Abstract
Background
Psoriasis is a chronic skin disorder manifested by recurrent episodes of scaly, red, itchy skin patches that occur within apparently normal skin.
Objectives
This study was performed to detect the expression of serum and tissue (lesion and non-lesion) LncRNA MALAT-1 and MiRNA-9 that might be used as biomarkers for psoriasis.
Methods
Blood samples were obtained from 60 psoriasis patients and 40 controls, as well as 4 mm punch biopsy from lesional and non lesional skin of psoriatic patient and normal skin of healthy controls. Expression of LncRNA MALAT-1 and miRNNA-9 in serum and tissues was detected by real time qRT-PCR.
Results
a statistically significant increase in the expression of MALAT-1 in lesional and non-lesional skin and serum of psoriatic patients in comparison to controls were detected. Moreover, there was statistically significant increase in serum MiRNA-9 in patients in comparison to controls, while its tissue level was significantly lower in patients.
Conclusion
This study highlights the dysregulation of LncRNA MALAT-1 and miRNA-9 in psoriasis. Elevated expression of MALAT-1 in lesional skin of psoriatic patients compared to non-lesional skin may possibly contribute to the development of psoriatic plaques.
Keywords: Psoriasis, MALAT-1, MiRNA-9, PCR
1. Introduction
Psoriasis vulgaris is a chronic immune mediated skin disorder influencing 1–2% of the white population. It is manifested by recurrent episodes of scaly, red, itchy skin patches that occur within apparently normal skin and is provoked by many factors as infection, stress, drugs and physical trauma to the skin [1]. Occurrence of autoimmune disorder is accompanied by genetic processes that regulate the gene networks in response to complex environmental factors [2]. Although causes of psoriasis still obscure, it is thought to be due to multiple factors involving genetic predisposition, environmental provokers as well as skin barrier perturbation and immune dysfunction [3].
A small number of long non coding RNAs have been determined in rheumatoid arthritis, systemic lupus erythematosus, and psoriasis. Though various studies have been carried out to detect LncRNAs, their biological and pathological functions are still controversial, and most transcriptome research in autoimmune disorders have only identified protein-coding transcripts [4]. Metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) is a significantly expressed nuclear long non coding RNA which is approximately 8000 nucleotides in length [5]. MALAT-1 plays a significant role in pathogenesis of systemic lupus erythematosus that regulates the expression of SIRT1and IL-21 in monocytes of patients which proposes a new relevance and a therapeutic implementation of this LncRNA in SLE [6].
MicroRNAs are non-coding small RNAs which organize gene expression at the post-transcriptional level via base-pairing mainly with a 3′-untranslated region of target mRNA, followed by degradation of mRNA or translational inhibition. MiRNAs have important role in regulation of the immune response that influence proliferation, differentiation, and activation of immune cells, in addition to formation of antibodies and secretion of inflammatory mediators. Disturbance of this regulatory process may result in occurrence of multiple pathological conditions, involving autoimmune inflammation [7]. Various microRNAs which have role in homeostasis and skin morphogenesis have been researched. For instance, the expression of miR-21 is increased in diseased skin, including psoriasis and squamous cell carcinoma [8]. Expression of MALAT-1 is regulated by direct binding with miR-9 which binds directly to two binding sites in the MALAT-1 molecule for degradation in the nucleus [9].
Our current study aims at assessing the expression of long non coding RNA MALAT-1 and miRNA-9 in lesional and non-lesional skin as well as serum of psoriatic patients. By this study, we hope to explore how these LncRNA and miRNA could share in the pathogenesis of psoriasis, thus pave the way for innovative treatment modalities for psoriasis targeting these LncRNA and miRNA.
2. Subjects and methods
This study was conducted at the Medical Biochemistry and Molecular Biology Department, Faculty of Medicine, Cairo University from March 2018 to June 2018, after approval of the Ethical Committee. A written informed consent had been obtained from each participant. 100 Egyptian adult subjects have been included in the study and were divided into 2 groups:
2.1. Patients' group
This group included 60 patients with psoriasis vulgaris presenting to the Dermatology outpatient clinic, Faculty of Medicine, Cairo University. Patients not receiving any relevant systemic therapy for at least four weeks or relevant local treatment for at least two weeks before initiation of our study.
-
•
Exclusion Criteria:
Pregnant and lactating females, erythrodermic or pustular psoriasis, patients with autoimmune diseases e.g. SLE, patients with hematological or solid malignancies e.g. leukemia, breast cancer. Patients with concurrent or recent history of systemic or cutaneous infections were also excluded.
