Abstract
Alveolar epithelial regeneration is essential for recovery from devastating lung diseases. This process occurs when type II alveolar pneumocytes (AT2) proliferate and transdifferentiate into type I alveolar pneumocytes (AT1). We used genome-wide analysis of chromatin accessibility and gene expression following acute lung injury to elucidate repair mechanisms. AT2 chromatin accessibility changed significantly following injury to reveal STAT3 binding motifs adjacent to genes that regulate essential regenerative pathways. Single-cell transcriptome analysis identified brain neurotrophic factor (Bdnf) as a STAT3 target gene with newly accessible chromatin in a unique population of regenerating AT2 cells. Furthermore, the BDNF receptor TrkB, was enriched on mesenchymal alveolar niche cells. Loss or blockade of AT2-specific Stat3, Bdnf or mesenchyme-specific TrkB compromised repair and reduced Fgf7 expression by niche cells. A TrkB agonist improved outcomes in vivo following lung injury. These data highlight the biological and therapeutic importance of the STAT3-BDNF-TrkB axis in orchestrating alveolar epithelial regeneration.
Introduction
Lung function depends on the integrity of the distal alveolar epithelium1,2. Damage to the alveolar epithelium, as occurs in pneumonia3, influenza4,5, and acute respiratory distress syndrome (ARDS)6,7, results in failure of gas exchange which may be fatal8. A subset of patients nonetheless survive these diseases, demonstrating that clinically meaningful lung regeneration is possible9-11. The current SARS-CoV-2 pandemic resulting in COVID-19 has demonstrated these principles on a worldwide scale12,13. This has led to an intense focus on understanding mechanisms of alveolar epithelial regeneration that could accelerate this process14,15.
Multiple cell types have been identified that function as progenitor cells to regenerate alveolar epithelium following acute lung injury14,15. These include type II alveolar pneumocytes (AT2)16-19, type I alveolar pneumocytes (AT1)20,21, bronchoalveolar stem cells22-24, airway stem cells25,26, and lineage negative progenitors27. This redundancy highlights the evolutionary importance of alveolar epithelial regeneration. Although the relative contribution of each pool of progenitors to alveolar epithelial regeneration remains to be determined, AT2 cells are indispensable for alveolar epithelial regeneration following sterile lung injury28. Furthermore, a subset of Axin2+ AT2 cells called alveolar epithelial progenitor (AEP) cells, marked by Tm4sf1 expression and FGF7 responsiveness, were shown to contribute to alveolar re-epithelialization following sterile and infectious lung injuries19. Efforts to elucidate the mechanisms of AT2 stem cell functions will undoubtedly enhance our understanding of alveolar epithelial regeneration. For example, reports of Krt5+ cell-mediated regeneration in areas where AT2-mediated regeneration fails27,29, suggest redundancy, even though Krt5+ cell-mediated regeneration may be less efficient at restoration of normal alveolar structure and function27,30.
Under homeostatic conditions, the AT2 cell is a quiescent surfactant producing cell31,32. Following acute lung injury, the AT2 cell functions as a progenitor cell that is capable of self-renewal and transdifferentiation into AT1 cells15,17,19, which are flat pneumocytes that facilitate gas exchange between the alveoli and adjacent capillaries33. Despite extensive research defining the stem cell functions of AT2 cells17-19, how changes in chromatin accessibility, transcription factor activation, and gene expression in AT2 cells influence interactions with adjacent mesenchymal niche cells is needed to improve our knowledge of how the alveolar epithelium regenerates. An integrated understanding of these biological processes is essential for developing therapeutics for devastating human lung diseases that injure alveolar epithelium15.
Discerning which factors and transcriptional targets are critical to regenerating epithelial cell functions34,35 requires genome-wide analyses36. To investigate this, we assayed chromatin architecture of AT2 cells for changes in accessibility following acid-induced lung injury using the assay for transposase accessible chromatin and next-generation sequencing (ATAC-seq)36. Studies of chromatin were coupled with single-cell transcriptomic analysis of AT2 cells and mesenchymal cells to improve our understanding of the cell signaling interactions between discrete populations of regenerating AT2 and niche cells. These genome-wide approaches converged on a single pathway in which the transcription factor STAT3 increases expression of growth factor BDNF in transdifferentiating AT2 cells. Using human and murine alveolar organoids37, we demonstrated that this is a conserved pathway in epithelial regeneration. Furthermore, drug treatments aimed at BDNF-TrkB signaling enhance repair following lung injury in mice, demonstrating that targeting this pathway may provide avenues for therapy of heretofore recalcitrant diseases38.
Results
The response to acid-induced lung injury is marked by changes in AT2 chromatin architecture.
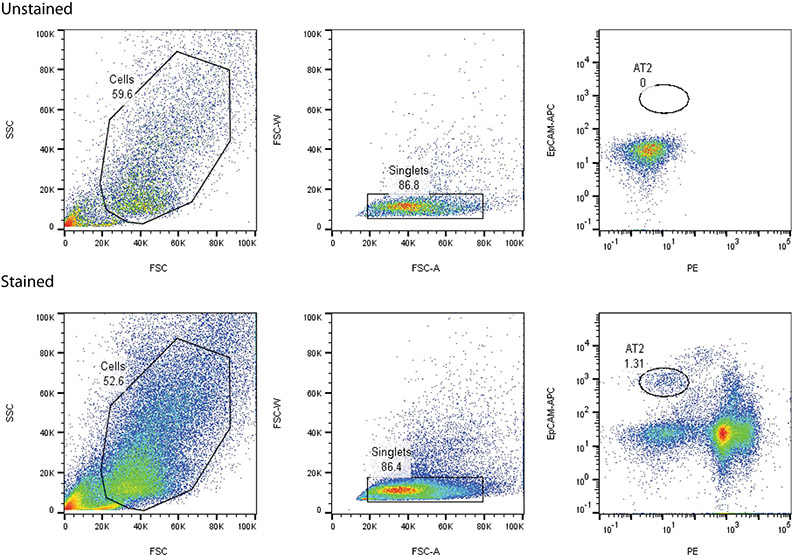
We previously described a model of alveolar lung injury and repair induced by acid aspiration in which AT2 cells proliferate within 24 hours following injury, and are the progenitors to AT1 cell reconstitution39. This unilateral injury model allows mice to survive an otherwise fatal injury, and has distinct inflammatory and regenerative phases39. We hypothesized that in response to epithelial loss, the AT2 cell adopts new stem cell functions not present at homeostasis, and that the regenerative function is facilitated by transcriptional changes resulting from alterations in chromatin architecture. We therefore isolated AT2 cells from mice 24 hours after acid instillation (Fig. 1A, 1B & Extended Figure 1), and compared them to AT2 cells from uninjured mice for chromatin accessibility using the ATAC-seq assay36 (Fig. 1C). We purposefully avoided Sftpc-lineage labeled mice for this experiment in order to capture cells from other progenitor types that may transiently adopt characteristics of AT2 cells following injury.
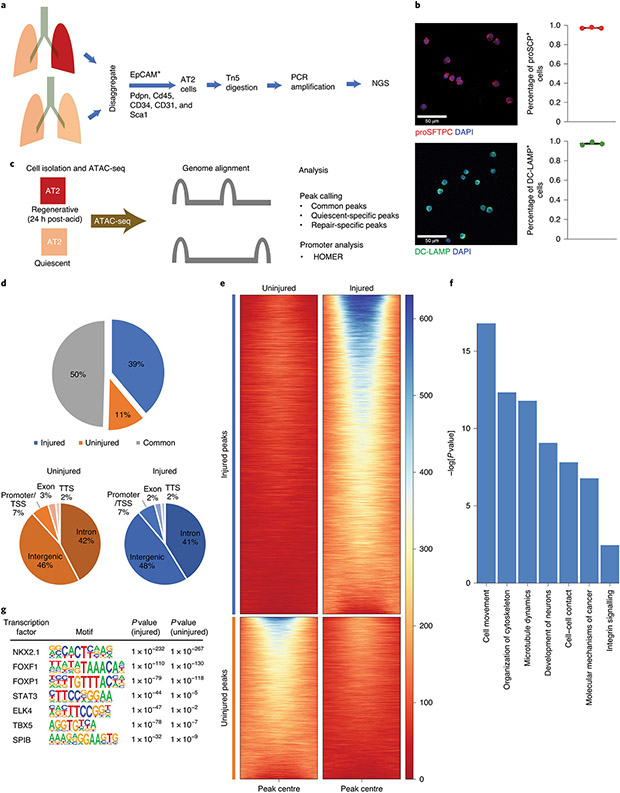
Figure 1 – The response to injury is marked by significant changes in AT2 chromatin architecture.
(A) AT2 cells were isolated from single cell suspensions of murine lung tissue (3 uninjured mice, 2 injured mice) using fluorescence activated cell sorting (FACS). Nuclei were isolated and incubated with Tn5 transpose followed by amplification of fragments and next-generation sequencing (NGS). (B) Staining of isolated cells for the AT2 markers proSFTPC and DC-LAMP confirmed >97% isolated cells are AT2 cells (100 cells per stain analyzed in n=3 independent experiments). (C) ATAC-seq was performed on AT2 cells from uninjured mice (quiescent) and from mice 24 hours post acid-induced lung injury (regenerative). NGS reads were aligned and uninjured and injured sequences were compared. (D & E) Quantitation of shared and unique regions of accessible chromatin. (F) Ingenuity pathway analyses of genes associated with differentially accessible chromatin following acute lung injury demonstrated newly accessible areas of the genome encode proteins important for alveolar epithelial regeneration. (G) HOMER analysis of accessible chromatin demonstrated common enrichment for alveolar epithelium-associated transcription factors including NKX2.1, FOXF1 and FOXP1. After injury STAT3, TBX5, and SPIB chromatin accessibility becomes increasingly enriched. For panel (B), data is shown as the mean +/− SEM. Statistical significance was determined using a two-tailed Student’s T-test (B) and with the findMotifGenome.pl script in the HOMER software package with adjustment for multiple comparisons (G). Scale bars, 50μm.
Regions of accessible chromatin were substantially different in AT2 cells from mice that underwent acid-induced lung injury when compared to those from uninjured mice (Fig. 1D & 1E). Using an unbiased pathway analysis (Fig. 1F), we identified areas of new chromatin accessibility that are associated with genes involved in cell movement and proliferation. In order to determine which transcription factors may occupy sites important for expression of these genes, we located binding sites with Hypergeometric Optimization of Motif EnRichment (HOMER) analysis40 (Fig. 1G). Examination of genes in areas of accessible chromatin demonstrated enrichment for lung development and alveolar epithelium-associated transcription factors including NKX2-141,42 and FOXP143,44. Binding site motifs that appear enriched following injury include motifs for STAT3, ELK445, TBX5, and SPIB. Thus, following acute lung injury the chromatin in AT2 cells has increased accessibility to specific transcription factor binding sites and genes involved in cell proliferation.
STAT3 is important for AT2-mediated recovery from sterile and infectious lung injuries.
