Abstract
Hepatocellular carcinoma (HCC) remains an extremely lethal disease worldwide. High-throughput methods have revealed global transcriptome dysregulation; however, a comprehensive investigation of the complexity and behavioral characteristics of the competing endogenous RNA (ceRNA) network in HCC is lacking. In this study, we extracted the transcriptome (RNA) sequencing data of 371 HCC patients from The Cancer Genome Atlas platform. With the comparison of the high Myc expression (Mychigh) tumor and low Myc expression (Myclow) tumor groups in HCC, we identified 1,125 differentially expressed (DE) mRNAs, 589 long non-coding RNAs (lncRNAs), and 93 microRNAs (miRNAs). DE RNAs predicted the interactions necessary to construct an associated Myc ceRNA network, including 19 DE lncRNAs, 5 miRNAs, and 72 mRNAs. We identified a significant signature (long intergenic non-protein-coding [LINC] RNA 2691 [LINC02691] and LINC02499) that effectively predicted overall survival and had protective effects. The target genes of microRNA (miR)-212-3p predicted to intersect with DE mRNAs included SEC14-like protein 2 (SEC14L2) and solute carrier family 6 member 1 (SLC6A1), which were strongly correlated with survival and prognosis. With the use of the lncRNA-miRNA-mRNA axis, we constructed a ceRNA network containing four lncRNAs (LINC02691, LINC02499, LINC01354, and NAV2 antisense RNA 4), one miRNA (miR-212-3p), and two mRNAs (SEC14L2 and SLC6A1). Overall, we successfully constructed a mutually regulated ceRNA network and identified potential precision-targeted therapies and prognostic biomarkers.
Keywords: TCGA, HCC, ceRNA, immune infiltration, methylation expression
Graphical abstract
Zhang et al. provide evidence that a ceRNA-based SEC14L2/SLC6A1 axis has an impact on the changes in the tumor immune microenvironment and might be a novel important prognostic factor associated with the diagnosis and prognosis of HCC.
Introduction
Liver cancer is categorized as primary hepatic cancer and metastatic hepatic cancer.1, 2, 3 Hepatocellular carcinoma (HCC) is the fifth-most common cancer worldwide and is a highly lethal malignancy, accounting for nearly 80% of all primary liver malignancies.4, 5, 6 According to 2018 Global Cancer statistics,7 there have been more than 100,000 new cases and deaths worldwide every year.
Although there have been great advancements in cancer therapy, the 5-year overall survival (OS) rate for HCC is still only 12.10%.8, 9, 10, 11 Surgical resection, liver transplantation, and chemotherapy are common therapies,12, 13, 14, 15 but they are appropriate only for patients with early-stage disease.16,17 Therefore, it is essential to understand the mechanisms underlying the pathogenesis of HCC and identify novel biomarkers.
In the past several years, the advancement of high-throughput technologies has provided more opportunities for biomarker identification.18, 19, 20, 21 Non-coding RNAs (ncRNAs), which include long ncRNAs (lncRNAs) and microRNAs (miRNAs), play a role in cancer progression and are potential biomarkers and therapeutic targets.22, 23, 24, 25 lncRNAs are transcripts with more than 200 nucleotides, which previously were thought to have no biological function. But in fact, lncRNAs are involved in several biological processes, including cell differentiation, cancer proliferation, and metastasis.26, 27, 28, 29 Competing endogenous RNA (ceRNA), including lncRNA, can interact with mRNA by competitively combining different miRNAs.30, 31, 32 miRNAs are a class of ncRNA molecules of 21–24 nucleotides that mediate negative post-transcriptional regulation.33, 34, 35 When the miRNA arm-imbalance mechanism is broken in the cell, dysregulation of downstream tumor-suppressor genes or oncogenes controlled by aberrant miRNAs occurs, leading to cancer development.36, 37, 38, 39, 40 For example, lncRNA LPP antisense RNA 2 has carcinogenic effects and promotes cell proliferation and metastasis through microRNA (miR)-7-5.41 The well-studied tumorigenic lncRNA, which is highly upregulated in liver cancer (HULC), serves as a ceRNA network through miR-372.42 AGAP2 antisense RNA 1, a competitive lncRNA, functions as an oncogene and upregulates annexin A11 expression through miR-16-5p, promoting proliferative capacity in liver cancer.43 These ceRNA networks provide a novel perspective and offer insight into undetected biomarkers for the early diagnosis and treatment of cancer.
The Myc gene is located on chromosome 8q24.21, which is dysregulated in most human neoplasia. This region is frequently genetically amplified in various human cancers.44 Moreover, MYC is overexpressed in more than 50% of tumors, including HCC. In this study, we first used the median expression level of MYC to divide the 371 HCC samples into two groups for subsequent analysis: high Myc expression (Mychigh) tumor and low Myc expression (Myclow) tumor. We used the transcriptome sequencing data of 371 lncRNAs, 371 mRNAs, and 367 miRNAs of HCC tumor tissue and adjacent normal tissue from The Cancer Genome Atlas (TCGA) platform to identify key differentially expressed (DE) RNAs, and we constructed an associated Myc ceRNA network to reveal the underlying mechanism in HCC carcinogenesis. We identified a prognostic signature (long intergenic non-protein-coding [LINC] RNA 2691 [LINC02691] and LINC02499) that effectively predicts OS and has protective effects.
We constructed a critical ceRNA network that included four lncRNAs (LINC02691, LINC02499, LINC01354, and NAV2 antisense RNA 4 [NAV2-AS4]), one miRNA (miR-212-3p), and two mRNAs (SEC14-like protein 2 [SEC14L2] and solute carrier family 6 member 1 [SLC6A1]). This study provided a better understanding of the pathogenesis of liver cancer from the perspective of polygenic association, thus offering novel insights into targeted combination therapies.
Results
Study process of transcriptome data
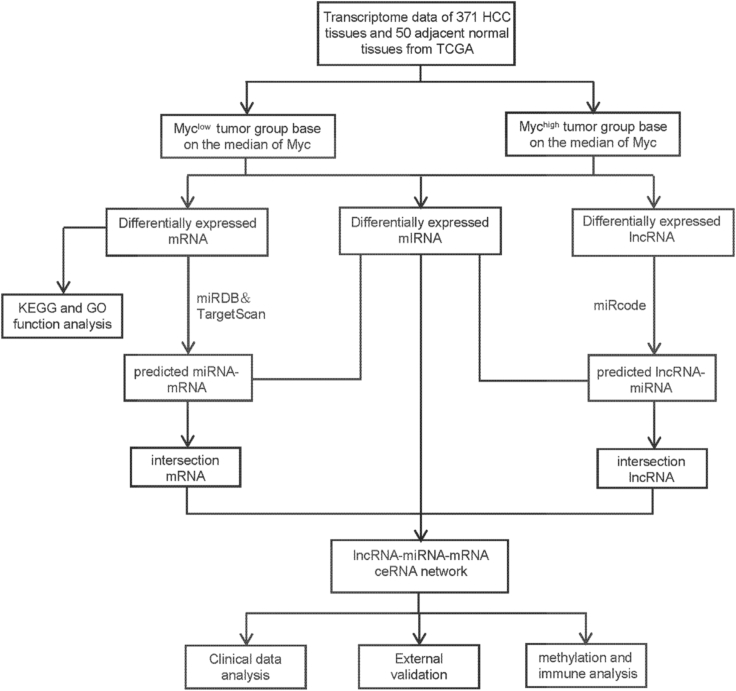
The study flow diagram is shown in Figure 1. We divided the transcriptome data of 371 HCC tissues into Mychigh tumor and Myclow tumor groups based on the standard median expression of Myc. We selected DE lncRNAs, miRNAs, and mRNAs from the Mychigh tumor group and Myclow tumor group. We considered lncRNA-miRNA to be a true interaction target by miRcode. We explored the miRDB database and TargetScan for miRNA-mRNA target prediction to construct a ceRNA network, followed by analyses of expression and survival.
Figure 1.
Research diagram of the Myc ceRNA network in HCC
Clinical analysis of Myc overexpression in HCC
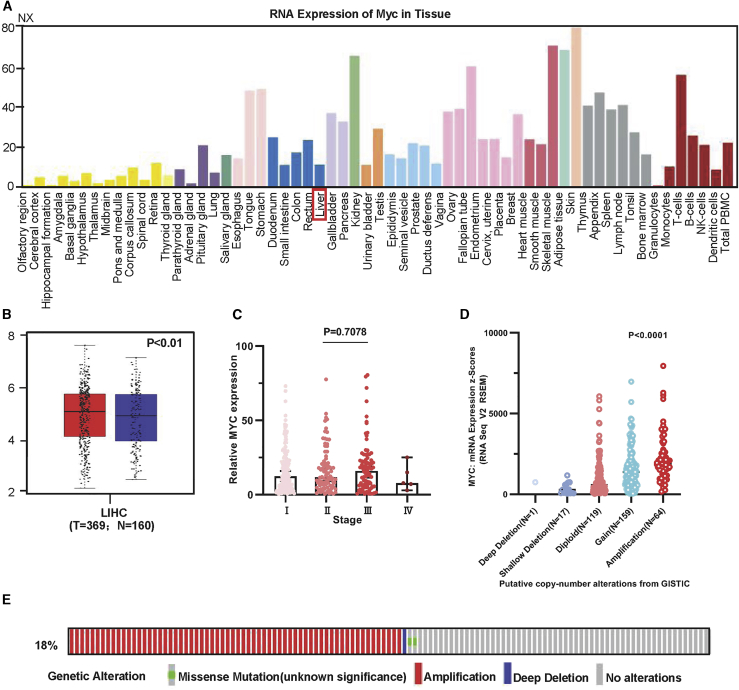
To explore whether Myc affects gene expression in liver cancer, we divided liver cancer patients into Mychigh tumor and Myclow tumor groups based on the median values of Myc in this study. The mRNA and protein levels of Myc were presented in various normal organs in the Human Protein Atlas (HPA) database (Figures 2A and S1A). The expression of Myc was high in cancer cell lines (Figure S1B). The region of Myc that is genomically altered in liver cancer was expressed through amplification and was found to be altered in 66 (18%) of 366 patients in HCC (Figures 2B and 2C). Myc expression was higher in HCC tissues (n = 369) compared with in normal liver tissues (n = 160, p < 0.01; Figure 2D). The expression of Myc increased as tumor invasion worsened (Figure 2E). Kaplan-Meier survival analysis showed the prognostic potential of Myc expression. These data suggested that Myc was upregulated in liver cancer samples.
Figure 2.
Functional analysis of Myc
(A) Transcription levels of Myc in various normal organs using the HPA database. (B and C) Frequency of genetic alterations associated with Myc. (D) Expression of Myc was analyzed between cancer tissues (n = 369) and normal tissues (n = 160) in HCC. (E) Distribution of Myc expression between TNM stages.
