Highlights
-
•
Angiogenesis-related genes are important in the aggressive behavior of dedifferentiated chondrosarcoma.
-
•
RNA sequencing shows different gene expression in the two components of dedifferentiated chondrosarcoma.
-
•
Differences in heat shock protein expression in dedifferentiated chondrosarcoma suggest relevant treatment targets.
Keywords: Chondrosarcoma, HSP70, HSP90, VEGF, pERK1/2, Angiogenesis
Abbreviations: CS, chondrosarcoma; DDCS, dedifferentiated chondrosarcoma; DD, dedifferentiated component of dedifferentiated chondrosarcoma (no matrix); WD, well differentiated chondroid matrix containing component of dedifferentiated chondrosarcoma; IPEX, immunohistochemistry
Abstract
Dedifferentiated chondrosarcomas (DDCS) are highly malignant bimorphic mesenchymal tumors with poor outcome and limited treatment options. Genes and proteins involved in angiogenesis play an important role in the development of invasion and metastasis. Immunohistochemical stains targeting HSP70, pERK1/2 and VEGFA were applied to a TMA containing 29 DDCS cases representing both tumor components. Higher expression of HSP70 and pERK1/2 was noted in the dedifferentiated component. RNA sequencing performed in 8 paired cases of DDCS comparing well differentiated and dedifferentiated components, showed higher expression of several HSP70 family members and HSP90 in the dedifferentiated component. Furthermore, high mobility group AT-hook 2 (HMAG2) and SET nuclear proto-oncogene demonstrated higher expression in the dedifferentiated component. Thus, the well differentiated and dedifferentiated components of DDCS are different, histologically and transcriptomically. The dedifferentiated component of DDCS shows higher expression of markers that are associated with malignant behavior. Some of these may represent future treatment targets.
1. Introduction
Dedifferentiated chondrosarcoma was first described by Dahlin and Beabout in 1971 [1]. Clinicopathologic characteristics of DDCS were delineated in 1986 [2]. This subtype of chondrosarcoma is histologically bimorphic with sharp demarcation between matrix containing grades 1, 2 and 3 (well differentiated, WD) chondrosarcoma and the adjacent high grade sarcoma devoid of cartilaginous matrix (dedifferentiated, DD). These chondrosarcomas represent approximately 10–15% of central chondrosarcomas and are associated with a very poor prognosis, up to 24% 5-year survival [3]. Treatment is mainly surgical, as conventional chemotherapy and radiation therapy regimens have been ineffective.
Understanding the molecular pathways operative in this neoplasm and, thus, determining appropriate targets for drug therapy, is paramount. The two clearly defined components of dedifferentiated chondrosarcoma share genetic alterations and demonstrate additional mutations in the high grade component. Pathways affected include those involving Rb, p53 and IDH mutations [4], [5].
Angiogenesis is a critical component in the development of numerous invasive malignancies and has been shown to be increased in chondrosarcomas, correlating with increasing grade [6], [7]. Angiogenesis is mediated by several cell processes and genes including the vascular endothelial growth factor (VEGF) family, members of the mitogen-activated protein kinase (MAPK) pathway and the heat shock protein (HSP) family, among others. In this study we attempt to identify expression of VEGF, phosphorylated extracellular signal-regulated kinase ½ (pERK1/2) and HSPs in a cohort of dedifferentiated chondrosarcomas utilizing immunohistochemistry and RNA sequencing.
2. Methods
The study was approved by the University of Pittsburgh Institutional Review Board (Study 20050109).
Cases: Three hundred and seventy six (376) conventional chondrosarcoma cases were identified from 1997 to 2018 in the UPMC Department of Pathology surgical pathology files. A subset of 29 primary dedifferentiated chondrosarcomas was procured from this original group to create a tissue microarray. Three cases from this group had sufficient material for RNA sequencing. Five additional dedifferentiated chondrosarcomas were identified in the same files from 2018 to 2020 for RNA sequencing.
TMA Construction: The histologic H&E stained slides of 29 dedifferentiated chondrosarcomas were reviewed by one pathologist (KES) and tissue blocks representing both well differentiated and dedifferentiated components of the tumors were selected. Targeted viable tissues were circled and corresponding tissue blocks were punched (2.0 mm cores) and re-embedded to create the TMA. Multiple tumor areas in each case were sampled (3–5 cores each). Twelve of the tissue blocks used in the TMA had been decalcified in Rapid Decalcifier RDO (Aurora, IL).
Antibodies for Immunohistochemistry (IPEX): Five micron thick unstained slides were cut from the TMA and used for immunohistochemistry. Antibodies utilized were VEGF, recognizing VEGFA only (VG1, Invitrogen, #MA1-16629, dilution 1:25, colon carcinoma positive control); pERK1/2 (pT202/pY204.22A, Santa Cruz Biotechnology, #sc-136521, dilution 1:50, normal brain positive control) and HSP70 (Santa Cruz Biotechnology, sc-24, dilution 1:300, normal lung positive control). Standard immunohistochemical methods were employed.