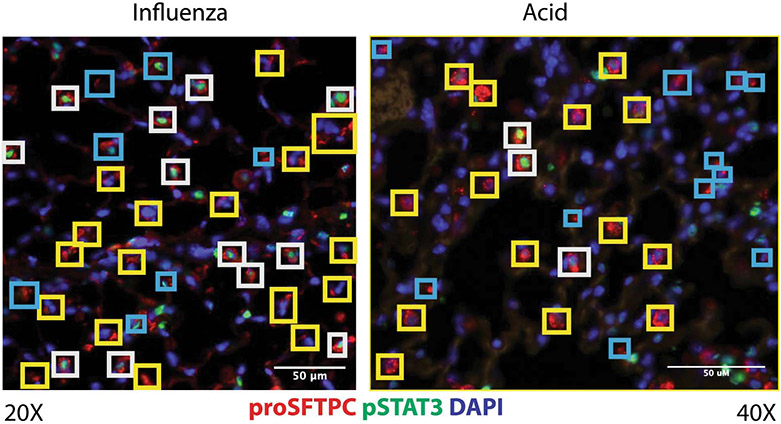
We focused on STAT3 since our previous work highlighted the importance of STAT3 activators granulocyte colony stimulating factor (G-CSF) and interleukin (IL-6) in alveolar epithelial regeneration39,46. Furthermore, we detected increased IL-6 in the BAL of mice 24h after acute lung injury, temporally corresponding to initiation of alveolar regeneration39 (Fig. 2A and Extended Figure 2). To determine whether altered accessibility and accumulation of activating cytokines47 accurately predicts activation of STAT3, we probed tissue sections obtained from acid-injured and control lungs for the presence of activated phosphorylated STAT348. Immunofluorescence staining 24 hours after acid-induced lung injury or 9 days after H1N1 influenza infection revealed activation of STAT3 in AT2 cells as well as other cell types (Fig. 2B, 2C and Extended Figure 2). We did not observe STAT3 activation in uninjured lungs (Fig. 2B & 2C). This observation was corroborated by evidence of STAT3 activation in AT2 cells of human explanted lungs with diffuse alveolar damage (Fig. 2D & 2E). Activation of STAT3 is thus an evolutionarily conserved response to lung injury.
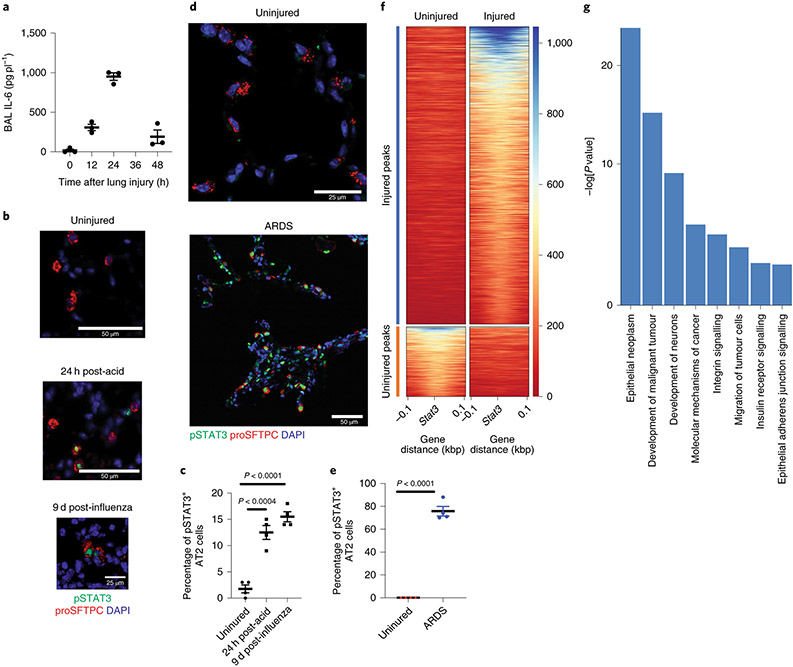
Figure 2 – Stat3 regulates key pathways in alveolar epithelial regeneration.
(A) IL-6 is undetectable in uninjured lungs. Levels of IL-6 in BAL fluid peak at 24h after acid-induced lung injury (n=3 mice per time point). (B & C) C57BL/6 mice that underwent acid-induced lung injury demonstrate activated pSTAT3 in AT2 cells (p=2.9x10−5 for 9 days post infection vs uninjured). There was minimal phosphorylated STAT3 in AT2 cells of uninjured mice (n= 4 mice per group). (D & E) Human samples with diffuse alveolar damage had increased AT2 specific pSTAT3 (p=1.8x10−6) that was not present in uninjured samples (n=4 patients per group). (F & G) Unbiased pathway analyses demonstrated newly accessible STAT3 binding sites adjacent to genes that control key biological mechanisms including proliferation and migration. For panels (A) (C) and (E), data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test (C & E). Scale bars 50μm (B, D) and 25μm (B, D).
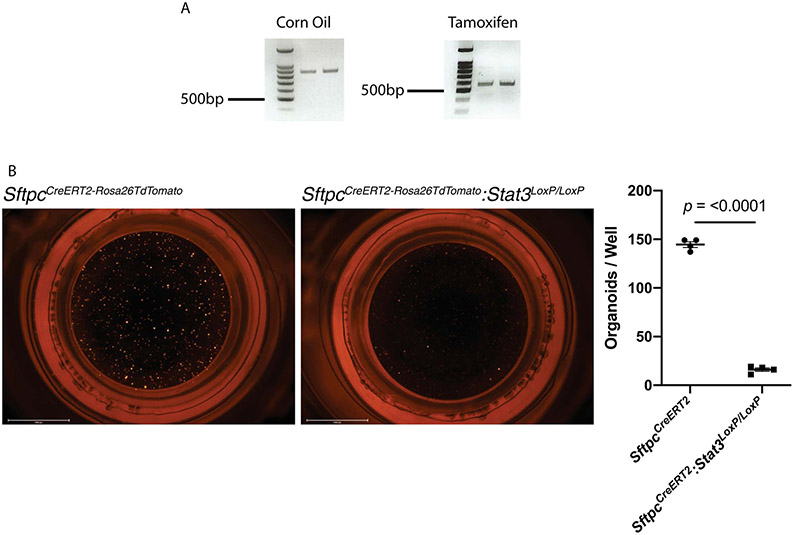
To determine downstream effectors of STAT3 activation, we performed an unbiased pathways analysis of genes in close proximity to newly accessible STAT3 binding sites. We identified multiple gene-expression pathways related to development, epithelial proliferation and insulin receptor signaling (Fig. 2F & G). In order to determine whether STAT3 activation is functionally critical for repair in AT2 epithelial cells, we generated SftpcCreERT2:Stat3LoxP/LoxP mice in which a critical element of Stat3 could be selectively deleted in AT2 cells upon administration of tamoxifen (Extended Figure 3A). Prior studies using organoids showed that IL-646 promoted organoid formation, and conversely STAT3 inhibition decreased organoid formation. These reports, however, did not delineate between the relative importance of AT2 specific STAT3 compared to mesenchymal specific STAT3. We therefore tested the importance of AT2-specific STAT3 signaling by assessing the ability of AT2 cells lacking Stat3 to form alveolar organoids when co-cultured with wild-type fibroblasts. We observed that AT2 cells lacking Stat3 formed significantly fewer organoids than those that retained Stat3 (Extended Figure 3B). Thus, AT2 specific STAT3 functionality is necessary for alveolar organoid formation.
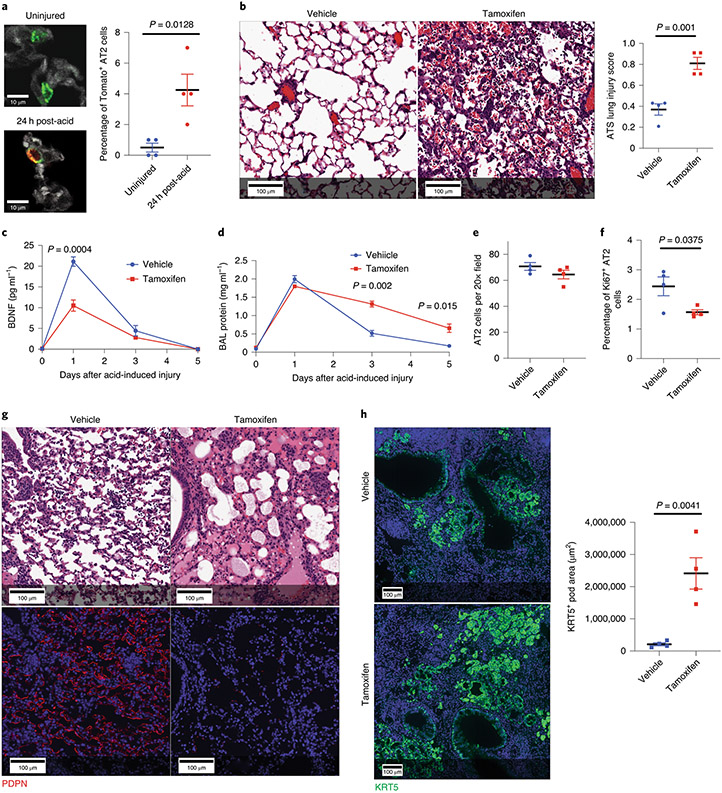
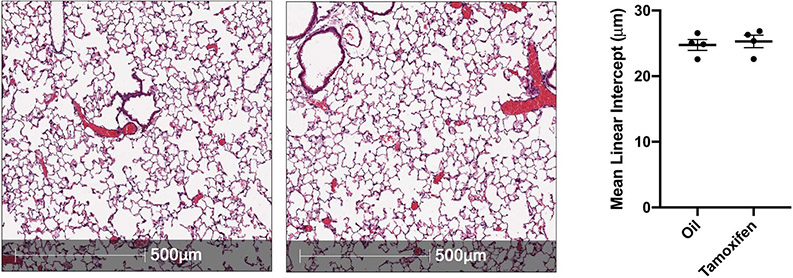
To test the hypothesis that AT2 cell-specific STAT3 promotes repair in vivo, SftpcCreERT2:Stat3LoxP/LoxP mice were administered tamoxifen or corn oil (vehicle control) 18 days prior to acid-induced lung injury. Distal alveoli of unchallenged lungs appeared histologically unaltered after deletion (Extended Figure 4). Whereas control mice showed normal rapid repair (Fig. 3A, upper panels), SftpcCreERT2:Stat3LoxP/LoxP mice had severe histologic changes consistent with repair failure after acid-induced lung injury (Fig. 3A, lower panels). This was accompanied by decreased staining for AT1 cells and significantly higher lung injury scores at later time points in mice lacking Stat3 specifically in AT2 cells (Fig. 3B & 3C).
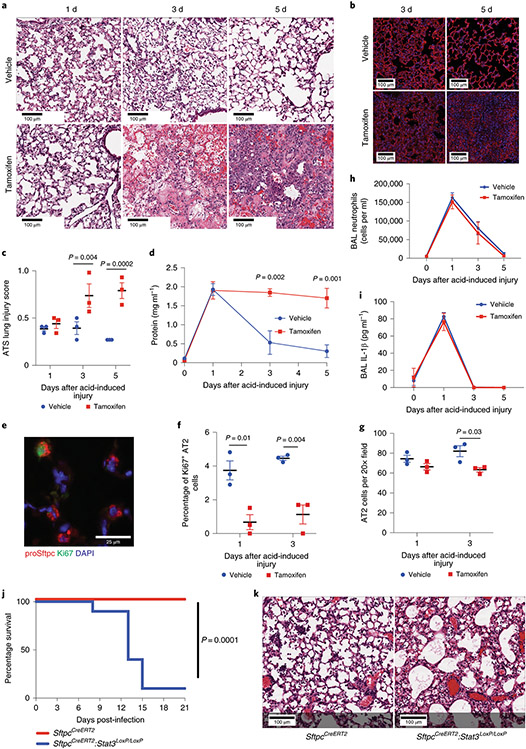
Figure 3 – Deletion of Stat3 in AT2 cells worsens outcomes following sterile and infectious lung injuries.
