Abstract
Background
Among the approximately 8000 Mendelian disorders, >1000 have cutaneous manifestations. In many of these conditions, the underlying mutated genes have been identified by DNA-based techniques which, however, can overlook certain types of mutations, such as exonic-synonymous and deep-intronic sequence variants. Whole-transcriptome sequencing by RNA sequencing (RNA-seq) can identify such mutations and provide information about their consequences.
Methods
We analyzed the whole transcriptome of 40 families with different types of Mendelian skin disorders with extensive genetic heterogeneity. The RNA-seq data were examined for variant detection and prioritization, pathogenicity confirmation, RNA expression profiling, and genome-wide homozygosity mapping in the case of consanguineous families. Among the families examined, RNA-seq was able to provide information complementary to DNA-based analyses for exonic and intronic sequence variants with aberrant splicing. In addition, we tested the possibility of using RNA-seq as the first-tier strategy for unbiased genome-wide mutation screening without information from DNA analysis.
Results
We found pathogenic mutations in 35 families (88%) with RNA-seq in combination with other next-generation sequencing methods, and we successfully prioritized variants and found the culprit genes. In addition, as a novel concept, we propose a pipeline that increases the yield of variant calling from RNA-seq by concurrent use of genome and transcriptome references in parallel.
Conclusions
Our results suggest that “clinical RNA-seq” could serve as a primary approach for mutation detection in inherited diseases, particularly in consanguineous families, provided that tissues and cells expressing the relevant genes are available for analysis.
Keywords: whole-transcriptome sequencing, RNA-seq, mutation detection, heritable skin diseases, epidermolysis bullosa, familial consanguinity
Introduction
Mendelian disorders comprise a heterogeneous group of diseases in which substantial progress has been made in understanding the genetic basis through identification of mutant genes and specific pathogenic sequence variants by next-generation sequencing (NGS) approaches. However, the genetic diagnosis has been made by DNA-based NGS methods in only 20%–50% of such cases and in many cases remains unknown, for several reasons (1). The challenges include difficulty in classifying the variants of unknown significance (VUS) and the inability of DNA-based NGS approaches to detect specific types of alterations, such as structural variants and repetitive sequence expansions and/or the disease-causing variants that are located in noncoding and regulatory regions of the genome (2, 3). The mutation-detection strategies have focused primarily on DNA analysis by Sanger sequencing and by using NGS approaches, including gene targeting, disease-specific NGS panels, whole-exome sequencing (WES), and whole-genome sequencing (WGS). These approaches have been assisted by genome-wide homozygosity mapping (HM) toward identification of associated mutant genes in consanguineous families (4).
More recently, RNA sequencing (RNA-seq) with appropriate bioinformatics analysis steps has been used for advanced mutation detection and for transcriptome quantitation and splicing profiling (5). During the genomic DNA analysis, mutations that potentially affect splicing events may be ignored and erroneously classified as synonymous changes or benign amino acid substitutions. RNA-seq is a multifaceted technique that can be used for variant calling, complementing the information derived from DNA-based analysis. In addition, it provides tools for prediction of the effects of some variants, such as exonic-synonymous nucleotide substitutions at exon/intron borders and deep-intronic variants affecting splicing. RNA-seq is also a robust technique to detect genes with monoallelic expression, particularly for diseases with recessive mode of inheritance that could not be captured by a single heterozygous rare variant identified by WES or WGS (6).
The importance of RNA-seq is emphasized by the notion that splicing defects are a major cause of 15%–60% of Mendelian disorders (7), and the pathogenic variants can be located in intronic sequences, not captured by WES (8). In addition, some regions in the genome are difficult to sequence, and RNA-seq can be helpful in finding rare causative variants in such regions. Consequently, “clinical RNA-seq” is a robust tool to facilitate genetic diagnosis of rare variants of Mendelian disorders complementing and extending the information available from DNA-based NGS approaches.
There are more than 1,000 Mendelian cutaneous disorders and, in some cases, the clinical findings are strictly limited to the skin and are thus nonsyndromic, whereas in many of the heritable skin diseases, the cutaneous findings are associated with extracutaneous manifestations and thus are syndromic (9). To fully investigate the applications of RNA-seq in Mendelian disorders, this study reports on a combination of previously unreported patients in our cohort and reanalyzes a few of our previously published cases with an algorithm developed in this study. The results demonstrated a high yield of mutation detection (88%), and in a subset of families, the findings were corroborated by DNA-based NGS analyses. In addition, in an unbiased approach, we interrogated RNA-seq as a first-tier genome-wide diagnostic tool for mutation detection in unknown heritable disorders. In particular, variant calling from high-throughput RNA-based sequencing techniques has been recently spotlighted in clinical diagnostic settings. Consequently, several novel computational workflows have been developed, such as VAP (Variant Analysis Pipeline), curated by Adetunji et al. (10), by using several RNA-seq splice-aware aligners to call single-nucleotide variants (SNVs) in nonhuman models using RNA-seq data only or Variant Detection in RNA (VaDiR), which integrates 3 variant callers: SNPiR, RVBoost, and MuTect2 (11–13). The VaDiR tool is a robust software to identify the mutations from RNA-seq data sets. Moreover, SNPiR is another tool to identify SNVs in RNA-seq data. Traditionally, RNA-seq is mostly used to measure the level of gene expression and to extract differentially expressed genes (DEGs), but it can also be an alternative to genomic methods to perform variant calling from RNA-seq with several applications. In this study, we developed and evaluated a pipeline for processing RNA-seq data from rare heritable skin diseases for variant calling. To improve the accuracy of variant-calling results, combinational analysis of genomic data sequencing was carried out with RNA-seq to broaden the discovery of genomic mutations.
Materials and Methods
Study Design
This study was approved by the Institutional Review Board of the Pasteur Institute of Iran, and the ethics committee of the Tarbiat Modares University, Tehran, Iran. All participants and the parents of children gave written informed consent to participate in research and to publish their images. From a total of about 1800 families referred to us in the past 6 years with different Mendelian disorders with cutaneous manifestations, we ascertained 2 groups of patients from whom samples were available for RNA-seq studies: (a) the patients were found to have VUS by genomic analysis or (b) they were subjected to RNA-seq as the first-tier strategy for unbiased genome-wide mutation screening without information from previous genomic data analyses. The criterion for study inclusion was a tentative diagnosis of a Mendelian skin disorder based on family history and clinical features consistent with the diagnosis. We analyzed samples from the probands of 40 families, 25 of them demonstrating familial consanguinity (Supplemental Table S1). In the current study, 4 types of NGS data, including targeted gene panel sequencing, WES, WGS, and RNA-seq, were analyzed (see details in the online Data Supplement).