All the patients were subjected to detailed history taking, clinical assessment including, (a) % body surface area affected (BSA) using the rule of 9 [10], (b) assessment of disease severity by the psoriasis area and severity index (PASI) score; which measures erythema, scaling, and thickness of lesions and is weighted by the area of involvement. The head, trunk, upper and lower extremities are the areas assessed. The PASI scale ranges from 0 to 72, calculated as the extent of involvement × (score for erythema + score for scaling + score for thickness) × area multiplier, where the extent of involvement is categorized as: 0 (0%), 1 (1–9%), 2 (10–29%), 3 (30–49%), 4 (50–69%), 5 (70–89%), or 6 (90–100%), and scores for erythema, scaling, and thickness are each rated on a scale of 0–4, while area multiplier is 0.1 for head and neck, 0.2 for upper extremities, 0.3 for trunk and 0.4 for lower extremities [11], and (c) the biopsied plaque severity score (BPSS) which is modified from Target Plaque Severity Score (TPSS), the target plaque was assessed separately for induration, scaling and erythema using a 5-point severity scale (0, none; 1, slight; 2, moderate; 3, marked; 4, very marked), and the scores summed to produce the TPSS sum score [13-point scale; maximum (most severe) score 12] [12].
Blood samples as well as two 4 mm punch skin biopsies were obtained from lesional and non lesional skin of psoriatic patients and stored in empty tubes at −80 °C till used for expression Lnc MALAT-1 and miRNA-9.
2.2. Control group
40 healthy subjects were included as control group. All recruits reported no history of chronic dermatological or systemic disease including renal, liver diseases or malnutrition. All subjects were subjected to detailed history taking, Blood sample and skin biopsy: the biopsies were taken from excess skin after abdominoplasty, breast reduction and brachioplasty operations.
Laboratory methods for detection of miRNA-9 and Lnc MALAT-1 in tissue and serum:
-
•
Sample collection and storage:
3 mL peripheral venous blood samples were collected from every participant by utilizing vacutainer system. Samples were collected and allowed to clot for 15 min, and then centrifuged at 4000×g for 10 min. Sera were separated and stored at −80 °C till the time of analysis. This sera were utilized in RNA extraction and detection of fold change of the 2 genes (microRNA −9 and long noncoding MALAT-1) using real time PCR.
Tissue is homogenized by grinding with QIAzol lysis buffer and the extraction of RNA is the same as the sera.
-
•
RNA extraction
RNA extraction from serum and tissues was performed utilizing miRNeasy mini kit and protocol for purification of serum total RNA, involving miRNA and long noncoding RNA (Qiagen, Valencia, CA, USA). RNA samples were exposed to RNA quantitation and purity assessment using the NanoDrop® (ND)-1000 spectrophotometer (NanoDrop Technologies, Inc. Wilmington, USA).
Reverse transcription and Quantitative Real-time PCR (qPCR) for Detection of Mature miRNA-9:
Reverse transcription was performed on total RNA in a final volume of twenty uL RT reactions utilizing the miScript II RT kit (Qiagen, Valencia, CA, USA).
Quantitative RT-PCR was performed utilizing miScript SYBR® Green PCR kit and protocol for mature miRNAs quantitative assessment (Qiagen, Valencia, CA, and USA) in a total volume of 25 μL per reaction volume utilizing the specific miRNA-9 primers (Cat No MS00003535).
2.3. Calculation of results
After finalization of the PCR cycles, melting curve analyses were carried out to confirm the specified production of the expected product of PCR. Because of absence of an endogenous reference housekeeping gene of miRNA in the serum, SNORD 68 was utilized to normalize the expression manner and for relative quantification of the studied miRNA. The expression level of miRNA-9 was estimated utilizing the ΔCt method. Control value was assumed equaled 1.
2.4. Quantitative Real-time PCR (qPCR) for detection of LncRNA MALAT-1
Serum expression levels of the studied lncRNA MALAT-1 was evaluated using GAPDH as internal control using ready-made primer for MALAT-1 and customized primer for GAPDH and Maxima SYBR Green PCR kit (Thermo, USA) according to the manufacturer's protocol. The primer sequences for GAPDH were as follows:
GAPDH - forward 5′-CCCTTCCCCGATTTCAACTT-3′, GAPDH-reverse, 5′-TGGCCCATGGTGATGAAATT-3′. In brief, real-time PCR was performed on 20 μl reaction mixture prepared by mixing 10 μl master mix, 1 μl forward primer, 1 μl reverse primer, 2.5 μl cDNA, and 5.5 μl RNAase-free water utilizing Rotor gene Q System (Qiagen) with the following conditions: 95 °C for 10 min, followed by 45 cycles at 95 °C for 15 s and 60 °C for 60s.
2.5. Statistical analysis
Descriptive analysis was performed in the form of numbers and percentages for qualitative data. Arithmetic means were calculated as central tendency measurement, while standard deviation as measure of dispersion for quantitative parametric data.
2.6. For quantitative parametric data
-
•
Independent student t-test was used to compare measure of 2-independent groups
-
•
One-way ANOVA test was used for comparing more than 2-independent groups with Benferroni Post-Hoc to test significance at p-value <0.05
For quantitative non-parametric data.