(A) SftpcCreERT2:Stat3LoxP/LoxP mice were given tamoxifen in corn oil or corn oil alone 18 days prior to acid-induced lung injury (3 mice per group). H&E staining shows that control mice had histological injury that resolved by day 5 post-injury. Mice that received tamoxifen which resulted in AT2 specific Stat3 deletion had persistent and pronounced cellular, proteinaceous and hemorrhagic infiltrates that obscured the alveolar structures. (B) PDPN staining revealed that mice lacking Stat3 had decreased AT1 populations at 3 and 5 days following acid-induced lung injury compared to controls (n=3 mice per group). (C) ATS lung injury scores were increased at later time points following acid-induced lung injury in mice lacking Stat3 (3 mice per group per time point ). (D) BAL protein from mice described in (A) showed non-resolving increase in alveolar protein following acid-induced lung injury in mice that lacked AT2 specific Stat3 (n=3 mice per group per time point). (E, F, G) The percent of Ki6+ AT2 cells was decreased in tamoxifen treated SftpcCreERT2:Stat3LoxP/LoxP mice following acid-induced lung injury (n=3 mice per group per time point). (H & I) BAL neutrophils and IL-1β levels following acid-induced lung injury were not changed by the presence or absence of functional AT2 specific Stat3 (n=3 mice per group per time point). (J) SfptcCreERT2 and SftpcCreERT2:Stat3LoxP/LoxP mice were given tamoxifen 18 days prior to intranasal infection with PR8 influenza (5x10−5 HAU/mouse). Mortality was significantly higher in the mice that lack AT2 specific STAT3 compared to controls (n=10 mice per group). (K) At 21 days post infection the mouse lacking AT2 specific Stat3 (1 mouse) had distorted distal alveolar lung architecture with cellular infiltrate and cystic structures that were not observed in controls (10 mice). For panels (C), (D), (F), (G), (H), and (I), data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test (C), (F), and (G), with a two-way ANOVA (D), and with the Log-rank (Mantel-Cox) test (J). Scale bars, 100μm (A, B, K) and 25μm (E).
Since the epithelial layer represents the major permeability barrier within the alveolus49,50, we measured broncholavelolar lavage (BAL) protein as an indicator of permeability, and confirmed that the epithelial permeability barrier fails to re-form in the absence of STAT3 (Fig. 3D). As alveolar epithelium regeneration is dependent on AT2 proliferation16,31,39, we quantified proliferation by Ki67 staining (Fig. 3E & 3F) and found significantly fewer proliferating AT2 cells in the absence of STAT3. We did not observe changes in the absolute number of AT2 cells one day after acid-induced lung injury in mice with an AT2 cell-specific Stat3 deletion (Fig. 3G), suggesting that changes in initial AT2 survival were not altered. Furthermore, no differences in the ATS lung injury scores (Fig. 3C), BAL protein (Fig. 3D), BAL neutrophils (Fig. 3H) or BAL IL-1β (Fig. 3I) were observed 24 hours after acid-induced lung injury, showing that loss of Stat3 in AT2 cells does not alter the acute inflammatory response.
In order to determine whether outcomes following chronic infectious injury would be similarly dependent on STAT3, we tested the effect of H1N1 Influenza infection on SftpcCreERT2:Stat3LoxP/LoxP and SftpcCreERT2 mice. Eighteen days after tamoxifen-pretreatment, mice were exposed to H1N1 (5x10−5 HAU/mouse, PR8 strain) and observed for 21 days post-infection. Mice with Stat3 deficient AT2 cells had significantly impaired survival beginning on day 8 after infection (Fig. 3J). Histologic analysis of lungs obtained at 14 days after infection revealed extensive damage when Stat3 was deleted from AT2 cells compared to controls (Fig. 3K). Thus, AT2 specific STAT3-activation and transcriptional activity is necessary for recovery from both sterile and infectious lung injuries.
Single-cell RNA-seq identifies BDNF as important for alveolar regeneration.
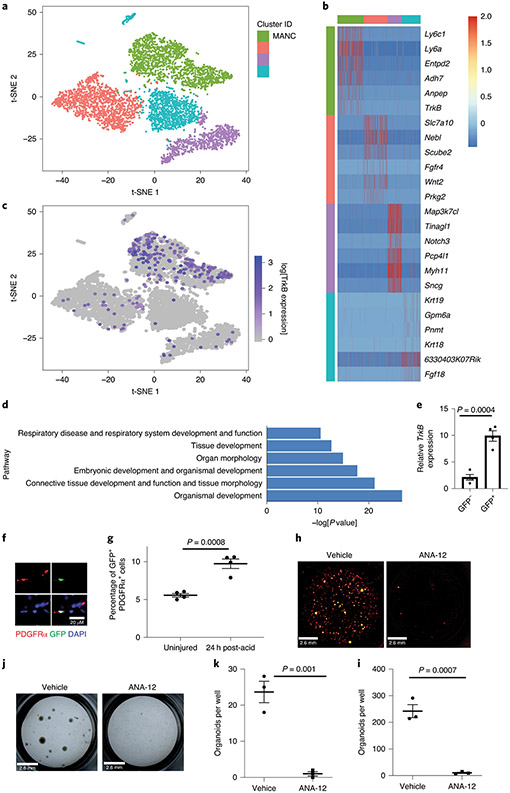
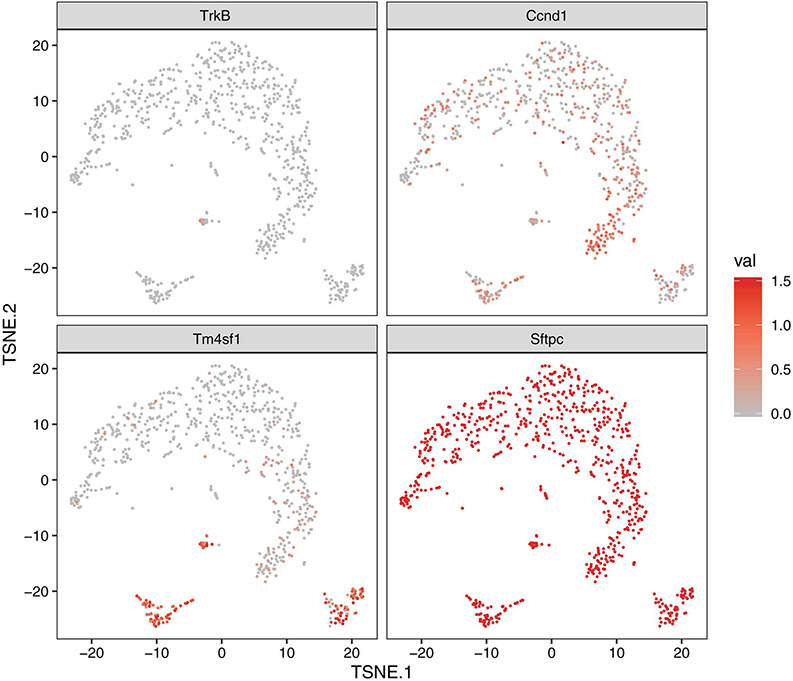
To identify relevant STAT3 target genes expressed after acute lung injury, we performed single-cell RNA-seq (scRNA-seq) to identify transcriptomic changes in AT2 cells after acid-induced lung injury (Fig. 4A and 4B). A recent report suggests that a specific subset of AT2 cells that express Tm4sf1 are important in alveolar epithelial regeneration19. Under the same conditions used for isolating cells for ATAC-seq (Fig. 1A), we observed discrete sub-populations of cells including those representing putative AEPs19 (Fig. 4C), following acid-induced lung injury. Cluster 2 (Fig. 4B), which we termed proliferating AT2 cell population, contains proliferating AT2 cells and Ccnd1 expression is restricted to this cluster (Fig. 4B, 4C, and Extended Figure 5). Furthermore, unbiased pathways analysis of this cluster showed enrichment for genes that regulate proliferation (Extended Figure 6). We focused our analysis on cluster 3 (Fig. 4B), which we termed transdifferentiating population. This sub-population is unique in its simultaneous expression of Tm4sf1, Sftpc and Pdpn (Fig. 4A-C and Extended Figure 5), similar to a previously reported population of transdifferentiating AT2 observed after lipopolysaccharide-induced lung injury51. These data effectively identified a sub-population of AT2 cells whose transcriptomic profile is most consistent with transdifferentiating alveolar epithelium.
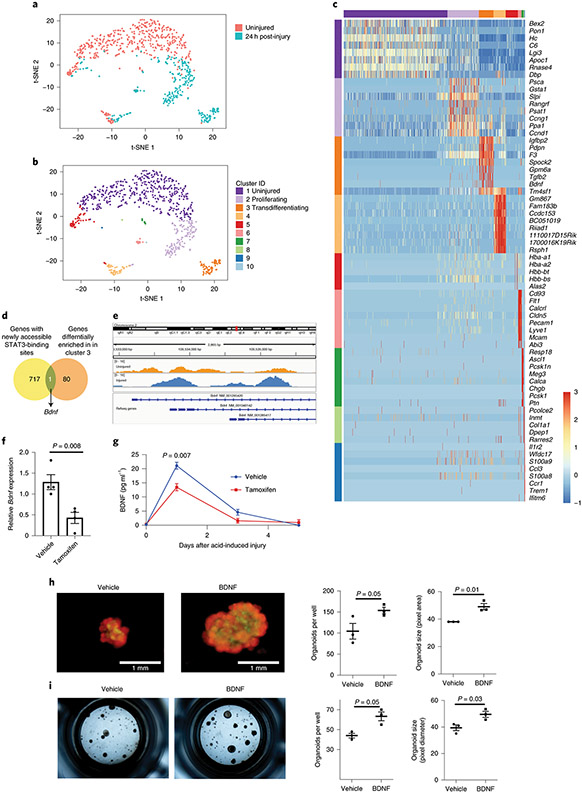
Figure 4 – BDNF identified as important for alveolar epithelial regeneration.
(A) AT2 cells were isolated from an uninjured C57BL/6 mouse and from a C57BL/6 mouse that underwent acid-induced lung injury 24 hours prior. Single-cell transcriptomic analysis was performed and cells were grouped into populations using t-SNE. There was little overlap between AT2 cells from uninjured and injured mice. (B) K-means analysis was used to sub-divide AT2 cells into groups based on their individual gene expression. (C) Genes whose expression was unique to each cluster are shown. Cluster 3 has gene expression from both AT2 and AT1 cells (Sftpc and Pdpn) and AEP marker Tm4sf1. (D) Bdnf is the only bona fide STAT3 target gene with newly accessible chromatin following acid-induced lung injury that is differentially expressed in cluster 3. (E) Data from ATAC-seq (shown in Fig. 1) demonstrate increased accessibility of the Bdnf promotor in cells isolated from mice 24 hours after acid-induced lung injury. (F) AT2 cells from tamoxifen-exposed SftpcCreERT2:Stat3LoxP/LoxP mice express less Bdnf than vehicle exposed mice (n=4 mice per group) (G) BAL fluid from SftpcCreERT2:Stat3LoxP/LoxP mice treated with either tamoxifen or corn oil (vehicle) prior to undergoing acid-induced lung injury shows that mice lacking AT2-specific Stat3 have significantly less BDNF in their BAL fluid (n=3 mice per group per time point). (H) AT2 cells from SftpcCreERT2-Rosa26TdTomato:Hopx3FlagGFP mice were co-cultured with PDGFRα+ mesenchymal cells from C57BL/6 mice in the presence of BDNF which increased alveolar organoid forming efficiency of murine AT2 cells. Right-hand panels show quantification from n=3 distinct cultures per condition. (I) Primary human AT2 cells were isolated and co-cultured with MRC5 fibroblasts for three weeks. We observed increased organoid size and forming efficiency when BDNF was added to the culture media. Right-hand panels show the average organoid forming efficiency and size from n=3 different donors. Individual points are shown with the mean and standard error of the mean depicted. For panels (F), (G), (H), and (I), data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test (F), (H), and (I), and with a two-way ANOVA (G). Scale bars, 1mm.