DE RNAs in HCC
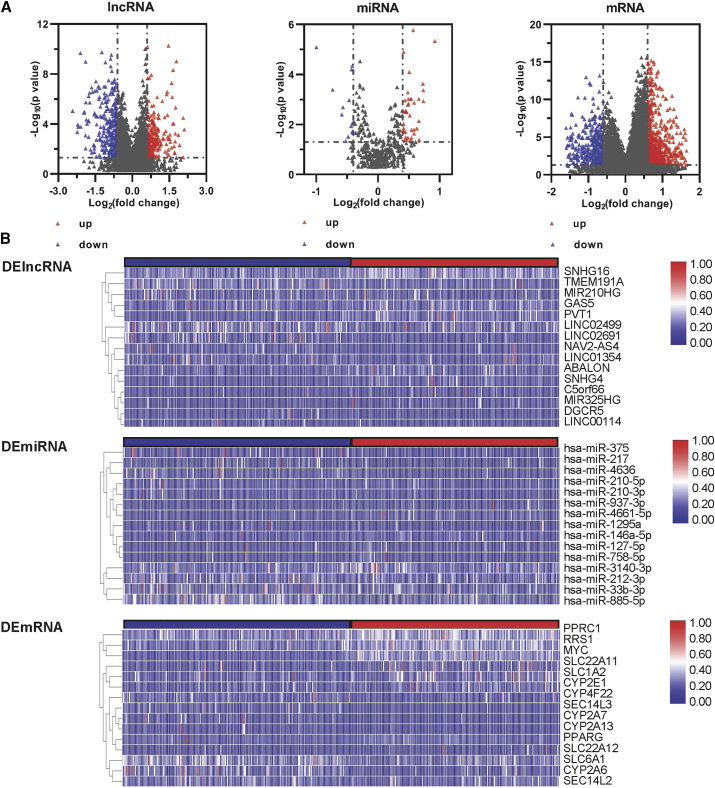
To better understand the relationship between DE RNAs and tumorigenesis-associated HCC, we downloaded gene-expression microarrays from TCGA database to search for DE RNAs. The gene-expression microarrays contained 371 samples (lncRNAs, mRNAs); 367 samples (miRNAs) were defined as Mychigh and Myclow based on the median expression levels of Myc.45 We analyzed the DE genes between the Mychigh and Myclow tumor groups with the standard of p < 0.05 and fold change (FC) ≥ 1.5. In total, we found 589 lncRNAs, 93 miRNAs, and 1,125 mRNAs of DE RNAs among the groups. The upregulated DE RNAs included 222 (38%) lncRNAs, 65 (70%) miRNAs, and 696 (62%) mRNAs. The downregulated DE RNAs included 367 (62%) lncRNAs, 38 (30%) miRNAs, and 429 (38%) mRNAs. In Figure 3, the volcano plots show the distribution of DE RNAs (Figure 3A), and the heatmap describes 15 significant DE RNAs (Figure 3B).
Figure 3.
Identification of DE genes
(A) Volcano maps of DE lncRNAs, miRNAs, and mRNAs between two groups: Mychigh and Myclow in HCC. (B) Heatmap of DE lncRNAs (top), miRNAs (middle), and mRNAs (bottom). Red and blue spots represent significantly upregulated and downregulated RNAs, respectively.
Functional enrichment analysis of DE mRNAs
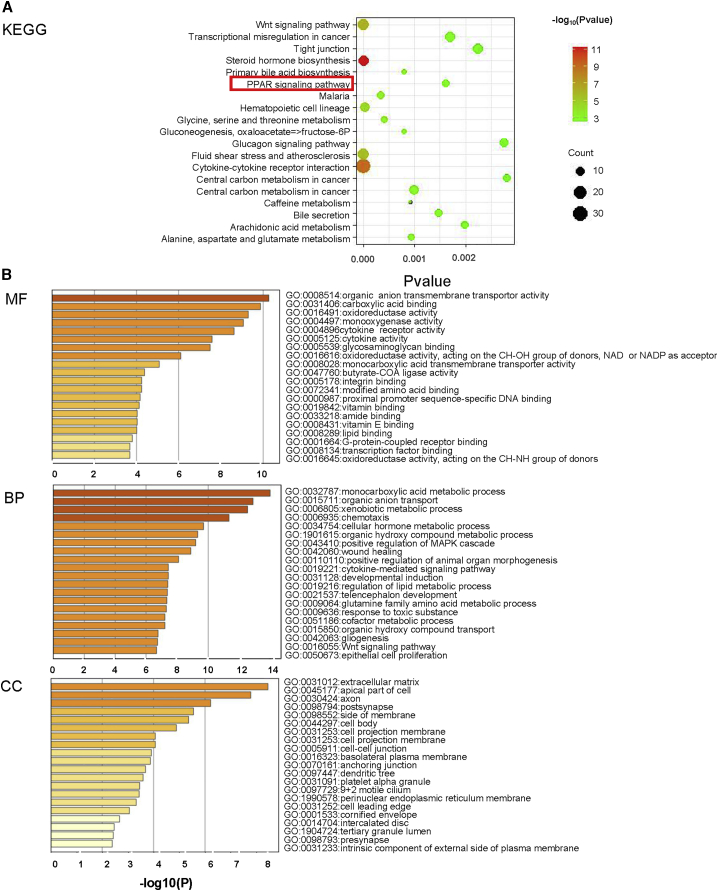
To better comprehend the mechanisms involved in HCC, we explored the function of 1,125 DE mRNAs from Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses using the Metascape database (Figures 4A and 4B). The most enriched GO terms in cellular component (CC), biological process (BP), and molecular function (MF) were “side of membrane,” “response to toxic substance,” and “organic anion transmembrane transporter activity,” respectively. We found the oxidation-reduction process, peroxisome proliferator-activated receptor (PPAR) signaling, and peroxisome pathway to participate in HCC, according to KEGG pathway enrichment analysis.
Figure 4.
Functional enrichment analysis of DE mRNAs
(A) KEGG pathway of DE mRNAs. (B) MF, BP, and CC of DE mRNAs.
Construction of the ceRNA network and identification of hub RNAs
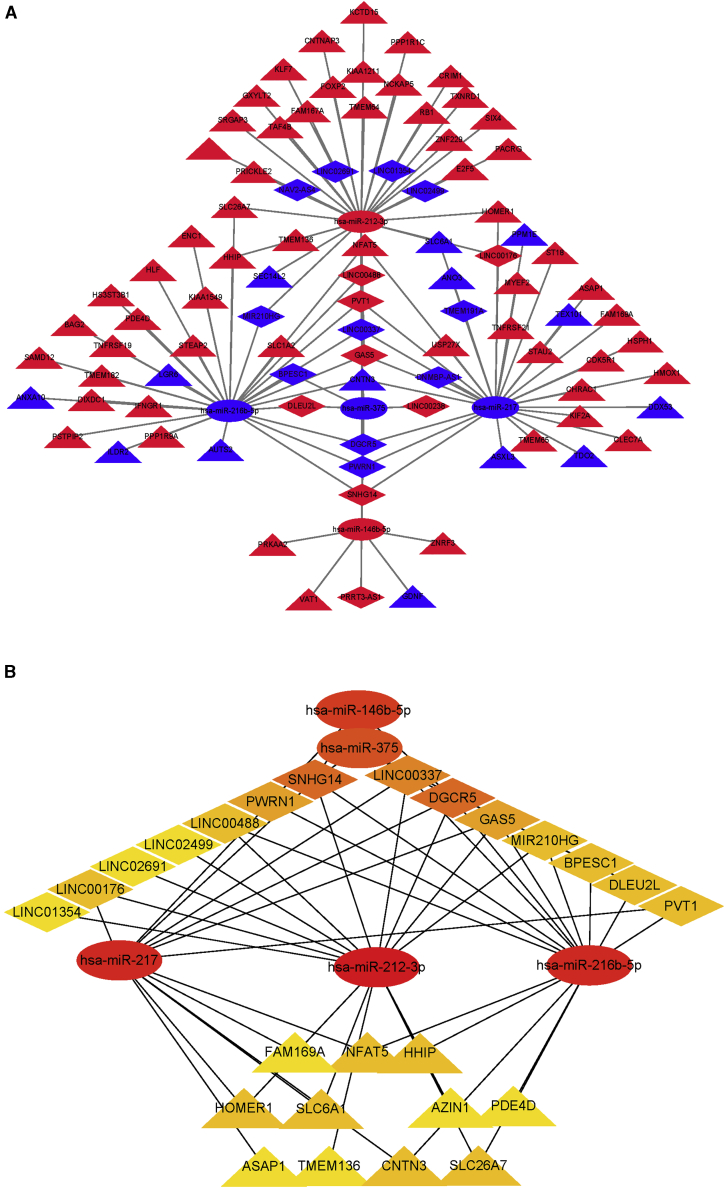
We used miRNA target informatic tools to identify DE RNAs in the ceRNA. The ceRNA network was constructed using Cytoscape 3.7 (Figure 5A) and included 19 lncRNAs, 5 miRNAs, and 72 mRNAs. The ceRNA network included 19 lncRNAs (11 downregulated and 8 upregulated) and 5 miRNAs (4 downregulated and 1 upregulated). In addition, 72 mRNAs (58 upregulated and 14 downregulated) and 5 miRNAs (4 downregulated and 1 upregulated) were used to construct the ceRNA network. We employed a large ceRNA network based on node-weighting arithmetic to recognize highly interacted hub clustering (Figure 5B). The hub RNAs contained 14 lncRNAs (6 upregulated and 8 downregulated), 5 miRNAs (2 upregulated and 3 downregulated), and 11 mRNAs (9 upregulated and 2 downregulated).
Figure 5.
Myc ceRNA network in HCC
(A) Blue rhombus represents downregulated lncRNAs, red ellipses represent upregulated miRNAs, and blue triangles represent downregulated mRNAs; red rhombus represents upregulated lncRNAs, blue ellipses represent downregulated miRNAs, and red triangles represent upregulated mRNAs. (B) The hub 30 genes in the ceRNA network.