Immunohistochemical expression was recorded as nuclear and/or cytoplasmic, semiquantitative intensity of staining [(0, no staining), (1+, greater than 0 up to 10% of cells, weak intensity), (2+, 11–50% of cells, weak to moderate intensity), (3 + greater than 50% of cells, moderate to strong intensity) and diffuse or focal. Results were analyzed using the student T-test, two tailed.
3. RNA sequencing
3.1. Sample acquisition, processing and RNA extraction
Paraffin embedded well differentiated and dedifferentiated tumor tissues were available in three cases of the original 29 cases comprising the TMA and an additional 5 cases found subsequently. These tissues were extracted for RNA sequencing. A pathologist (KES) reviewed H&E slides from each sample and marked regions of high tumor cell purity for RNA extraction. Subsequently, (5–7) 10 μM sections from the paraffin blocks were cut and areas of interest were macrodissected with a sterile, disposable No. 15 blade (Fisher Scientific, catalog 50-822-460). RNA extraction was performed with the Allprep DNA/RNA FFPE kit (Qiagen, catalog 80234) according to the manufacturer’s instructions under sterile RNase/DNase free conditions. RNA concentration was determined with the Qubit 4.0 Fluorometer (ThermoFisher Scientific). Quality RNA integrity number scores and fragment sizes (DV200 metrics) were obtained utilizing the Agilent 4200 TapeStation system.
3.2. Sequencing used NextSeq500 Platform, flowcell (1 mid output 150), read length 2 × 75, loading concentration (2.0 pM) and % Phi-X (1%)
SMART technology was used in this ligation-free protocol to preserve strand-of-origin information. Random priming allowed the generation of cDNA from all RNA fragments in the sample, including rRNA. When the SMARTScribe™Reverse Transcriptase (RT) reached the 5′ end of the RNA fragment, the enzyme’s terminal transferase activity added a few non-templated nucleotides to the 3′ end of the cDNA. The carefully designed LNA-TSO (included in the Template Switching Oligo Mix) base-paired with the non-templated nucleotide stretch, creating an extended template to enable the RT to continue replicating to the end of the oligonucleotide. The resulting cDNA contained sequences derived from the random primer and the LNA-TSO used in the reverse transcription reaction. In the next step, a first round of PCR amplification (PCR1) added full-length Illumina adapters, including barcodes. The Forward PCR Primer bound to the LNA-TSO sequence, while the Reverse PCR Primer bound to sequence associated with the random primer. The ribosomal cDNA (originating from rRNA) was then cleaved by ZapR in the presence of the mammalian-specific R-Probes. This process leaves the library fragments originating from non-rRNA molecules untouched, with priming sites available on both 5′ and 3′ ends for further PCR amplification. These fragments were enriched via a second round of PCR amplification (PCR2) using primers universal to all libraries. The final library contained sequences allowing clustering on an Illumina flow cell (Illumina) (TruSeq® HT) indexes as well as the regions recognized by sequencing primers.
4. RNA-Seq pipeline
The reverse stranded paired-end RNA-Seq reads, generated by SMARTer Stranded Total RNASeq Kit v2 – Pico Input Mammalian kit, were checked for presence of adapters and high-quality bases using FastQC (v 0.11.7). These high-quality reads were trimmed for the universal adapter using Cutadapt (v 1.18). The trimmed reads were later mapped against the Ensembl human reference genome (GRCh38 v 97) using the HISAT2 (v 2.1.0) mapping tool. The output file from HISAT2 was converted from SAM format to BAM format using SAMtools (v 1.9). Counts for expressed genes were generated using HT-Seq (v 0.11.2) and were output in text format. These count text files were then imported into the Bioconductor R package, edgeR (v 3.24.1). The package was utilized to calculate Counts per Million (CPM) and the average CPM of the two different groupings of samples. Counts per Million (CPM) is a unit of measurement where mapped reads or raw counts are normalized by library size, then multiplied by one million.
Data Analysis: Data analysis was performed using Edge R 4.0 and statistical significance was set at <0.05 (Fisher’s exact test). Principal Component Analysis (PCA) is a dimensionality-reduction statistical procedure, often used for processing large sets of data. This is used to reduce the number of variables within a project for simplicity and readability, while still preserving as much accuracy of the data as possible. PCA uses a technique called feature extraction, which combines input variables, but then drops the least important variables, while keeping the most valuable variables in the statistical model that best represent the data.