To understand the interdependence of chromatin changes after acute lung injury, STAT3-mediated gene expression and alveolar epithelial regeneration, we cross-referenced our ATAC-seq dataset with the transcriptomic profiles of the AT2 cells in cluster 3 (Fig. 4D). We identified brain-derived neurotrophic factor (Bdnf)52 as being the only bona fide STAT3 target gene53-56 whose chromatin is newly accessible after lung injury (Fig. 4D, Supplementary Table 1) and whose expression is restricted to the regenerating population of cells. Our ATAC-seq analysis revealed that the Bdnf locus becomes more accessible following injury (Fig. 4E), suggesting increased chromatin accessibility contributes to increased expression after injury. Furthermore, AT2 cells isolated from tamoxifen treated SftpcCreERT2:Stat3LoxP/LoxP had a significant decrease in Bdnf expression compared to mice that did not receive tamoxifen (Fig. 4F). We also observed that BDNF release following acid-induced injury was significantly attenuated in mice with an AT2-specific Stat3 deletion (Fig. 4G). Bdnf is thus the only STAT3-regulated gene whose expression is restricted to a transcriptionally-unique AT2 cell population which expresses Tm4sf1, as well as markers of both AT1 and AT2 cells.
The BDNF-TrkB axis promotes alveolar organoid formation.
To test whether BDNF might enhance alveolar repair, we used a system to generate alveolar organoids by co-culturing AT2 cells with pulmonary mesenchymal cells37. This reductionist approach facilitates the study of epithelial-mesenchymal cross-talk in alveolar epithelial regeneration. We show that the addition of recombinant BDNF to organoid media increased the efficiency and size of murine and human organoid formation (Fig. 4H & 4I).
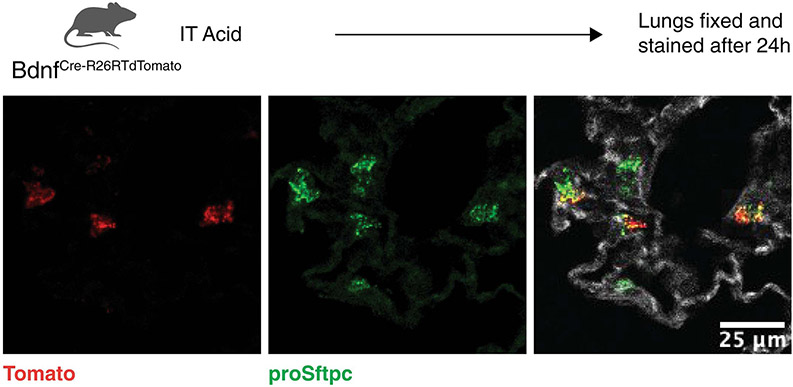
Using BdnfCre-R26lslTdTomato mice, we confirmed that a small percentage of AT2 cells express Bdnf after acid-induced lung injury (Fig. 5A and Extended Figure 7), as suggested by scRNA-seq and ATAC-seq (Fig. 4C and 4E). To characterize the impact of AT2 cell-specific Bdnf, we generated a SftpcCreERT2:BdnfLoxP/LoxP model. Mice lacking AT2 specific Bdnf had worse histological outcomes following acid-induced lung injury as reflected by significantly worse ATS Lung Injury Scores (Fig 5B). As expected, BAL levels of BDNF were significantly reduced in the tamoxifen-exposed SftpcCreERT2:BdnfLoxP/LoxP mice (Fig. 5C). Furthermore, tamoxifen-exposed SftpcCreERT2:BdnfLoxP/LoxP mice maintained significantly elevated BAL protein levels (Fig. 5D). Although overall AT2 cell numbers were unchanged following acid-induced lung injury in the absence of Bdnf (Fig. 5E), there was a significant decrease in AT2 proliferation 5 days after acid-induced lung injury (Fig. 5F). We saw similar histological outcomes 21 days post-influenza infection (Fig. 5G) and increased KRT5 pods (Fig. 5H) when Bdnf was selectively deleted in AT2 cells.
Figure 5 – Loss of AT2 specific BDNF worsens outcomes following sterile and infectious lung injuries.
(A) We quantified tomato positive AT2 cells in BdnfCre-R26TdTomao mice before and 24h after acid-induced lung injury and found a significant increase in co-positive cells following acute lung injury (n=4 mice per group). (B-F) H&E staining and ATS lung injury scores (B), BAL BDNF concentrations (C) BAL protein concentration (D) AT2 cell numbers (E) and number of proliferating AT2 cells (F) of tamoxifen-exposed and vehicle-exposed SftpcCreERT2:BdnfLoxP/LoxP mice 5 days after acid-included lung injury (n= 3 mice per time point for C & D, n=4 mice per group per time point for B, E, & F). (G) H&E and PDPN staining of lung tissue from tamoxifen-exposed and vehicle-exposed SftpcCreERT2:BdnfLoxP/LoxP mice that had been infected with intranasal PR8 influenza (5x10−5 HAU/mouse) (4 mice per group). (H) KRT5 staining of the mice described in (G) with quantification of KRT5+ pods in both groups (n=4 mice per group). For panels (A), (B), (C), (D), (E), (F) and (H), data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test (A), (B), (E), (F), and (H), and with a two-way ANOVA (C) and (D). Scale bars, 100μm (B, G, H) and 10μm (A).
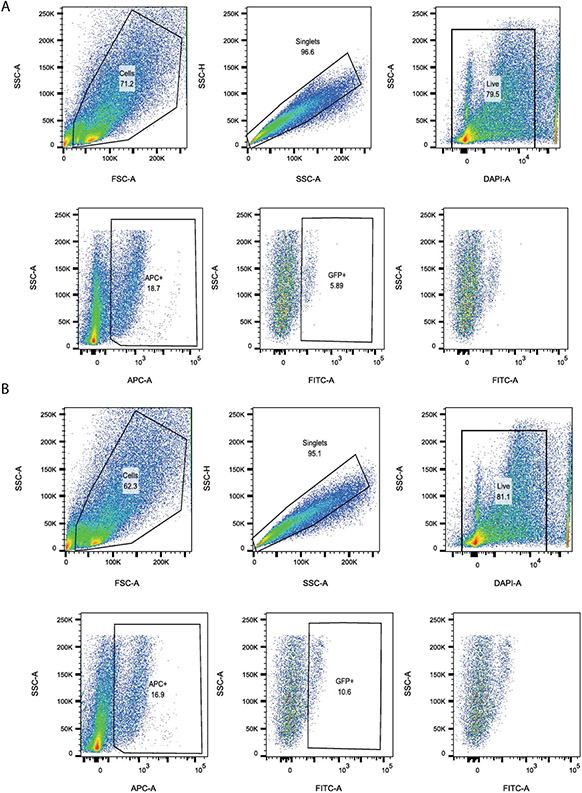
Since BDNF increased alveolar organoid formation, we sought to determine if TrkB, the receptor for BDNF, is expressed on AT2 and mesenchymal cells. Reanalysis of scRNA-seq data did not identify significant TrkB expression in AT2 cells (Extended Figure 5). However, analysis of scRNA-seq data from mesenchymal cells in the mouse lung46 revealed that TrkB is expressed in a subset of cells (Fig. 6A-C), previously identified as mesenchymal alveolar niche cells (MANCs), These cells have been identified as essential for organoid formation and alveolar regeneration after bleomycin-induced lung injury46. Unbiased pathways analysis of TrkB expressing mesenchymal cells showed enrichment for genes regulating respiratory function, organ morphology and embryonic development (Fig. 6D). Expression of TrkB on mesenchymal cells was confirmed using a TrkBCre-Rosa26TdTomato mouse, which successfully identified PDGFRα cells with increased TrkB expression (Fig. 6E & 6F). Using flow cytometry, we identified a significant expansion in TrkB expressing cells in the lung 24 hours after acid-induced lung injury (Fig. 6G and Extended Figure 8). Specifically, we found that 11.69% of MANC cells, and more broadly 1.25% of all non-MANC mesenchymal cells, expressed TrkB.
Figure 6 – BDNF-TrkB signaling promotes alveolar epithelial regeneration.
(A-C) Analysis of scRNA-seq of mesenchymal cells from uninjured mice showed that TrkB expression is enriched in mesenchymal alveolar niche cells. (D) Ingenuity pathways analysis show that mesenchymal cells expressing TrkB are enriched with expression of genes that control respiratory system development, organ development and tissue morphology. (E) Expression of TrkB in GFP+ and GFP− cells from TrkBEGFP mice (n=4 mice per group). (F) Expression of TrkB on mesenchymal cells was verified by staining lung tissue form TrkBEGFP mice for PDFRα. (G) Quantification of GFP and PDFRα co-positive cells in TrkBEGFP mice 24 hours after acid-induced lung injury (n=4 mice per group) The gating strategy is shown in Extended Figure 8. (H & I) AT2 cells isolated from a tamoxifen treated SftpcCreERT2-Rosa26mTmG mouse were co-cultured with PDGFRα+ mesenchymal cells for four weeks. ANA-12 abrogated organoid forming efficiency of AT2 cells (n=3 wells per condition). (J & K) The organoid forming capacity of primary human AT2 cells was abrogated by the addition of ANA-12 to the media. Cultures were grown for four weeks. Right-hand panels show average organoid forming efficiency and size from n=3 different donors. For panels (E), (G), (K) and (I), data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test (E), (G), (K) and (I). Scale bar, 2.6mm (H, J), 20μm (F).
These data suggest that BDNF derived from transdifferentiating AT2 cells influences mesenchymal cells to enhance alveolar repair. To test this hypothesis, we studied the impact of TrkB inhibition on alveolar organoid formation in vitro. Addition of ANA-12, a small molecule antagonist of TrkB57, completely abrogated organoid formation in both murine and human alveolar organoid systems (Fig. 6H-K).
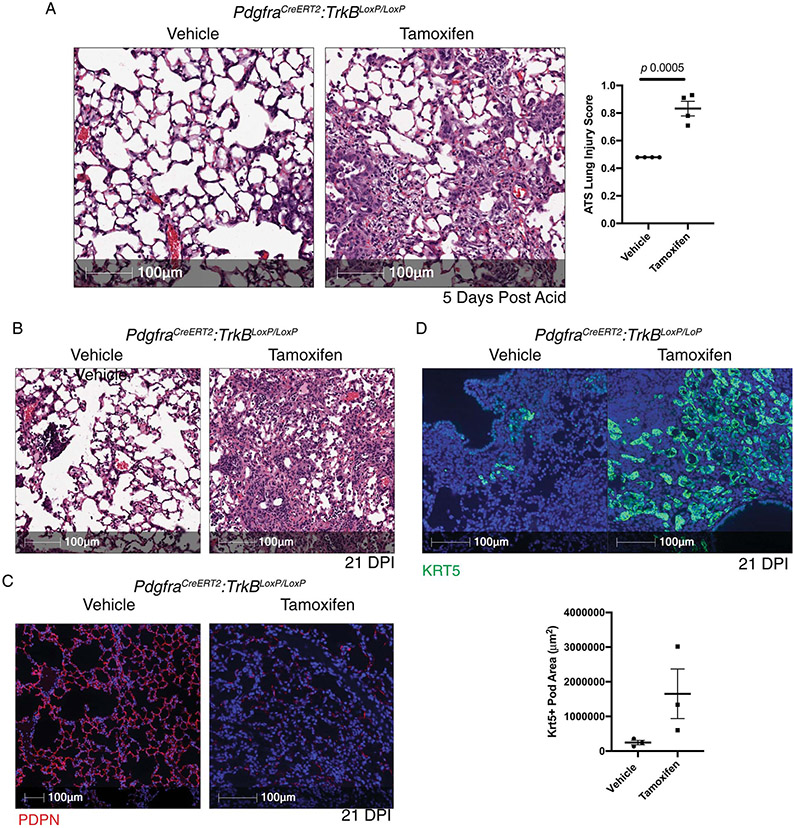
To test the importance of TrkB signaling in PDGFRα cells for alveolar repair, we generated a PdgfraCreERT2:TrkBLoxP/LoxP murine model. Mice lacking PDGFRα-cell specific TrkB had non-resolving pathology following our acid-induced lung injury model from which mice are usually able to recover (Extended Figure 9A). When tamoxifen-exposed PdgfraCreERT2:TrkBLoxP/LoxP mice were exposed to PR8 influenza, we observed worse histological outcomes, decreased PDPN staining (Extended Figure 9B & 9C) and increased Krt5 pods (Extended Figure 9D). These data corroborate the importance of BDNF-TrkB signaling in recovery from sterile and infectious lung injuries.