Comprehensive analysis of DE lncRNAs in the ceRNA network
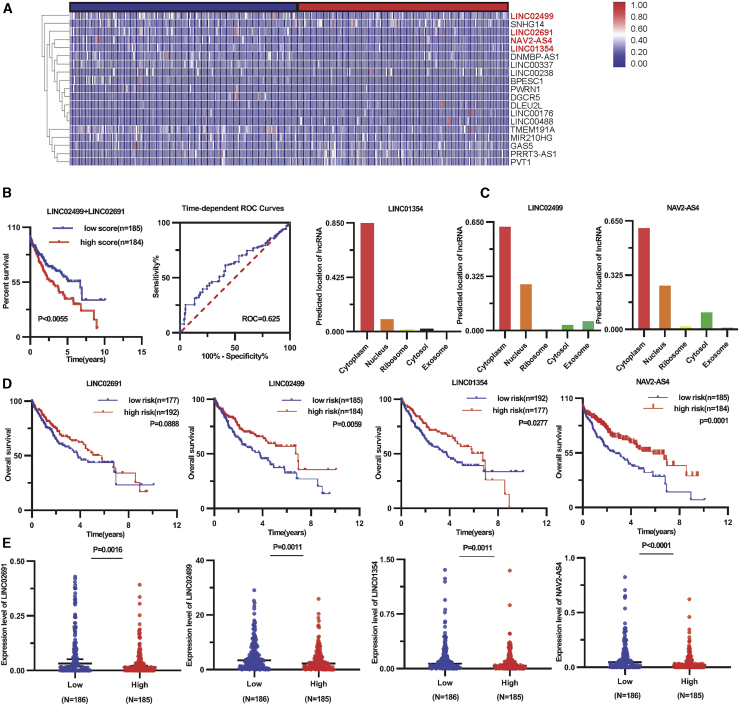
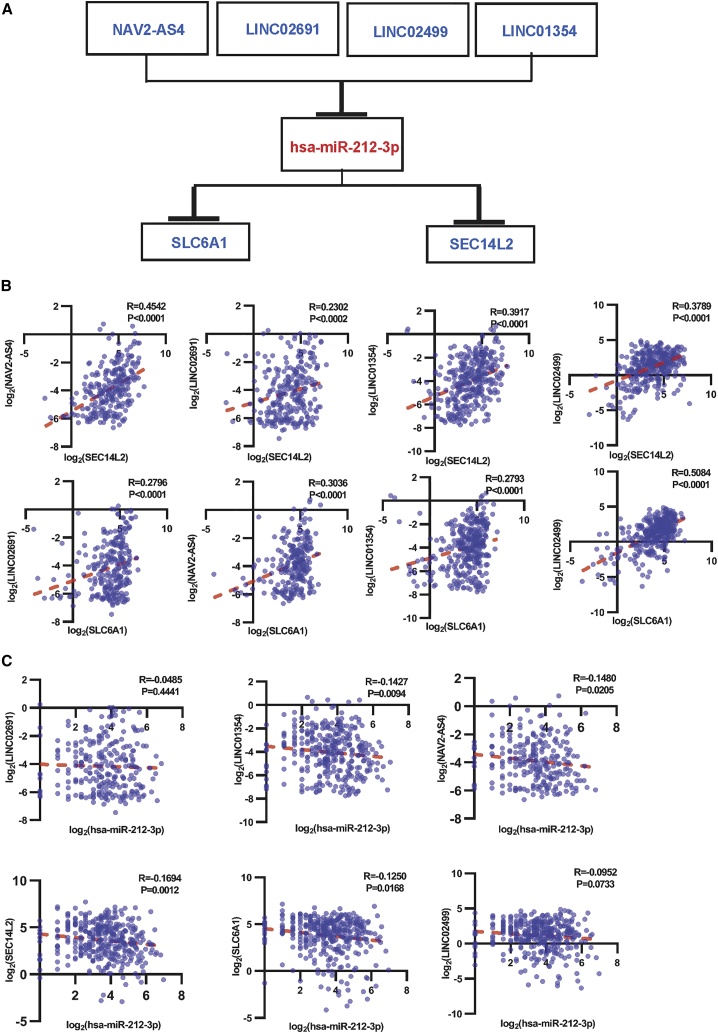
lncRNAs have an impact on the responses of miRNAs and mRNAs, commanding the upstream portion of the ceRNA network. The expression of lncRNAs is associated with OS in cancer patients.46, 47, 48, 49 Therefore, we analyzed 19 DE lncRNAs of ceRNA in this study and found differential expression in the Mychigh tumor group (n = 185) and Myclow tumor group (n = 186) using the heatmap (Figure 6A). Among the 19 DE lncRNAs, we screened potential prognosis-associated lncRNAs by univariate and multivariate Cox regression analyses. The final risk score method established that an lncRNA signal was a significant prognostic factor for HCC, as follows: prognostic index (PI) = (−0.6621 × expression level of LINC02691) + (−0.2047 × expression level of LINC02499). Notably, the classification of these two groups using the risk score improved the distinguishing ability and predictive power regarding OS based on Kaplan-Meier (p = 0.0055) and time-dependent receiver operating characteristic (ROC) curve analyses (area under the curve [AUC] = 0.625). LINC02691 and LINC02499 were found to be protective factors for HCC (Figure 6B). lncRNAs of the ceRNA network play a role in the cytoplasm and are involved in the regulation of downstream genes. Hence, we used the lncLocator online tool to predict the location of LINC02691, LINC02499, LINC01354, and NAV2-AS4. We found that the subcellular locations of LINC02691 did not possess a sequences’ record in LNCipedia, which was not predicted. The transcripts analysis of LINC02499, LINC01354, and NAV2-AS4 obtained from LNCipedia indicated their location in the cytoplasm using lncLocator online prediction (Figure 6C).
Figure 6.
Comprehensive analysis of DE lncRNAs in the ceRNA network
(A) Differential expression of 19 lncRNAs in the ceRNA network by a heatmap. (B) Validation of the two lncRNA signatures by Kaplan-Meier survival and ROC curves based on risk scores. (C) Prediction of lncRNA subcellular localization; a score ranging from 0 and 1 was given. (D) Expression of LINC02691, LINC02499, LINC01354, and NAV2-AS4 was analyzed in the Mychigh tumor group (n = 185) and Myclow tumor group (n = 186) in HCC. (E) Survival analysis.
To identify potentially significant RNAs regarding the survival of patients with HCC, we conducted DE analysis, clinical-stage analysis, and Kaplan-Meier survival curve for each RNA in the ceRNA network. A comparison of consequences follows: 4 lncRNAs (LINC02691, LINC02499, LINC01354, and NAV2-AS4) were suppressively expressed in the Mychigh tumor group (n = 185) relative to the Myclow tumor group (n = 186; Figure 6D); 4 lncRNAs were strongly correlated with OS (p < 0.05) (Figure 6E); and the remaining 15 lncRNAs were not significantly correlated with OS in HCC (Figure S2A).
We next explored the relationship of four lncRNAs according to clinical pathological stage. Following worsening tumor invasion, the expression of the four lncRNAs decreased (Figure S2B). As further verification of this finding, the difference in LINC02691, LINC02499, LINC01354, and NAV2-AS4 expression in liver cancer tissues (n = 371) was statistically significant (p < 0.05) compared with normal liver tissues (n = 50) (Figure S2C). The 50 paired cancer and paracancer tissues were employed to validate the results of DE lncRNAs (Figure S2D).
Functional analysis of miR-212-3p as a target
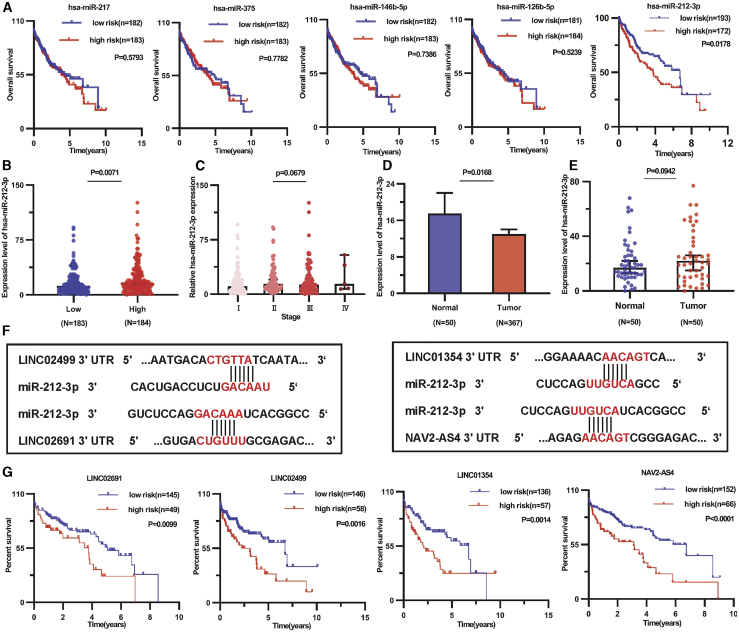
We searched for potentially significant miRNA in HCC. Five DE miRNAs (4 downregulated and 1 upregulated [miR-212-3p]) in the ceRNA network were associated with OS. We constructed a Kaplan-Meier survival curve for four miRNAs (Figure 7A). Moreover, patients with high miR-212-3p expression had a poor prognosis compared with those with low miR-212-3p expression. The results showed that miR-212-3p was a factor that promoted tumor growth. miR-212-3p was highly expressed in the Mychigh tumor group (n = 184) compared with the Myclow tumor group (n = 18; Figure 7B). The expression of miR-212-3p increased with high tumor-node-metastasis (TNM) stage in 371 liver cancers (Figure 7C).
Figure 7.
Expression and survival analysis of DE miRNAs
(A) Kaplan-Meier survival analysis of miR-212-3p, miR-217, miR-216b-5p, miR-375, and miR-146b-5p in 371 HCC tissues. (B) The miR-212-3p expressed in the Mychigh tumor group (n = 184) and Myclow tumor group (n = 183). (C) The miR-212-3p expression in the TNM stage in 371 liver cancer samples. (D) Expression level of miR-212-3p in 371 HCC tissues compared with 50 adjacent normal tissues. (E) In 50 pairs of HCC tissues, miR-212-3p was highly expressed in liver cancer. (F) A schematic diagram of the miR-212-3p of 3′ UTR in four lncRNA-binding sites. (G) The high-risk group had high miRNA expression and low lncRNA expression. The low-risk group had low miRNA expression and high lncRNA expression.
We performed further validation with cancer and paracancer data and compared adjacent normal tissues. We found that the expression level of miR-212-3p was significantly upregulated in 371 HCC tissues. In 50 paired HCC tissues, miR-212-3p was highly expressed in liver cancer relative to nontumor tissues (Figures 7D and 7E). In addition, through the informatics websites, we found that miR-212-3p included specific linking of the 3′ UTR region of the lncRNA gene. The 3′ UTR binding location of miR-212-3p of four lncRNAs is shown in Figure 7F. We combined miRNA and lncRNA to verify the ceRNA hypothesis. The high-risk group had high miRNA expression and low lncRNA expression, whereas the low-risk group had low miRNA expression and high lncRNA expression. We conducted survival analysis comparing the high-risk group with the low-risk group (Figure 7G).