5. Results
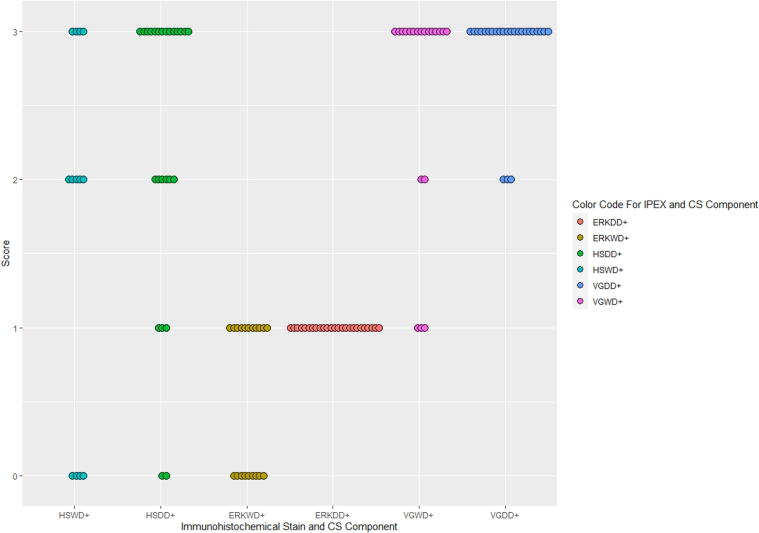
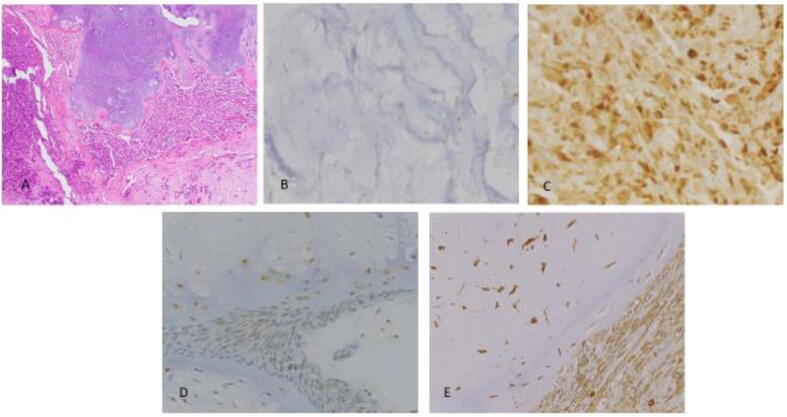
29 cases of primary dedifferentiated chondrosarcoma were identified, comprising 11 females and 18 males (age range: 32–91 years; mean: 62 years). Semiquantitative scoring of immunohistochemical stains against HSP70, pERK1/2 and VEGFA, as well as tumor location and size are recorded below (Table 1 and Fig. 1). While VEGFA expression was not statistically different in the two components, (p = 0.1), HSP70 demonstrated higher cytoplasmic expression in the dedifferentiated component as compared to the well-differentiated component, (p = 0.003). A statistically significant higher expression of pERK1/2 was also observed in the dedifferentiated component as compared to the well differentiated component (p = 0.02). Representative H&E and immunohistochemical features of HSP70, pERK1/2, and VEGFA expression are illustrated below (Fig. 2).
Table 1.
Site, size and immunohistochemical results of angiogenesis markers.
| SITE (SIZE, CM) | HSP70 |
pERK1/2 |
VEGF |
||||
|---|---|---|---|---|---|---|---|
| WD | DD | WD | DD | WD | DD | ||
| 1 | FEMUR (10.5) | – | N/C 2+ | – | C 3+ | – | 3+ |
| 2 | PELVIS (9.0) | 0 | N/C 2+ | N/C 3+ | C 3+ | 2+ | 3+ |
| 3 | R ILIUM (6.2) | 0 | N1+ | N/C 3+ | N/C 3+ | 3+ | 3+ |
| 4 | HUMERUS (7.0) | – | N1+ | 0 | C 3+ | 3+ | 3+ |
| 5 | TALUS (11.0) | – | – | – | – | – | – |
| 6 | PELVIS (6.5) | – | N1+ | – | N/C 3+ | – | 2+ |
| 7 | 3RD RIB (6.0) | N1+ | N/C 3+ | N/C 3+ | C 3+ | 3+ | 3+ |
| 8 | L FEMUR (11.0) | N1+ | N/C 3+ | N/C 3+ | N/C 3+ | 3+ | 3+ |
| 9 | HUMERUS (9.0) | – | N/C 2+ | – | C 3+ | – | 3+ |
| 10 | FEMUR(16.0) | – | N/C 2+ | N/C2+ | C 3+ | 1+ | 3+ |
| 11 | PELVIS (10.6) | – | N/C 2+ | C2+ | C 3+ | 2+ | 2+ |
| 12 | SCAPULA (10.6) | – | – | – | – | – | – |
| 13 | L HUMERUS (14.0) | N/C2+ | N/C 2+ | C3+ | C 3+ | 3+ | 3+ |
| 14 | L FEMUR (10.0) | – | N/C 2+ | N/C2+ | C 3+ | 1+ | 3+ |
| 15 | L FEMUR (20.5) | – | N/C 2+ | – | C 3+ | – | 3+ |
| 16 | PELVIS (18.0) | – | N/C 2+ | – | C 3+ | – | 3+ |
| 17 | L HUMERUS (9.5) | 0 | N 1+ | C 1+ | C 3+ | 1+ | 3+ |
| 18 | PELVIS (13.0) | – | 0 | C1+ | C 2+ | 3+ | 3+ |
| 19 | L PELVIS (12.0) | N1+ | N/C 2+ | N/C 3+ | N/C 3+ | 3+ | 3+ |
| 20 | R PELVIS (10.5) | – | – | – | – | – | – |
| 21 | R FEMUR (6.0) | 0 | N/C 2+ | N/C 3+ | N/C 3+ | 3+ | 3+ |
| 22 | TIBIA (9.5) | – | N/C 2+ | N/C 2+ | N/C 3+ | 3+ | 3+ |
| 23 | R FEMUR (7.0) | N1+ | 0 | N/C 3+ | N/C 3+ | 3+ | 3+ |
| 24 | FEMUR (11.5) | – | N/C 2+ | N 3+ | C 3+ | 3+ | 3+ |
| 25 | L FEMUR (17.5) | N/C2+ | N/C 3+ | N/C 3+ | N/C 3+ | 3+ | 3+ |
| 26 | R ILIAC (13.2) | N1+ | N 1+ | N 3+ | N 3+ | 3+ | 2+ |
| 27 | L FEMUR (33.0) | N/C2+ | N/C 2+ | C 3+ | C 3+ | 3+ | 3+ |
| 28 | RIB (10.0) | – | – | – | – | – | – |
| 29 | L FEMUR (21.2) | N1+ | N/C 2+ | – | C 2+ | 3+ | 3+ |
Note: Dashes reflect incomplete data due to TMA section dropout or disruption. N, nuclear; C, cytoplasmic.