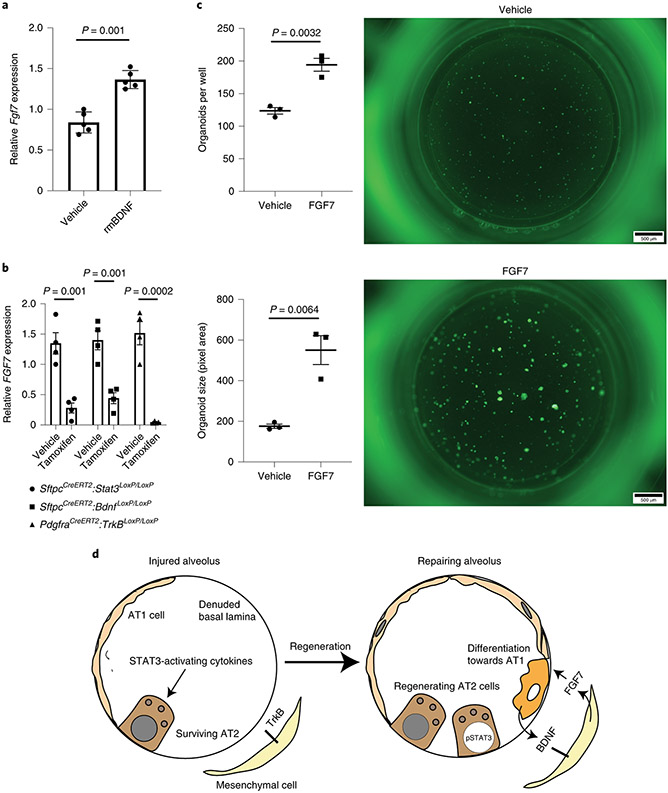
Since FGF7 has been identified as an essential MANC-derived cytokine19,46 promoting alveolar epithelial regeneration, we examined the impact of BDNF on FGF7 expression. Addition of recombinant BDNF to isolated mesenchymal cells from the organoid model shown in Figure 4H significantly increased expression of Fgf7 (Fig. 7A), providing a mechanistic basis for the observation that BDNF promotes alveolar epithelial regeneration. Similarly, PDGFRα cells from tamoxifen-exposed SftpcCreERT2:Stat3LoxP/LoxP, SftpcCreERT2:BdnfLoxP/LoxP, and PdgfraCreERT2:TrkBLoxP/LoxP mice 24 hours after exposure to acid-induced lung injury had reduced expression of Fgf7 in PDGFRα+ cells (Fig. 7B). Furthermore, we found that adding recombinant FGF7 to our organoid culture caused a significant increase in alveolar organoid forming efficiency and size (Fig. 7C). These data highlight the importance of the STAT3-BDNF-TrkB axis to promote regeneration, at least in part, by mediating release of FGF7 by mesenchymal niche cells (Fig. 7D).
Figure 7 – The STAT3-BDNF-TrkB axis modulates FGF7 expression by mesenchymal niche cells.
(A) Fgf7 mRNA expression in mesenchymal cells isolated from the organoid culture conditions described in Fig. 4H (n= 5 wells per condition). (B) Fgf7 expression in PDFRα cells isolated from tamoxifen or corn oil exposed SftpcCreERT2:Stat3LoxP/LoxP, SftpcCreERT2:BdnfLoxP/LoxP, and PdgfraCreERT2:TrkBLoxP/LoxP mice 24 hours after acid-induced lung injury (n=4 mice per group). (C) The addition of 0.1μg/mL of recombinant murine BDNF (rmBDNF) caused a significant increase in alveolar organoid forming efficiency and size (n=3 wells per condition). (D) In the setting of acute lung injury STAT3-activating cytokines accumulate in the alveolus and STAT3 becomes activated in the AT2 cell. Accompanying changes in chromatin accessibility within AT2 cells allows for expression of BDNF in transdifferentiating cells which then binds to TrkB on mesenchymal niche cells to stimulate expression of Fgf7, which promotes alveolar epithelial regeneration. For panels (A) (B) and (C), data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test (A) (B) and (C). Scale bars, 500μm.
A TrkB agonist improves lung histology following lung injury.
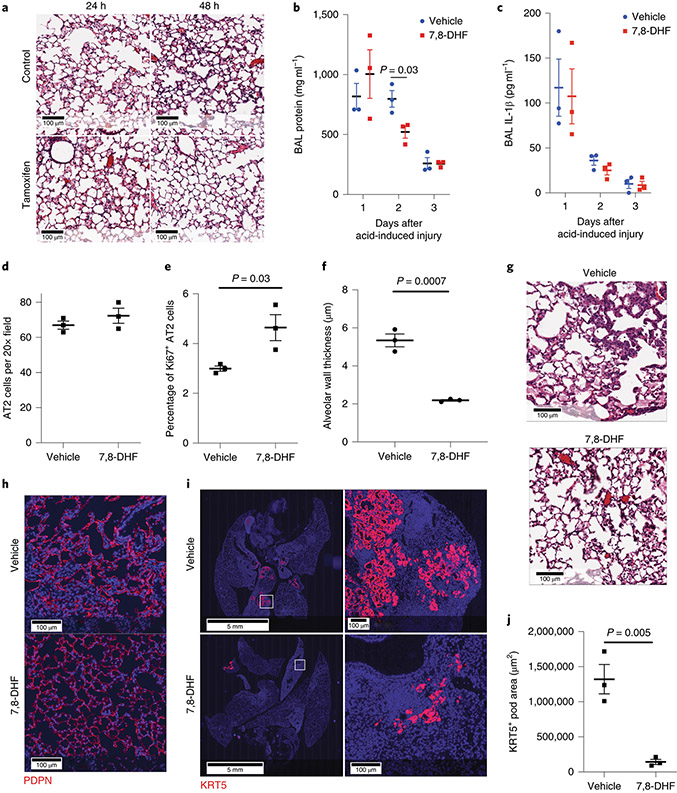
In order to determine if TrkB agonists alter lung repair after injury, we administered the TrkB agonist 7,8-Dihydroflavone (7,8-DHF)58 to mice that underwent acid-induced lung injury. We chose a 2-day time point to increase resolution of post-injury recovery in this model with rapid recovery39. 7,8-DHF attenuated the degree of histologic injury (Fig. 8A) and BAL protein levels 2 days after injury (Fig. 8B) without altering IL-1β levels (Fig. 8C). We also show increased AT2 cell proliferation 24h after injury and improved alveolar wall thickness 2 days after acid-induced lung injury (Fig. 8D- 8F).
Figure 8 – The TrkB agonist 7,8-dihydroxyflavone improves outcomes following sterile and infectious lung injury.
(A-E) C57BL/6 mice underwent acid-induced unilateral lung injury and were given an intraperitoneal injection of 7,8-DHF or vehicle control at the time of injury (n=3 mice per group). Mice that were euthanized at 48 hours received an additional intraperitoneal dose of 7,8-DHF or vehicle 24 hours after undergoing acid-induced lung injury. Mice that received 7,8-DHF had improved lung histology (A), significantly less BAL protein (B&C) and increased Ki67 and proSFTPC indicating increased AT2 cell proliferation (D&E) at 48 hours after lung injury. No differences in IL-1β or TNFα were observed. (F) Alveolar wall thickness was measured using 10 20x images per lung (means of n=3 mice per group). (G & H) H&E and PDPN staining of lung tissue from C57BL/6 mice infected with intranasal PR8 influenza (5x10−5 HAU/mouse). Intraperitoneal injections of 7,8-DHF or vehicle every other day (3 mice per group). (I&J) Quantification of KRT5+ pods from mice described in G & H (means of n=3 mice per group). For panels (B), (C), (D), (E), (F) and (J), data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test at each time point shown at (B), (C), (D), (E), (F) and (J). Scale bars, 100μm.
Since our model of acid-induced injury is rapidly repaired39, the effect of 7,8-DHF treatment to accelerate repair is likely to be inherently modest. In order to determine whether 7,8-DHF might affect disease characterized by continued injury and repair, we tested the effect of 7,8-DHF on influenza-induced injury. Histologic analysis of lungs 21 days post-infection revealed markedly improved architecture (Fig. 8G) and increased AT1 cells (Fig. 8H).
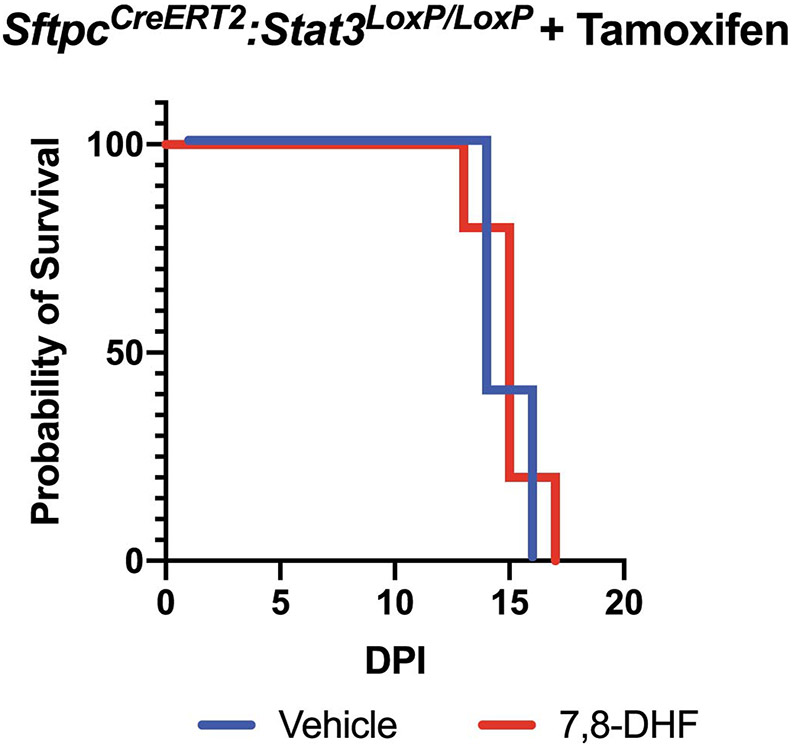
Since the presence of KRT5+ pods are known to negatively impact repair of severely damaged lung tissue27,30, we hypothesized that 7,8-DHF would enhance AT2-mediated repair and decrease dysmorphic responses. Quantitative histology of whole-lung sections revealed a significant decrease in KRT5+ pods in DHF treated murine tissue relative to controls (Fig. 8I and 8J). Interestingly, 7,8-DHF was unable to rescue influenza-exposed mice that lack AT2 specific Stat3 (Extended Figure 10), suggesting that other STAT3 targets, such as Cyclin D1, may also be important for the regenerative response to lung injury. These data demonstrate that 7,8-DHF, which pharmacologically enhances TrkB activation, is therefore a pathway to consider in developing treatments for enhancing recovery from sterile and infectious lung injuries.