RNA Sequencing
Along with genomic data sequencing, we used the RNA-seq method to investigate the mRNA content and processing in the skin or cultured cells. RNA-seq was initiated by the isolation of RNA either from a small (3-mm) whole-skin biopsy or cultured dermal fibroblasts or epidermal keratinocytes (Fig. 1). Sequencing was done at the Cancer Genomics facilities at Thomas Jefferson University and used 100 ng of total RNA with the TruSeq Stranded Total RNA Kit (Illumina), following the manufacturer's protocol. The mRNA capture and library preparation at 4-nmol/L concentration, including bar coding and sequencing on an Illumina NextSeq 500 machine, were performed according to standard procedures at the Thomas Jefferson University facility. With 150-bp paired-end chemistry, a sequencing depth of 20-100 million paired reads per sample was achieved.
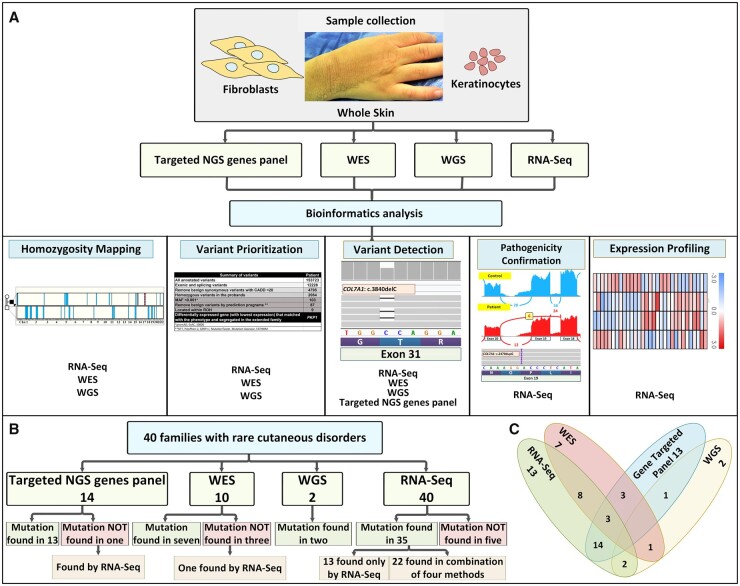
Fig. 1.
Schematic overview of the study outline. (A) Summary of NGS-based methods applied in this study and their potential applications to heritable skin diseases. The potential applications of RNA-seq data include HM, variant prioritization, variant detection, pathogenicity confirmation and expression profiling. As indicated, RNA-seq is complementary to exome sequencing in some of these functions, whereas some information is available only from RNA-seq. (B) Summary of the techniques applied to families with heritable skin disorders. Forty families with rare cutaneous disorders were analyzed by the combined analysis pipelines for variant calling by using genomic and transcriptomic data. (C) The overlap of methodologies utilized is illustrated by the Venn diagram. The genetic diagnostic yield by these techniques is indicated both in (B) and (C).
Preprocessing of RNA-Seq Data
The quality of raw reads was checked by the FastQC tool, and based on the quality-check results, the trimming process using Trimmomatic tool was applied to raw reads to remove the low-quality sequences. We proposed a novel bioinformatics pipeline that promotes the diagnostic yield in variant calling. In this regard, the high-quality sequences were mapped to human genome and transcriptome references by the Star and BWA/Bowtie2 tools, respectively (14).
Extraction and Visualization of Differentially Expressed Genes
Following the mapping phase with Star, the StringTie program was used to assemble the aligned sequences of 6 control and 40 patient samples with different genodermatoses. The outputs of the assembly were used to extract DEGs, and the edgeR package was applied. The obtained count matrix of all samples was normalized by trimmed mean of M values (TMM) method, and the t-test was carried out to identify the genes with differential expression levels between control and patient samples. The DEGs were selected based on a log fold-change >1 or less than −1 and a false discovery rate <0.05. In the following step, significantly meaningful DEGs were enriched by Enrichr. Although several types of rare skin diseases were investigated in this study, the DEGs were identified only between a specific type of disease and healthy samples. Heat maps were created using MeV (Multiple Experiment Viewer) version 4.9.0 (The Institute for Genomic Research (TIGR)).
RNA-Seq for Variant Calling
We used general genome mappers, such as BWA or Bowtie2, for mapping RNA-seq data to the reference transcriptome. Next, processing of the aligned reads along with variant calling were done by Picard and Samtools, respectively. Before combining the identified variants from this workflow with the ones obtained from mapping to the reference genome, the coordinates of called variants from the reference transcriptome were converted to their corresponding genomic coordinates. The discussed cases used GRCh37/hg19 from the University of California Santa Cruz (UCSC) Genome Browser Gateway as the reference genome and reference transcriptome, and the National Center for Biotechnology Information RefSeq track from the Genes and Gene Predictions group of the UCSC table browser was also used for transcriptomic to genomic coordinate conversion. Using the exons’ lengths and the available start and end positions, the coordinates of called variants using the reference transcriptome were translated to their corresponding coordinates on the reference genome. The variant calling from mapped reads to the reference genome was performed by GATK4. We included Filter_reads_with_N_cigar to have higher variant-calling sensitivity. The variant-calling region was set to the exons of all transcripts and surrounding 15 bp flanking intronic sequences. The next step of the analysis includes variant filtration and annotation by GATK and ANNOVAR (15), respectively, to combine the identified variants from both workflows. Final candidates were identified by overlapping the surviving variants with the regions of homozygosity (ROHs), as applicable to families with evidence of consanguinity.
Validation of the Utility of the Proposed Pipeline
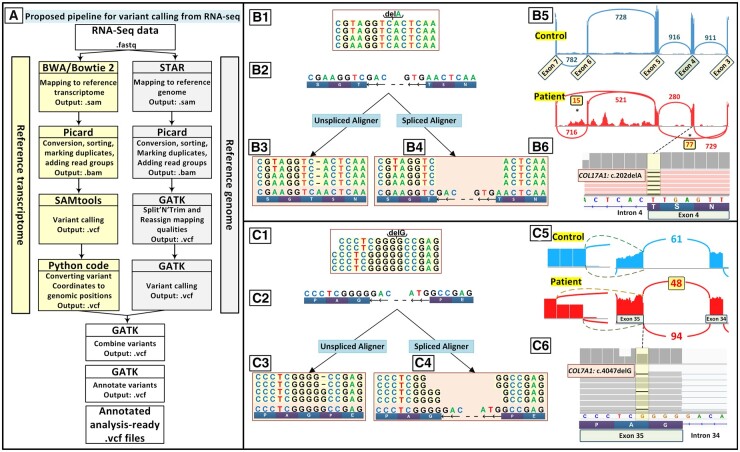
We implemented a novel approach of variant calling from RNA-seq data. To use this method to increase the diagnostic yield of variant calling in rare cutaneous diseases, we began by evaluating its accuracy and specificity based on the previously published GM12878 lymphoblastoid cell data set (GSM758559). This data set has been studied extensively, and SNVs detected in its genome have been continuously deposited into dbSNP, making it a good candidate set for evaluating the precision and sensitivity of our proposed pipeline (16, 17). As illustrated in Fig. 2, the parameters of the proposed pipeline were optimized to achieve the highly accurate variant calling from RNA-seq (see details in the online Data Supplement). Our pipeline is available at https://github.com/Vahidnezhad-Lab.
