Abstract
Although the successful clinical trials of immunotherapy show promising strategies for many cancers, its application in glioma has lagged in comparison with the progress seen in other cancers. Both isocitrate dehydrogenase (IDH) mutations and 1p/19q codeletions are critical molecular alterations affecting therapeutic response in lower-grade glioma (LGG). The systematic and comprehensive characterization of the immunological phenotypes with different molecular subtypes is key to improving our understanding and application of immunotherapies in LGG. Here, we collected the RNA-sequencing, somatic mutation, and clinical data from 1,052 patients from The Cancer Genome Atlas and Chinese Glioma Genome Atlas and stratified patients into three genetic subgroups: IDH mutations with 1p/19q codeletions (IDH mut-codel), IDH mutations without 1p/19q codeletions (IDH mut-noncodel), and IDH wild-type. Our evaluations revealed that IDH mutations and 1p/19q codeletions were associated with distinct immunological tumor microenvironments in LGG. In addition, immune cell infiltration, the expression of immune checkpoint and human leukocyte antigen (HLA) gene, and the activity of immune signaling pathways shared gradual increase from IDH mut-codel to IDH wild-type. We further constructed and validated an immune-related prognostic signature that presented high value in predicting the overall survival time in LGG. In conclusion, our study may provide valuable information for immunotherapy strategies in LGG patients.
Keywords: IDH mutations, 1p/19q codeletions, lower-grade glioma, tumor immune microenvironment, molecular subtypes, bioinformatics
Graphical abstract
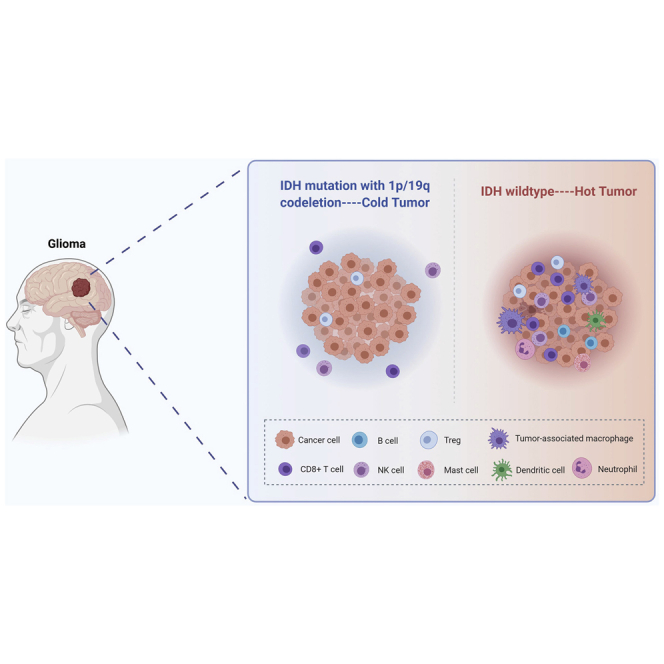
This study demonstrates that IDH mutation and 1p/19q codeletion status were associated with a distinct immunological tumor microenvironment in LGG, which may provide valuable information for immunotherapy strategies in LGG patients.
Introduction
Glioma is the most commonly diagnosed primary malignancy of the central nervous system and is characterized by aggressive malignant proliferation and invasion.1 Although grades II and III gliomas are often collectively described as diffuse lower-grade gliomas (LGGs),2 there are substantial heterogeneities among these tumors in terms of both biology and clinical outcome. Comprehensive and integrative genomic analysis has revealed hundreds of molecular alterations in LGG.2 Among these changes, mutations in the isocitrate dehydrogenase (IDH) gene, and codeletions of chromosome arms 1p and 19q (1p/19q codeletion) are of particular concern. According to the new classification by the World Health Organization (WHO), diffuse LGGs are divided into three molecular subtypes based on IDH mutation and 1p/19q codeletion status: (1) wild-type IDH, (2) IDH mutation without 1p/19q codeletion, and (3) IDH mutation with 1p/19q codeletion.3 Of these subtypes, IDH wild-type tumors are known to be the most clinically aggressive with the worst prognosis, followed by IDH mutation alone, and then IDH mutation with 1p/19q codeletion. This new classification has recently been demonstrated to mirror the biological characteristics of these LGGs.
Immunotherapy, especially immune checkpoint inhibitors (ICIs), has achieved great success in several extracranial cancers resulting in a clear motivation for their application in glioma. Responses to immunotherapy usually depend on interactions between tumor cells and the tumor immune microenvironment.4 Notably, the associations of specific molecular subtypes and their interaction with the tumor immune microenvironment have been reported for several tumor types. For example, HER-2 positive and triple-negative breast cancers are both reported to be associated with increased immune cell infiltration.5,6 In non-small cell lung cancer (NSCLC), Kirsten rat sarcoma 2 viral oncogene homolog (KRAS) mutations are associated with increased immune infiltration, whereas epidermal growth factor receptor (EGFR) mutations are essential to the generation of T regulatory (Treg) and tolerogenic dendritic cells (DCs).7,8
We hypothesized that the three glioma subtypes classified by IDH mutation and 1p/19q codeletion status may also produce with distinct tumor immune microenvironments. In this study, we aimed to characterize the immune profile of wild-type IDH, IDH mutation without 1p/19q codeletion, and IDH mutation with 1p/19q codeletion subtypes using bioinformatics-based evaluations of two independent datasets from The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) databases. We also aimed to determine the prognostic impact of the tumor-infiltrating immune cells on these subtypes and to develop and validate an immune-related risk signature, which might serve as an independent prognostic indicator during clinical evaluation.
Results
The immune landscape in LGG subtypes stratified according to IDH mutation and 1p/19q codeletion status
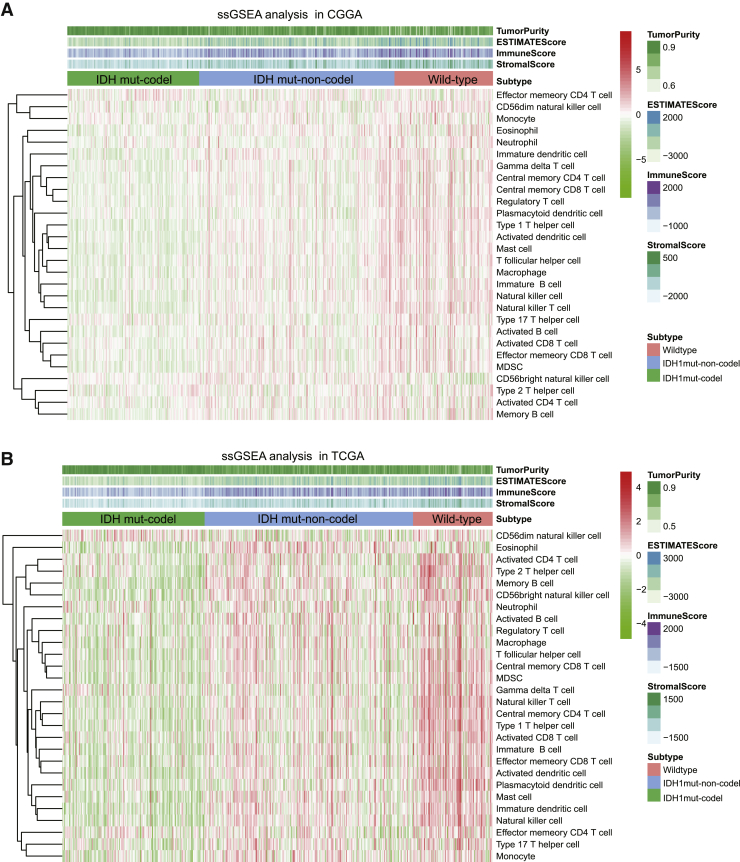
The composition of the immune cell population modulates a diverse range of immune responses and contributes to the anti-tumor effects of immunity by infiltrating tumors with immunoreactive cells. Accumulating evidence suggests that different molecular subtypes are associated with distinct immune cell infiltration patterns. In this study, a single sample gene set enrichment analysis (ssGSEA) was used to determine the immune heterogeneity among IDH wild-type, IDH mut-noncodel, and IDH mut-codel tumors. We generated a heatmap to visualize the relative abundance of 28 infiltrating immune cells in each tumor type and then used this data to guide our other evaluations. Overall, the IDH wild-type samples from the CGGA cohort were shown to present with a high degree of immune cell infiltration, suggesting that they adopt an immune-hot phenotype, whereas IDH mut-codel samples produced a low degree of immune cell infiltration suggesting that they have an immune-cold phenotype (Figure 1A). These observations were then validated in TCGA cohort (Figure 1B).