Discussion
Repair of lung tissue has long been known to underlie resolution of several devastating diseases for which no pharmacological therapies exist9,15,59-62. These principles have become increasingly important in light of the current SARS-CoV-2 pandemic that causes severe ARDS in some but not all patients12,13. During lung repair, the AT2 cell functions as a progenitor for new AT2 and AT1 cells16-19. We hypothesized that genome-wide changes in chromatin accessibility in AT2 cells following acute lung injury associated with transcriptomic analyses of AT2 and mesenchymal niche cells would yield important insights into distal alveolar epithelium regeneration. Here, we show that these analyses converge on a single important cell-signaling pathway. We propose that STAT3 activation induces expression of Bdnf, which acts through the receptor TrkB to increase mesenchymal expression of Fgf7 and support epithelial regeneration. Our data suggest that targeting this pathway with the TrkB agonist 7,8-7,8-DHF should be examined as a strategy for enhancing resolution of destructive lung diseases.
We identified changes in AT2 chromatin structure following acute lung injury using ATAC-seq on AT2 cells isolated from control mice compared to mice 24 hours after sustaining acid induced lung injury. The importance of these changes to alveolar regeneration was suggested by unbiased pathways analysis that classified genes within newly accessible chromatin as supporting regenerative processes. We focused on STAT3 because of our prior data implicating STAT3 activators in alveolar epithelial regeneration39,46 and because an unbiased pathway analysis of our ATAC-seq data identified that genes adjacent to newly accessible STAT3 binding sites are involved in regenerative pathways. Although mouse models have previously shown that STAT3 is important for maintaining alveolar epithelium following hyperoxia exposure63 and is important for host defense following Escherichia coli pneumonia64, none have specifically interrogated the function of STAT3 in alveolar epithelial regeneration. As such, no STAT3-target genes had yet been implicated in alveolar epithelial regeneration. Here, we demonstrate that changes in chromatin accessibility following acute lung injury portend a second function for STAT3 in the later phases of regeneration. It is important to note that as shown in Fig. 2B, STAT3 is activated in cell types other than AT2 cells65-67. Although we did not identify STAT3 binding sites near known Stat3 activators, we cannot rule out the possibility that STAT3 activity in non-AT2 cells is altered by the loss of STAT3 in AT2 cells.
Having identified the importance of STAT3 to alveolar epithelial regeneration, we next sought to identify important effectors using sc-RNA seq. We identified a unique cluster of cells, the transdifferentiating population, which emerges 24 hours after induced lung injury, that expresses Tm4sf1, the AT2 marker Sftpc and the AT1 marker Pdpn, despite having been selected against PDPN during isolation. By integrating ATAC-seq and scRNA-seq data, we identified Bdnf as the single gene that had a newly accessible STAT3 binding motif and was also differentially upregulated in these transdifferentiating AT2 cells. It is possible that using different selection criteria, we would have identified additional candidates. Although called “brain”-derived “neurotrophic” factor, the regulation of Bdnf by STAT3 has previously been reported in multiple cell types53-56. While BDNF has been detected in lung cancer lines68, it has not previously been implicated in alveolar epithelial regeneration. The importance of BDNF for alveolar epithelial regeneration was confirmed using organoids, a simplified model of alveolar development that effectively isolated the roles of epithelial and mesenchymal cells. Adding recombinant human BDNF to the organoid media significantly increased organoid size and forming efficiency of primary murine and human AT2 cells. Although we identified BDNF by isolating AT2 cells, its expression is seen only in transdifferentiating AT2 cells, and thus we cannot resolve whether this upregulation is part of the AT2 to AT1 transition or unique to the reparative state.
To understand the underlying cell-signaling mechanism through which BDNF promotes alveolar repair, we sought to identify which cell types express TrkB, the cognate receptor for BDNF69. Analysis of pulmonary mesenchymal cells using scRNA-seq indicated that TrkB expression was uniquely enriched in the mesenchymal alveolar niche cell population, which expands during regeneration46. Likewise, we find that the number of TrkB and PDGFRα co-positive cells significantly increases following acute lung injury, further suggesting the importance of BDNF-responsive niche cells as a part of the regenerative response. Adding the TrkB antagonist, ANA-12, to organoid media abrogated alveolar organoid formation, demonstrating that TrkB signaling is necessary for alveolar organoid development. These data suggest that activating the TrkB receptor could be beneficial to recovery from acute lung injury. Administering the recently-developed TrkB agonist 7,8-DHF58 led to faster recovery from acid-induced injury and H1N1 influenza infection. While we implicated a role for FGF7, further work is necessary to determine the impact of BDNF and TrkB agonists, during homeostasis and regeneration on FGF7 and other potential mediators as well. further studies are needed to see if the impact of 7,8-DHF on outcomes following lung injury are due to differences in inflammation or regeneration alone. Our studies using BDNF and ANA-12 in vitro strongly suggest that at least part of the effect we observed with 7,8-DHF treatment is due to increased alveolar epithelial regeneration.
In summary, these data demonstrate that effective regeneration of the alveolar epithelium requires bidirectional communication between epithelial and mesenchymal cells. In the epithelial cells, our data shows that activation of STAT3 in AT2 cells plays a central role in repair. At least one of the downstream effects of STAT3 activation appears to be induction of Bdnf, which activates TrkB-expressing mesenchymal niche cells to modulate expression of Fgf7, which subsequently interacts with epithelial cells to play an essential regenerative role (Fig. 8D). From a translation standpoint, this process can be enhanced pharmacologically by administration of a TrkB agonist. Future studies are needed to elucidate how chromosomal architecture is regulated in AT2 cells after acute lung injury, how STAT3 becomes activated in AT2 cells, and which other cells may respond to BDNF in vivo. Nevertheless, the contribution of the STAT3-BDNF-TrkB axis in orchestrating alveolar epithelial regeneration, might open up opportunities for drug discovery, precision targets and therapeutic interventions.
Materials and Methods
Mice
All mice were housed in specific pathogen free conditions in an animal facility at the Children’s Hospital of Philadelphia. The mice are housed at an ambient temperature of 70°F +/− 1°F with humidity levels kept at 60%. Lighting in the rooms is on a 12-hour cycle such that they are on from 6:15 AM to 6:15 PM and are otherwise kept off. All mouse protocols were approved by the IACUC at the Children’s Hospital of Philadelphia. The study is compliant with all relevant ethical regulations regarding animal research. Mice aged 10-12 weeks were used for experiments (18-25g). We strived to use male and female mice in equal proportions. Unless otherwise specified, we used a minimum of 3 mice per time point per treatment condition in each experiment. Mice were housed and used in accordance with institutional and AALAC guidelines. SftpcCreERT2 mice70 were generously donated by Harold A. Chapman at the University of California, San Francisco. TrkbEGFP mice71 were generously donated by David D. Ginty at Harvard University. TrkBLoxP mice72 were generated by Louis F. Parada, currently at Memorial Sloan Kettering Cancer Center, and were shared with us by James O. McNamera at Duke University. All other mice were obtained from the Jaxon Laboratory – C57BL/6 (stock #000664), BdnfCre (stock #030189)73, BdnfLoxP (stock #004339)74, Hopx3FlagGFP (stock #029271)75, PdgfraCreERT2 (stock #018280)76, R26RTdTomato (stock #007914)77, and Stat3LoxP/LoxP (stock #016923)78.
Acid-Induced Lung Injury
Sedated mice were intubated using a 20G angiocatheter (BD Biosciences catalog #381434) using a previously described technique79. The mice were then placed in the right lateral recumbent position and a polytehylene 10 (PE-10) catheter (BD catalog #427400) was directed into the right main stem bronchus as previously described80. Injury was induced by instilling 2μL/g of osmotically balanced 0.1N HCl into the right lung through the PE-10 catheter. Where indicated, mice received intraperitoneal injections of 10mg/kg 7,8-Dihydroxyflavone (Sigma-Aldrich catalogue #D5446) in DMSO and PBS (1:1 ratio) on the day of acid-instillation followed by daily intraperitoneal injections. Mice in the control group received the identical volume of DMSO and PBS at the same time points.
Influenza Lung Injury
PR8 H1N1 influenza was a generous gift of Dr. Carolina B. Lopez at the University of Pennsylvania. For infection, virus was diluted in PBS and a dose of 5x10−5 HAU per mouse was administered via intranasal instillation. Following infection, animals were weighed and monitored three times per week (Monday, Wednesday and Friday) for up to 21 days. Animals that lost >30% of starting weight or were moribund were euthanized humanely. Where indicated, mice received intraperitoneal injections of 10mg/kg 7,8-Dihydroxyflavone (Sigma-Aldrich catalogue #D5446) or vehicle control on the day of infection and on subsequent Mondays, Wednesdays and Fridays. Mice in the control group received the identical volume of DMSO and PBS at the same time points.
Isolation of Murine AT2 and Mesenchymal Cells
Murine lung tissue was digested into a single-cell suspension as previously described using dispase (Corning catalog #354235), collagenase (Roche catalog #10103578001), and DNase (Roche catalog #10104159001). When isolating AT2 cells from SftpcCreERT2:R26RTdTomato mice, the animals received 200μg/gm tamoxifen (Sigma-Aldrich catalog #T55648) in corn oil (Sigma-Aldrich catalog #C8267) via gavage 3 days prior to being euthanized. Tomato+ cells were isolated using flow cytometry (MoFlow Astrios with Summit v 6.3.0.16900 software). To isolate AT2 cells from mice we stained single-cell suspensions of murine lung tissue and gated based on the following criteria: positive for EpCAM-APC (BioLegend catalog #118213) and negative for CD31-PE (eBioscience catalog #12-0311-81), CD45-PE (eBioscience catalog #12-0451-81), podoplanin-PE (eBioscience catalog #12-5381-80), Sca1-PE (eBioscience catalog #12-5981-81), CD24-PE (BioLegend catalog #119307), and DAPI (BioLegend catalog #422801) as previously described81. To isolate mesenchymal cells we sorted for cells that were positive for CD140a (BioLegend catalog #135907) and negative for and DAPI (BioLegend catalog #422801). Antibody concentrations are in Supplementary Table 2.
Isolation of Human AT2 Cells
Samples of uninjured, de-identified human lungs were obtained from non-utilized lungs donated for organ transplantation via an established protocol for the Prospective Registry of Outcomes in Patients Electing Lung Transplant Study approved by University of Pennsylvania Institutional Review Board with informed consent in accordance with institutional procedures. A 2x2cm piece of distal lung tissue was obtained, pleura and large airways were carefully dissected away, and tissue was processed into a single cell suspension using the same combination of dispase, collagenase I, and DNase used for mouse lungs. A Miltenyi gentleMACS dissociator was used for mincing and incubation for 35 minutes at 37°C. Cells were washed, passed over 70μM and 40μM filters, and RBCs were lysed with Ammonium – Chloride - Potassium (ACK) lysis buffer. After a single-cell suspension was obtained, cells were sorted using the MACS multisort kit, MACs LS columns, and the following antibodies: HT2-280 (mouse IgM), a gift of Leland Dobbs at the University of California San Francisco, anti-mouse IgM microbeads (Miltenyi catalog #130-047-302). The full protocol for digestion and sorting of human lung epithelial cells, and their propagation as alveolar organoids, has been made available via Nature Protocol Exchange82.
Organoid Assay
Clonal alveolar organoid assays were performed as described previously with some modifications from the original protocol17,37,81. In brief, 5 × 103 AT2 cells were isolated as described above and mixed with 5 x104 lung PDGFRα+ isolated cells from adult wild type mice as previously described19 for mouse or MRC5 cells (ATCC CCL-171, tested negative for mycobacterial contamination, at no greater than passage 7) for human cultures. For the first two days of culture, ROCK inhibitor Y27632 (Sigma-Aldrich catalog #Y0503) was added to the media (SAGM; Lonza catalog #CC-3118). After two days of culture, Y276632 was removed and ligand treatments of organoids were performed using the following reagents at the indicated concentrations: ANA-12 0.02μg/mL (Alomone Labs catalog #A-215), recombinant human BDNF 0.05μg/mL (Alomone Labs catalog #B-250) and recombinant murine FGF7 0.01μg/mL (R&D Systems catalog #5028-KG). DMSO was used as a control for the ANA-12 and 2% BSA was used as a control for the BDNF and FGF7. Media was changed every 48 hours, and fresh ligands were included at each media change. After 21 days (or 28 days where indicated) organoids were imaged using an Olympus MVX10 microscope or EVOS FL microscope (Life Technologies).