Fig. 2.
Parallel use of a reference genome and a reference transcriptome improves variant calling from RNA-seq data for Mendelian disorders. (A) Workflow of our proposed pipeline for variant calling from RNA-seq data with 2 parallel processes for mapping of RNA-seq data to the reference genome and reference transcriptome. (B) Mapping to the reference genome detected the mutation c.5422C>T in EXPH5 (data not shown) in a patient with EB with 2 homozygous mutations but not a deletion mutation c.202delA in COL17A1, which was detected by mapping to the reference transcriptome. (B.1) Schematic representation of RNA sequenced reads for the homozygous mutation c.202delA in COL17A1. (B.2) Reference genome corresponding to the location of sequenced reads with a negative-strand orientation. (B.3) RNA sequenced reads when mapped to the reference genome by a splice aligner tool such as STAR. (B.4) RNA sequence reads were mapped with the reference transcriptome by unspliced aligners tools. (B.5) A sashimi plot revealed that the c.202delA mutation at the end of exon 4 leads to complex aberrant splicing of COL17A1 pre-mRNA, including skipping of exons 4 and 6 in 77 and 15 sequencing reads, respectively. (B.6) Screenshot of the genomic sequence visualized by IGV demonstrating the homozygous mutation COL17A1: c.202delA. (C.1) Schematic representation of sequenced reads harboring the heterozygous deletion of c.4047delG in COL7A1 by RNA sequencing in a patient affected by DEB with compound heterozygous deletion mutations of c.3840delC and c.4047delG. (C.2) Reference genome corresponding to the location of sequenced reads with a negative-strand orientation. (C.3) Mapped reads to the reference genome show a lower number of reads for the first 2 nucleotides of an exon and extra reads for the last nucleotide of the adjacent exon and first nucleotide at the beginning of the intron. (C.4) Sequences mapped to the reference transcriptome clearly show the heterozygous deletion. (C.5) A sashimi plot revealed that c.4047delG in exon 35 of COL7A1 leads to aberrant splicing. (C.6) Screenshot of IGV demonstrating the heterozygous mutation COL7A1:4047delG.
Results
Study Outline
In a combination of NGS-based technologies, patient samples (RNA and/or DNA) were analyzed with appropriate bioinformatics pipelines. RNA-seq shared applications with DNA-based NGS techniques, such as HM, variant prioritization, variant detection, and pathogenicity confirmation (Fig. 1A). Overall, we analyzed 14 patients by disease-targeted gene panels, 10 by WES, 2 by WGS, and 40 by RNA-seq either alone or in combination (see Venn diagram in Fig. 1C). The definitive diagnosis was made in 13 patients based exclusively on RNA-seq results, emphasizing the role of this method in increasing the diagnostic yield. Five cases (12%) remain genetically unresolved, even after WES was done for 2 of them. Collectively, 35 of 40 families (88%) were genetically diagnosed. In all cases, the mutations were confirmed by Sanger sequencing. Accordingly, we exploited both genomic and transcriptomic data for the variant-calling process particularly in rare skin diseases (Fig. 1B). This success in identifying the genetic basis of rare heritable skin diseases reflects the combined use of whole-transcriptome sequencing by RNA-seq with DNA-based analyses using a gene-targeted panel, WES, WGS and HM.
Concurrent Use of Reference Genome and Reference Transcriptome Improves Variant Calling from RNA-Seq
We propose a novel pipeline to analyze the result files of whole-transcriptome sequencing with both reference transcriptome and reference genome for variant detection and prioritization. Although the application of a reference transcriptome has been mentioned previously in the literature, it has not been reported to be applied for variant calling before this study. The reference transcriptome is routinely used in recently developed software programs, such as Sailfish, kallisto, Salmon-SMEM, and Salmon-Quasi, with the aim of gene expression analysis. Therefore, in the current study, the high-quality sequences were mapped to human genome and transcriptome references by STAR and BWA/Bowtie2 tools, respectively. The workflow of these mapping pipelines is shown in Fig. 2A. On several occasions, mapping of the variant data to the reference genome alone according the GATK pipeline was not able to detect the mutation. We applied our proposed pipeline to a previously published case in which we found 2 homozygous mutations, a nonsense mutation EXPH5: c.5422C>T and a deletion mutation COL17A1:c.202delA, by manual interrogation of BAM files (4). The homozygous deletion mutation COL17A1:c.202delA in the middle of the reads of RNA-seq is shown in Fig. 2B.1; the reference genome corresponding to the location of the sequenced reads with a negative strand orientation is shown in Fig. 2B.2. When the transcriptome reads were mapped to reference genome, it was noted that because of the canonical splice sequence right before the second mutation, the most common splice-aware mapping tools (splice aligners such as STAR or HISAT2) (18) were unable to map this mutation, as the remaining part of the sequence reads was mapped to the adjacent exon and this space was considered as an unread nucleotide (Fig. 2B.3). However, when mapping tools were used with reference transcriptome by unspliced aligner programs, instead of the reference genome, the variant was successfully detected (Fig. 2B.4). A sashimi plot of RNA-seq revealed that deletion of the nucleotide A at the end of exon 4 led to complex aberrant splicing of COL17A1 pre-mRNA and skipping of exons 4 and 6 in 77 and 15 sequencing reads, respectively (Fig. 2B.5). Fig. 2B.6 depicts the screenshot of the genomic sequence visualized by Integrative Genomics Viewer (IGV) demonstrating the homozygous mutation.
Another advantage of using reference transcriptome is given by examination of a patient affected by dystrophic epidermolysis bullosa (DEB) with compound heterozygous COL7A1 mutations, c.4047delG (Fig. 2C.1) and c.3840delC; reference genome corresponding to the location of the sequenced reads with a negative-strand orientation is shown (Fig. 2C.2). Mapped reads to the reference genome (splice aligner) showed a lower number of reads for the 2 first nucleotides of an exon and extra reads for the last nucleotide of the adjacent exon and the first nucleotide at the beginning of the intron (Fig. 2C.3). In contrast, mapped sequences to the reference transcriptome (unspliced aligner) clearly showed the heterozygous deletion (Fig. 2C.4). This deletion can be mistakenly assumed as the correct map with lower number of reads. However, when the same data were mapped to reference transcriptome using WES/WGS mapping tools, the correct one-nucleotide heterozygous deletion was detected (Fig. 2C.4). Sashimi plot of RNA-seq revealed that c.4047delG in exon 35 of COL7A1 leads to aberrant splicing (Fig. 2C.5). Fig. 2C.6 shows the screenshot of IGV demonstrating heterozygous mutation of COL7A1:c.4047delG.