Figure 1.
The immune cell landscape in different molecular subtypes of LGG
(A) Heatmap visualization of the relative abundance of 28 infiltrating immune cell types in the CGGA database data. (B) These results were further validated in TCGA database. Each small grid represents each immune cell, and the shade of color represents the infiltration level of this immune cell. The larger infiltration level is, the darker color will be (red is upregulated, and green is downregulated).
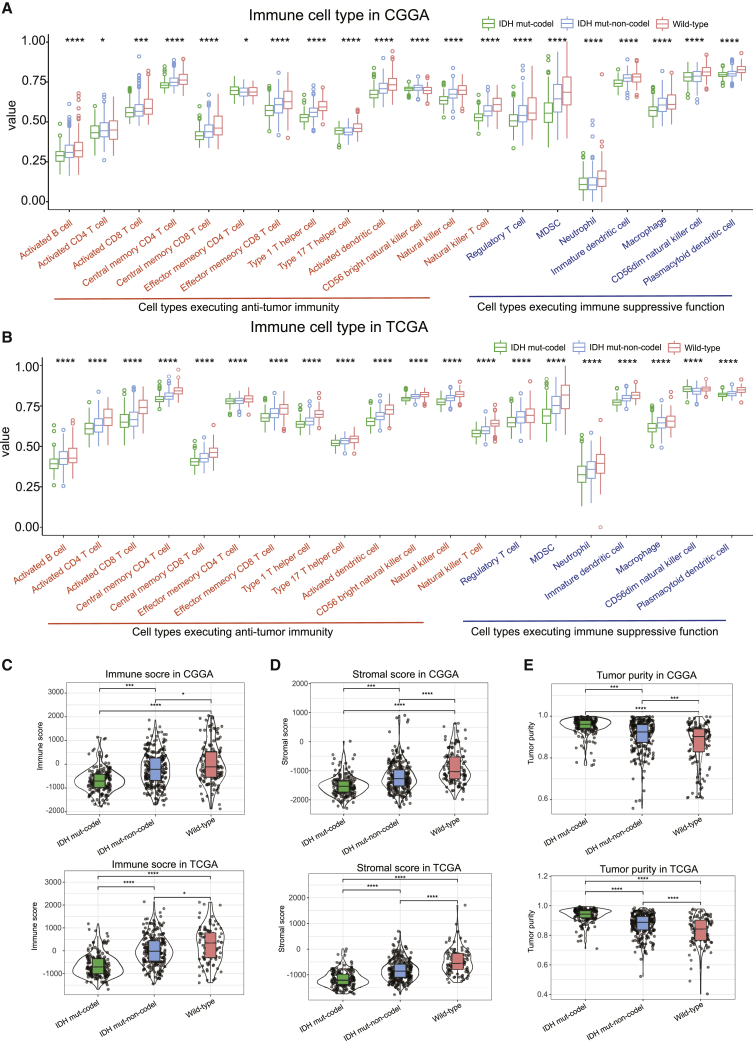
We then evaluated the proportion of immunoreactive and immunosuppressive cells in each of these populations. Interestingly, although the IDH wild-type was shown to present with an immune-hot phenotype, the infiltrating cells were shown to be largely associated with immune suppression (regulatory T cell, MDSC, neutrophil, macrophage, etc.) as opposed to immune activation (e.g., activated B cell, activated CD4 T cell, activated CD8 T cell, effector memory CD8 T cell, natural killer cell, etc.). However, the opposite was true for the IDH mut-noncodel and IDH mut-codel subtypes (Figures 2A and 2B).
Figure 2.
The proportion of immune cells and immune score in each subtype
(A and B) Boxplots represent the differential distribution of immunoreactive and immunosuppressive cells in the various LGG subtypes identified in CGGA (A) and TCGA (B) databases. (C–E) Violin plots show the median, quartile, and kernel density estimations for each immune score (C), stromal score (D), and tumor purity score (E). The black horizontal line represents the median score for each subtype, and the edges in the box represent the upper and lower quartiles in the dataset. The violin graph can also reflect the data density, where a more concentrated dataset yields a broader graph. p values were determined using the Wilcoxon or Kruskal test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001).
In addition, the immune and stromal scores both shared a gradual increase between the IDH mut-codel to the IDH wild-type subtypes, whereas tumor purity demonstrated a gradual decrease (Figures 2C–2E).
GSEA identified a variety of changes in the immune-related signaling pathway activations between IDH wild-type and IDH mut-codel subtype tumors
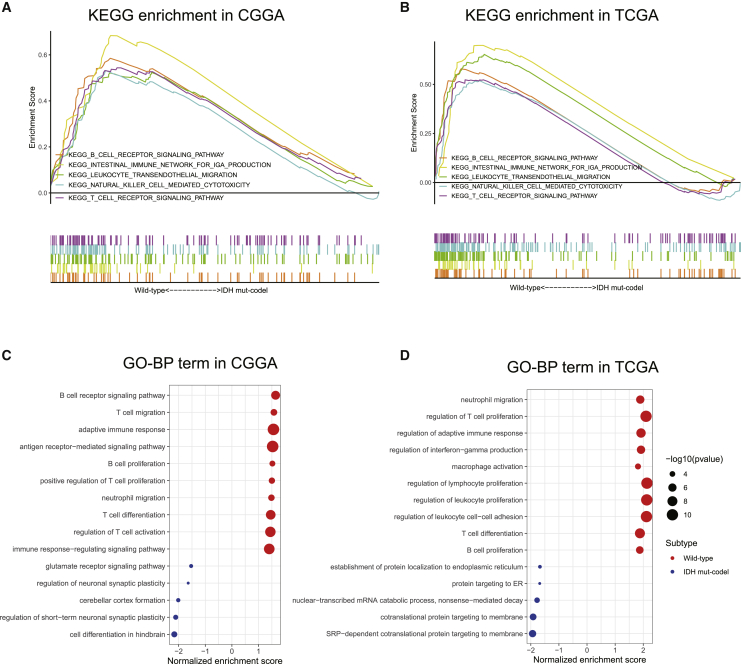
We next evaluated the differences in the signaling pathways activated in the IDH wild-type and IDH mut-codel subtype samples using GSEA. These evaluations revealed that various gene sets were differentially enriched in the IDH wild-type samples within the CGGA database and that these gene sets were closely associated with the regulation of immune signaling pathways, such as the B cell receptor signaling pathway, the leukocyte transendothelial migration pathway, the natural killer cell-mediated cytotoxicity, and the T cell receptor signaling pathways (Figure 3A). These results were further confirmed in TCGA database (Figure 3B).