Fig. 1.
Immunohistochemistry scores for HSP70, pERK1/2 and VEGFA in both components of DDCS. Immunohistochemistry scores for WD and DD components where ERK = pERK1/2; HS = HSP70 and VG = VEGFA.
Fig. 2.
HSP70, pERK1/2, and VEGFA Expression in Both Components of DDCS: A: H&E depicting both components of DDCS (100X) B: HSP70 immunohistochemistry in well differentiated component of dedifferentiated chondrosarcoma (400X); C: HSP70 immunohistochemistry in dedifferentiated component of dedifferentiated chondrosarcoma (400X); D: pERK1/2 in both components (400X); E: VEGFA immunohistochemistry in both components (400X).
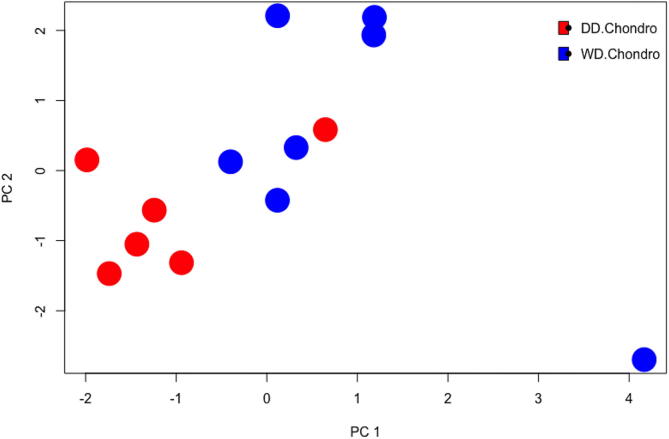
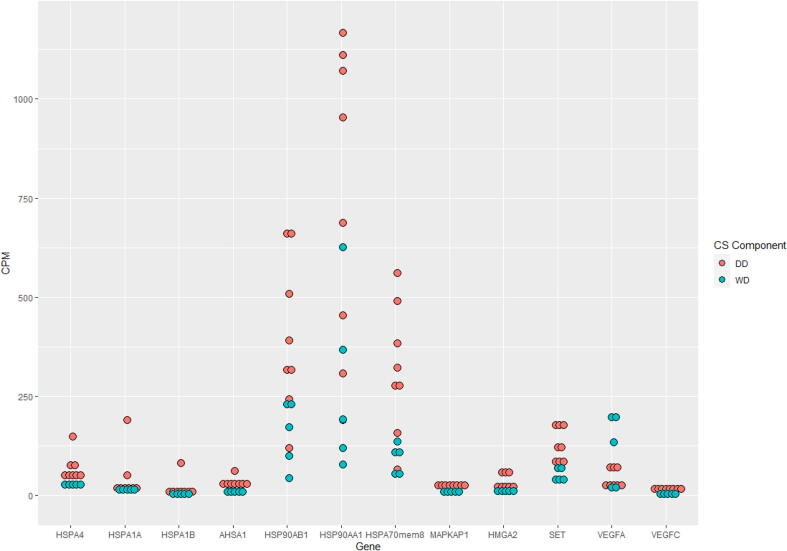
Moreover, RNA sequencing results, performed on 8 cases, demonstrated differences in RNA expression between the two components of dedifferentiated chondrosarcoma (Fig. 3). These results also independently confirmed the presence of VEGF family members, MAPK members, and HSP members including HSP70 and HSP90. Higher expression of VEGFA was seen in the well-differentiated component, while VEGFC was higher in the dedifferentiated component. Overexpression of HSP70 and HSP90 family members was observed in the dedifferentiated component. Of interest, high mobility group AT-hook 2 (HMGA2) and SET nuclear proto-oncogene also showed higher expression in the dedifferentiated component (Fig. 4 and Table 2).
Fig. 3.