Comprehensive analysis of DE mRNAs in the ceRNA network
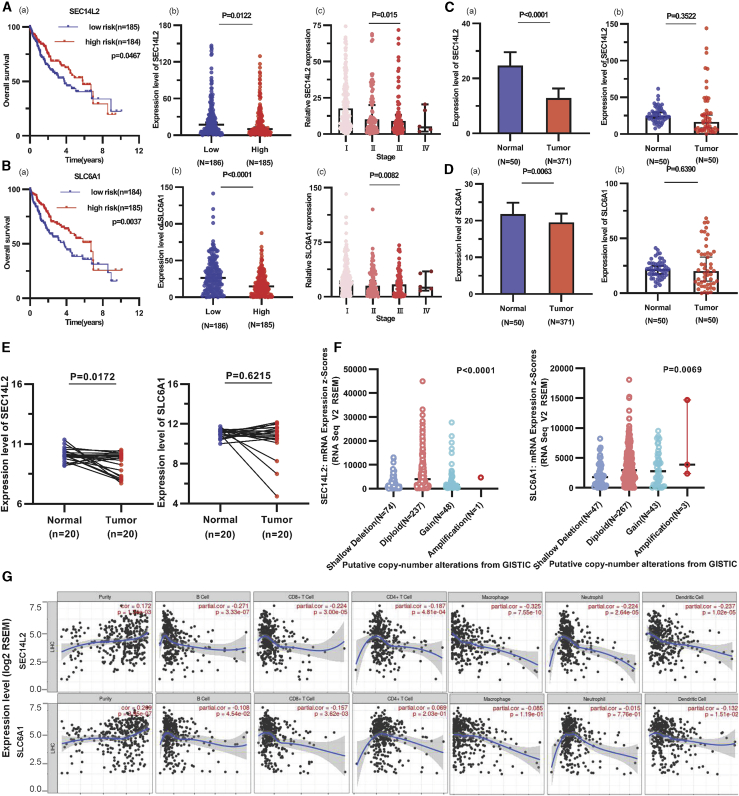
To further understand the biological functions, we explored the prognostic values of the 72 DE mRNAs involved in the ceRNA network. In the ceRNA network, we found that SEC14L2 and SLC6A1 were downstream target genes of miR-212-3p. According to Kaplan-Meier survival analysis, SEC14L2 and SLC6A1 were significantly associated with prolonged prognosis (p < 0.05), whereas the remaining the mRNAs in the ceRNA network had no survival significance in HCC (Figure S3). SEC14L2 and SLC6A1 were overexpressed in the Myclow tumor group and were associated with low-grade TNM stage (Figures 8A and 8B). Further validation showed that the expression of SEC14L2 and SLC6A in 371 liver cancers was lower than that in 50 paracancer samples; 50 samples were paired to explain the expression of the genes in cancer and paracancer tissues (Figures 8C and 8D). No genetic alterations were found in SEC14L2 and SLC6A1 in HCC; however, SEC14L2 was altered in 1 (0.3%) of 366 patients, and SLC6A1 was altered in 5 (1.4%) of 366 patients with liver cancer in the cBioPortal database. In addition, SEC14L2 and SLC6A1 had a lower expression in liver cancer (n = 10) compared with paired-normal tissue samples (n = 10) taken from the Gene Expression Omnibus (GEO) profiles (Figures 8E and 8F).
Figure 8.
Analysis of SEC14L2 and SLC6A1
(A and B) Comparison of SEC14L2 and SLC6A1 expression in neoplasm development and association with prolonged prognosis. (C and D) Verification of SEC14L2 and SLC6A1 expression in normal tissues and HCC tissues in liver. (E and F) Verification of SEC14L2 and SLC6A1 in HCC relative to paired normal tissue samples from GEO profiles. (G) Immune cell infiltration of SEC14L2 and SLC6A in HCC using the TIMER database.
Expression of SEC14L2 and SLC6A1 in various cancers
We analyzed the transcription and protein levels of SEC14L2 and SLC6A1 in various organ tissues using the HPA database (Figures S4 and S5). To further evaluate SEC14L2 and SLC6A1 expression in human cancer, we used RNA sequencing data to examine SEC14L2 and SLC6A1 expression between tumor and normal tissues. The expression of SEC14L2 was significantly lower in some cancers, including bladder urothelial carcinoma, cholangiocarcinoma, kidney chromophobe, lung adenocarcinoma, and HCC. SLC6A1 expression was lower in most cancers, except renal cell clear carcinoma and HCC (Figure S6). The expression of SEC14L2 and SLC6A1 was low in various cancer cell lines (Figure S7).
Association of immune infiltration with SEC14L2 and SLC6A1 expression in HCC
To explore the role of SEC14L2 and SLC6A1 in immune infiltration in HCC, we evaluated the relationship between differential expression gene and immune cell infiltration by the TIMER platform. A positive correlation between SEC14L2 expression and immune cell infiltration (Cor = 0.172, p = 1.34e-03). The most positive correlation of SEC14L2 and SLC6A1 expression was with B cells (Cor = −0.271, p = 3.33e-07), CD8+ T cells (Cor = −0.224, p = 3.00e-05), macrophages (Cor = −0.325, p = 7.55e-10), neutrophils (Cor = −0.224, p = 2.64e-05), and dendritic cells (Cor = −0.237, p = 1.02e-05) (Figure 8G). We researched the correlation between SEC14L2 and SLC6A1 expression and gene biomarkers of immune cells. The results were most strongly correlated with T cells and T cell exhaustion (Table S1). The correlation between SEC14L2 and gene biomarkers of immune cells in tumor and normal samples is shown in Table S2.
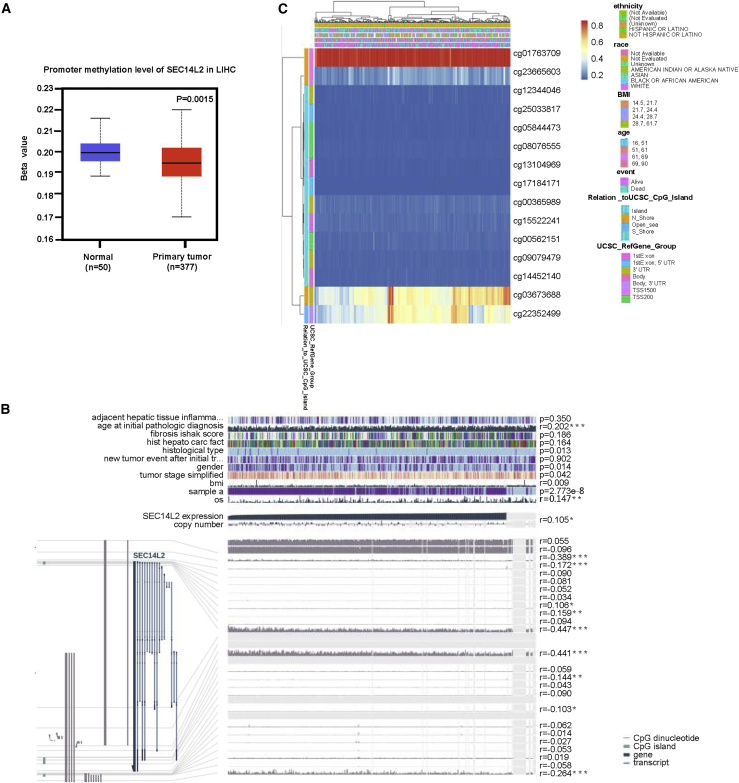
Correlation between methylation and SEC14L2 expression in HCC
The UALCAN database showed that SEC14L2 is highly methylated in the HCC tissue database (Figure 9A). We analyzed the relationship between SEC14L2 methylation and clinical information using the MEXPRESS database. We found significant methylation of SEC14L2 in various clinical factors, including new tumor events after initial treatment, histological type, gender, tumor stage, and OS. The methylation of SEC14L2 occurred on multiple sites, including cg03673688, cg22352499, and cg23665603 (r = 0.441, 0.447, and 0.389, respectively) (Figure 9B). We described the relationship between the methylation sites (cg03673688, cg22352499, and cg23665603) of SEC14L2 and clinical factors of patients using the MethSurv database (Figure 9C).
Figure 9.
Methylation of SEC14L2
(A) Methylation of SEC14L2 in HCC tissues was described using the UALCAN database. (B) Relationship between methylation of SEC14L2 and clinical factors by the MEXPRESS database. (C) Methylation sites (cg03673688, cg22352499, and cg23665603) of SEC14L2 described by the MethSurv database.
Construction of the lncRNA-miRNA-mRNA network
To verify the ceRNA mechanism in HCC, we constructed a lncRNA-miRNA-mRNA network after analysis, which included four lncRNAs, one miRNA, and two mRNAs (Figure 10A). We analyzed the correlation between Myc and these key RNAs (Figure S8A). We found that miR-212-3p is a connecting linker gene that participates in ceRNA pathways, whereas lncRNAs indirectly regulated mRNA expression by preferentially encompassing the miRNA response region. We analyzed LINC02691, LINC02499, LINC01354, and NAV2-AS4 and found that lncRNAs were positively correlated with SEC14L2 and SLC6A1 through the same miRNA, miR-212-3p (Figure 10B). Spearman correlation analysis showed that miR-212-3p was negatively regulated in lncRNAs and mRNAs of HCC (Figure 10C). The correlation among lncRNAs, miRNA, and mRNAs is shown (Figure S8B).
Figure 10.
Correlation of linear regression analysis between DE lncRNAs and mRNAs
(A) The identified lncRNA-miRNA-mRNA axis is integrated into a circuit map. (B) Relationship of lncRNAs and mRNAs. (C) Correlation between hsa-miR-212-3p and lncRNAs and mRNAs.
Relationship between key RNAs and clinical features
We analyzed the relationship between key RNAs (lncRNAs, miRNAs, and mRNAs) and clinical information, including age, gender, body mass index (BMI), race, TNM stage, distant metastasis, lymph node metastasis, diameter, and prior malignancy (Table S3). SEC14L2 and SLC6A1 had significant differences regarding BMI, tumor stage, and diameter (p < 0.05). TNM, diameter, lymph node metastasis, distant metastasis, and prior malignancy were significantly related to HCC prognosis (p < 0.05). LINC02499 was the lncRNA most significantly correlated with the clinical factors (Table S4).
Discussion
HCC is the fifth-most common cancer, with the second-highest mortality rate worldwide.50, 51, 52 The incidences of liver cancer and mortality have been increasing in the past few years, particularly in Asia. Traditional surgery is no longer the preferred treatment for HCC, so new therapeutic directions need to be explored.17,53, 54, 55 Molecular targeted therapy is an important topic for the treatment of HCC; however, the precise molecular mechanisms remain unknown. The development of high-throughput transcriptome sequencing technologies and the ceRNA hypothesis have been proposed, in which lncRNAs and mRNAs interact with each other through shared miRNAs.56, 57, 58, 59 Recently, studies have been conducted to construct a ceRNA network between HCC tissues and adjacent nontumor liver tissues.60 The molecular mechanisms of ceRNA associated with Myc remain unclear in HCC.
Myc is an oncogene that is dysregulated in >50% of tumors.61, 62, 63, 64 In this study, we first analyzed the large cohort of associated Myc transcriptome profiling with HCC using TCGA database. We selected DE lncRNAs, miRNAs, and mRNAs by comparing the Mychigh tumor tissues with Myclow tumor tissues based on Myc expression, and we constructed a ceRNA network that identified 1125 DE mRNAs, 589 DE lncRNAs, and 93 DE miRNAs.