Figure 3.
Functional enrichment between wild-type and IDH mut-codel LGG data
(A and B) KEGG signaling pathway enrichment analysis of CGGA (A) and TCGA datasets (B). (C and D) Gene Ontology (GO) enrichment analysis of CGGA (C) and TCGA datasets (D). The x axis represents normalized enrichment score, and the y axis is GO term. The size of the dot represents –log10(p value), and the color of the dot represents different subtypes.
In line with Kyoto Encyclopedia of Genes and Genomes (KEGG) results, gene ontology (GO) functional analysis also revealed that the IDH wild-type was associated with the immune-related signaling pathways (Figures 3C and 3D). In the biological process group, the gene sets were mainly aggregated in the B cell receptor signaling pathway, T cell migration, adaptive immune response, neutrophil migration, T cell differentiation, and the antigen receptor-mediated signaling pathway.
Prognostic impact of changes in immune checkpoint expression and immunosuppressive cell infiltration in LGG patients
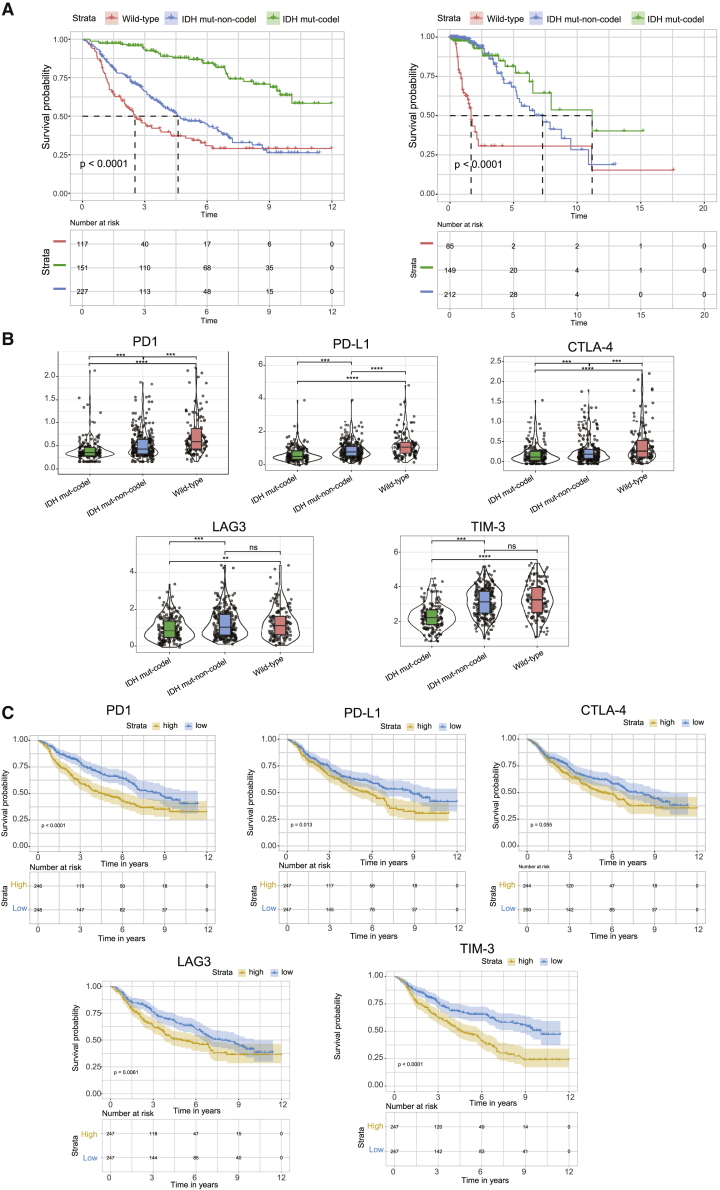
Clinical data show that IDH wild-type gliomas are clinically aggressive and present with the worst prognosis, followed by IDH mut-noncodel and IDH mut-codel.3 Survival data from both CGGA and TCGA cohorts were consistent with these findings (Figure 4A).
Figure 4.
The prognostic impact of changes in the immune checkpoint on LGG patients
(A) Kaplan-Meier curves describing the overall survival (OS) for different molecular subtypes. (B) Violin plots demonstrating the differential expression of the immune checkpoint proteins in these subtypes. (C) Kaplan-Meier curves showing the prognostic impact of changes in the immune checkpoint on LGG patients. p values were determined using a Wilcoxon test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗p < 0.0001).
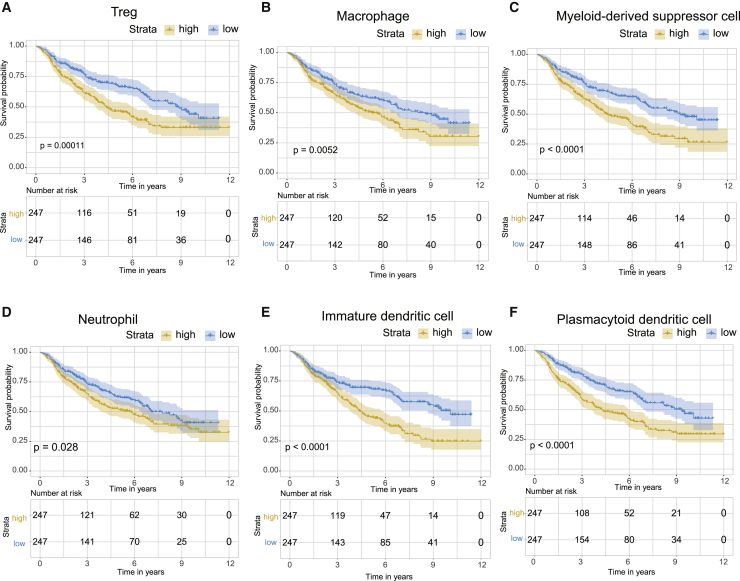
Numerous studies have demonstrated a clear link between high immune cell infiltration and favorable clinical outcomes in several cancers. Notably, our study identified that IDH wild-type samples present with an immune-hot phenotype but have a poor prognosis, whereas IDH mut-codel subtype samples present with an immune-cold phenotype but have more favorable clinical outcomes. We hypothesized that the abundance of immune checkpoints and suppressor cells contributed to the poor prognosis of the IDH wild-type tumors. Indeed, the immune checkpoints (e.g., PD-L1, PD1, CTLA-4, LAG3, and TIM-3) in the CGGA dataset were shown to share a gradual increase in expression from the IDH mut-codel to the IDH wild-type samples (Figure 4B). Moreover, overexpression of immune checkpoints was correlated with poorer prognosis in LGG (Figure 4C). In line with the immune checkpoint data, the proportion of immunosuppressive cells was shown to be enriched in IDH wild-type samples when compared to the IDH mut-noncodel and IDH mut-codel subtypes (Figures 2A and 2B). A high proportion of these immunosuppressive cell types (e.g., Treg, macrophage, myeloid-derived suppressor cell, neutrophil, immature dendritic cell, and plasmacytoid dendritic cells) was associated with poorer clinical outcomes (Figures 5A–5F).
Figure 5.
The prognostic impacts of changes in immunosuppressive cell content on LGG patients
(A–F) A high proportion of Treg (A), macrophage (B), myeloid-derived suppressor cell (C), neutrophil (D), immature dendritic cell (E), and plasmacytoid dendritic cells (F) were shown to be closely associated with reduced OS in LGG patients.