-
•
Kruskalwallis test was used to compare more than 2-independent groups
-
•
Mann-whitney test was used to test significance between more than 2-independent groups
For qualitative data.
-
•
Bivariate Pearson correlation test to find out the association between different groups with a two-tailed to test the significance
-
•
Sensitivity and specificity test were generated for testing a new test with ROC Curve (Receiver Operating Character)
-
•
P-value<0.05 was considered as a cutoff value for significance
3. Results
This study was conducted on 60 psoriatic patients and 40 healthy controls with no statistically significant differences regarding age and sex between patients and controls (Table 1). Forty-eight patients (80%) had negative family history of psoriasis, 12 patients were hypertensive (20%) and only 6 patients had diabetes mellitus (10%). Extent of disease, PASI score and BPSS among psoriatic patients are shown in(Table 2).
Table 1.
Demographic characters (age and sex) of patients and controls.
| Variables | Controls (n = 40) Mean ± SD |
Patients (n = 60) Mean ± SD |
P-value |
|---|---|---|---|
| Age (year) | 40.50 ± 12.19 | 41.80 ± 12.25 | 0.69 |
| Sex | |||
| Males Females |
28 (70%) 12 (30%) |
44 (73%) 16 (27%) |
0.823 |
Table 2.
Description of disease characters among psoriatic group.
| Variables | Minimum | Maximum | Mean | ±SD |
|---|---|---|---|---|
| Extent of disease (%) | 7 | 80 | 37.7 | 22.7 |
| PASI score | 5.6 | 56.8 | 22.2 | 12.2 |
| BPSS | 4 | 12 | 7.2 | 2.4 |
3.1. MiRNA-9 expression in psoriatic patients and controls
-
-
MiRNA-9 expression ranged from 0.002 to 5.7 folds in lesional skin, with mean of 0.61 and this difference was significantly lower than controls (p = 0.001) (Table 3).
-
-
MiRNA-9 expression ranged from 0.006 to 6.8 folds in non-lesional skin, with a mean of 0.65, and this difference was significantly lower than controls (p = 0.001) (Table 3).
-
-
Serum MiRNA-9 expression among patients ranged from 0.11 to 32.8 folds, with a mean of 5.1 and this difference was significantly higher than controls (p = 0.001) (Table 3).
Table 3.
Description of fold change of miRNA-9 among study groups.
| Variables | Cases (n = 60) |
controls (n = 40) |
p-value | Sig. | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | |||||
| Lesion | 0.61 | 0.32 | 0.99 | 0.01 | 0.001 | HS | ||
| Non lesion | 0.65 | 0.35 | 0.99 | 0.01 | 0.001 | HS | ||
| Serum | 5.1 | 1.8 | 0.99 | 0.02 | 0.001 | HS | ||
3.2. Long noncoding MALAT-1 expression in psoriatic patients and controls
Among patients, the fold change of long noncoding MALAT-1 in lesional skin ranged between 0.05 and 47.1 with a mean of 7.5, while in non-lesional skin, it ranged between 0 and 66.7 with a mean of 5.8 and its serum values ranged between 0.02 and 79.4 with a mean of 9.3. These fold changes were significantly higher among patients in comparison to controls as regard lesional, non-lesional and serum levels of MALAT1 (p = 0.001, 0.02, and 0.001 respectively) (Table 4).
Table 4.
Description of fold change of MALAT-1 among study groups.
| Variables | Cases (n = 60) |
controls (n = 40) |
p-value | Sig. | |||
|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | ||||
| Lesion | 7.5 | 3.3 | 1.1 | 0.01 | 0.001 | HS | |
| Non lesion | 5.8 | 3.6 | 1.1 | 0.01 | 0.02 | S | |
| Serum | 9.3 | 4.9 | 0.99 | 0.01 | 0.001 | HS | |
Moreover, lesional MALAT-1 fold change was significantly higher among females in comparison to male patients in the psoriasis group (p < 0.05) (Table 5).
Table 5.
Comparisons of different markers in different genders.