Utility of RNA-Seq Data for Mutation Detection as a First-Tier Method
After variant calling through mapping the RNA sequencing result files with either genome reference or transcriptome reference, the comma-separated values (csv) output file was annotated. Variant prioritization toward identification of candidate genes and pathogenic mutations initially focused on exonic sequence variants followed by removal of benign synonymous variants with a Combined Annotation Dependent Depletion (CADD) score of <20 when rare heritable diseases are studied. The variants were filtered by including only those with minor allele frequency <0.001, followed by removal of benign variants based on bioinformatics prediction programs. In case of consanguineous families with autosomal recessive disorders, the loci of putative pathogenic mutant genes were aligned with ROHs determined by HM, which resulted in significant reduction in the number of variants to be considered as causative (19–21). The specific applications of RNA-seq used in this study are described for challenging and illustrative cases (cases 1–9). The first 2 cases exemplify RNA-seq application as a first-tier method for mutation detection of the single nucleotide substitution at a position in intron/exon borders.
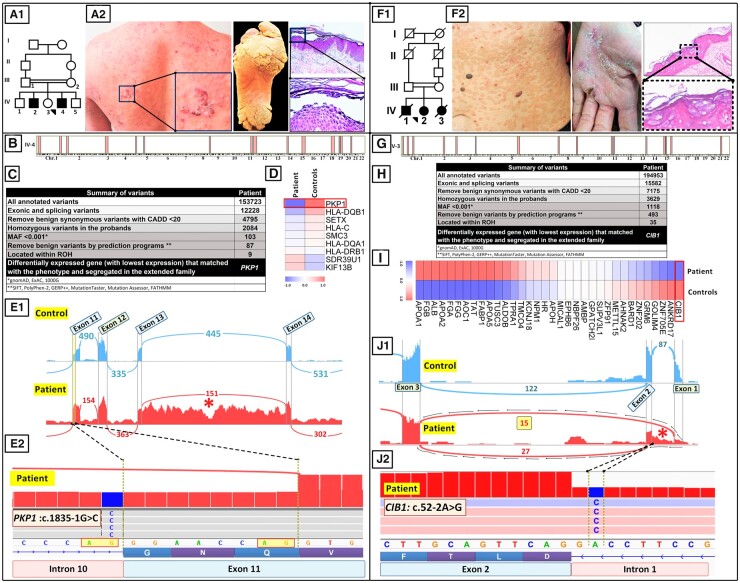
Case 1 describes a 32-year-old male born to a family with consanguineous parents and an older affected brother (Fig. 3A.1). The patient demonstrated superficial blistering and erosions on the trunk and hands, extensive plantar hyperkeratosis, and oral leukokeratosis. Histopathology of skin demonstrated suprabasal intraepidermal blistering with acanthotic cells, consistent with a skin fragility disorder with hyperkeratosis—a group of diseases associated with mutations in >60 distinct genes (Fig. 3A.2) (22). RNA-seq data-based HM revealed 15 ROHs >2.0 Mb that collectively contained 647 genes (Fig. 3B). After performing the required steps explained for variant detection by RNA-seq, 87 of 153 723 variants survived and were then overlapped with the 15 ROHs (Fig. 3C). Nine of the 87 variants aligned with 3 ROHs and were shared with the 2 affected individuals (IV-2 and IV-4) but were absent in the clinically unaffected obligate heterozygous parents (III-1 and III-2). Among the 9 genes listed in Fig. 3D, only one of them, PKP1, was known to be associated with skin fragility and hyperkeratosis phenotype. In addition, gene expression analysis of those 9 genes showed the lowest expression level for PKP1 (Fig. 3D). Analysis of the RNA-seq data identified a homozygous variant in the patient, PKP1: IVS10: c.1835-1G>C, which resided in the intron 10/exon 11 border affecting the last nucleotide in the intron 10 in the canonical splice site sequence agGG (Fig. 3E.2). Indeed, RNA-seq confirmed the pathogenicity of the canonical splice site mutation that was detected in PKP1, as shown in Fig. 3E. A Sashimi plot revealed abnormal splicing of intron 11 and retention of intron 13 sequences that were predicted to result in frameshift and synthesis of a truncated protein (Fig. 3, E.1 and E.2). Thus, whole-transcriptome analysis confirmed the pathogenicity of this sequence variant.
Fig. 3.
RNA-seq as a first-tier method of mutation detection in skin diseases. (A–F) Case 1. Family pedigree (A.1) and clinical features (A.2) in a patient with a skin fragility disorder with hyperkeratosis. (B) HM based on RNA-seq data revealed 15 ROHs in the proband. (C) Bioinformatics filtering steps of annotated variants that were used to narrow the candidate gene down to PKP1, which has been associated previously with a skin fragility syndrome. (D) A heat map of extracted DEGs revealed that the expression of PKP1 was most downregulated among the 9 genes coinciding with the ROHs. (E.1) A Sashimi plot revealed that this mutation resulted in aberrant splicing of exon 11 and retention of intron 13 sequences (asterisk). (E.2) IGV visualization of the genomic sequence revealed a homozygous mutation, PKP1:c.1835-1G>C affecting the last nucleotide in intron 10. (F–J) Case 2. Family pedigree (F.1) and clinical features (F.2) and histopathology of a patient with EV. (G) RNA-seq–based HM revealed 17 ROHs in the proband. (H) Stepwise bioinformatics filtering and coalignment of the putative mutant genes with ROHs identified a pathogenic variant in CIB1. (I) CIB1 was the most downregulated gene among the 35 variants coinciding with ROHs. (J.1, J.2) Variant calling from RNA-seq data identified a homozygous mutation, CIB1:c.52-2A>G. The Sashimi plot revealed complex aberrant splicing, including partial intron 1 retention and exon 2 skipping (asterisk).
Case 2 shows a 41-year-old female (IV-2) born to consanguineous parents (Fig. 3F.1). The proband presented with widespread distribution of flat warts on the trunk and palmar hyperkeratosis, and histopathology of the skin lesions revealed epidermal hyperkeratosis with keratinocytes showing coarse keratohyalin granules, perinuclear halo, and blue-gray pallor, consistent with viral infection (Fig. 3F.2). An older brother and a younger sister had died with similar clinical presentations. The clinical diagnosis of epidermodysplasia verruciformis (EV) was made, confirmed by human papillomavirus (HPV) typing, which revealed the presence of HPV5 and HPV8 (results not shown). HM, based on RNA-seq data, revealed 17 ROHs >2 Mb, and collectively, these regions of the genome contained 2859 genes (Fig. 3G). RNA-seq identified 194 953 sequence variants (Fig. 3H). Stepwise bioinformatics filtering, as shown in Fig. 3H, reduced the total number of possible pathogenic variants to 493. When these variants were overlapped with the 17 ROHs, the number of potentially pathogenic variants was reduced to 35. Gene expression analysis of those 35 genes showed CIB1 as the most downregulated gene in the patient (Fig. 3I); mutations in this gene have been recently shown to be associated with the EV phenotype (23, 24). The RNA-seq data identified a homozygous mutation, CIB1:c.52-2A>G affecting the canonical splice site at the intron 1/exon 2 border (Fig. 3J.2). A Sashimi plot from the patient in comparison to the controls revealed complex aberrant splicing of the CIB1 pre-mRNA, including deletion of exon 2 and partial retention of intron 1 sequences (Fig. 3J.1). The deletion of exon 2 was confirmed by reverse transcription PCR amplification followed by Sanger sequencing (data not shown). These data demonstrate efficient variant detection by whole-transcriptome sequencing in identification of homozygous pathogenic mutations and validation of their pathogenic consequences at the RNA level.