Differences in MHC class I gene expression and tumor mutation burden (TMB) in samples with IDH wild-type, IDH mut-noncodel, and IDH mut-codel subtypes
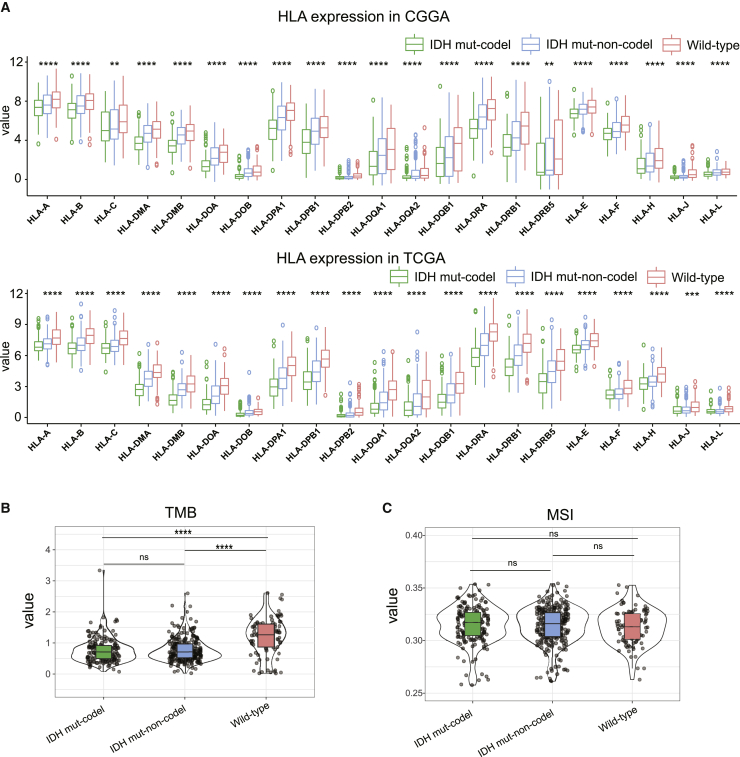
Tumor-related antigen presentation via the major histocompatibility complex (MHC) class I complexes, TMB, and microsatellite instability (MSI) are all prerequisites for effective immune surveillance and are essential for the clinical outcomes associated with immune checkpoint blockade therapies. Given this, we evaluated the expression of the human leukocyte antigen (HLA) genes encoding the MHC class I proteins in the different molecular subtypes in the CGGA-LGG cohort and then validated these data with an independent evaluation of TCGA cohort. All analyzed HLA genes shared a gradual decrease in expression between the IDH wild-type and IDH mut-codel subtypes (Figure 6A), indicating that patients with IDH wild-type might benefit more from immune checkpoint blockade therapy.
Figure 6.
Differences in MHC class I gene expression and tumor mutation burden in different subtypes of LGG
(A) Boxplots show the differential HLA expression between different LGG subtypes. (B) Violin plots represent differential TMB expression in these subtypes. (C) Violin plots display the differential expression of MSI in each of these subtypes. p values were determined using a Wilcoxon or Kruskal test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
This assumption was further supported by the increase in TMB in IDH wild-type samples compared to the IDH mut-codel subtype samples (Figure 6B). However, there were no differences in MSI in any of these subtypes (Figure 6C).
Somatic mutations in IDH wild-type, IDH mut-noncodel, and IDH mut-codel subtypes
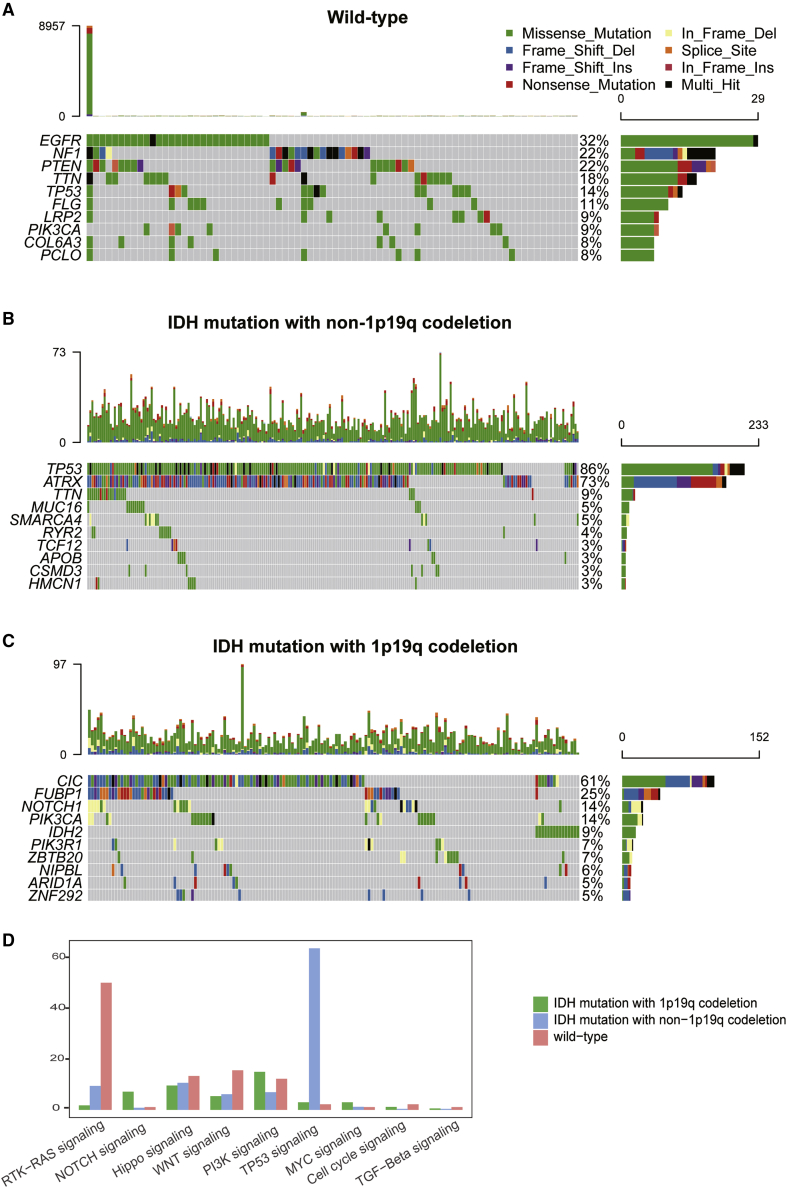
Given our previous data, we determined the genomic alterations associated with each of these tumor subtypes using the somatic mutations data from TCGA database. We noted an increase in the relative frequencies for EGFR, NF1, and PTEN mutations in IDH wild-type samples when compared to the IDH mut-noncodel and IDH mut-codel subtypes (Figure 7A). IDH mut-noncodel subtype samples demonstrated an increase in the frequency of TP53 and ATRX mutations, accounting for 86% and 73% of the total, respectively (Figure 7B). The three most commonly mutated genes in the IDH mut-codel samples were CIC, FUBP1, and NOTCH1 (Figure 7C).
Figure 7.
Comparison of somatic mutations among different LGG subtypes
(A–C) Oncoprint visualization of the top ten most frequently mutated genes in IDH wild-type (A), IDH mut-noncodel (B), and IDH mut-codel subtypes (C). (D) The mutation frequencies of nine common oncogenic pathways in each of these three subtypes.