Principal Component Analysis Plot of DDCS Components. This principal component analysis compares the two components of dedifferentiated chondrosarcoma in a statistical model that best represents the RNA sequencing data. Red dots represent the DD component of DDCS and blue dots represent the WD component of DDCS. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
HSP, MAPK, HMGA2, SET and VEGF Expression in DDCS Components by RNA Sequencing X axis shows HSP family members, MAPK, HMGA2, SET and VEGF gene expression in well differentiated (green) and dedifferentiated (red) components of dedifferentiated chondrosarcoma. Y axis shows RNA expression in counts per million (CPM, mapped reads normalized by library size multiplied by 1 million). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Mean CPM Values of HSP, VEGF, MAPK, HMGA2 and SET in DDCS Components.
| Gene | WD Component | DD Component | P value |
|---|---|---|---|
| HSPA4 | 28.67 | 67.36 | 0.04 |
| HSP70member8 | 92.64 | 317.03 | 0.007 |
| HSPA1A | 9.83 | 39.94 | 0.06 |
| HSPA1B | 3.07 | 16.15 | 0.02 |
| AHSA1 | 8.42 | 31.26 | 0.0006 |
| HSP90AB1 | 155.11 | 402.51 | 0.04 |
| HSP90AA1 | 276.8 | 742.74 | 0.05 |
| VEGFA | 113.9 | 41.56 | 0.004 |
| VEGFC | 3.05 | 11.19 | 0.02 |
| MAPKAP1 | 10.24 | 21.05 | 0.06 |
| HMGA2 | 6.01 | 34.72 | 0.01 |
| SET | 48.96 | 128.85 | 0.01 |
CPM = Counts per million (RNA sequencing mapped reads normalized by library size multiplied by 1 million).
6. Discussion
To date dedifferentiated chondrosarcoma is treated surgically with scant benefit shown for adjuvant chemoradiation therapy [8]. Prognosis is bleak with a 5 year overall survival of up to 24% [3], [9], [10], [11].
In an effort to elucidate alternative treatment targets in chondrosarcoma, various researchers have identified specific molecular pathways active in these tumors, including p13K/Akt/mTOR, p53, Rb [12], [13], [14], [15], HIF1a [14], PTHR1 [16] and BCL2 [15], [16]. Increased levels of Hedgehog pathway factors are also expressed in high grade chondrosarcomas [15]. As a result of the multiple mechanisms involved in the development and behavior of chondrosarcoma, crosstalk between pathways may represent a factor in resistance to therapy [12]. Other mediators of treatment resistance include p-glycoprotein [12] and the BCL2 family [15], [16].
Angiogenesis is critical to the growth, invasion and metastatic properties of chondrosarcoma [17], [18], [19]. Antiangiogenic therapy has been proposed for treatment of high grade chondrosarcoma based on evidence of increased microvessel density and intracartilaginous vascularity in chondrosarcoma [6], [20]. VEGF is upregulated in chondrosarcoma and is mediated by several factors and pathways such as Adiponectin, p13K/Akt/mTOR, HIF1a and hypoxic conditions in the microenvironment [15], [21], [22]. Increased VEGF expression is present in grades 2 and 3 chondrosarcomas [6], [7]. In turn, ERK1/2 regulates endothelial cell migration and proliferation in response to VEGF [23]. Furthermore, IDH mutations in chondrosarcoma increase D2- hydroxyglutarate, stabilizing HIF1a and promoting angiogenesis.
Drugs targeting angiogenic pathways have been developed. The antiangiogenic drug, Sorafanib, for example, induces hypoxia, reduces endothelial cell proliferation and downregulates pERK [24]. Antiangiogenic drugs, Pazopanib and Ramucirumab, were used in the treatment of ten patients with advanced chondrosarcoma and demonstrated a mean progression free survival of 22.6 months as compared to 4.7 months for chemotherapy [10].
HSP70, by contrast, has diverse cellular roles and overexpression is associated with high grade malignancy, inhibition of apoptosis, tumor invasion, resistance to therapy and reduced host survival [25], [26], [27], [28], [29], [30]. On the other hand, down regulation of HSP70 leads to the demise of cancer cells. HSP70 is present in all of the subcellular compartments of nucleated cells including cell membranes [31]. As an angiogenesis regulator, HSP70 can activate ERK [27]. Tumor cells also harbor membrane bound exosomes containing HSP70. These exosomes can be harnessed as part of an immune response to neoplastic cells [32].
To our knowledge, matched well differentiated and dedifferentiated components of dedifferentiated chondrosarcoma have not been studied and compared with regard to expression of angiogenesis factors. By immunohistochemistry, VEGFA showed similar expression in the two components, although RNA sequencing demonstrated higher VEGFA expression in the well differentiated component, while the dedifferentiated component showed higher VEGFC expression. In the study by Ayala, et al, VEGF expression was seen almost exclusively in high grade (2 and 3) conventional chondrosarcomas as compared to enchondromas and grade 1 chondrosarcomas. One dedifferentiated case in their cohort showed immunohistochemical expression of VEGF in both components [6]. The family members of VEGF have been studied in solid tumors and VEGFA promotes blood vessel formation, while VEGFC fosters lymphatic vessel formation [33]. Although angiogenesis has not been studied extensively in dedifferentiated chondrosarcoma, it is plausible that the absence of cartilage matrix in the dedifferentiated component may facilitate the development of abnormal, complex, lymphovascular networks, leading to hypoxia and promoting aggressive behavior.