Applications of RNA-Seq in Identification of Unusual Mutations in Heritable Skin Diseases
Other applications of RNA-seq, in combination with DNA-based NGS approaches, are illustrated in unusual cases with heritable skin diseases.
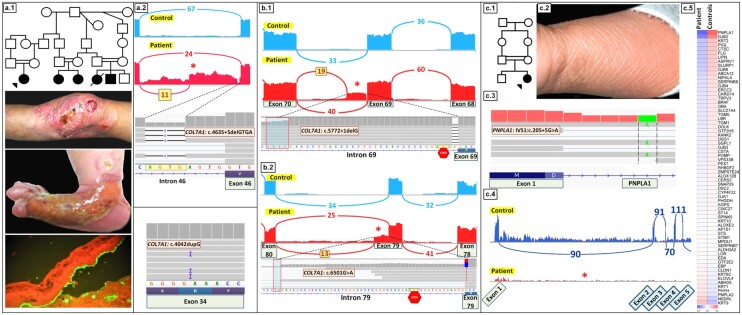
Case 3 depicts a female born to a family with extensive consanguinity (Fig. 4A.1). The patient had lifelong history of erosions and scarring of the skin, and she was born with aplasia cutis congenita on the right foot (Fig. 4A.1). Immune epitope mapping showed sub–lamina densa tissue separation, as demonstrated with immunostaining with an anti–type IV collagen antibody, consistent with DEB diagnosis (Fig. 4A.1). Four additional children in the same generation had similar cutaneous features, and the parents were clinically normal. Given extensive consanguinity in the family, we expected to identify a homozygous mutation in COL7A1, but RNA-seq identified 2 heterozygous COL7A1 mutations in the proband, including noncanonical splice site mutation in intron 46, COL7A1:c.4635 + 5delGTGA, and an exonic insertion in exon 34, COL7A1:c.4042dupG (Fig. 4A.2). A Sashimi plot revealed that the noncanonical splice site VUS at position +5 created a cryptic splice site inside intron 46, leading to partial retention of the intron. This retention, consisting of 379 nucleotides, results in frameshift and premature termination codon, predicting synthesis of a truncated, nonfunctional type VII collagen protein. The c.4042dupG mutation results in frameshift and generates premature codon 12 nucleotides downstream (Fig. 4A.2, lower panel).
Fig. 4.
Applications of RNA-seq for mutation detection in challenging cases of genodermatoses. (A) Case 3. (A.1) Family pedigree, clinical features and immune epitope mapping of the blistering led to diagnosis of recessive DEB. (A.2) RNA-seq revealed compound heterozygous mutations in COL7A1, a noncanonical splice site insertion mutation c.4635 + 5delGTGA in intron 46 and an insertion mutation c.4042dupG in exon 34. The noncanonical intronic mutation at the +5 position resulted in aberrant splicing, including partial retention of intron 46 (asterisk). (B) Case 4. (B.1, B.2) RNA-seq identified compound heterozygous mutations in COL7A1 in a patient with recessive DEB, namely, deletion mutation in canonical donor splice site, c.5772 + 1delG in intron 69 and a synonymous mutation, c.6501G>A, in exon 79. These mutations resulted in aberrant splicing, leading to partial retention of intron 69 (B.1, asterisk), and creation of an alternative donor splice site resulting in partial retention of intron 79 sequences (B.2, asterisk). (C) Case 5. Family pedigree (C.1) and clinical features (C.2) of a patient with congenital ichthyosiform erythroderma manifesting with mild generalized erythema and scaly skin lesions. (C.3) IGV visualization of genomic sequence identified a homozygous mutation, PNPLA1:IVS1, c.205 + 5G>A. (C.4) A Sashimi plot revealed that this mutation resulted in the absence of PNPLA1 transcripts (lower panel, red, asterisk) compared with an age- and sex-matched control (upper panel, blue). (C.5) Differential expression of PNPLA1 was also demonstrated by heat map analysis of the patient’s RNA in comparison to the average of 6 healthy controls.
Case 4 was clinically diagnosed as recessive DEB, and RNA-seq identified compound heterozygous mutations, including a nucleotide deletion in canonical donor splice site in intron 69, COL7A1:c.5772 + 1delG, and a synonymous exonic VUS COL7A1:c.6501G>A (Fig. 4, B.1 and B.2). The c.5772 + 1delG mutation results in partial use of a newly created donor splice site within intron 69 and retention of 33 nucleotides. The second heterozygous mutation, c.6501G>A, is synonymous but abolishes the normal donor site at the end of exon 79, resulting in partial use of an alternative donor site within intron 79. Use of this alternative donor site results in a transcript with a stop codon 12 nucleotides downstream from the exon 79/intron 79 border (Fig. 4B.2).
Case 5 describes a 1.5-year-old female born to consanguineous parents (Fig. 4C.1). She manifested with mild generalized erythema and scaly skin and was diagnosed with congenital ichthyosiform erythroderma (Fig. 4C.2). Initial application of a 38-gene ichthyosis targeted sequencing panel revealed a homozygous VUS, PNPLA1:IVS1, c.205 + 5G>A (Fig. 4C.3). Mutations in this gene have been previously associated with patients with autosomal recessive congenital ichthyosis (25, 26). A Sashimi plot of RNA-seq data revealed that this homozygous VUS in intron 1 resulted in complete absence of PNPLA1 transcript in comparison to an age- and sex-matched control (Fig. 4C.4). Heat map analysis of the patient’s RNA in comparison to the average of 6 healthy controls showed that PNPLA1 had the lowest gene expression level among 67 genes known to be associated with different forms of ichthyosis (Fig. 4C.5). The absence of PNPLA1 transcript could reflect nonsense-mediated decay of mRNA or other mechanisms, leading to transcript instability and shortened RNA half-life.
