Abstract
MiR-22 has been demonstrated to inhibits tumor growth in several cancers. However, its function in the tumor microenvironment is still unclear, especially for T cell differentiation. Here, miR-22 expression in the circulating T cells from hepatocellular carcinoma (HCC) patients and healthy controls was analyzed with quantitative polymerase chain reaction (qPCR). Diethylnitrosamine (DEN)/phenobarbital (PB)-mediated primary HCC and Hepa1-6 subcutaneous tumor mouse models were established and subjected to lenti-miR-22 injection. Mice immunoreconstituted with miR-22-overexpressing T cells were employed to investigate the antitumor effect of miR-22 in mice. Luciferase assay, immunofluorescent staining, in vitro Th17 cell differentiation assay, and rescue experiments were employed to investigate the mechanism underlying the miR-22-mediated regulation of Th17 cell differentiation and liver tumor growth. Results confirmed the dramatic downregulation of miR-22 expression in malignant tissues and circulating T cells from patients with HCC. MiR-22 expression correlated with good prognosis of patients. Overexpression of miR-22 impaired the DEN/PB-induced primary HCC formation and the growth of Hepa1-6 subcutaneous tumors by promoting Th17 differentiation. Injection of miR-22-overexpressing T cells in irradiated mice resulted in the inhibition of Hepa1-6 subcutaneous tumor growth via Th17 differentiation promotion. MiR-22 could directly bind to Jarid2, which played an important role during the miR-22-mediated regulation of Th17 differentiation. Taken together, our study expands the understanding of miR-22 function and provides a therapy target for HCC.
Keywords: HCC, miR-22, T cell differentiation, Jarid2, Th17
Introduction
Hepatocellular carcinoma (HCC), usually accompanied with poor prognosis, is the third leading cause of cancer-associated deaths [1]. In particular, the incidence of HCC in China is 5-10 times higher than that in developed countries [2]. Several factors contribute to the risk of HCC, including diabetes, hepatitis virus B or C, genetic metabolic diseases, and alcoholic or nonalcoholic steatohepatitis, [3-6]. Diverse tremendous, including surgery, chemotherapy, radiation therapy, and immunotherapy have developed for HCC treatment, but the patients show bad prognosis and high rates of postoperative recurrence, which was the main reason for the survival rate less than 50% [7]. Thus, it’s important to develop new therapeutic targets and identify prognostic biomarkers for HCC.
Various tumor-supporting cells and tumor epithelium comprised the tumor microenvironment, which is complex and dynamic and has attracted the attention of amount of cancer researchers [8,9]. Immune cells and immunosuppressive cells are the main effector of tumor-supporting cells [8,9]. CD4+ Th17 cells is a subset of CD4+ T cells and provide protective immunity against infections but are also involved in inflammation [10]. In tumor immunity, Th17 cells take a complex and controversial role and exhibit a fluctuating identity in cancer [11,12]. By promoting the secretion of interleukin (IL)-1β, IL-6, IL-23, and transforming growth factor (TGF)-β1 and other inflammatory cytokines, Th17 cells facilitate contact between tumor cells and tumor-derived fibroblasts and consequently promote tumorigenesis [13,14]. However, Th17 cells also cause antitumor immune responses via activating effector CD8+ T cells, or producing interferon (IFN)-γ [15]. Thus, it is necessary to clarifying the underlying mechanism of Th17 function and differentiation.
MiR-22 serves as a biomarker for the prognosis of several cancers and acts as a tumor suppressor. Downregulation of miR-22 in the serum and malignant tissues was shown to correlate with bad prognosis of patients with breast cancer [16,17] and pancreatic cancer [18]. Overexpression of miR-22 could suppress tumorigenesis in breast and gastric cancers [19,20], inhibit epithelial-mesenchymal transition (EMT) in lung and bladder cancers [21,22], and enhance radiosensitivity in small-cell lung and breast cancers [19,23]. In HCC, high miR-22 expression correlated with good prognosis of patients [24]. Serum miR-22 level could be used as an early detection biomarker for HCC [25]. Previous functional investigations have indicated the antitumor effect of miR-22 on the proliferation, tumorigenesis, and metastasis of HCC via targeting YWHAZ, CD147, galectin-1, CCNA2, and Sp1 [26-30]. Upregulation in the expression of miR-22 with berberine and waltonitone was also found to inhibit the proliferation of HCC cells [26,29]. However, the function of miR-22 in the tumor microenvironment and T cell differentiation is indistinct.
In our study, we aimed to investigate the miR-22 expression in the circulating T cells from health people and HCC patients by qPCR. DEN/PB-mediated primary HCC and Hepa1-6 subcutaneous tumor mouse models were established and subjected to lenti-miR-22 treatment. Mice immunoreconstituted with miR-22-overexpressing T cells were used to investigate the function of miR-22 in vivo. Luciferase assay, immunofluorescent staining, in vitro Th17 cell differentiation assay, and rescue experiments were used to investigate the potential mechanism of miR-22-mediated regulation of T cell differentiation and liver tumor growth. The results of our study may expand the understanding of miR-22 function and supply a therapy target for HCC.