| Variables | Males (n = 44) |
Females (n = 16) |
p-value | Sig. | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | |||||
| MiRNA-9 | ||||||||
| Lesion | 0.51 | 0.38 | 0.88 | 0.62 | 0.6 | NS | ||
| Non lesion | 0.84 | 0.46 | 0.057 | 0.008 | 0.4 | NS | ||
| Serum | 5.6 | 2.4 | 3.6 | 1.7 | 0.7 | NS | ||
| MALAT 1 | ||||||||
| Lesion | 3.8 | 1.9 | 18.7 | 11.3 | 0.04 | S | ||
| Non lesion | 7.4 | 4.7 | 0.70 | 0.41 | 0.4 | NS | ||
| Serum | 10.9 | 6.5 | 4.2 | 2.4 | 0.5 | NS | ||
3.3. Correlations between MiRNA- 9 and other variables among patients
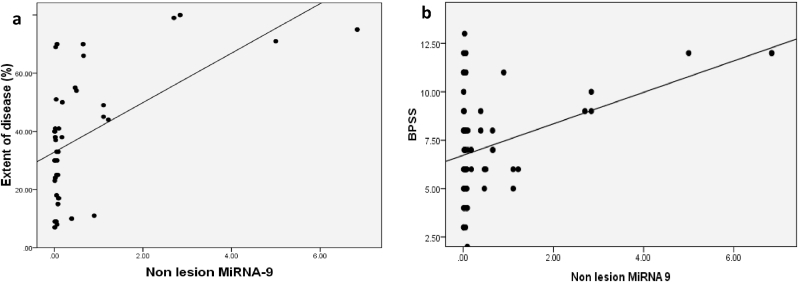
Significant positive correlations were calculated, between non-lesion level of MiRNA-9 and each of extent of disease and BPSS score (p = 0.007 and 0.02 respectively) (Fig. 1).
Fig. 1.
(a) Correlation between extent of disease and non-lesional MiRNA-9 among patients. (b) Correlation between BPSS and non-lesional MiRNA-9 among patients.
3.4. Correlations between MALAT- 1 and other variables among patients
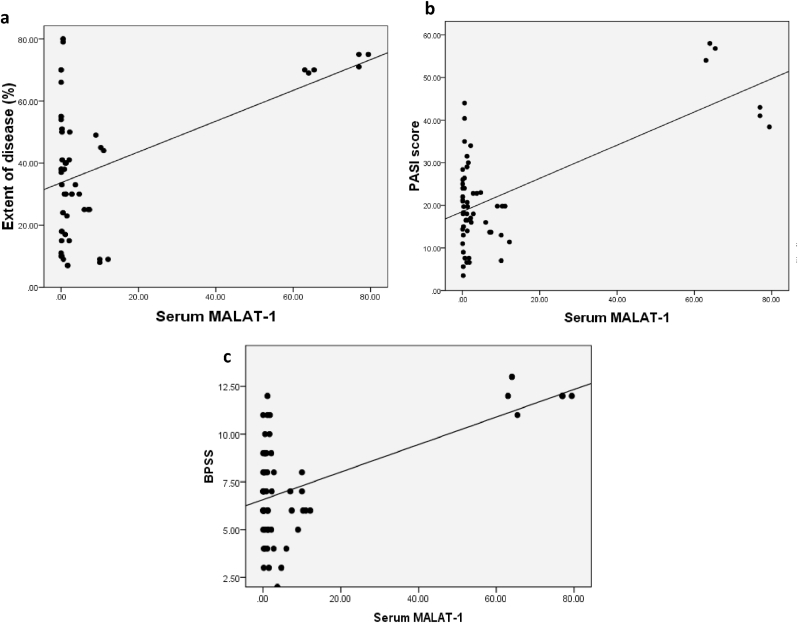
There were positive correlations with p-values of 0.02, 0.002 and 0.007 between serum level of MALAT-1 and extent of disease, PASI and BPSS score respectively (Fig. 2).
Fig. 2.
(a) Correlation between extent of disease and serum MALAT-1 among patients.(b) Correlation between PASI score and serum MALAT-1 among patients. (c)Correlation between BPSS and serum MALAT-1 among patients.
3.5. Correlations between MiRNA- 9 and MALAT-1 in different samples among patients
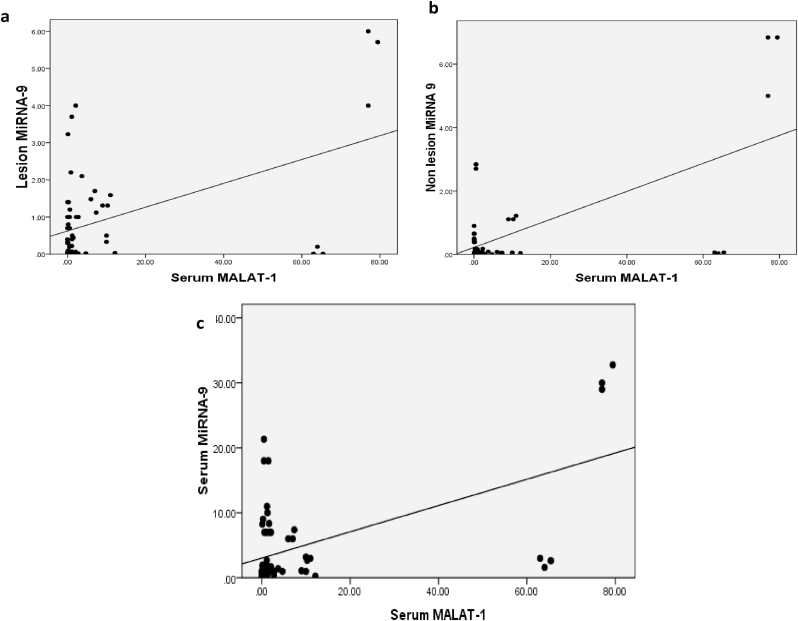
Serum level of MALAT-1 among cases correlated positively with lesional, non-lesional, as well as serum levels of MiRNA-9 (p = 0.004, 0.002 and 0.01 respectively) (Fig. 3).