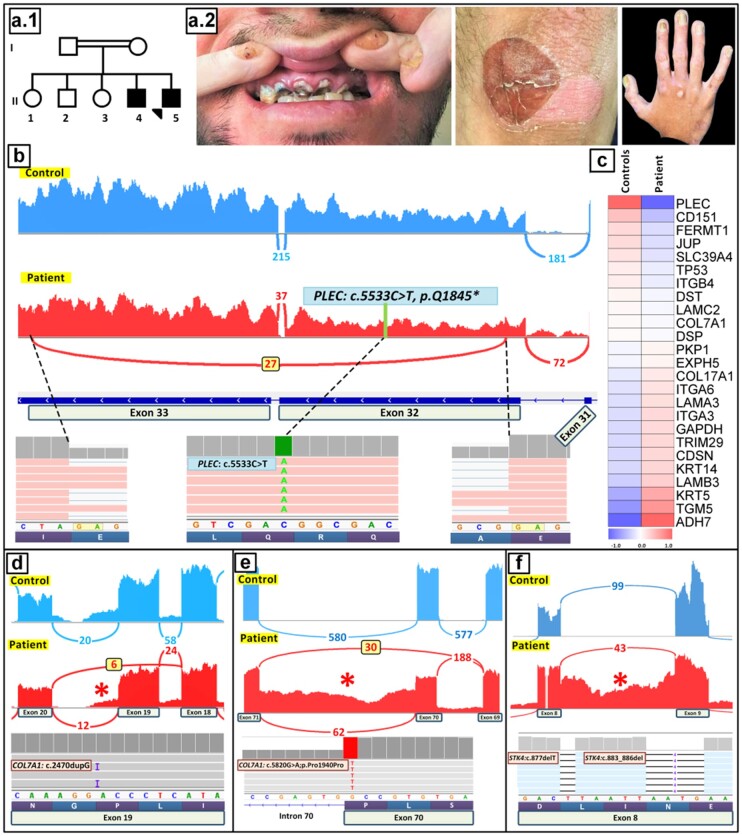
Case 6 depicts a 27-year-old male (II-5) and his older brother (II-4) who were born to consanguineous parents (Fig. 5A.1). The patients manifested with erosions in the skin and nail dystrophy, as well as dental anomalies (Fig. 5A.2), and were diagnosed with a form of autosomal recessive epidermolysis bullosa (EB) simplex with relatively mild blistering phenotype. Filtering of the annotated variants identified by RNA-seq by the pipeline detailed in cases 1 and 2 (Fig. 3) reduced the variants under consideration from 195 990 to 74 before applying the ultimate ROH filtration step, and alignment with ROHs reduced the number of candidate variants to 11. Phenotypic correlation and cosegregation analysis in the family identified a homozygous mutation in the plectin gene, PLEC:c.5533C>T, p. Q1845*, within exon 32 (Fig. 5B). The pathogenicity of this sequence variant was further supported by heat map analysis, which revealed that PLEC had the lowest expression level among the 21 EB-associated genes tested (Fig. 5C). A sashimi plot showed abnormal splicing extending from exon 32 to exon 33, which affected approximately 50% of the corresponding transcripts. This aberrant splicing resulted in elimination of 6567 nucleotides encoding part of plectin, a critical adhesion molecule associated with skin fragility disorders in the spectrum of EB simplex with late-onset muscular dystrophy (27). Importantly, this aberrant splicing event eliminated the stop-codon mutation within exon 32, a correction response known as nonsense-associated altered splicing, that is triggered by the premature termination codon. This deletion was in-frame in the coding region, predicting synthesis of a truncated plectin polypeptide. Interestingly, this same mutation in PLEC has been previously associated with a lethal form of EB simplex (28). In our patients, the relatively mild phenotype could be explained by nonsense-associated altered splicing, which eliminated the mutation, and the truncated plectin molecule may have retained partial function as an adhesion molecule.
Fig. 5.
Identification of aberrant splicing patterns due to unusual noncanonical splice site mutations. (A–C) Case 6. Family pedigree (A.1) and clinical findings (A.2) in a patient with EB simplex. (B) Variant calling identified a homozygous mutation PLEC:c.5533C>T, p. Q1845*. A Sashimi plot revealed partial skipping of the stop codon in exon 32 by nonsense-associated altered splicing, a correction triggered by a premature termination codon. (C) PLEC also showed the lowest level of gene expression among the EB-associated genes by heat map analysis. (D) Case 7. IGV screenshot showed a homozygous insertion in COL7A1 exon 19, c.2470dupG in a patient affected by DEB. This frameshift mutation led to exon 19 skipping, which occurred in 6 reads in the patient (red) compared with the controls (blue), as illustrated by Sashimi plots. (E) Case 8. In a patient with 2 coexisting heritable diseases, recessive DEB and acrodermatitis enteropathica, WES revealed a homozygous synonymous mutation at the border of exon/intron 70 in COL7A1:c.5820, p.Pro1940Pro. A Sashimi plot of the mutant COL7A1 mRNA revealed complex splicing, including partial skipping of exon 70 and retention of intron 70 sequences (asterisk). (F) Case 9. Transcriptome analysis of a case with atypical form of epidermodysplasia verruciformis revealed 2 frameshift mutations: c.877delT and c.883_886del in exon 8 of the STK4 gene. A Sashimi plot demonstrated retention of intron 8 sequences in the patient (red, asterisk) compared with the controls.
Case 7 shows a patient diagnosed as recessive DEB with a homozygous insertion mutation in the type VII collagen gene, COL7A1:c.2470dupG (Fig. 5D). This mutation resulted in skipping of exon 19, causing a frameshift and predicting synthesis of shortened, nonfunctional protein.
Case 8 describes a patient diagnosed as recessive DEB coexisting with another heritable disorder, acrodermatitis enteropathica, caused by homozygous mutations in COL7A1 and SLC39A4, respectively (29). The COL7A1 mutation, c.5820G>A, p. Pro1940Pro was initially eliminated from consideration as a pathogenic variant because of the synonymous nature of the consequence of the mutation. However, a Sashimi plot of the transcriptome profile of the mutant COL7A1 mRNA revealed complex splicing, including partial skipping of exon 70 in 30 reads of splicing events and retention of intron 70 sequences in 62 reads (Fig. 5E).
Case 9 involves atypical EV, and transcriptome analysis revealed 2 frameshift mutations in the STK4 gene, c.877delT and c.883_886del in exon 8 (Fig. 5F). The pathogenicity of these mutations was validated, and a Sashimi plot demonstrated retention of intron 8.
Collectively, these unusual cases of heritable skin diseases illustrate the power of NGS in identifying the molecular bases in these families. Table S1 indicates the presence or absence of consanguinity in any given family and lists the combinations of different methods (gene-targeted panel, WES, WGS, and RNA-seq) applied for mutation detection in each case. Specifically, cases 1, 2, and 9 were consanguineous and analyzed by the RNA-seq method only. In additional cases from consanguineous families (cases 3, 5, 6, and 8), the mutation detection utilized a combination of DNA-based sequencing approaches and RNA-seq, as indicated in Supplemental Table S1. Finally, cases 4 and 7 showed no evidence of familial consanguinity, and these cases were analyzed by a combination of gene-targeted sequencing panel and RNA-seq. As indicated in Supplemental Table S1, the utility of a targeted gene panel was unyielding in case 6, but the homozygous mutation was identified by a combination of WGS and RNA-seq. These data indicate that RNA-seq can be successfully utilized for mutation detection, with concomitant demonstrations of the effects of the mutations at the RNA level—namely, aberrant splicing and differential gene expression—shown in Sashimi plots and heat map analyses, respectively.