Numerous studies have reported that lncRNAs play a critical role in different cancers.65, 66, 67, 68 lncRNA MIR503HG inhibits cell proliferation and promotes apoptosis in triple-negative breast cancer (TNBC) cells via the miR-224-5p/HOXA9 axis.69 lncITPF (long non-coding idiopathic pulmonary fibrosis) promotes pulmonary fibrosis by targeting hnRNP-L depending on its host gene ITGBL1.70 lncRNA LINC00858 functions through the miR-153-3p/Rabl3 axis, which promotes cell proliferation and infiltration in HCC.71 Therefore, we analyzed 19 DE lncRNAs in the ceRNA network by multidirectional analysis and calculated risk scores through univariate and multivariate Cox regression analyses. Our results showed that LINC02691, LINC02499, LINC01354, and NAV2-AS4 were significant in predicting OS in HCC, especially LINC02691 and LINC02499. Therefore, the prognostic signature of lncRNAs might act as a prognostic and diagnostic marker for HCC.
Among the prognostic signature lncRNAs, the oncogenic function of LINC01354 has been studied in a variety of cancers—in particular, in colorectal carcinoma, gastric carcinoma, lung carcinoma, and neck squamous cell carcinoma—in which LINC01354 accelerates the proliferation response, migration, and infiltration of cancer cells.72, 73, 74, 75, 76 This study found suppression of LINC01354 expression in Mychigh tumor tissues compared with Myclow tumor tissues. LINC02499 is highly expressed in HCC and inhibits cell-proliferation capacity, migration, and immune cell infiltration in HCC77 and was first analyzed in the associated Myc ceRNA network. Therefore, we successfully analyzed a series of lncRNAs with functions in the associated Myc ceRNA network. lncRNAs, including LINC02691 and LINC01354, were found to be strongly negatively regulated in HCC through the miR-212-3p/SEC14L2 axis.
miRNA connects lncRNA and mRNA in various cancers, including HCC, acting as a bridge for the ceRNA network.78, 79, 80, 81 miR-96 reduces HCC migration, functioning as a therapeutic target in this disease.82 miR-221-3p promotes HCC by downregulating the expression of O6-methylguanine-DNA methyltransferase.83 In this study, we analyzed five miRNAs in the ceRNA network, and only miR-212-3p overexpression was correlated with poor OS. The ceRNA network illustrates that hsa-miR-212-3p may promote cancer cell migration and invasion.84, 85, 86, 87
In this comprehensive study, we screened a ceRNA axis, including the downstream coding gene of SEC14L2 and SLC6A1. We analyzed downstream genes through GO enrichment analysis and KEGG pathway analysis. We found that the GO terms of the dysregulated mRNAs (SEC14L2 and SLC6A1) in HCC could be classified into MF, CC, and BP to further explain the pathways involved. In terms of MF, organic anion transmembrane transporter activity and vitamin E binding suggested that HCC may be a multigene-related disease. During this study, the PPAR signaling pathway was the most important KEGG pathway in the Metascape database, which also has been found in several other cancer types.88, 89, 90, 91, 92, 93, 94
SEC14L2-like phosphatidylinositol transfer proteins SEC14L3/SEC14L2 mediated Wnt/Ca2+ signaling by acting as GTPase proteins.95 SLC6A1 is overexpressed in prostate cancer and is associated with drug resistance and a poor prognosis96 but is downregulated in HCC.97 SEC14L2 and SLC6A1 mRNAs serve as novel transcriptional targets related to Myc and suppress HCC progression and thus may be a promising target for the treatment of Myc-driven HCC. Our results show that Kaplan-Meier survival analysis of ceRNA-correlated genes demonstrated that 19 of the 72 mRNAs had a statistically significant impact on prognosis. Many genes significantly influenced the OS of HCC patients; however, only SEC14L2 and SLC6A1 participated in establishing a complete endogenous regulation network.
This study had some limitations. Because of the lack of other HCC-associated samples, we did not perform further in vitro and in vivo experiments on clinical samples. Additionally, some exploratory experiments remain necessary to identify the functions of the unreported RNAs (lncRNAs, miRNAs, and mRNAs) in this study.
In summary, we introduced a Myc-ceRNA network from genome-wide transcriptome data by various bioinformatics analyses to provide a comprehensive analysis. This approach identified some key RNAs that were significantly associated with prognosis and that provide potential prognostic and diagnostic biomarkers for HCC.
Materials and methods
Data processing and analysis of DE proteins in HCC
We downloaded the transcriptome sequencing data of 421 lncRNAs and mRNAs and 417 miRNAs from 50 patients with HCC through TCGA data portal (https://portal.gdc.cancer.gov/). The 421 RNAs (417 miRNAs) were from 371 tumor tissues and 50 normal tissues. For external validation, we used 10 liver cancer samples and paracancer samples as validation sets from GEO profiles. To study the mechanism underlying the pathogenesis of liver cancer, we defined tumor samples as Mychigh tumor and Myclow tumor with standard median expression levels of MYC. We obtained DE mRNAs, lncRNAs, and miRNAs by comparing the Mychigh and Myclow tumor groups, with thresholds of p < 0.05 and a FC of ≥1.5.98
Functional enrichment analysis of DE mRNA
Metascape (http://metascape.org)99 is an informatics functional annotation tool that integrates many dominating databases to comprehensively analyze genes. We analyzed the enriched function of DE mRNAs using the GO and KEGG pathways in the Metascape website, with the restrictions of p < 0.01, a minimum count of 3, and an enrichment factor of >1.5. We visualized enriched GO pathways by Metascape and presented KEGG analyses using the “ggplot2” package of the R platform.
Construction of the ceRNA network and identification of hub RNAs
Cytoscape is a visualized software for network data, which provides users with a more picturesque biological process network.100 We used lncRNA as a true interaction miRNA target explored by miRcode (http://www.mircode.org/) and used miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/) for miRNA-mRNA target gene prediction. We employed these predicted relationship pairs to construct an endogenous competitive network based on the predicted miRNA expression relationship, which was visualized by Cytoscape version 3.7.0 software. We calculated the densely connected degree of gene nodes in a ceRNA network, and we employed the top 30 genes with the highest confidence scores as hub genes.
Prediction of the subcellular location of lncRNAs
The subcellular localization of lncRNA has an impact on its function. Therefore, we predicted the sequences of the significant lncRNA biomarkers obtained from the LNCipedia (https://lncipedia.org/). Then we used the sequences of lncRNAs to predict the subcellular localization in lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/). A score for each potential subcellular localization of lncRNA included the cytoplasm, nucleus, ribosome, cytosol, and exosome. We analyzed the final results using GraphPad Prism 8.3.
cBioPortal database analysis
cBioPortal (http://www.cbioportal.org/) is an online source application, which provides information on somatic mutations, altered copy number, and mRNA expression, used to visualize cancer genomics data.101, 102, 103, 104 In this study, we used cBioPortal to show SEC14L2 and SLC6A1 genetic changes in HCC.
Immune infiltrate analysis of SEC14L2 and SLC6A1
The TIMER database (http://cistrome.org/TIMER/) is a compositive resource that provides systematic analysis of the abundance of immune cells in the infiltrate of various cancers.105 We analyzed the expression of SEC14L2 and SLC6A1 in a variety of cancers, as well the correlation between their expression levels and the abundance of the immune cells. In addition, we evaluated the correlation of SEC14L2 and SLC6A1 expression with the biomarkers of immune cells. Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/) is an open database that contains tumor and normal samples from TCGA and Genotype-Tissue Expression databases. We used this database to confirm the correlation between DE mRNA and markers of immune cells in tumor and normal samples.
Methylation analysis of SEC14L2
The UALCAN database (http://ualcan.path.uab.edu/) is a data-mining platform, in which the methylation of DE mRNA in tumors can be queried.106, 107, 108 In this study, we used the UALCAN database to analyze SEC14L2 methylation in liver cancer tissues and paracancerous tissues. The MethSurv database (https://biit.cs.ut.ee/methsurv/) is an open website used to obtain CpG methylation data,109 and it contains significant information about a single CpG. We screened DE mRNA using the MethSurv database and then verified the most important methylated site associated with HCC patient outcomes. MEXPRESS (https://mexpress.be/) is a data-visualization tool used to visualize TCGA expression and the relationship between methylation expression and clinical information.110, 111, 112
The HPA database
The HPA database (https://www.proteinatlas.org/) was initiated in 2003 to map all human proteins in cells, tissues, and organs. In this study, we assembled the protein and RNA expression levels of SEC14L2 and SLC6A1 in various cancers tissues using this database.113, 114, 115
Cancer Cell Line Encyclopedia (CCLE) database analysis
CCLE (https://portals.broadinstitute.org/ccle) is an open-source website, which consists of a large amount of human cancer cell lines, including genomic data, gene expression, copy number, and abundant sequencing data. We analyzed the expression of SEC14L2 and SLC6A1 in HCC cell lines using the CCLE database.
Relationship between key RNAs and clinical features
We selected key RNAs according to the previously described analyses, which are presented as the median ± standard deviation. We conducted nonparametric tests to determine whether the expression of RNAs was correlated with the following clinical features: age (≥60 years versus <60 years), gender (female versus male), tumor stage (I + II versus III + IV); BMI (≤18.5, 18.6–23.9, 24–27.9, and ≥28), race (Asian versus White), diameter (≥5 cm versus <5 cm), lymph node and distant metastases (positive versus negative), and prior malignancy (yes versus no). p < 0.05 was used as the cutoff value.
Statistical analysis
We compared differential expression between the Mychigh and Myclow tumor groups using the nonparametric test. The correlation and survival analysis of the relative expression of the important lncRNA-miRNA-mRNA network were processed using GraphPad Prism 8.3. We explored potential lncRNAs using Cox regression analyses in the R package version 4.0. We completed all differential expression analyses using nonparametric tests. p < 0.05 was considered statistically significant.
Acknowledgments
We would like to thank Prof. Li-Ping Gu for data analysis and critical discussion of the manuscript. This study was supported partly by grants from the National Natural Science Foundation of China (81972214, 81772932, 81472202, and 81302065); Shanghai Natural Science Foundation (20ZR1472400); Natural Science Foundation of Hunan Province of China (2020WK2020); Construction of Clinical Medical Centre for Tumor Biological Samples in Nantong (HS2016004); Jiangsu 333 Program (BRA2017205); and Wu Jieping Medical Foundation (320.6750.14326).
Author contributions
Y.-S.M., D.F., and W.-J.Z. designed the study and contributed to study materials and consumables. D.-D.Z., Y.S., J.-B.L., X.-L.Y., R.X., H.-M.W., P.-Y.W., C.-Y.J., Y.-S.M., D.F., and W.-J.Z. conducted the study. D.-D.Z., Y.S., J.-B.L., X.-L.Y., R.X., Y.-S.M., and D.F. collected data. D.-D.Z., Y.S., D.F., and Y.-S.M. performed the statistical analyses and interpreted the data. D.-D.Z., Y.-S.M., and D.F. wrote the manuscript. All authors contributed to the final version of the manuscript and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.04.019.
Contributor Information
Wen-Jie Zhang, Email: zhangwj82@yahoo.com.
Yu-Shui Ma, Email: mayushui2006@126.com.
Da Fu, Email: fu800da900@126.com.