We then analyzed the mutation frequencies in nine common oncogenic pathways in each of these subtypes. Mutations in the RTK-RAS, Hippo, WNT, cell-cycle, and transforming growth factor-β (TGF-β) signaling pathways were most frequently detected in the wild-type tumors, whereas the TP53 signaling pathways exhibited the highest mutation frequency in the IDH mut-noncodel samples. The mutation frequencies for the PI3K and MYC pathways were highest in the IDH mut-codel tumors (Figure 7D).
Identification of differentially expressed immune-related genes in different IDH subtypes
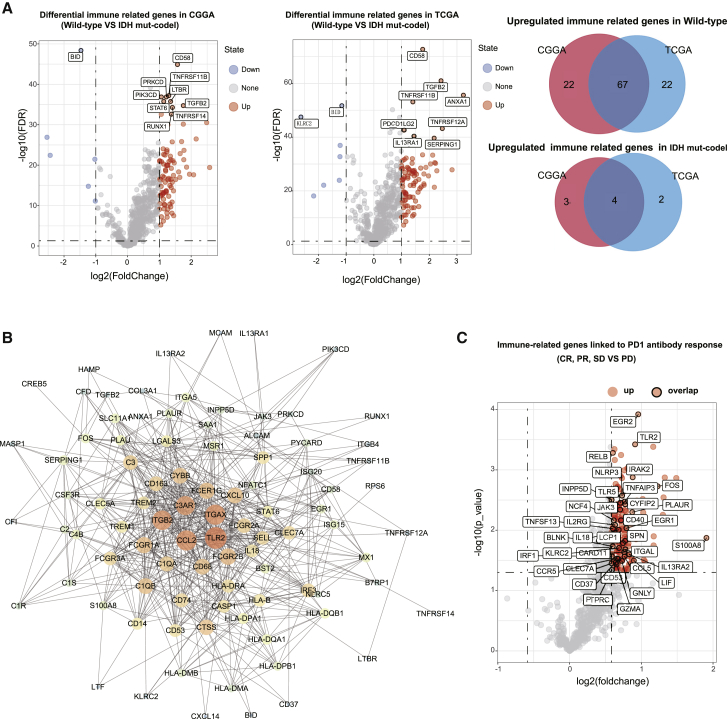
As the wild-type samples presented with an immune-hot phenotype and the IDH mut-codel samples presented with an immune-cold phenotype, we identified the key differentially regulated genes in each subtype in an effort to understand the differences in immune regulation in these LGG tumors. We identified a total of 95 dysregulated immune-related genes in the CGGA cohort and 96 dysregulated immune-related genes in TCGA cohort (Figure 8A). Most of the dysregulated genes were overexpressed in the wild-type samples, whereas only seven genes in the CGGA and six genes in TCGA data were shown to be upregulated in the IDH mut-codel samples. The interactions between these differentially expressed genes was further evaluated using a protein-protein interaction network constructed using the STRING database. The 20 key immune-related genes with the highest degree of interaction were identified and included TLR2, ITGAX, CCL2, ITGB2, C3AR1, FCER1G, CXCL10, FCGR2B, FCGR2A, SELL, CD68, CD163, CYBB, C1QA, HLA-DRA, CASP1, HLA-B, C1QB, CD74, and interleukin-18 (IL-18), suggesting their important role in regulating the tumor immune microenvironment (Figure 8B).
Figure 8.
Identification of the key immune-related genes undergoing differential expression when comparing the wild-type and IDH mut-codel LGG subtypes
(A) Volcano plots identify the differentially expressed immune related genes (wild-type versus IDH mut-codel) in both CGGA and TCGA databases; the Venn diagram summarizes the co-regulated genes in both databases. (B) Protein-protein interaction network analysis revealed the key immune-related genes in both the wild-type and IDH mut-codel subtypes. (C) Volcano plots represent the significant differentially expressed genes among progressive disease (PD), complete response (CR), partial response (PR), and stable disease (SD), and the red dots with black outlines indicate the overlapping genes in the wild-type LGG data.
To determine whether these differentially expressed genes were associated with changes in the response to ICIs, we analyzed the expression data for various immune gene panels in both melanoma and NSCLC patients treated with anti-PD1 antibodies. Patients with progressive disease were assigned to the non-response group, whereas those with complete response, partial response, and stable disease were assigned to the response group. These evaluations identified 153 immune genes that were differentially expressed, with a significantly higher level of expression in the response group (fold change > 0.5 and p value < 0.05). Notably, most of these immune genes overlapped with the upregulated genes in the IDH wild-type data (Figure 8C) suggesting that patients with IDH wild-type tumors might benefit from ICI therapy.
Construction and validation of the immune-related prognostic signature
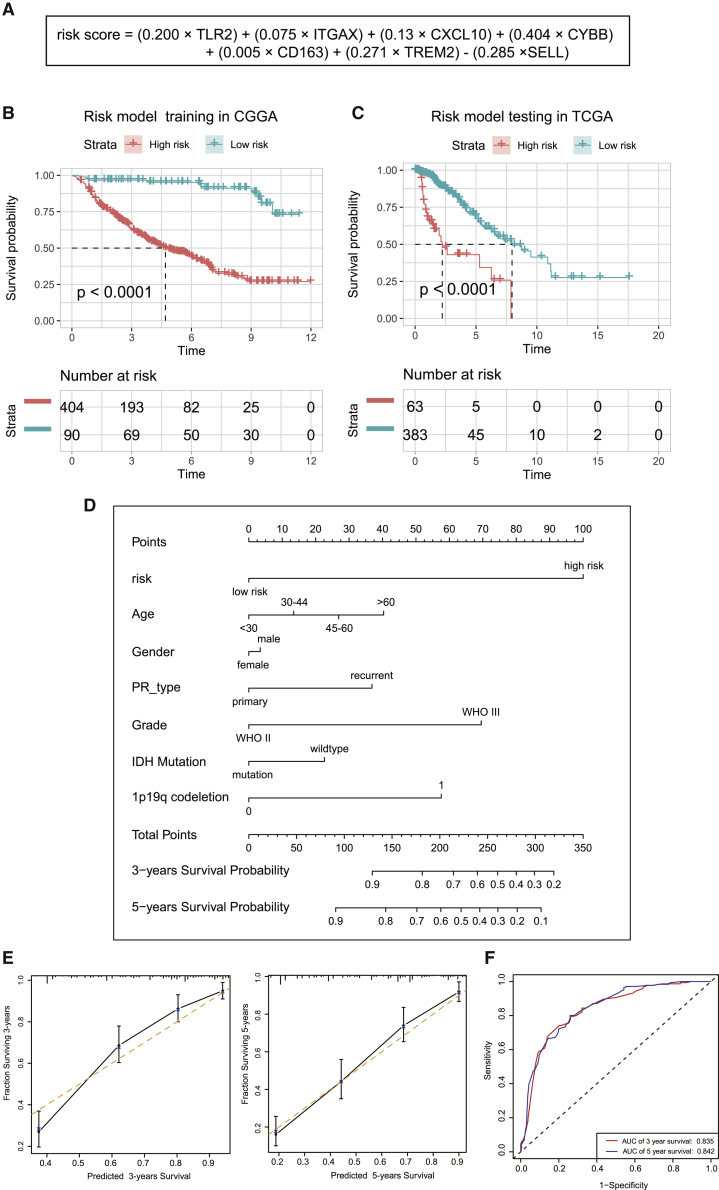
Finally, we developed a prognostic model based on the differential immune-gene expression profile of wild-type and IDH mut-codel subtype tumors. Data were first evaluated using a least absolute shrinkage and selection operator (LASSO) regression analysis which identified seven of the 71 immune-related genes as critical to prognosis and then used these to construct the prognostic model. A risk-score model was established as follows: risk score = (0.200 × TLR2) + (0.075 × ITGAX) + (0.13 × CXCL10) + (0.404 × CYBB) + (0.005 × CD163) + (0.271 × TREM2) – (0.285 × SELL) (Figure 9A). Kaplan-Meier analysis was then used to determine the prognostic value of this risk signature in LGG. A high-risk score was shown to correlate with poor overall survival (OS) in the CGGA cohort, which was further validated by a similar outcome in TCGA cohort (Figures 9B and 9C). We then constructed a nomogram model in which the immune-related prognostic signature was integrated with six clinical characteristics (age, gender, primary recurrence type, grade, IDH mutation status, and 1p19q codeletion status) and then used to predict clinical outcome (Figure 9D). To evaluate the efficiency of this nomogram model in predicting the 3- and 5-year survival rates of patients, we generated a series of calibration plots and a receiver operating characteristic (ROC) curve. The calibration plots describing observed versus predicted probabilities of 3- and 5-year OS demonstrated excellent concordance, and the area under the ROC curve was 0.835 at 3 years and 0.842 at 5 years, respectively, indicating a high predictive value (Figures 9E and 9F).