The authors also speculate that VEGF may be upregulated in the well differentiated component of dedifferentiated chondrosarcoma as compared to low grade (grade 1) chondrosarcoma/atypical cartilaginous tumor (not analyzed in this study). Additional work is required to substantiate this hypothesis.
By contrast, immunohistochemistry for pERK1/2 and HSP70 demonstrated higher expression in the dedifferentiated component of dedifferentiated chondrosarcoma. Likely, multiple pathways with crosstalk between VEGF, pERK1/2 and HSP70, among others, are involved in angiogenesis and malignancy in dedifferentiated chondrosarcoma. These factors interact and allow for evasive tumor behavior when confronted with chemotherapeutic agents. That HSP70 shows higher expression in the dedifferentiated component of chondrosarcoma also serves to emphasize the aggressive behavior of this neoplasm. A study by Trieb [34], et al, demonstrated differential expression of HSPs in chondrosarcoma and chondroma by immunohistochemistry. Their study showed decreased expression of HSP72 with increasing grade of chondrosarcoma and greater expression in chondroma. Dedifferentiated chondrosarcoma was not studied, however [34]. In a later study, Trieb, et al showed overexpression of HSP73 and HSP90 by immunohistochemistry in locally recurrent chondrosarcoma [35].
The presence of HSP70, HSP90, pERK1/2, and VEGF was substantiated in our dedifferentiated chondrosarcoma (DDCS) cases by RNA sequencing. Higher expression of HSP family members: AHSA1, HSPA4, HSPA1B, HSP70 member 8, and HSP90AB1 was seen in the dedifferentiated (DD) component. HSPA1B, a member of the HSP70 family, is known to participate in processes promoting aggressive tumor behavior, and is involved in antigen presentation and the MAPK pathway. HSP90 also induces angiogenesis and is involved in regulation of key oncoproteins [30], [36], [37]. Given that HSP90′s client proteins include EGFR, MET, HIFa and AKT, pharmacologic targeting of HSP90 could impact these proteins, as well [30]. That HMGA2 exhibits higher expression in the dedifferentiated component may relate to misexpression of HMGA2 in mesenchymal tumorigenesis. Its association with chemotherapeutic drug resistance and metastasis is known [38]. The overexpression of SET nuclear proto-oncogene in the dedifferentiated component may have a protective effect for the tumor in that apoptosis is inhibited and autophagy is promoted [39]. Utilizing multi-drug/modality combination therapies is likely the best way of overcoming treatment resistance.
Potential applications for HSP expression in dedifferentiated chondrosarcomas are broad. It is possible to target HSPs directly or utilize them in immune therapy as demonstrated in other malignancies such as carcinomas [30], [32], [40]. Further studies are needed to better define the tumor microenvironment and identify therapeutic targets in dedifferentiated chondrosarcoma.
CRediT authorship contribution statement
Karen Schoedel: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Writing - original draft, Writing - review & editing. Virginia Miller: Data curation, Investigation. David Osei-Hwedieh: Writing - review & editing. Rebecca Watters: Methodology, Writing - review & editing. Anette Duensing: Writing - review & editing. Ivy John: Writing - review & editing. Uma Chandran: Formal analysis, Software. Alexander Chang: Formal analysis, Data curation, Software. Vishal Soman: Formal analysis, Software. Kurt Weiss: Resources, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the Pittsburgh Sarcoma Research Collaborative (PSaRC) without which this work would not be possible. We also acknowledge the mentorship of the late Dr. Uma N. M. Rao of UPMC Pathology.
References
- 1.D. Dahlin, J. Beabout, Dedifferentiation of low grade chondrosarcomas. 1971 Cancer; 28:461. doi: 10.1002/1097-0142(197108)28:2<461::aid-cncr2820280227>3.0.co;2-u. [DOI] [PubMed]
- 2.S. Johnson, B. Tetu, A. Ayala, S. Chawla, Chondrosarcoma with additional mesenchymal component (dedifferentiated chondrosarcoma) a clinicopathologic study of 26 cases. 1986 Cancer 58:278. doi: 10.1002/1097-0142(19860715)58:2<278::aid-cncr2820580213>3.0.co;2-6. [DOI] [PubMed]
- 3.WHO Classification of Tumours of Soft Tissue and Bone, 5th ed. 2020 (IARC, Lyon).
