylate oxidase during leaf ontogeny in white clover. Plant Physiol. 1999;120:131–142. doi: 10.1104/pp.120.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang W, et al. The involvement of cytokinin and nitrogen metabolism in delayed flag leaf senescence in a wheat stay-green mutant, tasg1. Plant Sci. 2019;278:70–79. doi: 10.1016/j.plantsci.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 145.Kang K, Shim Y, Gi E, An G, Paek N-C. Mutation of ONAC096 enhances grain yield by increasing panicle number and delaying leaf senescence during grain filling in rice. Int. J. Mol. Sci. 2019;20:5241. doi: 10.3390/ijms20205241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Janeckova H, et al. Exogenous application of cytokinin during dark senescence eliminates the acceleration of photosystem II impairment caused by chlorophyll b deficiency in barley. Plant Physiol. Biochem. 2019;136:43–51. doi: 10.1016/j.plaphy.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 147.Talla SK, et al. Cytokinin delays dark-induced senescence in rice by maintaining the chlorophyll cycle and photosynthetic complexes. J. Exp. Bot. 2016;67:1839–1851. doi: 10.1093/jxb/erv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang, Y., Yin, S., Tu, Y., Mei, H. & Yang, Y. A novel microRNA, SlymiR208, promotes leaf senescence via regulating cytokinin biosynthesis in tomato. Physiol. Plant. 10.1111/ppl.13068 (2020). [DOI] [PubMed]
- 149.Xu J, et al. Crosstalk between cytokinin and ethylene signaling pathways regulates leaf abscission in cotton in response to chemical defoliants. J. Exp. Bot. 2019;70:1525–1538. doi: 10.1093/jxb/erz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim J, et al. Comparative transcriptome analysis in Arabidopsis ein2/ore3 and ahk3/ore12 mutants during dark-induced leaf senescence. J. Exp. Bot. 2018;69:3023–3036. doi: 10.1093/jxb/ery137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kant S, et al. Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.) PLoS ONE. 2015;10:e0116349. doi: 10.1371/journal.pone.0116349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McCabe, M. S. et al. Effects of P(SAG12)-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol. 127, 505–516, https://www.ncbi.nlm.nih.gov/pubmed/11598225, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC125086/pdf/pp1001000505.pdf (2001). [PMC free article] [PubMed]
- 153.Ori, N. et al. Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell11, 1073–1080, https://www.ncbi.nlm.nih.gov/pubmed/10368178, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC144237/pdf/111073.pdf (1999). [PMC free article] [PubMed]
- 154.Swartzberg D, Dai N, Gan S, Amasino R, Granot D. Effects of cytokinin production under two SAG promoters on senescence and development of tomato plants. Plant Biol. (Stuttg.) 2006;8:579–586. doi: 10.1055/s-2006-924240. [DOI] [PubMed] [Google Scholar]
- 155.Masferrer A, et al. Overexpression of Arabidopsis thaliana farnesyl diphosphate synthase (FPS1S) in transgenic Arabidopsis induces a cell death/senescence-like response and reduced cytokinin levels. Plant J. 2002;30:123–132. doi: 10.1046/j.1365-313X.2002.01273.x. [DOI] [PubMed] [Google Scholar]
- 156.Rivero RM, et al. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl Acad. Sci. USA. 2007;104:19631–19636. doi: 10.1073/pnas.0709453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim HJ, et al. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl Acad. Sci. USA. 2006;103:814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zwack PJ, Robinson BR, Risley MG, Rashotte AM. Cytokinin response factor 6 negatively regulates leaf senescence and is induced in response to cytokinin and numerous abiotic stresses. Plant Cell Physiol. 2013;54:971–981. doi: 10.1093/pcp/pct049. [DOI] [PubMed] [Google Scholar]
- 159.Cortleven A, et al. Cytokinin regulates the etioplast-chloroplast transition through the two-component signaling system and activation of chloroplast-related genes. Plant Physiol. 2016;172:464–478. doi: 10.1104/pp.16.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Raines T, et al. The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 2016;85:134–147. doi: 10.1111/tpj.13097. [DOI] [PubMed] [Google Scholar]
- 161.Mothes K, Engelbrecht L. On the activity of a kinetin-like root factor. Life Sci. 1963;2:852–857. doi: 10.1016/0024-3205(63)90098-5. [DOI] [Google Scholar]
- 162.Balibrea Lara ME, et al. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell. 2004;16:1276–1287. doi: 10.1105/tpc.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tang GQ, Luscher M, Sturm A. Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell. 1999;11:177–189. doi: 10.1105/tpc.11.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Goetz M, et al. Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl Acad. Sci. USA. 2001;98:6522–6527. doi: 10.1073/pnas.091097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Roitsch T, Balibrea ME, Hofmann M, Proels R, Sinha AK. Extracellular invertase: key metabolic enzyme and PR protein. J. Exp. Bot. 2003;54:513–524. doi: 10.1093/jxb/erg050. [DOI] [PubMed] [Google Scholar]