Supplemental information
References
- 1.Liu Z., Lin Y., Zhang J., Zhang Y., Li Y., Liu Z., Li Q., Luo M., Liang R., Ye J. Molecular targeted and immune checkpoint therapy for advanced hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019;38:447. doi: 10.1186/s13046-019-1412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Y.S., Huang T., Zhong X.M., Zhang H.W., Cong X.L., Xu H., Lu G.X., Yu F., Xue S.B., Lv Z.W., Fu D. Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol. Cancer. 2018;17:139. doi: 10.1186/s12943-018-0890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhen L., Zhao Q., Lü J., Deng S., Xu Z., Zhang L., Zhang Y., Fan H., Chen X., Liu Z. miR-301a-PTEN-AKT Signaling Induces Cardiomyocyte Proliferation and Promotes Cardiac Repair Post-MI. Mol. Ther. Nucleic Acids. 2020;22:251–262. doi: 10.1016/j.omtn.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Wang D., Liu B., Jin X., Wang X., Pan J., Tu W., Shao Y. IMP3 accelerates the progression of prostate cancer through inhibiting PTEN expression in a SMURF1-dependent way. J. Exp. Clin. Cancer Res. 2020;39:190. doi: 10.1186/s13046-020-01657-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Sun L.L., Xiao L., Du X.L., Hong L., Li C.L., Jiao J., Li W.D., Li X.Q. MiR-205 promotes endothelial progenitor cell angiogenesis and deep vein thrombosis recanalization and resolution by targeting PTEN to regulate Akt/autophagy pathway and MMP2 expression. J. Cell. Mol. Med. 2019;23:8493–8504. doi: 10.1111/jcmm.14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho D.W., Tsui Y.M., Sze K.M., Chan L.K., Cheung T.T., Lee E., Sham P.C., Tsui S.K., Lee T.K., Ng I.O. Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and stemness-related subpopulations in liver cancer. Cancer Lett. 2019;459:176–185. doi: 10.1016/j.canlet.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 8.Yu F., Chen B., Dong P., Zheng J. HOTAIR Epigenetically Modulates PTEN Expression via MicroRNA-29b: A Novel Mechanism in Regulation of Liver Fibrosis. Mol. Ther. 2020;28:2703. doi: 10.1016/j.ymthe.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchings C., Phillips J.A., Djamgoz M.B.A. Nerve input to tumours: Pathophysiological consequences of a dynamic relationship. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188411. doi: 10.1016/j.bbcan.2020.188411. [DOI] [PubMed] [Google Scholar]
- 10.Siemers N.O., Holloway J.L., Chang H., Chasalow S.D., Ross-MacDonald P.B., Voliva C.F., Szustakowski J.D. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS ONE. 2017;12:e0179726. doi: 10.1371/journal.pone.0179726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J., Dong Y., Ding L., Dong Y., Wu Z., Wang W., Shen M., Duan Y. Local delivery of arsenic trioxide nanoparticles for hepatocellular carcinoma treatment. Signal Transduct. Target. Ther. 2019;4:28. doi: 10.1038/s41392-019-0062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y., Zhang Y.N., Wang K.T., Chen L. Lenvatinib for hepatocellular carcinoma: From preclinical mechanisms to anti-cancer therapy. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188391. doi: 10.1016/j.bbcan.2020.188391. [DOI] [PubMed] [Google Scholar]
- 13.Jasirwan C.O.M., Hasan I., Sulaiman A.S., Lesmana C.R.A., Kurniawan J., Kalista K.F., Nababan S.H., Gani R.A. Risk factors of mortality in the patients with hepatocellular carcinoma: A multicenter study in Indonesia. Curr. Probl. Cancer. 2020;44:100480. doi: 10.1016/j.currproblcancer.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z., Yu W., Qiang Y., Xu L., Ma F., Ding P., Shi L., Chang W., Mei Y., Ma X. LukS-PV Inhibits Hepatocellular Carcinoma Progression by Downregulating HDAC2 Expression. Mol. Ther. Oncolytics. 2020;17:547–561. doi: 10.1016/j.omto.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei L., Wang X., Lv L., Liu J., Xing H., Song Y., Xie M., Lei T., Zhang N., Yang M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol. Cancer. 2019;18:147. doi: 10.1186/s12943-019-1086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Li G., Tao T., Kang X., Liu C., Zhang X., Wang C., Li C., Guo X. The μ-opioid receptor (MOR) promotes tumor initiation in hepatocellular carcinoma. Cancer Lett. 2019;453:1–9. doi: 10.1016/j.canlet.2019.03.038. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y., Jo M., Schmidt J., Luo X., Prakash T.P., Zhou T., Klein S., Xiao X., Post N., Yin Z., MacLeod A.R. Enhanced Potency of GalNAc-Conjugated Antisense Oligonucleotides in Hepatocellular Cancer Models. Mol. Ther. 2019;27:1547–1557. doi: 10.1016/j.ymthe.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong Z., Yu J., Yang S., Lai P.B.S., Chen G.G. FOX transcription factor family in hepatocellular carcinoma. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188376. doi: 10.1016/j.bbcan.2020.188376. [DOI] [PubMed] [Google Scholar]
- 19.Yuan F., Zhang Y., Ma L., Cheng Q., Li G., Tong T. Enhanced NOLC1 promotes cell senescence and represses hepatocellular carcinoma cell proliferation by disturbing the organization of nucleolus. Aging Cell. 2017;16:726–737. doi: 10.1111/acel.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q., Song G., Yao L., Liu Y., Liu M., Li S., Tang H. miR-3928v is induced by HBx via NF-κB/EGR1 and contributes to hepatocellular carcinoma malignancy by down-regulating VDAC3. J. Exp. Clin. Cancer Res. 2018;37:14. doi: 10.1186/s13046-018-0681-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Ping Y., Zhou Y., Hu J., Pang L., Xu C., Xiao Y. Dissecting the Functional Mechanisms of Somatic Copy-Number Alterations Based on Dysregulated ceRNA Networks across Cancers. Mol. Ther. Nucleic Acids. 2020;21:464–479. doi: 10.1016/j.omtn.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mu M., Niu W., Zhang X., Hu S., Niu C. LncRNA BCYRN1 inhibits glioma tumorigenesis by competitively binding with miR-619-5p to regulate CUEDC2 expression and the PTEN/AKT/p21 pathway. Oncogene. 2020;39:6879–6892. doi: 10.1038/s41388-020-01466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu W., Jing D., Meng Z., Hu B., Zhong B., Deng X., Jin X., Shao Z. FGD1 promotes tumor progression and regulates tumor immune response in osteosarcoma via inhibiting PTEN activity. Theranostics. 2020;10:2859–2871. doi: 10.7150/thno.41279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng L.-L., Cheng H.-H., Yeh T.-S., Huang S.-C., Syu Y.-Y., Chuu C.-P., Yuh C.-H., Kung H.-J., Wang W.-C. Targeting the histone demethylase PHF8-mediated PKCα-Src-PTEN axis in HER2-negative gastric cancer. Proc. Natl. Acad. Sci. USA. 2020;117:24859–24866. doi: 10.1073/pnas.1919766117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai L., Ye Y., Jiang Q., Chen Y., Lyu X., Li J., Wang S., Liu T., Cai H., Yao K. Author Correction: Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat. Commun. 2020;11:3437. doi: 10.1038/s41467-020-17048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X.Z., Cheng T.T., He Q.J., Lei Z.Y., Chi J., Tang Z., Liao Q.X., Zhang H., Zeng L.S., Cui S.Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol. Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Q., Li J., Xiong J., Song Z. Long noncoding RNA LINC00514 accelerates pancreatic cancer progression by acting as a ceRNA of miR-28-5p to upregulate Rap1b expression. J. Exp. Clin. Cancer Res. 2020;39:151. doi: 10.1186/s13046-020-01660-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Man X., Piao C., Lin X., Kong C., Cui X., Jiang Y. USP13 functions as a tumor suppressor by blocking the NF-kB-mediated PTEN downregulation in human bladder cancer. J. Exp. Clin. Cancer Res. 2019;38:259. doi: 10.1186/s13046-019-1262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi X., Xu R., Hong S., Xu T., Zhang W., Liu M. M2 Macrophage-Derived Exosomal lncRNA AFAP1-AS1 and MicroRNA-26a Affect Cell Migration and Metastasis in Esophageal Cancer. Mol. Ther. Nucleic Acids. 2020;22:779–790. doi: 10.1016/j.omtn.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z., Hu H. Identification of prognosis biomarkers of prostatic cancer in a cohort of 498 patients from TCGA. Curr. Probl. Cancer. 2019;43:100503. doi: 10.1016/j.currproblcancer.2019.100503. [DOI] [PubMed] [Google Scholar]
- 31.Tang G.D., Luo L.Y., Zhang J.L., Zhai D.F., Huang D.Q., Yin J., Zhou Q., Zhang Q., Zheng G.P. LncRNA LINC01057 promotes mesenchymal differentiation by activating NF-κB signaling in glioblastoma. Cancer Lett. 2021;498:152–164. doi: 10.1016/j.canlet.2020.10.047. [DOI] [PubMed] [Google Scholar]
- 32.Tong Y., Yang L., Yu C., Zhu W., Zhou X., Xiong Y., Wang W., Ji F., He D., Cao X. Tumor-Secreted Exosomal lncRNA POU3F3 Promotes Cisplatin Resistance in ESCC by Inducing Fibroblast Differentiation into CAFs. Mol. Ther. Oncolytics. 2020;18:1–13. doi: 10.1016/j.omto.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Huang Y.X., Wang D.L., Yang B., Yan H.Y., Lin L.H., Li Y., Chen J., Xie L.M., Huang Y.S. LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network. Theranostics. 2020;10:10823–10837. doi: 10.7150/thno.47830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou R., Sun H., Zheng S., Zhang J., Zeng D., Wu J., Huang Z., Rong X., Bin J., Liao Y. A stroma-related lncRNA panel for predicting recurrence and adjuvant chemotherapy benefit in patients with early-stage colon cancer. J. Cell. Mol. Med. 2020;24:3229–3241. doi: 10.1111/jcmm.14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin L., Que Y., Lu P., Li H., Xiao M., Zhu X., Li D. Chidamide Inhibits Acute Myeloid Leukemia Cell Proliferation by lncRNA VPS9D1-AS1 Downregulation via MEK/ERK Signaling Pathway. Front. Pharmacol. 2020;11:569651. doi: 10.3389/fphar.2020.569651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng W., Zhang C., Peng J., Huang Y., Peng C., Tan Y., Ji D., Zhang Y., Zhang D., Tang J. Lnc-FAM84B-4 acts as an oncogenic lncRNA by interacting with protein hnRNPK to restrain MAPK phosphatases-DUSP1 expression. Cancer Lett. 2020;494:94–106. doi: 10.1016/j.canlet.2020.08.036. [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Yin Z., Fan J., Zhang S., Yang W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct. Target. Ther. 2019;4:47. doi: 10.1038/s41392-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu Y., Tang Q., Feng H., Zhu L., Wang Y. lncRNA KTN1-AS1 promotes glioma cell proliferation and invasion by negatively regulating miR-505-3p. Oncol. Rep. 2020;44:2645–2655. doi: 10.3892/or.2020.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aleksakhina S.N., Kashyap A., Imyanitov E.N. Mechanisms of acquired tumor drug resistance. Biochim. Biophys. Acta Rev. Cancer. 2019;1872:188310. doi: 10.1016/j.bbcan.2019.188310. [DOI] [PubMed] [Google Scholar]