Fig. 3.
(a) Correlation between lesional MiRNA-9 and serum MALAT-1 among patients. (b) Correlation between non-lesional MiRNA-9 and serum MALAT-1 among patients. Correlation between serum MiRNA-9 and serum MALAT-1 among patients.
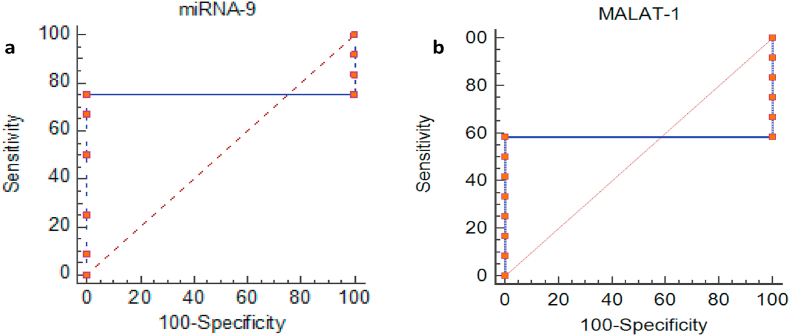
Statistics showed that Lesional MiRNA-9 and MALAT-1 fall in Area Under Curve 0.75 and 0.97 respectively with cutoff value 1.31 and 2.79. The sensitivity of MiRNA-9 is 91% and specificity of 100%. While the sensitivity for MALAT-1 is 75% and specificity of 100% (Fig. 4, Table 6).
Fig. 4.
ROC Curves for Lesional (a) MiRNA-9 and (b) MALAT-1.
Table 6.
Sensitivity and Specificity percentages for Lesional MiRNA-9 and MALAT-1 in Psoriasis patients.
| Lesional Biomarkers | AUC | Cut-off value | Sensitivity | Specificity |
|---|---|---|---|---|
| MiRNA-9 | 0.75 | 1.31 | 91% | 100% |
| MALAT-1 | 0.97 | 2.79 | 75.0% | 100% |
Area Under Curve for non Lesional MiRNA-9 and MALAT-1 are 0.917 and 0.50 respectively with sensitivity 91% and specificity 100% for MiRNA-9and 50% and 100% for MALAT-1. The cutoff values for MiRNA-9 and MALAT-1 are 1.11 and 2.16 respectively (Fig. 5, Table 7).
Fig. 5.
ROC Curves for Non Lesional (a) MiRNA-9 and (b) MALAT-1.
Table 7.
Sensitivity and Specificity percentages for non Lesional MiRNA-9 and MALAT-1 in Psoriasis patients.
| Non-lesional Biomarkers | AUC | Cut-off value | Sensitivity | Specificity |
|---|---|---|---|---|
| MiRNA-9 | 0.917 | 1.11 | 91% | 100% |
| MALAT-1 | 0.50 | 2.16 | 50% | 100% |
Area Under Curve for serum MiRNA-9 and MALAT-1 are 0.50 for both with sensitivity 55% and specificity of 100% forMiRNA-9 and 58% and 100% for MALAT-1. The cutoff values for MiRNA-9 and MALAT-1 are 1.95 and 1.22 respectively. (Fig. 6, Table 8).
Fig. 6.
ROC Curves for serum (a) MiRNA-9 and (b) MALAT-1.
Table 8.
Sensitivity and Specificity percentages for serum MiRNA-9 and MALAT-1 in Psoriasis patients.
| Serum Biomarkers | AUC | Cut-off value | Sensitivity | Specificity |
|---|---|---|---|---|
| MiRNA-9 | 0.50 | 1.95 | 55% | 100% |
| MALAT-1 | 0.50 | 1.22 | 58% | 100% |
4. Discussion
Psoriasis is an autoimmune disease in which inflammation is mediated by cells and molecules of the adaptive and innate immune systems. The immune pathways which operate in psoriasis constitute exaggeration of immune tracks that occur as fundamental or stimulatory pathways in normal skin of human [13]. Keratinocytes are major contributors in innate immunity calling up T cells to the skin, and T cells are essential in maintaining disease activity.