Discussion
Over the past 6 years, our laboratory has focused on mutation detection in a cohort of about 1800 families with different Mendelian skin disorders referred to us, including >500 families with EB, about 300 with various forms of keratinization disorders (ichthyosis and keratoderma), and about 600 with pseudoxanthoma elasticum, an ectopic mineralization disorder. More recently, we have analyzed rare diseases, such as EV manifesting with chronic cutaneous HPV infection due to genetic mutations in genes contributing to the skin immunity. Our initial mutation detection utilized DNA-based strategies, but over the past several years, the focus has shifted to RNA-based transcriptome sequencing.
This study demonstrates the utility of whole-transcriptome sequencing, either as a primary approach or complementary to DNA-based analyses, in the identification of pathogenic sequence variants and their consequences in patients with rare heritable skin diseases. Of 40 enrolled families in this study, mutation detection was successful in 35 families, providing a diagnostic yield of 88% (35/40). Some particularly informative cases were highlighted in this study, and they collectively illustrate the utility of whole-transcriptome sequencing by RNA-seq as a platform to confirm and extend information derived from DNA-based mutation-detection strategies, being in part complementary but also providing information not available from DNA analysis. Specifically, RNA-seq has demonstrated its usefulness in identifying consequences of putative splice junction mutations. The Sashimi plots visualize the pattern of aberrant splicing in qualitative and semiquantitative terms and/or the lack of gene expression at the mRNA level as a result of promoter mutations or large gene deletions, which could be confirmed at the protein level by immunofluorescence or Western blot analysis. Transcriptome data were also used for quantitation of DEGs visualized by heat maps, and reduced expression profiles could direct attention to the most likely pathogenic variants when patient and control tissues were compared in parallel. Another utility of RNA-seq data in the context of single-gene disorders is pathway analysis, which is useful for understanding the pathomechanisms of such diseases, with identification of potential targets for therapy. Although no pathway analysis was reported in this study, such analyses have been shown in other studies applying RNA-seq to a number of diseases (see Supplemental Table S2).
It should be noted that our filtering pipeline for identification of pathogenic mutations includes alignment of the candidate variants with ROH as the ultimate step. This filtering step is most applicable to consanguineous families with anticipation of homozygous mutations. In this context, it is important to note that up to 20% of all families in the world demonstrate some degree of consanguinity (30), and 1.1 billion people live in countries with customary consanguineous marriages (31), rendering our pipeline, including the ROH filtering step, applicable to a huge number of families in the global setting. Our pipeline is also applicable to outbred populations with rare autosomal recessive disorders with homozygous mutations inherited in the mode of identity by descent (IBD) (32). Nevertheless, even without application of the ROH alignment step, our pipeline was highly efficient in filtering the initially annotated variants. Specifically, the preceding filtering steps eliminated the overwhelming majority of the variants initially identified by RNA-seq (mean: 99.88%; median: 99.95%), and application of ROH reduced them to a few candidate genes that were interrogated by phenotypic correlations and cosegregation in the family to reach the causative genetic variant.
This study of Mendelian heritable skin diseases demonstrated the validity of our approach in identifying pathogenic variants in a number of cutaneous diseases, including EB, ichthyosis, and EV. These approaches are applicable not only to genodermatoses but also to different kinds of Mendelian disorders with a spectrum of manifestations and extensive genotypic/phenotypic heterogeneity. Whole-transcriptome sequencing has been applied to cohorts with other disorders, including neuromuscular, mitochondrial, and undiagnosed diseases, but with much lower diagnostic yield than obtained in our study, possibly reflecting the tremendous heterogeneity of those disorders (Supplemental Table S2). The prerequisite for the RNA-seq studies is the availability of tissues and cells expressing the relevant genes. In case of heritable skin disorders, skin biopsies or cultured cells, such as epidermal keratinocytes and dermal fibroblasts, are readily available in most cases. It is important to note that RNA is stable in appropriate transport media, such as RNAlater, for at least 2 weeks at room temperature, allowing shipping of skin biopsies or cultured cells to the laboratories performing the RNA-seq analysis. Although some of the information derived from RNA-seq, such as variant calling, is complementary to the data derived from DNA-based NGS, RNA-seq extends the repertoire of identified mutations, thus expanding the databases to include mutations that can be overlooked by NGS, especially those having an impact on splicing events. The results suggest that “clinical RNA-seq” could serve as the primary approach for mutation detection, provided that tissues and cells expressing the relevant genes are available for analysis.
Collectively, transcriptome profiling by RNA-seq was shown to facilitate genetic diagnosis of rare variants of Mendelian disorders by providing information complementing and extending the data available from DNA-based NGS approaches.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
L. Youssefian, statistical analysis, provision of study material or patients; A.H. Saeidian, statistical analysis; F. Palizban, statistical analysis; A. Bagherieh, statistical analysis, provision of study material or patients; S. Sotoudeh, provision of study material or patients; N. Mozafari, provision of study material or patients; H. Mahmoudi, provision of study material or patients; S. Zeinali, provision of study material or patients; P. Fortina, financial support, administrative support; J.C. Salas-Alanis, administrative support, provision of study material or patients; A.P. South, provision of study material or patients; H. Vahidnezhad, statistical analysis, administrative support, provision of study material or patients; J. Uitto, financial support, statistical analysis, administrative support, provision of study material or patients.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
P. Fortina, Clinical Chemistry, AACC.
Consultant or Advisory Role
None declared.
Stock Ownership
None declared.
Honoraria
None declared.
Research Funding
National Institutes of Health grant R01 AI143810 and DEBRA International.
Expert Testimony
None declared.
Patents
None declared.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments
The authors thank the patients and their families for participation in this study. Dr. Masoomeh Faghankhani performed genetic counseling, and Ali Jazayeri assisted in next-generation sequencing analysis. Carol Kelly assisted in manuscript preparation.