ATAC-seq
Individual ATAC-seq libraries were generated from sorted AT2 cells using methods outlined above and previously described83. Briefly, 5x104 cells were sorted into media, washed, and lysed to obtain nuclei. Nuclei were exposed to Tn5 transposase (Illumina catalogue #FC-121-1030), and fractionated DNA was used for amplification and library preparation. Libraries were then purified and underwent paired-end sequencing (100bp) using the Illumina HiSeq 4000. After sequencing the adapters were trimmed with attack (version 0.1.5, https://atactk.readthedocs.io/en/latest/index.html) raw reads were aligned against the mouse reference genome (mm9) using the bowtie-1.1.2 aligner with the following flags: “--chunkmbs 2000 --sam --best --strata -m1 -X 2000”84. Samtools and Picard-tools were used to exclude duplicate reads and reads mapping to ChrM were excluded from further analysis. We used MACS285 for peak-calling with a q cutoff of 0.05. Downstream analysis and visualization was done using HOMER40 and deepTools286.
Single-cell RNA-seq
Murine AT2 cells were isolated using the above protocols. Single cells were captured using the 10x Genomics microfluidic platform. RNA was isolated and libraries prepared using 10X Genomics reagents according to the manufacturer’s protocol. Sequencing was performed on an Illumina HiSeq 2500 using asymmetric reads, as suggested by 10x Genomics. We downloaded data on the sc-RNA expression of pulmonary mesenchymal cells from a prior publication46 whose data was deposited in GEO (GSE99714). Fastq files were assessed for quality control using the FastQC program (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). Fastq files were aligned against the mouse reference genome (mm9) using the Cell Ranger environment (10X Genomics). T-SNE clustering and differential expression analysis was done using Seurat87.
Pathways Analysis
The differential genes between clusters were identified using the R package ‘cellrangerRkit’. These lists were then analyzed using IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). The final heatmap was visualized using the gplots R package.
Histology
At the time of tissue harvest, mice were euthanized by CO2 inhalation. The chest cavity was exposed and the lungs cleared of blood by perfusion with cold PBS via the right ventricle. Lungs were inflated with 4% paraformaldehyde under constant pressure of 30 cm water and allowed to fix for 24 hours. Tissue was then dehydrated, paraffin embedded, and sectioned. Hematoxylin and eosin staining was performed to examine morphology, and to score regions based on the severity of injury using the ATS-ERS criteria88. Immunohistochemistry was used to detect protein expression using the following antibodies on paraffin sections: DC-LAMP (Novus Biologicals catalog #DDX0191P-100), GFP (Molecular Probes catalog #A1122), Ki67 (Abcam catalog #ab16667, KRT5 (BioLegend catalog #905501 and #905903), PDGFRα (Cell Signaling Technology catalog #3177), PECAM-1 (Dianova catalog #DIA-310), Podoplanin (University of Iowa Developmental Studies Hybridoma Bank clone 8.1.1), pSTAT3 (Cell Signaling Technology catalog #9145), proSFTPC (MilliporeSigma catalog #AB3786), and RFP (Rockland Immunochemicals catalog #600-401-379). Antibody concentrations are in Supplementary Table 2.
Image Acquisition
Confocal immunofluorescent images were obtained using the Leica TCS SP8 scanning confocal microscope using the Leica LAS X software (version 3.0.13). Images of H&E stained sections were obtained using the Aperio Image Scope (version 12.4).
Quantification of pSTAT3 in AT2 Cells
Histological specimens were obtained and stained as outlined above. Random confocal images were randomly obtained by an operator who was unaware of the experimental conditions or hypothesis. The digital sections were then reviewed by a blinded investigator who identified cells are co-positive for proSFTPC and pSTAT3 as well as cells that are positive for proSFTPC but not pSTAT3. The blinded investigator also identified several areas where they did not feel confident making a determination. Reasons for uncertainly included autofluorescence of red blood cells, unclear staining patterns or adjacent nuclei being too close to each other to accurately distinguish adjacent cells.
Quantitative PCR
RNA was isolated from sorted mesenchymal cells or AT2 cells using the RNeasy Mini Kit (Qiagen catalog #74104) and reverse transcribed using the High Capacity RNA to cDNA Kit (Applied Biosystems catalog #4387406). Quantitative PCR was performed using primers specific for Bdnf, Fgf7, Hprt1, and TrkB with the PowerSYBR Green PCR Master Mix (Applied Biosystems catalog #4367659). Quantitative PCR was performed using the ViiA 7 Real-Time PCR System (Applied Biosystems). Primer sequences are in Supplementary Table 3.
PCR
RNA was isolated from sorted AT2 cells using the RNeasy Mini Kit (Qiagen catalog #74104) and reverse transcribed using the High Capacity RNA to cDNA Kit (Applied Biosystems catalog #4387406). PCR was performed with the primers specific for STAT3. Expected product is 807bp without Cre-mediated recombination and 519bp after Cre-mediated recombination. Primer sequences are in Supplementary Table 4.
ELISAs and Protein Assay
BAL samples were obtained and cell counts were analyzed as previously described89. Levels of IL-1β were measured using an ELISA kit (eBioscience catalog #501129749). Levels of IL-6 were measured using an ELISA kit (eBioscience catalog #88-7064-88). Protein assays were done using the BCA Microplate BCA Protein Assay Kit (Thermo Scientific catalog #23252). All plates were read on the Spectra Max 250 spectrophotometer (Molecular Devices).
ATS Lung Injury Scoring
A blinded reviewer examined random H&E sections from 3 or 4 mice per time point per condition and scored each mouse according to published guidelines88.
Calculation of alveolar septal thickness and mean linear intercepts
We used the Matlab 2018a (RRID:SCR_001622) image processing and statistical toolboxes to perform high-throughput alveolar wall thickness analysis and mean length intercept analysis of H&E stained sections taken under a brightfield microscope with a 20x objective. We obtained 10 random images from each influenza infected mouse and 5 random images from mice that underwent acid-induced lung injury. We obtained fewer images from the acid injured mice because the acid injury is unilateral. The program performs binarization using automated thresholding based on Otsu's method, it performs erosion, dilation to fill the holes in the alveolar walls and inverts the image such that the alveolar spaces are black and the walls are white. Subsequently, the program uses the Laplacian operator to extract the boundaries of the alveolar spaces, and using the bwboundaries function, detects alveolar spaces as distinct objects. The object detection enables computation of the minimum distance between each alveolar space in an iterative manner where distances between each alveolar space with the neighboring spaces are compared and minimum distances corresponding to the narrowest segments are extracted. About 100-200 measurements per field are taken and the average value representing the mean interseptal wall thickness. For the mean linear intercept, the dimensions of the image were extracted and based on the vertical dimension, and an equal spaced line grid consisting of 20 lines was created. The number of times the lines intersect with the walls and calculate the length of the intercepts for each of lines based on dimensions of the image were counted. The mean length of the intercept for each mouse is reported.
Statistics and Reproducibility
Statistical analysis was performed in Prism for Mac (version 8.4.2) and R. P values were obtained usng two-tailed Student’s t-test for comparison of two data sets or by analysis and variance (ANOVA), where appropriate. Statistical analysis of transcription binding motifs in Fig. 1G was done using the findMotifGenome.pl script in the HOMER software package with adjustment for multiple comparisons Generation of odds ratios for distribution of ATAC regions near genes was evaluated using Fisher’s exact test and contingency table analysis. Statistical data was considered significant if p < 0.05. Center values of all plots represent means and error bars represent the standard error of the mean. Results were reproducible and conducted with established internal controls. When feasible, experiments were repeated three or more times and yielded similar results. We have indicated the n used for analysis in each figure legend. Cell cultures were routinely screened for mycoplasma.
Extended Data
Extended Data Fig. 1. Sorting Strategies for Isolating AT2 cells.
Lungs from C57BL/6 mice were digested into a single-cell suspension. We collected EpCAM positive cells (APC) that were PDPN, CD45, CD34, CD31 and SCA1 negative (all PE).
Extended Data Fig. 2. STAT3 is activated following sterile and infectious lung injuries.
Cells staining positive for proSFTPC+ and pSTAT3+ are highlighted with a white box while proSFTPC + pSTAT3− cells are highlighted with a yellow box. Areas that were felt to be indeterminate for cell type or pSTAT3 status are highlighted in blue. Scale bars, 50μm.
Extended Data Fig. 3. Loss of Stat3 diminished alveolar organoid formation.
(A) PCR shows the expected 802bp product when corn oil is administered to SftpcCreERT2:Stat3LoxP/LoxP mice and the recombined 514bp product when tamoxifen is administered to SftpcCreERT2:Stat3LoxP/LoxP mice. The gels in this figure represent n=2 mice per condition (corn oil and tamoxifen). This experiment was repeated 4 times with similar results. (B) AT2 cells from tamoxifen treated SftpcCreERT2-Rosa26tdTomato and SftpcCreERT2-Rosa26tdTomato:Stat3LoxP/LoxP mice were isolated and co-cultured with PDGFRα mesenchymal cells in transwells containing Matrigel for four weeks (n=4 wells per condition). Organoid forming efficiency was significantly decreased in the absence of STAT3 signaling in AT2 cells (p=2.2x10−8). Data is shown as the mean +/− SEM. Statistical significance was determined using a two-tailed Student’s t-test. Scale bars, 2.5mm.
Extended Data Fig. 4. Loss of Stat3 in AT2 cells does not alter distal lung morphology.
SftpcCreERT2:Stat3LoxP/LoxP mice were given tamoxifen or vehicle and euthanized three months later. Representative H&E stained histological specimens are shown. The Mean linear intercept was unchanged by the loss of Stat3 in these unchallenged mice (n=4 mice per group). Data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test. Scale bars, 500μm.
Extended Data Fig. 5. Single-cell expression of TrkB, Ccnd1, Tm4sf1 and Sftpc in isolated AT2 cells.
Plots show relative expression of TrkB, Ccnd1, Tm4sf1 and Sftpc in AT2 cells isolated isolated from uninjured mice and mice 24h after acid-induced lung injury.
Extended Data Fig. 6. Pathways analysis of isolated AT2 cells reveals multiple subsets of cells with multiple functions.
Pathways analysis of differentially expressed genes in the clusters shown in Fig. 4B.
Extended Data Fig 7. AT2 cells express Bdnf after acid-induced lung injury.
BdnfCre-Rosa26tdTomato were subjected to acid-induced lung injury (n=4 mice per group). One day after injury the mice were euthanized, and the lungs were probed for Tomato (red) and proSFTPC (green). Autofluorescence delineates tissue structure. Scale bar, 25μm.
Extended Data Fig. 8. Loss of TrkB in mesenchymal cells worsens outcomes following sterile and infectious lung injuries.
Lungs from PdgfraEGFP mice were digested into a single-cell suspension either before or 24h after acid-induced lung injury (n=4/group). We first gated on PDGFR α + and then quantified the percent of GFP+ cells within that population.
Extended Data Fig. 9. Loss of TrkB in mesenchymal cells worsens outcomes following sterile and infectious lung injuries.