Inflammatory myeloid dendritic cells (DCs) secrete IL-12 and IL-23 to activate Th1 cells, Th22 cells, and IL-17–secreting T cells to release plentiful psoriatic cytokines IL-22, IL-17, TNF-α, and IFN-γ [13,14].
The role of microRNAs in regulation of gene expression at the post-transcriptional level is deemed an essential genetic mechanism, and there were many proofs revealing that microRNAs have significant role in the pathogenesis of psoriasis [15].
The regulatory effect of long non coding RNAs in immune system has been a pivotal field in research toward personalized medicine. Research on LncRNAs in immune cells detected that LncRNAs play important role in immune cells development, differentiation and activation [16].
The aim of this study was to examine the role of miRNA- 9 and Lnc MALAT-1 in the pathogenesis of psoriasis and their relation to disease severity. To the best of our knowledge, this is the first study to check both miRNA-9 and MALAT-1 in psoriatic patients’ lesionl and non-lesional skin as well as serum.
In the present study, the expression of miRNA-9 and lncMALAT-1 were detected by qRT-PCR in psoriatic patients and compared to controls.
Regarding miRNA-9, its serum expression was significantly increased in patients than controls, while its tissue levels were significantly decreased in patients than controls. Liang et al., deduced that microRNAs exist in blood in a stabilized format and their levels are modified in diseases [17].
Majd et al., revealed that microRNA-9 in increased in plasma of autoimmune MS patients in relapsing phase compared to healthy controls [18]. O'Connell et al., reported that MicroRNA-9 participate in regulation of cell activation by many loops of feedback inhibition working at the level of NFKB1, which is a transcriptional factor greatly take part in the inflammatory response [19]. Thus, in lesional psoriasis; another immune mediated disease, one may hypothesize that the downregulation of miR-9 we detected in comparison to controls may possibly be one of the factors that contribute to the upregulation of NFKB1. Puzzlingly, Bazzoni et al., state that, the proinflammatory cytokines TNF-α, Toll-IL-1R (TIR) and IL-1β; which are all upregulated in psoriasis, causes upregulation of miR-9 levels in both PMN and monocytes [20].
Adding to the confusion, miRNA-9 is upregulated by NFKB1 itself via LPS and MyD88 interactions [21]. However, it seems that such upregulation signals are dampened in lesional skin, by a yet an unidentified mechanism which does not seem to affect its serum levels which were upregulated in patients’ serum as we demonstrate.
Lesional, non-lesional and serum expression of long noncoding MALAT- 1 were significantly higher in patients than controls. Moreover, the increased expression of MALAT-1 in lesional skin in comparison to non-lesional skin may provide an assertion of the involvement of this lncRNA in disease progression where non-lesional skin may be considered pre-psoriatic skin that may express the disease phenotype in a genetically predisposed individual.
MALAT-1 is a long noncoding RNA whose importance in immune regulation stays mostly obscure. Several research have revealed that LncRNAs can be used as biomarkers for diagnosis and prognosis of diseases. Lately, abnormal expression of LncRNAs has been detected in rheumatoid arthritis and many autoimmune disorders [22].
In the present study, MALAT-1 was significantly increased in psoriatic patients, lesional, non lesional and serum samples compared to controls. Dysregulation of MALAT-1 is reported to be NF-ƘB- dependent. NF-ƘB is a major transcription factor which is important for promoter activity of MALAT-1 [23] and has repeatedly been demonstrated as an essential component for psoriasis development [24].
Furthermore, Puthanveetil et al., displayed that hyperglycemia initiates an inflammatory response cascade through MALAT-1 mediated up-regulation of serum amyloid antigen [25], thus inducing the release of inflammatory markers TNF-α and IL-6, both of which are major regulators in pathogenesis of psoriasis [26].
A good example of abnormal expression of MALAT-1 in autoimmune disease is reported by Yang et al., who showed that MALAT-1 was upregulated in PBMCs from autoimmune disease (SLE) patients compared to that in healthy controls, and the major regulatory role of MALAT-1 in the pathogenesis of systemic lupus via SIRT1signaling pathway [6]. Rasheed et al., detected dysregulation of SIRT1 in psoriasis patients in comparison to healthy controls [27] and Yang et al., reported the role of MALAT-1 in regulation of SIRT1 signaling pathway[6].
In the present study, results showed that lesion level of miRNA-9 and MALAT-1 was higher in females than males. Our result was confirmed by Zandman et al., and Greer et al., who detected that various stimuli may differentially impact females and males, making the higher potential of autoimmune diseases among females [28,29].
Alonso and Hernan revealed that for epigenetic modifications to explain sexual dimorphism in autoimmunity, it is important to assume that epigenetic factors are more commonly occurred in one gender than in the other, or that individuals of one sex are more susceptible than the other [30]. While, in view of the higher incidence of some autoimmune diseases among females, but not males, over the last 100 years, and the main social changes that mostly influenced females over that time, it is an appealing supposition.