Glossary
Nonstandard Abbreviations
- NGS
next generation sequencing
- VUS
variants of unknown significance
- WES
whole exome sequencing
- WGS
whole genome sequencing
- HM
homozygosity mapping
- RNA-seq
RNA sequencing
- SNV
single-nucleotide variant
- DEG
differentially expressed gene
- ROH
region of homozygosity
- IGV
Integrative Genomics Viewer
- DEB
dystrophic epidermolysis bullosa
- EV
epidermodysplasia verruciformis
- HPV
human papillomavirus
- EB
epidermolysis bullosa
References
- 1. Wright CF, McRae JF, Clayton S, Gallone G, Aitken S, FitzGerald TW, et al. ; DDD Study. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet Med 2018;20:1216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith ED, Blanco K, Sajan SA, Hunter JM, Shinde DN, Wayburn B, et al. A retrospective review of multiple findings in diagnostic exome sequencing: half are distinct and half are overlapping diagnoses. Genet Med 2019;21:2199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Truty R, Paul J, Kennemer M, Lincoln SE, Olivares E, Nussbaum RL, Aradhya S.. Prevalence and properties of intragenic copy-number variation in Mendelian disease genes. Genet Med 2019;21:114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vahidnezhad H, Youssefian L, Saeidian AH, Touati A, Sotoudeh S, Jazayeri A, et al. Next generation sequencing identifies double homozygous mutations in two distinct genes (EXPH5 and COL17A1) in a patient with concomitant simplex and junctional epidermolysis bullosa. Hum Mutat 2018;39:1349–54. [DOI] [PubMed] [Google Scholar]
- 5. Saeidian AH, Youssefian L, Vahidnezhad H, Uitto J.. Research techniques made simple: whole-transcriptome sequencing by RNA-seq for diagnosis of monogenic disorders. J Invest Dermatol 2020;140:1117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonorazky HD, Naumenko S, Ramani AK, Nelakuditi V, Mashouri P, Wang P, et al. Expanding the boundaries of RNA sequencing as a diagnostic tool for rare mendelian disease. Am J Hum Genet 2019;104:1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park E, Pan Z, Zhang Z, Lin L, Xing Y.. The expanding landscape of alternative splicing variation in human populations. Am J Hum Genet 2018;102:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaz-Drago R, Custodio N, Carmo-Fonseca M.. Deep intronic mutations and human disease. Hum Genet 2017;136:1093–111. [DOI] [PubMed] [Google Scholar]
- 9. Vahidnezhad H, Youssefian L, Saeidian AH, Uitto J.. Phenotypic spectrum of epidermolysis bullosa: the paradigm of syndromic versus non-syndromic skin fragility disorders. J Invest Dermatol 2019;139:522–7. [DOI] [PubMed] [Google Scholar]
- 10. Adetunji MO, Lamont SJ, Abasht B, Schmidt CJ.. Variant analysis pipeline for accurate detection of genomic variants from transcriptome sequencing data. PLoS One 2019;14:e0216838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neums L, Suenaga S, Beyerlein P, Anders S, Koestler D, Mariani A, Chien J.. VaDir: an integrated approach to variant detection in RNA. Gigascience 2018;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Piskol R, Ramaswami G, Li JB.. Reliable identification of genomic variants from RNA-seq data. Am J Hum Genet 2013;93:641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang C, Davila JI, Baheti S, Bhagwate AV, Wang X, Kocher JP, et al. RVboost: RNA-seq variants prioritization using a boosting method. Bioinformatics 2014;30:3414–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang K, Li M, Hakonarson H.. Annovar: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quinn EM, Cormican P, Kenny EM, Hill M, Anney R, Gill M, et al. Development of strategies for SNP detection in RNA-seq data: application to lymphoblastoid cell lines and evaluation using 1000 genomes data. PLoS One 2013;8:e58815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brison O, El-Hilali S, Azar D, Koundrioukoff S, Schmidt M, Nahse V, et al. Transcription-mediated organization of the replication initiation program across large genes sets common fragile sites genome-wide. Nat Commun 2019;10:5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim D, Langmead B, Salzberg SL.. Hisat: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vahidnezhad H, Youssefian L, Zeinali S, Saeidian AH, Sotoudeh S, Mozafari N, et al. Dystrophic epidermolysis bullosa: COL7A1 mutation landscape in a multi-ethnic cohort of 152 extended families with high degree of customary consanguineous marriages. J Invest Dermatol 2017;137:660–9. [DOI] [PubMed] [Google Scholar]
- 20. Vahidnezhad H, Youssefian L, Jazayeri A, Uitto J.. Research techniques made simple: genome-wide homozygosity/autozygosity mapping is a powerful tool to identify candidate genes in autosomal recessive genetic diseases. J Invest Dermatol 2018;138:1893–900. [DOI] [PubMed] [Google Scholar]
- 21. Vahidnezhad H, Youssefian L, Saeidian AH, Zeinali S, Abiri M, Sotoudeh S, et al. Genome-wide single nucleotide polymorphism-based autozygosity mapping facilitates identification of mutations in consanguineous families with epidermolysis bullosa. Exp Dermatol 2019;28:1118–21. [DOI] [PubMed] [Google Scholar]
- 22. Uitto J, Youssefian L, Saeidian AH, Vahidnezhad H.. Molecular genetics of keratinization disorders—what's new about ichthyosis. Acta Derm Venereol 2020. Mar 25;100(7)adv00095.doi:10.2340/00015555-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Jong SJ, Matos I, Crequer A, Hum D, Gunasekharan V, Lorenzo-Diaz L, et al. The human CIB1-EVER1-EVER2 complex governs keratinocyte-intrinsic immunity to beta-papillomaviruses. J Exp Med 2018;215:2289–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vahidnezhad H, Youssefian L, Saeidian AH, Mansoori B, Jazayeri A, Azizpour A, et al. A CIB1 splice-site founder mutation in families with typical epidermodysplasia verruciformis. J Invest Dermatol 2019;139:1195–8. [DOI] [PubMed] [Google Scholar]
- 25. Boyden LM, Craiglow BG, Hu RH, Zhou J, Browning J, Eichenfield L, et al. Phenotypic spectrum of autosomal recessive congenital ichthyosis due to PNPLA1 mutation. Br J Dermatol 2017;177:319–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vahidnezhad H, Youssefian L, Saeidian AH, Zeinali S, Mansouri P, Sotoudeh S, et al. Gene-targeted next generation sequencing identifies PNPLA1 mutations in patients with a phenotypic spectrum of autosomal recessive congenital ichthyosis: the impact of consanguinity. J Invest Dermatol 2017;137:678–85. [DOI] [PubMed] [Google Scholar]
- 27. McMillan JR, Akiyama M, Rouan F, Mellerio JE, Lane EB, Leigh IM, et al. Plectin defects in epidermolysis bullosa simplex with muscular dystrophy. Muscle Nerve 2007;35:24–35. [DOI] [PubMed] [Google Scholar]
- 28. Koss-Harnes D, Hoyheim B, Jonkman MF, de Groot WP, de Weerdt CJ, Nikolic B, et al. Life-long course and molecular characterization of the original Dutch family with epidermolysis bullosa simplex with muscular dystrophy due to a homozygous novel plectin point mutation. Acta Derm Venereol 2004;84:124–31. [DOI] [PubMed] [Google Scholar]
- 29. Vahidnezhad H, Youssefian L, Sotoudeh S, Liu L, Guy A, Lovell PA, et al. Genomics-based treatment in a patient with two overlapping heritable skin disorders: epidermolysis bullosa and acrodermatitis enteropathica. Hum Mutat 2020;41:906–12. [DOI] [PubMed] [Google Scholar]
- 30. Modell B, Darr A.. Science and society: genetic counselling and customary consanguineous marriage. Nat Rev Genet 2002;3:225–9. [DOI] [PubMed] [Google Scholar]
- 31. Hamamy H, Antonarakis SE, Cavalli-Sforza LL, Temtamy S, Romeo G, Kate LP, et al. Consanguineous marriages, pearls and perils: Geneva International Consanguinity Workshop report. Genet Med 2011;13:841–7. [DOI] [PubMed] [Google Scholar]
- 32. Schuurs-Hoeijmakers JH, Hehir-Kwa JY, Pfundt R, van Bon BW, de Leeuw N, Kleefstra T, et al. Homozygosity mapping in outbred families with mental retardation. Eur J Hum Genet 2011;19:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.