- 40.Zhang P.F., Pei X., Li K.S., Jin L.N., Wang F., Wu J., Zhang X.M. Correction to: Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol. Cancer. 2020;19:21. doi: 10.1186/s12943-020-1131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X., Niu W., Mu M., Hu S., Niu C. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J. Exp. Clin. Cancer Res. 2020;39:196. doi: 10.1186/s13046-020-01695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaker O., Mahfouz H., Salama A., Medhat E. Long Non-Coding HULC and miRNA-372 as Diagnostic Biomarkers in Hepatocellular Carcinoma. Rep. Biochem. Mol. Biol. 2020;9:230–240. doi: 10.29252/rbmb.9.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z., Wang Y., Wang L., Yao B., Sun L., Liu R., Chen T., Niu Y., Tu K., Liu Q. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019;38:194. doi: 10.1186/s13046-019-1188-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Le Grand M., Mukha A., Püschel J., Valli E., Kamili A., Vittorio O., Dubrovska A., Kavallaris M. Interplay between MycN and c-Myc regulates radioresistance and cancer stem cell phenotype in neuroblastoma upon glutamine deprivation. Theranostics. 2020;10:6411–6429. doi: 10.7150/thno.42602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas Research Network. Electronic address. wheeler@bcm.edu. Cancer Genome Atlas Research Network Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pang B., Zhu Y., Ni J., Thompson J., Malouf D., Bucci J., Graham P., Li Y. Extracellular vesicles: the next generation of biomarkers for liquid biopsy-based prostate cancer diagnosis. Theranostics. 2020;10:2309–2326. doi: 10.7150/thno.39486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min L., Zhu S., Wei R., Zhao Y., Liu S., Li P., Zhang S. Integrating SWATH-MS Proteomics and Transcriptome Analysis Identifies CHI3L1 as a Plasma Biomarker for Early Gastric Cancer. Mol. Ther. Oncolytics. 2020;17:257–266. doi: 10.1016/j.omto.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heller L., Thinard R., Chevalier M., Arpag S., Jing Y., Greferath R., Heller R., Nicolau C. Secretion of proteins and antibody fragments from transiently transfected endothelial progenitor cells. J. Cell. Mol. Med. 2020;24:8772–8778. doi: 10.1111/jcmm.15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Zhu S., Meng N., He Y., Lu R., Yan G.R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol. Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Y.S., Chu K.J., Ling C.C., Wu T.M., Zhu X.C., Liu J.B., Yu F., Li Z.Z., Wang J.H., Gao Q.X. Long Noncoding RNA OIP5-AS1 Promotes the Progression of Liver Hepatocellular Carcinoma via Regulating the hsa-miR-26a-3p/EPHA2 Axis. Mol. Ther. Nucleic Acids. 2020;21:229–241. doi: 10.1016/j.omtn.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Chen Z., Zheng Z., Feng L., Huo Z., Huang L., Fu M., Chen Q., Ke Y., Yang J., Hou B. Overexpression of miR-382 Sensitizes Hepatocellular Carcinoma Cells to γδ T Cells by Inhibiting the Expression of c-FLIP. Mol. Ther. Oncolytics. 2020;18:467–475. doi: 10.1016/j.omto.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teng F., Zhang J.X., Chang Q.M., Wu X.B., Tang W.G., Wang J.F., Feng J.F., Zhang Z.P., Hu Z.Q. LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2020;39:235. doi: 10.1186/s13046-020-01739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fergusson D.A., Wesch N.L., Leung G.J., MacNeil J.L., Conic I., Presseau J., Cobey K.D., Diallo J.S., Auer R., Kimmelman J. Assessing the Completeness of Reporting in Preclinical Oncolytic Virus Therapy Studies. Mol. Ther. Oncolytics. 2019;14:179–187. doi: 10.1016/j.omto.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma Y.S., Wang X.F., Yu F., Wu T.M., Liu J.B., Zhang Y.J., Xia Q., Jiang Z.Y., Lin Q.L., Fu D. Inhibition of USP14 and UCH37 deubiquitinating activity by b-AP15 as a potential therapy for tumors with p53 deficiency. Signal Transduct. Target. Ther. 2020;5:30. doi: 10.1038/s41392-020-0143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu H., Chen G.F., Ma Y.S., Zhang H.W., Zhou Y., Liu G.H., Chen D.Y., Ping J., Liu Y.H., Mou X., Fu D. Hepatic Proteomic Changes and Sirt1/AMPK Signaling Activation by Oxymatrine Treatment in Rats With Non-alcoholic Steatosis. Front. Pharmacol. 2020;11:216. doi: 10.3389/fphar.2020.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao B., Ke K., Wang Y., Wang F., Shi Y., Zheng X., Yang X., Liu X., Liu J. HIF-1α and HDAC1 mediated regulation of FAM99A-miR92a signaling contributes to hypoxia induced HCC metastasis. Signal Transduct. Target. Ther. 2020;5:118. doi: 10.1038/s41392-020-00223-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao Y., Najeeb R.M., Ma D., Yang K., Zhong Q., Liu Q. Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms. J. Exp. Clin. Cancer Res. 2019;38:458. doi: 10.1186/s13046-019-1444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tam B.Y., Chiu K., Chung H., Bossard C., Nguyen J.D., Creger E., Eastman B.W., Mak C.C., Ibanez M., Ghias A. The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett. 2020;473:186–197. doi: 10.1016/j.canlet.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 59.Li M., Shao J., Guo Z., Jin C., Wang L., Wang F., Jia Y., Zhu Z., Zhang Z., Zhang F. Novel mitochondrion-targeting copper(II) complex induces HK2 malfunction and inhibits glycolysis via Drp1-mediating mitophagy in HCC. J. Cell. Mol. Med. 2020;24:3091–3107. doi: 10.1111/jcmm.14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y., Zhao H., Li H., Feng X., Tang H., Qiu C., Zhang J., Fu B. LINC01234/MicroRNA-31-5p/MAGEA3 Axis Mediates the Proliferation and Chemoresistance of Hepatocellular Carcinoma Cells. Mol. Ther. Nucleic Acids. 2020;19:168–178. doi: 10.1016/j.omtn.2019.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Shao Y., Song X., Jiang W., Chen Y., Ning Z., Gu W., Jiang J. MicroRNA-621 acts as a tumor radiosensitizer by directly targeting SETDB1 in hepatocellular carcinoma. Mol. Ther. 2019;27:355–364. doi: 10.1016/j.ymthe.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng C., Liu S., Lu S., Yu X., Lai J., Wu Y., Chen S., Wang L., Yu Z., Luo G., Li Y. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol. Cancer. 2018;17:130. doi: 10.1186/s12943-018-0884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van den Ende T., van den Boorn H.G., Hoonhout N.M., van Etten-Jamaludin F.S., Meijer S.L., Derks S., de Gruijl T.D., Bijlsma M.F., van Oijen M.G.H., van Laarhoven H.W.M. Priming the tumor immune microenvironment with chemo(radio)therapy: A systematic review across tumor types. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188386. doi: 10.1016/j.bbcan.2020.188386. [DOI] [PubMed] [Google Scholar]
- 64.Wang H.L., Liu P.F., Yue J., Jiang W.H., Cui Y.L., Ren H., Wang H., Zhuang Y., Liu Y., Jiang D. Somatic gene mutation signatures predict cancer type and prognosis in multiple cancers with pan-cancer 1000 gene panel. Cancer Lett. 2020;470:181–190. doi: 10.1016/j.canlet.2019.11.022. [DOI] [PubMed] [Google Scholar]
- 65.Cheng J., Zhuo H., Wang L., Zheng W., Chen X., Hou J., Zhao J., Cai J. Identification of the Combinatorial Effect of miRNA Family Regulatory Network in Different Growth Patterns of GC. Mol. Ther. Oncolytics. 2020;17:531–546. doi: 10.1016/j.omto.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian X., Yu H., Li D., Jin G.J., Dai S.D., Gong P.C., Kong C.C., Wang X.J. The miR-5694/AF9/Snail axis provides metastatic advantages and a therapeutic target in basal-like breast cancer. Mol. Ther. 2021;29:1239–1257. doi: 10.1016/j.ymthe.2020.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hewes A.M., Sansbury B.M., Barth S., Tarcic G., Kmiec E.B. gRNA Sequence Heterology Tolerance Catalyzed by CRISPR/Cas in an In Vitro Homology-Directed Repair Reaction. Mol. Ther. Nucleic Acids. 2020;20:568–579. doi: 10.1016/j.omtn.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bao W.D., Zhou X.T., Zhou L.T., Wang F., Yin X., Lu Y., Zhu L.Q., Liu D. Targeting miR-124/Ferroportin signaling ameliorated neuronal cell death through inhibiting apoptosis and ferroptosis in aged intracerebral hemorrhage murine model. Aging Cell. 2020;19:e13235. doi: 10.1111/acel.13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S.M., Pang J., Zhang K.J., Zhou Z.Y., Chen F.Y. lncRNA MIR503HG inhibits cell proliferation and promotes apoptosis in TNBC cells via the miR-224-5p/HOXA9 axis. Mol. Ther. Oncolytics. 2021;21:62–73. doi: 10.1016/j.omto.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song X., Xu P., Meng C., Song C., Blackwell T.S., Li R., Li H., Zhang J., Lv C. lncITPF Promotes Pulmonary Fibrosis by Targeting hnRNP-L Depending on Its Host Gene ITGBL1. Mol. Ther. 2019;27:380–393. doi: 10.1016/j.ymthe.2018.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qi W.Y., Mao X.B., He Y.B., Xiao C.H. Long non-coding RNA LINC00858 promotes cells proliferation and invasion through the miR-153-3p/Rabl3 axis in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020;24:9343–9352. doi: 10.26355/eurrev_202009_23017. [DOI] [PubMed] [Google Scholar]
- 72.Cui J., Wang L., Zhong W., Chen Z., Chen J., Yang H., Liu G. Identification and validation of methylation-driven genes prognostic signature for recurrence of laryngeal squamous cell carcinoma by integrated bioinformatics analysis. Cancer Cell Int. 2020;20:472. doi: 10.1186/s12935-020-01567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang Y., Luo Y. LINC01354 Promotes Osteosarcoma Cell Invasion by Up-regulating Integrin β1. Arch. Med. Res. 2020;51:115–123. doi: 10.1016/j.arcmed.2019.12.016. [DOI] [PubMed] [Google Scholar]