Figure 9.
Prognostic value of the immune-related risk signatures in LGG
(A) Establishment of an immune-related risk formula. (B and C) Kaplan-Meier OS curves for patients with high or low risk scores. (D) Construction of a nomogram model for predicting the probability of 1-, 3-, and 5-year OS. (E) Calibration plots of the nomogram for 3-year and 5-year survival. The x axis shows the nomogram-predicted probability, and the y axis represents the observed probability. (F) ROC curves showing the predictive value of the nomogram model for 1-, 3-, and 5-year survival rates.
Discussion
Immunotherapies, including antibodies, chimeric antigen receptor T cells, and ICIs, have received tremendous attention in their application to cancer treatment.9, 10, 11 However, apart from some exciting clinical trials, the general application of immunotherapies has seen a highly variable response with some patients even proving to be resistant to these interventions.12, 13, 14 Therefore, the characterization of tumor immunologic profile, which stratifies patients with a high or low sensitivity, will help to maximize the efficacy of immunotherapy. Here, we present a comprehensive characterization of the tumor immune microenvironment for LGG tumors and reveal a significant association between these immunological parameters and the three genetic subgroups of the LGG tumor. This information may prove valuable in evaluating the outcomes of precision immunotherapy in LGG patients.
Glioma is highly heterogeneous with multiple subtypes. The identification of several key molecular markers, most notably IDH mutations, has allowed for a precise method with which to categorize glioma with clear prognostic implications. Apart from IDH mutation, recent evaluations have identified a second important mutation in these tumors, the 1p/19q codeletion, affecting their clinical outcome. The initial retrospective series and subsequent retrospective analyses of large randomized trials have validated 1p/19q deletion as a strong prognostic and predictive marker in LGG.15 In the revised fourth edition of the WHO Classification of central nervous system tumors published in 2016, classification of diffuse gliomas has fundamentally changed: a large subset of glioma is now defined based on IDH mutation and 1p/19q codeletion status and can be stratified into three genetic subgroups (IDH wild-type, IDH mutation and 1p/19q-noncodeletion, and IDH mutation and 1p/19q-codeletion).3,16 This stratification based on IDH mutation and 1p/19q deletion status presents a more precise stratification in natural histories, molecular profiling, prognosis, and responses to treatment when compared to IDH stratification alone, and many genome studies of glioma are based on the three subgroups.17, 18, 19, 20, 21, 22 Given the importance of the IDH and 1p/19q codeletion mutations in LGG, we stratified our LGG data based on their molecular subtype, producing three groups: (1) wild-type IDH, (2) IDH mutation without 1p/19q codeletion, and (3) IDH mutation with 1p/19q codeletion.
Recently, IDH mutation status has been reported to be associated with PD-L1 expression and tumor-infiltrating lymphocyte (TIL) infiltration in diffuse gliomas.23, 24, 25, 26, 27 Our study was largely in agreement with available data but differed in several important aspects. We integrated IDH mutation and 1p/19q codeletion status and conducted a comprehensive and systematic analysis of 28 immune cell types and their infiltration in these subtypes; the expression of the immune checkpoint components in each subtype was assessed and TMB, MSI, and MHC class I gene expression was categorized in these subtypes. Our evaluations also included descriptions of somatic mutations and changes in the molecular pathways associated with the immune response in these subtypes. All our evaluations used bioinformatics-based scrutiny of genomic and transcriptomic data from two independent databases (TCGA and CGGA). Nevertheless, it should be noted that our findings require further validation in vitro or in vivo. Our findings should be interpreted with this limitation in mind.
Hegde et al.28 summarized the top 10 challenges in anti-tumor immunotherapy. One of these challenges is the determination of the dominant drivers of cancer immunity. Tumor immunological phenotypes are usually stratified as immune-hot or immune-cold tumors and are associated with survival and prediction of response to immunotherapy.4,29 Accumulating evidence has identified the biomarkers of the immune-hot type beyond the increase in the number of TILs, including the expression of immune checkpoint proteins in tumor or tumor-associated immune cells, genomic instability as defined by MSI or TMB, and intact antigen presentation.14 In contrast, apart from low immune cell infiltration, immune-cold tumors are also characterized by low immune checkpoint expression, low mutational burden, and defects in antigen presentation. Our study showed that IDH wild-type tumors were infiltrated with the highest diversity of immune cells and were associated with high immune checkpoint expression, TMB, and MHC class I gene expression, indicating an immune-hot phenotype. Conversely, IDH mut-noncodel and IDH mut-codel subtypes were infiltrated by a lower number of immune cells and were shown to correlate with reduced immune checkpoint expression, TMB, and MHC class I gene expression, indicating an immune-cold phenotype.
It has been reported that IDH wild-type gliomas present with the worst prognosis, when compared to the IDH mut-noncodel and IDH mut-codel subtypes.3 One of the proposed mechanisms of this poor prognosis is likely to involve the relative radio-resistance of IDH wild-type tumors when compared to the other two subtypes.30 Here, we reveal high degrees of immunosuppressive cell infiltration and immune checkpoint expression in the IDH wild-type tumors, both of which may contribute to its poor prognosis. This could be another important mechanism underlying the differences in clinical outcomes between the subtypes following treatment.
The aberrant activation of the EGFR signaling pathway is likely responsible for the immunosuppression of the tumor microenvironment.31 This mechanism may involve the aberrant regulation mediated by mutated EGFR in the generation of Treg and tolerogenic DCs.32,33 Our study went on to evaluate other somatic mutations in these three tumor subtypes and showed that both the receptor tyrosine kinase-Ras pathway and the EGFR pathway mutations were more common in the IDH wild-type, which may account for its enhanced immunosuppressive cell infiltration.
In conclusion, our study highlights the associations of the IDH mutation and 1p/19q codeletion with changes in the immunological tumor microenvironment in LGG. These observations may benefit future immune therapy-based interventions for LGG patients. We also established and validated an immune-related prognostic signature, which demonstrated significant value in predicting OS time in LGG patients.