- 4.J. Bovee, A.M. Cleton-Jansen, C. Rosenberg, et al. Molecular genetic characterization of both components of a dedifferentiated chondrosarcoma, with implications for its histogenesis. 1999 J. Pathol 189; 454. doi: 10.1002/(SICI)1096-9896(199912)189:4<454::AID-PATH467>3.0.CO;2-N. [DOI] [PubMed]
- 5.Meijer D., de Jong D., Pansuriya T.C., van den Akker B.E., Picci P., Szuhai K., Bovée J.V.G.M. Genetic characterization of mesenchymal, clear cell and dedifferentiated chondrosarcoma 2012 Genes. Genes Chromosom. Cancer. 2012;51(10):899–909. doi: 10.1002/gcc.v51.1010.1002/gcc.21974. [DOI] [PubMed] [Google Scholar]
- 6.G. Ayala, C. Liu, R. Nicosia, et al. Microvasculature and VEGF expression in cartilaginous tumors. 2000 Human Pathol. 31;3:341. doi: 10.1016/s0046-8177(00)80248-8. [DOI] [PubMed]
- 7.R. McGough, C. Lin, P. Meitner, et al. Angiogenic cytokines in cartilage tumors. 2002 Clinical Orthopedics and Related Research 397:62 doi: 10.1097/00003086-200204000-00009. [DOI] [PubMed]
- 8.A. Van Maldegem, J. Bovee, H. Gelderblom, Comprehensive analysis of published studies involving systemic treatment for chondrosarcoma of bone between 2000 and 2013. 2014 Clin. Sarcoma Res. 4:11 doi: 10.1186/2045-3329-4-11. [DOI] [PMC free article] [PubMed]
- 9.P. Strotman, T. Reif, S. Kliethermes, et al. Dedifferentiated chondrosarcoma: a survival analysis of 159 cases for the SEER database (2001–2011). 2017 J Surg Oncol 116:252. doi: 10.1002/jso.24650. [DOI] [PubMed]
- 10.R. Jones, D. Katz, E. Loggers, et al. Clinical benefit of antiangiogenic therapy in advanced and metastatic chondrosarcoma 2017 Med Oncol 34:167. doi: 10.1007/s12032-017-1030-2. [DOI] [PMC free article] [PubMed]
- 11.H. Lee, C. Lin, J. Shih, et al. Adiponectin promotes VEGF-A dependent angiogenesis in human chondrosarcoma through p13K, Akt, mTOR and HIF-a pathway. 2015 Oncotarget 6;34:36746. doi: 10.18632/oncotarget.5479. [DOI] [PMC free article] [PubMed]
- 12.B. Mery, S. Espenel, J. Guy, et al. Biological aspects of chondrosarcomas: leaps and hurdles. 2018 Crit. Rev. Oncol./Hematol. 126:32. doi: 10.1016/j.critrevonc.2018.03.009. [DOI] [PubMed]
- 13.J. Bovee, P. Hogendoorn, J. Wunder, B. Alman, Cartilage tumours and bone development:molecular pathology and possible therapeutic targets. 2010 Nat. Rev. Cancer 10:481. doi: 10.1038/nrc2869. [DOI] [PubMed]
- 14.Y. Zhang, J. van Oosterwijk, et al. Functional profiling of receptor tyrosine kinases and downstream signaling in human chondrosarcomas identifies pathways for rational targeted therapy. 2013 Clin Cancer Res; 19(14); 3796. doi: 10.1158/1078-0432.CCR-12-3647. [DOI] [PubMed]
- 15.F. Speetjens, Y. de Jong, H. Gelderblom, J. Bovee, Molecular oncogenesis of chondrosarcoma:impact for targeted treatment. 2016 Current Opin Oncol 28:314. doi: 10.1097/CCO.0000000000000300. [DOI] [PubMed]
- 16.J.G. Van Oosterwijk, D. Meijer, M. van Ruler, et al. Screening for potential targets for therapy in mesenchymal, clear cell, and dedifferentiated chondrosarcoma reveals BCL2 family members and TGFB as potential targets. 2013 Am. J. Pathol. 182 (4): 1347. 10.1016/j.ajpath.2012.12.036. [DOI] [PubMed]
- 17.Yang W.-H., Chang A.-C., Wang S.-W., Wang S.-J., Chang Y.-S., Chang T.-M., Hsu S.-K., Fong Y.-C., Tang C.-H. Leptin promotes VEGF-C production and induces lymphangiogenesis by suppressing miR-27b in human chondrosarcoma cells. Sci. Rep. 2016;6(1) doi: 10.1038/srep28647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. Wu, P. Huang, M. Hsieh, et al. Endothelin-1 promotes epithelial-mesenchymal transition in human chondrosarcoma cells by repressing miR-300. 2016 Oncotarget 7(43): 70232. doi: 10.18632/oncotarget.11835. [DOI] [PMC free article] [PubMed]
- 19.K. Boehme, S. Schleicher, F. Traub, B. Rolauffs, Chondrosarcoma: A rare misfortune in aging human cartilage? The role of stem and progenitor cells in proliferation, malignant degeneration and therapeutic resistance. 2018 Int. J. of Molecular Sciences 19:311doi: 10.3390/ijms19010311. [DOI] [PMC free article] [PubMed]
- 20.F. Cintra, M. Etchebehere, J. Goncalves, et al. Analysis of angiogenic factors and cyclooxygenase-2 expression in cartilaginous tumors- clinical and histological correlation. 2011 Clinics 66(9):1591. doi: 10.1590/s1807-59322011000900015. [DOI] [PMC free article] [PubMed]