Moreover, the significant positive correlation between non-lesion miRNA-9 and both extend of disease and BPSA affected by psoriasis, suggests an important relationship between this miRNA and the disease initiation and activity, and may encourage further investigations to explore the possibility of using this miRNA as one of the markers of psoriasis severity.
Interestingly, serum MALAT-1 expression was positively correlated with disease extension, PASI score and BPSS among cases, which indicated that the increase in these variables was associated with increase in serum level of MALAT-1. Such observations may suggest a direct participation of MALAT-1 in the development and severity of psoriasis.
In the current study, a significant positive correlation between serum level of MALAT-1 and each of lesion, non-lesion, and serum level of MiRNA-9 among cases with p-value <0.05, which indicated that the increase in serum level of MALAT-1 was associated with increase in lesion, non-lesion and serum level of micro RNA- 9.
These results are opposing the result of Leucci et al., who reported that miRNA-9 overexpression resulted in a significant downregulation of MALAT-1 both in the cytoplasm and the nucleus of L428 Hodgkin lymphoma cells[9]. Psoriasis and lymphoma are 2 diverse disorders with very different pathogenesis and immune dysregulations. The relation of miRNA-9 with MALAT-1 cannot be simply restricted to a dual interaction when a vast number of cytokines involved in psoriasis are dysregulated; e.g. NFKB, TIR-IL1 and TNF, and are shown to be direct regulators of miRNA-9 and or MALAT-1.
5. Conclusion
This study highlights the contribution of MALAT-1and miRNA-9 in the pathogenesis of psoriasis. We detected that the expression of MALAT-1 in lesional skin of psoriatic patients is elevated in comparison to non-lesional skin which might be an important factor in the development of psoriatic plaques. The high serum expression of MALAT-1 and miRNA-9 indicate that such circulating markers can serve as potential markers for diagnosis of psoriasis and need to be compared with the papulosquamous disorders. These results could also pave the way for the development of a potential therapy for psoriasis by manipulating these markers, which also seem to be involved in co-morbidities associated with psoriasis.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
All authors contribute equally in conception, design, practical work, supervision, manuscript writing, editorial help, collection of data and data analysis through the whole study.
Ethics approval
Done.
Consent to participate
Done.
Consent for publication
Done.
Availability of data and material
Available.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
Not applicable.
Contributor Information
Azza M. Elamir, Email: azzaelamer2013@gmail.com, ama27@fayoum.edu.eg.
Olfat G. Shaker, Email: olfatshaker@yahoo.com.
Mohamed HM. El-Komy, Email: m_elkomy@kasralainy.edu.eg.
Mai Mahmoud sharabi, Email: mmm29@fayoum.edu.eg.
Nesreen M. Aboraia, Email: Nesreen_aboraia@yahoo.com.
References
- 1.Rongioletti F., Fiorucci C., Parodi A. Psoriasis induced or aggravated by drugs. J. Rheumatol. Suppl. 2009;83:59–61. doi: 10.3899/jrheum.090227. [DOI] [PubMed] [Google Scholar]
- 2.Tsokos G.C. Systemic lupus erythematosus. N. Engl. J. Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 3.Raychaudhuri S.K., Maverakis E., Raychaudhuri S.P. Diagnosis and classification of psoriasis. Autoimmun. Rev. 2014;13(4–5):490–495. doi: 10.1016/j.autrev.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Dolcino M., Pelosi A., Fiore P.F., Patuzzo G., Tinazzi E., Lunardi C., Puccetti A. Long non-coding RNAs play a role in the pathogenesis of psoriatic arthritis by regulating MicroRNAs and genes involved in inflammation and metabolic syndrome. Front. Immunol. 2018;9:1533. doi: 10.3389/fimmu.2018.01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei L., Zeng Q., Lu J., Ding S., Xia F., Kang J., Tan L., Gao L. MALAT1 participates in ultraviolet B-induced photo-aging via regulation of the ERK/MAPK signaling pathway. Mol. Med. Rep. 2017;15:3977–3982. doi: 10.3892/mmr.2017.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H., Liang N., Wang M., Fei Y., Sun J., Li Z., Xu Y., Guo C. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget. 2017;8(44):77400–77406. doi: 10.18632/oncotarget.20490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baulina N.M., Kulakova O.G., Favorova O.O. MicroRNAs: the role in autoimmune inflammation. ACTA NATURAE. 2016;8(28):21–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Ambica B., William G., Diana D., Amelia S., Elizabeth G., Zhenquan Y. The Grainy head transcription factor Grhl3/Get1, suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. NIH Public Access. 2013:1497–1507. doi: 10.1038/onc.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.E1 Leucci, Patella F., Waage J., Holmstrøm K., Lindow M., Porse B., Kauppinen S. MicroRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 2013;3:1–6. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livingston E.H., Lee S. Percentage of burned body surface area determination in obese and nonobese patients. J. Surg. Res. 2000;91(2):106–110. doi: 10.1006/jsre.2000.5909. [DOI] [PubMed] [Google Scholar]