(A) H&E staining and ATS lung injury scores of tamoxifen and corn oil exposed PdgfrαCreERT2:TrkBLoxP/LoxP mice 24 hours after acid-induced lung injury (n=4 mice per group). (B-D) H&E (B), PDPN (C) and Krt5 (D) staining of tamoxifen and corn oil exposed PdgfrαCreERT2:TrkBLoxP/LoxP mice 21 days after infection with PR8 influenza (n=3 mice per group). Statistical analysis was performed with a two-tailed Student’s t-test or ANOVA, where appropriate. For panels (A) and (D), data is shown as the mean +/− SEM. Statistical significance was determined with a two-tailed Student’s t-test (A) and (D). Scale bars, 100μm.
Extended Data Fig. 10. The TrkB agonist 7,8-DHF is unable to rescue mice with an AT2-specific Stat3 deletion.
Survival curves for Tamoxifen-exposed SftpcCreERT2:Stat3LoxP/LoxP mice that had been infected with intranasal PR8 influenza (5x10−5 HAU/mouse) and were given intraperitoneal injections of 7,8-DHF or vehicle every other day (n=5 mice per group). The data analyzed using the Log-rank (Mantel-Cox) test and is not statistically significant.
Supplementary Material
Acknowldedgements
The authors wish to acknowledge Steven M. Albelda, MD, Jacob S. Brenner, MD, PhD, and Andrew E. Vaughan, PhD for their help in preparing this manuscript as well as the following cores at the Children’s Hospital of Philadelphia Research Institute: Pathology, Flow Cytometry, Nucleic Acid and PCR, and Center for Applied Genomics. This work was supported by the Parker B. Fellowship Program (AJP) and multiple grants from the National Institutes of Health – K08 HL136698 (AJP), R01 AI121321 (MDW), R01 AI099479 (GSW), R01 DK114054 (GSW). The authors also wish to thank the patients and families who donated lung tissue to support this research.
Footnotes
Competing Interests
The authors have applied for a provisional patent in the United States related to this work. AJP and GSW are listed as co-inventors on this application.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Data Availability
ATAC-seq and scRNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE132535.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Matthay MA, Robriquet L & Fang X Alveolar epithelium: role in lung fluid balance and acute lung injury. Proc Am Thorac Soc 2, 206–213 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Guillot L et al. Alveolar epithelial cells: master regulators of lung homeostasis. Int J Biochem Cell Biol 45, 2568–2573 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Tyrrell C, McKechnie SR, Beers MF, Mitchell TJ & McElroy MC Differential alveolar epithelial injury and protein expression in pneumococcal pneumonia. Exp Lung Res 38, 266–276 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Herold S, Becker C, Ridge KM & Budinger GR Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur Respir J 45, 1463–1478 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Herold S et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. The Journal of experimental medicine 205, 3065–3077 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthay MA, Ware LB & Zimmerman GA The acute respiratory distress syndrome. The Journal of clinical investigation 122, 2731–2740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson BT, Chambers RC & Liu KD Acute Respiratory Distress Syndrome. The New England journal of medicine 377, 1904–1905 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Martin TR, Hagimoto N, Nakamura M & Matute-Bello G Apoptosis and epithelial injury in the lungs. Proc Am Thorac Soc 2, 214–220 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herridge MS et al. One-year outcomes in survivors of the acute respiratory distress syndrome. The New England journal of medicine 348, 683–693 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Matthay MA & Wiener-Kronish JP Intact epithelial barrier function is critical for the resolution of alveolar edema in humans. The American review of respiratory disease 142, 1250–1257 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Ingbar DH & Matthay RA Pulmonary sequelae and lung repair in survivors of the adult respiratory distress syndrome. Crit Care Clin 2, 629–665 (1986). [PubMed] [Google Scholar]
- 12.Guan WJ et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatraju PK et al. Covid-19 in Critically Ill Patients in the Seattle Region - Case Series. The New England journal of medicine (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konigshoff M, Saglani S, Marsland BJ & Eickelberg O Rebuilding a diseased lung: repair and regeneration. Eur Respir J 41, 497–499 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Kotton DN & Morrisey EE Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med 20, 822–832 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans MJ, Cabral LJ, Stephens RJ & Freeman G Renewal of alveolar epithelium in the rat following exposure to NO2. The American journal of pathology 70, 175–198 (1973). [PMC free article] [PubMed] [Google Scholar]
- 17.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. The Journal of clinical investigation 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA & Desai TJ Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zacharias WJ et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain R et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat Commun 6, 6727 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y et al. Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proceedings of the National Academy of Sciences of the United States of America 115, 2407–2412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim CF et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Liu Q et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat Genet 51, 728–738 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Salwig I, et al. Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. The EMBO journal (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo W, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 517, 616–620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray S, et al. Rare SOX2+ Airway Progenitor Cells Generate KRT5+ Cells that Repopulate Damaged Alveolar Parenchyma following Influenza Virus Infection. Stem Cell Reports 7, 817–825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaughan AE, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nolen-Walston RD, et al. Cellular kinetics and modeling of bronchioalveolar stem cell response during lung regeneration. American journal of physiology. Lung cellular and molecular physiology 294, L1158–1165 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yee M, et al. Alternative Progenitor Lineages Regenerate the Adult Lung Depleted of Alveolar Epithelial Type 2 Cells. Am J Respir Cell Mol Biol 56, 453–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xi Y, et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat Cell Biol 19, 904–914 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason RJ & Williams MC Type II alveolar cell. Defender of the alveolus. The American review of respiratory disease 115, 81–91 (1977). [DOI] [PubMed] [Google Scholar]
- 32.Fehrenbach H Alveolar epithelial type II cell: defender of the alveolus revisited. Respir Res 2, 33–46 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crapo JD, et al. Morphometric characteristics of cells in the alveolar region of mammalian lungs. The American review of respiratory disease 128, S42–46 (1983). [DOI] [PubMed] [Google Scholar]
- 34.Takahashi K & Yamanaka S Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Rodolfa KT & Eggan K A transcriptional logic for nuclear reprogramming. Cell 126, 652–655 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barkauskas CE et al. Lung organoids: current uses and future promise. Development 144, 986–997 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadyk LC, DeWitt ND & Gomperts B Proceedings: Regenerative Medicine for Lung Diseases: A CIRM Workshop Report. Stem Cells Transl Med 6, 1823–1828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paris AJ et al. Neutrophils promote alveolar epithelial regeneration by enhancing type II pneumocyte proliferation in a model of acid-induced acute lung injury. American journal of physiology. Lung cellular and molecular physiology, ajplung 00327 02016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura S et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 10, 60–69 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Yuan B et al. Inhibition of distal lung morphogenesis in Nkx2.1(−/−) embryos. Dev Dyn 217, 180–190 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Li S et al. Foxp1/4 control epithelial cell fate during lung development and regeneration through regulation of anterior gradient 2. Development 139, 2500–2509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shu W, et al. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development 134, 1991–2000 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Ghahary A & Ghaffari A Role of keratinocyte-fibroblast cross-talk in development of hypertrophic scar. Wound Repair Regen 15 Suppl 1, S46–53 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Zepp JA, et al. Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell 170, 1134–1148.e1110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schutte H, et al. Bronchoalveolar and systemic cytokine profiles in patients with ARDS, severe pneumonia and cardiogenic pulmonary oedema. Eur Respir J 9, 1858–1867 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Zhong Z, Wen Z & Darnell JE Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264, 95–98 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Zemans RL & Matthay MA Bench-to-bedside review: the role of the alveolar epithelium in the resolution of pulmonary edema in acute lung injury. Crit Care 8, 469–477 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matthay MA, Folkesson HG & Clerici C Lung epithelial fluid transport and the resolution of pulmonary edema. Physiol Rev 82, 569–600 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Riemondy KA, et al. Single cell RNA sequencing identifies TGFbeta as a key regenerative cue following LPS-induced lung injury. JCI Insight 5(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leibrock J, et al. Molecular cloning and expression of brain-derived neurotrophic factor. Nature 341, 149–152 (1989). [DOI] [PubMed] [Google Scholar]
- 53.Chen B, et al. Autocrine activity of BDNF induced by the STAT3 signaling pathway causes prolonged TrkB activation and promotes human non-small-cell lung cancer proliferation. Sci Rep 6, 30404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang QP, et al. STAT3 signal that mediates the neural plasticity is involved in willed-movement training in focal ischemic rats. J Zhejiang Univ Sci B 17, 493–502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang JX, et al. Unique genome-wide map of TCF4 and STAT3 targets using ChIP-seq reveals their association with new molecular subtypes of glioblastoma. Neuro Oncol 15, 279–289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hixson KM, Cogswell M, Brooks-Kayal AR & Russek SJ Evidence for a non-canonical JAK/STAT signaling pathway in the synthesis of the brain's major ion channels and neurotransmitter receptors. BMC Genomics 20, 677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cazorla M, et al. Identification of a low-molecular weight TrkB antagonist with anxiolytic and antidepressant activity in mice. The Journal of clinical investigation 121, 1846–1857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang SW, et al. A selective TrkB agonist with potent neurotrophic activities by 7,8-dihydroxyflavone. Proceedings of the National Academy of Sciences of the United States of America 107, 2687–2692 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hogan B Stemming Lung Disease? The New England journal of medicine 378, 2439–2440 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Weiss DJ Concise review: current status of stem cells and regenerative medicine in lung biology and diseases. Stem Cells 32, 16–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J & Jia Z Cell-based therapy in lung regenerative medicine. Regen Med Res 2, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang M & Thebaud B Stem cell biology and regenerative medicine for neonatal lung diseases. Pediatric research 83, 291–297 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Hokuto I, et al. Stat-3 is required for pulmonary homeostasis during hyperoxia. J Clin Invest 113, 28–37 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quinton LJ, et al. Alveolar epithelial STAT3, IL-6 family cytokines, and host defense during Escherichia coli pneumonia. Am J Respir Cell Mol Biol 38, 699–706 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Severgnini M, et al. Activation of the STAT pathway in acute lung injury. American Journal of Physiology-Lung Cellular and Molecular Physiology 286, L1282–L1292 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Liu X STAT3 activation inhibits human bronchial epithelial cell apoptosis in response to cigarette smoke exposure. Biochem Biophys Res Commun 353, 121–126 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Tadokoro T, et al. IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proceedings of the National Academy of Sciences of the United States of America 111, E3641–3649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giard DJ, et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst 51, 1417–1423 (1973). [DOI] [PubMed] [Google Scholar]
- 69.Chao MV Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci 4, 299–309 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Chapman HA, et al. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest 121, 2855–2862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell 147, 1615–1627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luikart BW, Nef S, Shipman T & Parada LF In vivo role of truncated trkb receptors during sensory ganglion neurogenesis. Neuroscience 117, 847–858 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Tan CL, et al. Warm-Sensitive Neurons that Control Body Temperature. Cell 167, 47–59 e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rios M et al. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol Endocrinol 15, 1748–1757 (2001). [DOI] [PubMed] [Google Scholar]
- 75.Takeda N et al. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development 140, 1655–1664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y et al. Thermal stress induces glycolytic beige fat formation via a myogenic state. Nature 565, 180–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moh A, et al. Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab Invest 87, 1018–1028 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Das S, MacDonald K, Chang HY & Mitzner W A simple method of mouse lung intubation. J Vis Exp, e50318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paris AJ, et al. Using selective lung injury to improve murine models of spatially heterogeneous lung diseases. PLoS One 14, e0202456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alder JK, et al. Telomere dysfunction causes alveolar stem cell failure. Proceedings of the National Academy of Sciences of the United States of America 112, 5099–5104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zacharias W & Morrisey E Isolation and culture of human alveolar epithelial progenitor cells. Nature Protocol Exchange (2018). [Google Scholar]
- 83.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21.29.21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matute-Bello G, et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol 44, 725–738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nick JA, et al. Selective suppression of neutrophil accumulation in ongoing pulmonary inflammation by systemic inhibition of p38 mitogen-activated protein kinase. Journal of immunology 169, 5260–5269 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ATAC-seq and scRNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE132535.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.