- 21.M. Wu, C. Huang, J. Lin, et al. Endothelin-1 promotes vascular endothelial growth factor-dependent angiogenesis in human chondrosarcoma cells. 2014 Oncogene 33:1725doi: 10.1038/onc.2013.109. [DOI] [PubMed]
- 22.A. Olsson, A. Dimberg, J. Kreuger, L. Claesson-Welsh, VEGF receptor signalling-in control of vascular function. 2006 Nat. Rev. Mol. Cell Biol. 7:359. doi: 10.1038/nrm1911. [DOI] [PubMed]
- 23.R. Srinivasan, T. Zabuawala, H. Huang, et al. Erk1 and Erk2 regulate endothelial cell proliferation and migration during mouse embryonic angiogenesis. 2009 Plos One 4(12):e8283doi: 10.1371/journal.pone.0008283. [DOI] [PMC free article] [PubMed]
- 24.D. Murphy, S. Makonnen, W. Lassoued, et al. Inhibition of tumor endothelial ERK activation, angiogenesis and tumor growth by Sorafenib. 2006 Am J Pathol 169:1875doi: 10.2353/ajpath.2006.050711. [DOI] [PMC free article] [PubMed]
- 25.K. Rashmi, H. Atreya, M. Raj, et al. A pyrrole-based natural small molecular mitigates HSP90 expression in MDA-MB-231 cells and inhibits tumor angiogenesis in mice by inactivating HSF-1. 2017 Cell Stress and Chaperones. 22:751. DOI: 10.1007/s12192-017-0802-0. [DOI] [PMC free article] [PubMed]
- 26.M. Sherman, V. Gabai, Hsp70 in cancer: back to the future. 2015 Oncogene 34:4153. doi: 10.1038/onc.2014.349. [DOI] [PMC free article] [PubMed]
- 27.T. Kim, H. Na, W. Lee, M. Jeoung, S. Lee, Heat shock protein 70-1A is a novel angiogenic regulator. 2016 Biochem. Biophys. Res. Commun. 469:222. doi: 10.1016/j.bbrc.2015.11.125. [DOI] [PubMed]
- 28.D. Dimas, C. Perlepe, T. Sergentanis, et al. The prognostic significance of HSP70/HSP90 expression in breast cancer: a systematic review and meta-analysis. 2018 Anticancer Research 38: 1551. DOI: 10.21873/anticanres.12384. [DOI] [PubMed]
- 29.C. Boudesco, S. Cause, G. Jego, C. Garrido, Ch. 27 HSP70: A cancer target inside and outside the cell in Calderwood S and Prince T, eds, Chaperones: Methods and Protocols, Methods in Molecular Biology, v.1709: 371 (2018 Springer Science+Business media) doi: 10.1007/978-1-4939-7477-1_27. [DOI] [PubMed]
- 30.Wu J., Liu T., Rios Z., Mei Q., Lin X., Cao S. Heat shock proteins and cancer. Trends Pharm. Sci. 2017;38(3):226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 31.M. Shevtsov, G. Huile, G. Multhoff, Membrane heat shock protein 70: A theranostic target for cancer therapy. 2017 Phil. Trans. R. Soc. B 373: 20160526. doi: 10.1098/rstb.2016.0526. [DOI] [PMC free article] [PubMed]
- 32.R. Binder, Functions of heat shock proteins in pathways of the innate and adaptive immune system. 2014 J. Immunol. 193: 5765 doi: 10.4049/jimmunol.1401417. [DOI] [PMC free article] [PubMed]
- 33.Majidpoor J., Mortezaee K. Angiogenesis as a hallmark of solid tumors-clinical perspectives. Cell. Oncol. 2021 doi: 10.1007/s13402-021-00602-3. [DOI] [PubMed] [Google Scholar]
- 34.K. Trieb, R. Kohlbeck, S. Lang, et al. Heat shock protein 72 expression in chondrosarcoma correlates with differentiation. 2000 J. Cancer Res. Clin. Oncol.; 126:667-670. doi: 10.1007/s004320000167. [DOI] [PubMed]
- 35.Trieb K., Sulzbacher I., Kubista B. Recurrence rate and progression of chondrosarcoma is correlated with heat shock protein expression. Oncol. Lett. 2016;11:521. doi: 10.3892/ol.2015.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatterjee S., Burns T. Targeting heat shock proteins in cancer: a promising therapeutic approach. Int. J. Mol. Sci. 2017;18:1–39. doi: 10.3390/ijms18091978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.E. Taha, K. Ono, T. Eguchi, Roles of extracellular HSPs as biomarkers in immune surveillance and immune evasion. 2019 Int. J. Mol. Sci. 20:4588. DOI: 10.3390/ijms20184588 [DOI] [PMC free article] [PubMed]
- 38.U. Unachukwu, K. Chada, J. Armiento, High mobility group AT-hook2 oncogenicity in mesenchymal and epithelial neoplasia. 2020 Int. J. Mol. Sci., 21: 3151. DOI: 10.3390/ijms21093151 [DOI] [PMC free article] [PubMed]
- 39.A.T. Ouchida, V. Uyemura, A. Queiroz, et al. SET protein accumulation prevents cell death in head and neck squamous cell carcinoma through regulation of redox state and autophagy. 2019 BBA-Molecular cell research 1866: 623. doi: 10.1016/j.bbamcr.2019.01.005. [DOI] [PubMed]
- 40.G. Polychronidou, V. Karavasilis, S. Pollack, et al. Novel therapeutic approaches in chondrosarcoma. 2017 Future Oncol. 13(7): 637 doi: 10.2217/fon-2016-0226. [DOI] [PubMed]