- 11.Wittkowski K.M., Leonardi C., Gottlieb A., Menter A., Krueger G.G., Tebbey P.W. International Psoriasis Council. Clinical symptoms of skin, nails, and joints manifest independently in patients with concomitant psoriasis and psoriatic arthritis. PloS One. 2011;6(6) doi: 10.1371/journal.pone.0020279. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ports W.C., Khan S., Lan S., Lamba M., Bolduc C., Bissonnette R. A randomized phase 2a efficacy and safety trial of the topical Janus kinase inhibitor tofacitinib in the treatment of chronic plaque psoriasis. Br J Dermatol. Jul. 2013;169(1):137–145. doi: 10.1111/bjd.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowes M.A., Suarez-Farinas M., Krueger J.G. Immunology of psoriasis. Annu. Rev. Immunol. 2014;32:227–255. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perera G.K., Di Meglio P., Nestle F.O. Psoriasis. Annu. Rev. Pathol. 2012;7:385–422. doi: 10.1146/annurev-pathol-011811-132448. [DOI] [PubMed] [Google Scholar]
- 15.Chandra A., Ray A., Senapati S., Chatterjee R. Genetic and epigenetic basis of psoriasis pathogenesis. Mol. Immunol. 2015;64(2):313–323. doi: 10.1016/j.molimm.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Atianand M.K., Fitzgerald K.A. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014;20(11):623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y., Pan H.F., Ye D.Q. MicroRNAs function in CD8þT cell biology. J. Leukoc. Biol. 2015;97:487–497. doi: 10.1189/jlb.1RU0814-369R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majd M., Hosseini A., Ghaedi K., Kiani-Esfahani A., Tanhaei S., Shiralian-Esfahani H., Rahnamaee S., Nasr-Esfahani M. MiR-9-5p and miR-106a-5p dysregulated in CD4+ T-cells of multiple sclerosis patients and targeted essential factors of T helper17/regulatory T-cells differentiation. Iran J Basic Med Sci. 2018;21(3):277–283. doi: 10.22038/ijbms.2018.25382.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Connell R.M., Taganov K.D., Boldin M.P., Cheng G., Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bazzoni F., Roseate M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M.A., Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to pro inflammatory signals. Proc. Natl. Acad. Sci. U. S. A. 2009;106(13):5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., Sun Z.X., Allgayer H., Yang H. Downregulation of E-cadherin is an essential event in activating beta-catenin/Tcf-dependent transcription and expression of its target genes in Pdcd4 knockdown cells. Oncogene. 2010;29:128–138. doi: 10.1038/onc.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Li, Li Xingye, Jiang C., William K., Qian W., Matthew T. Long non‐coding RNAs in rheumatoid arthritis. Cell Prolif. 2018;51(1):1–6. doi: 10.1111/cpr.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gui Z., Zhenyi S., Dan S., Mao Y., Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-ƘB. Federation of European Biochemical Societies (FEBS) 2016;590:2884–2895. doi: 10.1002/1873-3468.12315. [DOI] [PubMed] [Google Scholar]
- 24.Goldminz A.M., Au S.C., Kim N., Gottlieb A.B., Lizzul P.F. NF-κB: an essential transcription factor in psoriasis. J. Dermatol. Sci. 2013;69(2):89–94. doi: 10.1016/j.jdermsci.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Puthanveetil P., Chen S., Feng B., Gautam A., Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell Mol. Med. 2015;19:1418–1425. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blauvelt A. IL-6 differs from TNF-α: unpredicted clinical effects caused by IL-6 blockade in psoriasis. J. Invest. Dermatol. 2017;137(3):541–542. doi: 10.1016/j.jid.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Rasheed H., El-Komy M.H., Shaker O.G., Hegazy R.A., Gawdat H.I., AlOrbani A.M. Expression of sirtuins 1, 6, tumor necrosis factor, and interferon-γ in psoriatic patients. Int. J. Immunopathol. Pharmacol. 2016;29(4):764–768. doi: 10.1177/0394632016662475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zandman-Goddard G., Peeva E., Shoenfeld Y. Gender and autoimmunity. Autoimmun. Rev. 2007;6(6):366–372. doi: 10.1016/j.autrev.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Greer J.M., McCombe P.A. Role of gender in multiple sclerosis: clinical effects and potential molecular mechanisms. J. Neuroimmunol. 2011;234(1–2):7–18. doi: 10.1016/j.jneuroim.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Alonso A., Hernán M.A. Temporal trends in the incidence of multiple sclerosis: a systematic review. Neurology. 2008;71(2):129–135. doi: 10.1212/01.wnl.0000316802.35974.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Available.