Materials and methods
Datasets
The RNA sequencing, somatic mutation, and corresponding clinical data from 536 LGG patients were obtained from the CGGA (http://www.cgga.org.cn/) database.34,35 Similar data was also collected for the 516 LGG patients in TCGA (http://cancergenome.nih.gov/) database to act as a validation set. LGG patients were divided into wild-type IDH, IDH mutation without 1p/19q codeletion (IDH mut-noncodel), and IDH mutation with 1p/19q codeletion (IDH mut-codel) subtypes, and the clinical information for each cohort is summarized in Table 1.
Table 1.
Clinical characteristic
| Clinical characteristics | Total (n = 1,052) |
|
|---|---|---|
| CGGA (n = 536) | TCGA (n = 516) | |
| Age | ||
| <45 | 371 | 267 |
| ≥45 | 164 | 190 |
| N/A | 1 | 59 |
| Gender | ||
| Female | 224 | 201 |
| Male | 312 | 256 |
| N/A | 0 | 59 |
| IDH mutation status | ||
| Mutant | 412 | 419 |
| Wild-type | 124 | 94 |
| N/A | 0 | 3 |
| 1p19q codeletion status | ||
| Codel | 166 | 169 |
| Noncodel | 370 | 347 |
| Grade | ||
| G2 | 254 | 216 |
| G3 | 282 | 241 |
| N/A | 0 | 59 |
Transcriptome expression data describing a predefined immune gene panel and the corresponding clinical outcome for patients treated with anti-PD1 antibodies were downloaded from GEO: GSE93157.36
ssGSEA
ssGSEA was used to quantify the 28 types of immune cells infiltrating each LGG sample and was completed using the “GSVA” and “GSEABase” packages in R.37 The ssGSEA enrichment score was then used as the measure of immune cell infiltration in each sample. Gene signatures for each immune cell type were obtained from previously published data.38
Somatic mutation analysis
Somatic mutation data for each of the of the LGG samples were downloaded from TCGA GDC Data Portal in “maf” format (VarScan2 Variant Aggregation and Masking; https://portal.gdc.cancer.gov). Waterfall plots were then constructed using the “Maftools” package in R software, which facilitated the visualization and summarization of the mutated genes and aberrant signaling pathways in each of the three LGG subtypes.39
Functional enrichment analysis
GSEA was used to identify the significantly altered signaling pathways in each subtype, as identified by their enrichment in the MSigDB Collection (c2.kegg.v7.1 symbols.gmt; c5.bp.v7.1 symbols.gmt). Gene set permutations were set at 1,000 repeats for each analysis. The results were then visualized using gglpot2, grid, and gridExtra for R.
Differential expression of immune-related genes
The differential expression of immune-related genes was identified using the “limma” R package. Adj. p < 0.05, and |fold change| > 1.5 were used as the threshold values.
Construction of the immune-related prognostic signature
The statistically significant immune-related genes identified in the univariable Cox regression analysis were then subjected to a LASSO cox regression analysis to calculate the specific coefficient values for each association. LASSO is a regression analysis method that performs both variable selection and regularization in order to enhance the prediction accuracy and interpretability of the resulting statistical model. Thus, LASSO cox regression is well-suited for the construction of prognostic model based on gene-expression profile.40, 41, 42, 43, 44
These values were then used to develop the following formula:
OS for the low and high-risk groups was compared using Kaplan-Meier analysis completed using the Survminer and survival packages for R. A nomogram model was then established in which the immune-related prognostic signature, which integrates the age, gender, primary recurrence type, grade, IDH mutation status, and 1p19q codeletion status, was then produced. The calibrations plot and survival ROC curve were then generated to validate the predictive value of this nomogram model in patients’ OS using the survivalROC, rms, and foreign packages in R.
Statistical analysis
The OS among the different groups was compared via Kaplan-Meier analysis using the survival and survminer packages in R. Differences among the subtypes were tested using the Wilcoxon signed-rank test or Kruskal-Wallis test. Univariate Cox analysis was applied to identify potential prognostic factors. A ROC curve was performed to validate the accuracy of the risk model in predicting the patients’ OS via the survivalROC R package. All statistical analyses were performed using the R software (version 3.5.2).
Acknowledgments
Our results were generated using some of the basic data from TCGA Research Network: https://www.cancer.gov/tcga. Differential expression analysis was performed using the Sangerbox tools, a free online platform for data analysis (http://www.sangerbox.com/tool). This work was supported by the National Key Research and Development Program of China (project number 2018YFC0115700). The graphical abstract was created with BioRender.
Author contributions
Conceptualization and methodology, W.L.; writing, X.Q. and W.L.; statistical calculation and validation, P.S. and Y.Y.; review and approval of concept/methodology, J.J.L. and L.K.; and project administration and funding acquisition, J.J.L., and L.K. All authors read and approved the manuscript for publication.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Lin Kong, Email: lin.kong@sphic.org.cn.
Jiade J. Lu, Email: jiade.lu@sphic.org.cn.
References
- 1.Ostrom Q.T., Gittleman H., Farah P., Ondracek A., Chen Y., Wolinsky Y., Stroup N.E., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro-oncol. 2013;15:ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brat D.J., Verhaak R.G., Aldape K.D., Yung W.K., Salama S.R., Cooper L.A., Rheinbay E., Miller C.R., Vitucci M., Morozova O., Cancer Genome Atlas Research Network Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glajcar A., Szpor J., Hodorowicz-Zaniewska D., Tyrak K.E., Okoń K. The composition of T cell infiltrates varies in primary invasive breast cancer of different molecular subtypes as well as according to tumor size and nodal status. Virchows Arch. 2019;475:13–23. doi: 10.1007/s00428-019-02568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karn T., Jiang T., Hatzis C., Sänger N., El-Balat A., Rody A., Holtrich U., Becker S., Bianchini G., Pusztai L. Association Between Genomic Metrics and Immune Infiltration in Triple-Negative Breast Cancer. JAMA Oncol. 2017;3:1707–1711. doi: 10.1001/jamaoncol.2017.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X.C., Wang J., Shao G.G., Wang Q., Qu X., Wang B., Moy C., Fan Y., Albertyn Z., Huang X. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat. Commun. 2019;10:1772. doi: 10.1038/s41467-019-09762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu S., Liu D., Shen B., Shi M., Feng J. Immunotherapy strategy of EGFR mutant lung cancer. Am. J. Cancer Res. 2018;8:2106–2115. [PMC free article] [PubMed] [Google Scholar]
- 9.Restifo N.P., Dudley M.E., Rosenberg S.A. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat. Rev. Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumeister S.H., Freeman G.J., Dranoff G., Sharpe A.H. Coinhibitory Pathways in Immunotherapy for Cancer. Annu. Rev. Immunol. 2016;34:539–573. doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 11.Kerr W.G., Chisholm J.D. The Next Generation of Immunotherapy for Cancer: Small Molecules Could Make Big Waves. J. Immunol. 2019;202:11–19. doi: 10.4049/jimmunol.1800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emens L.A., Ascierto P.A., Darcy P.K., Demaria S., Eggermont A.M.M., Redmond W.L., Seliger B., Marincola F.M. Cancer immunotherapy: Opportunities and challenges in the rapidly evolving clinical landscape. Eur. J. Cancer. 2017;81:116–129. doi: 10.1016/j.ejca.2017.01.035. [DOI] [PubMed] [Google Scholar]