- 74.Li R., Yang Y.E., Yin Y.H., Zhang M.Y., Li H., Qu Y.Q. Methylation and transcriptome analysis reveal lung adenocarcinoma-specific diagnostic biomarkers. J. Transl. Med. 2019;17:324. doi: 10.1186/s12967-019-2068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J., He M., Xu W., Huang S. LINC01354 interacting with hnRNP-D contributes to the proliferation and metastasis in colorectal cancer through activating Wnt/β-catenin signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:161. doi: 10.1186/s13046-019-1150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang G., Yang C., She Y., Shen Z., Gao P. LINC01354 enhances the proliferation and invasion of lung cancer cells by regulating miR-340-5p/ATF1 signaling pathway. Artif. Cells Nanomed. Biotechnol. 2019;47:3737–3744. doi: 10.1080/21691401.2019.1667816. [DOI] [PubMed] [Google Scholar]
- 77.Ma X., Mo M., Tan H.J.J., Tan C., Zeng X., Zhang G., Huang D., Liang J., Liu S., Qiu X. LINC02499, a novel liver-specific long non-coding RNA with potential diagnostic and prognostic value, inhibits hepatocellular carcinoma cell proliferation, migration, and invasion. Hepatol. Res. 2020;50:726–740. doi: 10.1111/hepr.13491. [DOI] [PubMed] [Google Scholar]
- 78.Hu J., Chen Z., Bao L., Zhou L., Hou Y., Liu L., Xiong M., Zhang Y., Wang B., Tao Z., Chan K. Single-Cell Transcriptome Analysis Reveals Intratumoral Heterogeneity in ccRCC, which Results in Different Clinical Outcomes. Mol. Ther. 2020;28:1658–1672. doi: 10.1016/j.ymthe.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kawamura E., Maruyama M., Abe J., Sudo A., Takeda A., Takada S., Yokota T., Kinugawa S., Harashima H., Yamada Y. Validation of Gene Therapy for Mutant Mitochondria by Delivering Mitochondrial RNA Using a MITO-Porter. Mol. Ther. Nucleic Acids. 2020;20:687–698. doi: 10.1016/j.omtn.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elsedawy N.B., Nace R.A., Russell S.J., Schulze A.J. Oncolytic Activity of Targeted Picornaviruses Formulated as Synthetic Infectious RNA. Mol. Ther. Oncolytics. 2020;17:484–495. doi: 10.1016/j.omto.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gupta S.C., Awasthee N., Rai V., Chava S., Gunda V., Challagundla K.B. Long non-coding RNAs and nuclear factor-κB crosstalk in cancer and other human diseases. Biochim. Biophys. Acta Rev. Cancer. 2020;1873:188316. doi: 10.1016/j.bbcan.2019.188316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iwai N., Yasui K., Tomie A., Gen Y., Terasaki K., Kitaichi T., Soda T., Yamada N., Dohi O., Seko Y. Oncogenic miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in hepatocellular carcinoma. Int. J. Oncol. 2018;53:237–245. doi: 10.3892/ijo.2018.4369. [DOI] [PubMed] [Google Scholar]
- 83.Chen Z., Xiang B., Qi L., Zhu S., Li L. miR-221-3p promotes hepatocellular carcinogenesis by downregulating O6-methylguanine-DNA methyltransferase. Cancer Biol. Ther. 2020;21:915–926. doi: 10.1080/15384047.2020.1806642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J., Cui R., Liu Y. MicroRNA-212-3p inhibits paclitaxel resistance through regulating epithelial-mesenchymal transition, migration and invasion by targeting ZEB2 in human hepatocellular carcinoma. Oncol. Lett. 2020;20:23. doi: 10.3892/ol.2020.11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghosh S., Bhowmik S., Majumdar S., Goswami A., Chakraborty J., Gupta S., Aggarwal S., Ray S., Chatterjee R., Bhattacharyya S. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int. J. Cancer. 2020;147:2934–2947. doi: 10.1002/ijc.33111. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y., Cao Y., Cai W., Wu L., Zhao P., Liu X.G. Aberrant expression of two miRNAs promotes proliferation, hepatitis B virus amplification, migration and invasion of hepatocellular carcinoma cells: evidence from bioinformatic analysis and experimental validation. PeerJ. 2020;8:e9100. doi: 10.7717/peerj.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X., Liao X., Huang K., Zeng X., Liu Z., Zhou X., Yu T., Yang C., Yu L., Wang Q. Clustered microRNAs hsa-miR-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for hepatocellular carcinoma. J. Cancer. 2019;10:2520–2533. doi: 10.7150/jca.29207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen S.M., Ji Y., Zhang C., Dong S.S., Yang S., Xiong Z., Ge M.K., Yu Y., Xia L., Guo M. Nuclear PTEN safeguards pre-mRNA splicing to link Golgi apparatus for its tumor suppressive role. Nat. Commun. 2018;9:2392. doi: 10.1038/s41467-018-04760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen J., Liu A., Lin Z., Wang B., Chai X., Chen S., Lu W., Zheng M., Cao T., Zhong M. Downregulation of the circadian rhythm regulator HLF promotes multiple-organ distant metastases in non-small cell lung cancer through PPAR/NF-κb signaling. Cancer Lett. 2020;482:56–71. doi: 10.1016/j.canlet.2020.04.007. [DOI] [PubMed] [Google Scholar]
- 90.Huang Z., Yu H., Du G., Han L., Huang X., Wu D., Han X., Xia Y., Wang X., Lu C. Enhancer RNA lnc-CES1-1 inhibits decidual cell migration by interacting with RNA-binding protein FUS and activating PPARγ in URPL. Mol. Ther. Nucleic Acids. 2021;24:104–112. doi: 10.1016/j.omtn.2021.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zeng J., Tang Z., Zhang Y., Tong X., Dou J., Gao L., Ding S., Lu J. Ozonated autohemotherapy elevates PPAR-γ expression in CD4+ T cells and serum HDL-C levels, a potential immunomodulatory mechanism for treatment of psoriasis. Am. J. Transl. Res. 2021;13:349–359. [PMC free article] [PubMed] [Google Scholar]
- 92.Yu Q., Cheng P., Wu J., Guo C. PPARγ/NF-κB and TGF-β1/Smad pathway are involved in the anti-fibrotic effects of levo-tetrahydropalmatine on liver fibrosis. J. Cell. Mol. Med. 2021;25:1645–1660. doi: 10.1111/jcmm.16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen H.X., Li M.Y., Jiang Y.Y., Hou H.T., Wang J., Liu X.C., Yang Q., He G.W. Role of the PPAR pathway in atrial fibrillation associated with heart valve disease: transcriptomics and proteomics in human atrial tissue. Signal Transduct. Target. Ther. 2020;5:4. doi: 10.1038/s41392-019-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miao Y., Zheng Y., Geng Y., Yang L., Cao N., Dai Y., Wei Z. The role of GLS1-mediated glutaminolysis/2-HG/H3K4me3 and GSH/ROS signals in Th17 responses counteracted by PPARγ agonists. Theranostics. 2021;11:4531–4548. doi: 10.7150/thno.54803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Z., Lou Y., Tian G., Wu J., Lu A., Chen J., Xu B., Shi J., Yang J. Discovering master regulators in hepatocellular carcinoma: one novel MR, SEC14L2 inhibits cancer cells. Aging (Albany NY) 2019;11:12375–12411. doi: 10.18632/aging.102579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen C., Cai Z., Zhuo Y., Xi M., Lin Z., Jiang F., Liu Z., Wan Y., Zheng Y., Li J. Overexpression of SLC6A1 associates with drug resistance and poor prognosis in prostate cancer. BMC Cancer. 2020;20:289. doi: 10.1186/s12885-020-06776-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kudo M., Han K.H., Ye S.L., Zhou J., Huang Y.H., Lin S.M., Wang C.K., Ikeda M., Chan S.L., Choo S.P. A Changing Paradigm for the Treatment of Intermediate-Stage Hepatocellular Carcinoma: Asia-Pacific Primary Liver Cancer Expert Consensus Statements. Liver Cancer. 2020;9:245–260. doi: 10.1159/000507370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gagan J., Van Allen E.M. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7:80. doi: 10.1186/s13073-015-0203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li S., Cui Z., Gu J., Wang Y., Tang S., Chen J. Effect of porcine corneal stromal extract on keratocytes from SMILE-derived lenticules. J. Cell. Mol. Med. 2021;25:1207–1220. doi: 10.1111/jcmm.16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Han H.J., Sung J.Y., Kim S.H., Yun U.J., Kim H., Jang E.J., Yoo H.E., Hong E.K., Goh S.H., Moon A. Fibronectin regulates anoikis resistance via cell aggregate formation. Cancer Lett. 2021;508:59–72. doi: 10.1016/j.canlet.2021.03.011. [DOI] [PubMed] [Google Scholar]
- 104.Cai Y., Wu G., Peng B., Li J., Zeng S., Yan Y., Xu Z. Expression and molecular profiles of the AlkB family in ovarian serous carcinoma. Aging (Albany NY) 2021;13:9679–9692. doi: 10.18632/aging.202716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li B., Severson E., Pignon J.C., Zhao H., Li T., Novak J., Jiang P., Shen H., Aster J.C., Rodig S. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V.S.K., Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zheng Y., Tang L., Chen G., Liu Z. Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma. Technol. Cancer Res. Treat. 2020;19 doi: 10.1177/1533033820983284. 1533033820983284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo J., Zhu J., Wang Q., Wang J., Jia Y. Comparative Efficacy of Seven Kinds of Chinese Medicine Injections in Acute Lung Injury and Acute Respiratory Distress Syndrome: A Network Meta-analysis of Randomized Controlled Trials. Front. Pharmacol. 2021;12:627751. doi: 10.3389/fphar.2021.627751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu C., Hao X., Zhang S., Hu W., Li J., Sun J., Zheng M. Characterization of the prognostic values of the NDRG family in gastric cancer. Therap. Adv. Gastroenterol. 2019;12 doi: 10.1177/1756284819858507. 1756284819858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koch A., De Meyer T., Jeschke J., Van Criekinge W. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics. 2015;16:636. doi: 10.1186/s12864-015-1847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koch A., Jeschke J., Van Criekinge W., van Engeland M., De Meyer T. MEXPRESS update 2019. Nucleic Acids Res. 2019;47(W1):W561–W565. doi: 10.1093/nar/gkz445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li J.Y., Li C.J., Lin L.T., Tsui K.H. Multi-Omics Analysis Identifying Key Biomarkers in Ovarian Cancer. Cancer Contr. 2020;27 doi: 10.1177/1073274820976671. 1073274820976671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mulder J., Wernérus H., Shi T.J., Pontén F., Hober S., Uhlén M., Hökfelt T. Systematically generated antibodies against human gene products: high throughput screening on sections from the rat nervous system. Neuroscience. 2007;146:1689–1703. doi: 10.1016/j.neuroscience.2007.02.054. [DOI] [PubMed] [Google Scholar]
- 114.Dammeyer P., Arnér E.S. Human Protein Atlas of redox systems - what can be learnt? Biochim. Biophys. Acta. 2011;1810:111–138. doi: 10.1016/j.bbagen.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 115.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.