- 13.Farkona S., Diamandis E.P., Blasutig I.M. Cancer immunotherapy: the beginning of the end of cancer? BMC Med. 2016;14:73. doi: 10.1186/s12916-016-0623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambi M., Bagheri L., Szewczuk M.R. Current Challenges in Cancer Immunotherapy: Multimodal Approaches to Improve Efficacy and Patient Response Rates. J. Oncol. 2019;2019:4508794. doi: 10.1155/2019/4508794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R., Smith-Cohn M., Cohen A.L., Colman H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics. 2017;14:284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louis D.N., Perry A., Burger P., Ellison D.W., Reifenberger G., von Deimling A., Aldape K., Brat D., Collins V.P., Eberhart C., International Society Of Neuropathology--Haarlem International Society Of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24:429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz V.Y., Praska C.E., Armstrong G., Kollmeyer T.M., Yamada S., Decker P.A., Kosel M.L., Eckel-Passow J.E., Lachance D.H., Bainbridge M.N., Gliogene Consortium Molecular subtyping of tumors from patients with familial glioma. Neuro-oncol. 2018;20:810–817. doi: 10.1093/neuonc/nox192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otani R., Uzuka T., Higuchi F., Matsuda H., Nomura M., Tanaka S., Mukasa A., Ichimura K., Kim P., Ueki K. IDH-mutated astrocytomas with 19q-loss constitute a subgroup that confers better prognosis. Cancer Sci. 2018;109:2327–2335. doi: 10.1111/cas.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauber C., Klink B., Seifert M. Comparative analysis of histologically classified oligodendrogliomas reveals characteristic molecular differences between subgroups. BMC Cancer. 2018;18:399. doi: 10.1186/s12885-018-4251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi T., Sonoda Y., Shibahara I., Saito R., Kanamori M., Kumabe T., Tominaga T. Impact of gross total resection in patients with WHO grade III glioma harboring the IDH 1/2 mutation without the 1p/19q co-deletion. J. Neurooncol. 2016;129:505–514. doi: 10.1007/s11060-016-2201-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhou H., Chang K., Bai H.X., Xiao B., Su C., Bi W.L., Zhang P.J., Senders J.T., Vallières M., Kavouridis V.K. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J. Neurooncol. 2019;142:299–307. doi: 10.1007/s11060-019-03096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barthel F.P., Johnson K.C., Varn F.S., Moskalik A.D., Tanner G., Kocakavuk E., Anderson K.J., Abiola O., Aldape K., Alfaro K.D., GLASS Consortium Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019;576:112–120. doi: 10.1038/s41586-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berghoff A.S., Kiesel B., Widhalm G., Wilhelm D., Rajky O., Kurscheid S., Kresl P., Wöhrer A., Marosi C., Hegi M.E., Preusser M. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro-oncol. 2017;19:1460–1468. doi: 10.1093/neuonc/nox054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cejalvo T., Gargini R., Segura-Collar B., Mata-Martínez P., Herranz B., Cantero D., Ruano Y., García-Pérez D., Pérez-Núñez Á., Ramos A. Immune Profiling of Gliomas Reveals a Connection with IDH1/2 Mutations, Tau Function and the Vascular Phenotype. Cancers (Basel) 2020;12:3230. doi: 10.3390/cancers12113230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson L.G., Nieman L.T., Stemmer-Rachamimov A.O., Zheng X.S., Stafford K., Nagashima H., Miller J.J., Kiyokawa J., Ting D.T., Wakimoto H. IDH-mutant gliomas harbor fewer regulatory T cells in humans and mice. OncoImmunology. 2020;9:1806662. doi: 10.1080/2162402X.2020.1806662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amankulor N.M., Kim Y., Arora S., Kargl J., Szulzewsky F., Hanke M., Margineantu D.H., Rao A., Bolouri H., Delrow J. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 2017;31:774–786. doi: 10.1101/gad.294991.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Rao A., Sette P., Deibert C., Pomerantz A., Kim W.J., Kohanbash G., Chang Y., Park Y., Engh J. IDH mutant gliomas escape natural killer cell immune surveillance by downregulation of NKG2D ligand expression. Neuro-oncol. 2016;18:1402–1412. doi: 10.1093/neuonc/now061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegde P.S., Chen D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity. 2020;52:17–35. doi: 10.1016/j.immuni.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Galon J., Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 30.Wang X.W., Ciccarino P., Rossetto M., Boisselier B., Marie Y., Desestret V., Gleize V., Mokhtari K., Sanson M., Labussière M. IDH mutations: genotype-phenotype correlation and prognostic impact. BioMed Res. Int. 2014;2014:540236. doi: 10.1155/2014/540236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L., Li A., Lei Q., Zhang Y. Tumor-intrinsic signaling pathways: key roles in the regulation of the immunosuppressive tumor microenvironment. J. Hematol. Oncol. 2019;12:125. doi: 10.1186/s13045-019-0804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan H.Y., Wang N., Lam W., Guo W., Feng Y., Cheng Y.C. Targeting tumour microenvironment by tyrosine kinase inhibitor. Mol. Cancer. 2018;17:43. doi: 10.1186/s12943-018-0800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson K.G., Stromnes I.M., Greenberg P.D. Obstacles Posed by the Tumor Microenvironment to T cell Activity: A Case for Synergistic Therapies. Cancer Cell. 2017;31:311–325. doi: 10.1016/j.ccell.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Qian T., You G., Peng X., Chen C., You Y., Yao K., Wu C., Ma J., Sha Z. Localizing seizure-susceptible brain regions associated with low-grade gliomas using voxel-based lesion-symptom mapping. Neuro-oncol. 2015;17:282–288. doi: 10.1093/neuonc/nou130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Z., Meng F., Wang W., Wang Z., Zhang C., Jiang T. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci. Data. 2017;4:170024. doi: 10.1038/sdata.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prat A., Navarro A., Paré L., Reguart N., Galván P., Pascual T., Martínez A., Nuciforo P., Comerma L., Alos L. Immune-Related Gene Expression Profiling After PD-1 Blockade in Non-Small Cell Lung Carcinoma, Head and Neck Squamous Cell Carcinoma, and Melanoma. Cancer Res. 2017;77:3540–3550. doi: 10.1158/0008-5472.CAN-16-3556. [DOI] [PubMed] [Google Scholar]
- 37.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 39.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tibshirani R. The lasso method for variable selection in the Cox model. Stat. Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Sauerbrei W., Royston P., Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat. Med. 2007;26:5512–5528. doi: 10.1002/sim.3148. [DOI] [PubMed] [Google Scholar]
- 42.Bøvelstad H.M., Nygård S., Størvold H.L., Aldrin M., Borgan Ø., Frigessi A., Lingjaerde O.C. Predicting survival from microarray data--a comparative study. Bioinformatics. 2007;23:2080–2087. doi: 10.1093/bioinformatics/btm305. [DOI] [PubMed] [Google Scholar]
- 43.Hu X., Martinez-Ledesma E., Zheng S., Kim H., Barthel F., Jiang T., Hess K.R., Verhaak R.G.W. Multigene signature for predicting prognosis of patients with 1p19q co-deletion diffuse glioma. Neuro-oncol. 2017;19:786–795. doi: 10.1093/neuonc/now285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei J.-H., Feng Z.-H., Cao Y., Zhao H.-W., Chen Z.-H., Liao B., Wang Q., Han H., Zhang J., Xu Y.-Z. Predictive value of single-nucleotide polymorphism signature for recurrence in localised renal cell carcinoma: a retrospective analysis and multicentre validation study. Lancet Oncol. 2019;20:591–600. doi: 10.1016/S1470-2045(18)30932-X. [DOI] [PubMed] [Google Scholar]