Materials and methods
Clinical sample
The HCC patients who underwent hepatectomy and without radiotherapy or chemotherapy prior to the hepatectomy at the Affiliated Eighth People’s Hospital, Jiangsu University between 2013 and 2015 were included in our study. After pathologic diagnosis, the liver tissues samples were used in the present study. The forward tissues of HCC patients were used as normal samples. Informed consent for the collection of tissues for the present study was signed by the patients. The protocols for collection of liver tissue samples was approved by the Medical Ethics Committee of the Jiangsu University.
Animal study
C57BL/6 mice (female, 8- to 10-week-old) were purchased from Hfkbio (Beijing, China) and housed at a SPF (specific pathogen free) condition, with free access to water and food. For primary HCC mice model, C57BL/6 mice received intraperitoneal injections of DEN (20 mg/kg, Sigma, MA, USA), and feeded with sterile containing 0.05% PB. Between 6 to 12 weeks post DEN injection, lentivirus-based miR-22 overexpression system and lenti-control were intravenously injected one time a week. At 52 weeks later, mice were sacrificed and tumors were collected. The liver weight and tumors in each liver were analyzed. For subcutaneous tumor model, 1 × 107 Hepa1-6 cells (dilution in 100 μl sterile PBS) were injected into the subcutaneous of right ventral. Between 11 to 15 days post tumor cell injection, lentivirus-based miR-22 overexpression system and lenti-control were intravenously injected one time a day for five times.
Cultivation of spleen-derived T cells
For T cell isolation, spleens of C57BL/6 mice (8-weeks old) were collected for preparing single-cell leukocyte suspensions. Then, flow cytometry was employed to isolate naive CD4+ T cells. The isolated naive CD4+ T cells were cultured as previous study indicated [31]. Briefly speaking, isolated naive CD4+ T cells were resuspended in 0.5 ml conditional medium (complete RPMI 1640 medium containing 10% FCS (Invitrogen, MA, USA), 200 mmol/L L-glutamine (Invitrogen, MA, USA), 100 U/mL penicillin/streptomycin (Invitrogen, MA, USA), and 4.5 g/L glucose (Invitrogen, MA, USA), 10 mg/mL plate-bound anti-mouse a-CD3 (17A2) and 2 mg/mL soluble a-CD28) and cultured in 48-well flat-bottom plates. CD4+ T cell were infected with lenti-miR-22 over expression system with a MOI of 20.
Th17 differentiation assay
For Th17 differentiation, cultured naive CD4+ T cells were stimulated with 1 mg/mL anti-mouse CD3 (BioLegend), 5 mg/ml anti-mouse CD28 (eBioscience), 10 ng/ml recombinant IL-2 (Biolegend) in RPMI-1640 medium with 10% fetal bovine serum (FBS) with 50 ng/mL IL-6 (BioLegend), 10 ng/mL IL-1β (eBioscience), 10 ng/mL IL-23 (BioLegend), 10 ng/mL TGF-β (eBioscience), 10 μg/mL anti-mouse IFN-γ (eBioscience) and 10 μg/ml anti-mouse IL-4 (BioLegend). After culture for 4 days, the cells were collected for flow cytometry analysis.
Immunoreconstituted mouse model
Immunoreconstituted mice were established as previous study indicated [32]. In short, RS-200 Biological Irradiator was used to irradiate the recipient mice (8- to 10-week-old C57BL/6) with 7.0 Gray. Then 2 × 106 T-miR-22 cells and T-control cells were intravenously injected into the irradiated mice. At 8 weeks after T cells injection, mice blood was collected for miR-22 determination by qPCR.
Quantitative PCR
Liver tissues and cells were collected for total RNA extraction by Trizol (Invitrogen, MA, USA) following the instruction of producer. cDNA was prepared with reverse transcription kit with gDNA eraser (Takara, Tokyo, Japan) was used to synthesize cDNA and SYBR Green (Applied Biosystems; Thermo Fisher Scientific, Inc.) kit was used for qPCR analysis.
The qPCR conditions: denaturation at 94°C for 2 min, amplification at 94°C for 30 s for 40 cycles, annealing at 58°C for 30 s and extension at 72°C for 60 s, followed by a terminal elongation step for 10 min at 72°C. All of the data was calculated following the 2-ΔΔCq method after normalization to U6 [33]. U6 was determined as a internal control. The primer of miR-22 and U6 were purchased from Ruibio (Guangzhou, China) and the sequence was not supplied according to the rule of the company.
H&E staining and immunofluorescent staining
Liver tissues and tumors were fixed with 4% paraformaldehyde for 48 hours before processing into paraffin-embedded blocks. After cutting into 4 μm sections, the slides were then stained with H&E kit (Beyotime, Beijing China). The tumor area in each slide were calculated based on the image of H&E staining. For immunofluorescent staining, the sections were performed for antigen retrieval for 3 mins in citrate under high pressure, after dewaxing and hydration. Then the slides were incubated with primary antibody against Jarid2 (abcam, CA, UK) for overnight at 4°C. The sections were washed with PBS for 3 times and incubated with tetramethylrhodamine-conconjugated secondary antibody (Invitrogen, MA, USA) at 37°C for 1 hour. 4’,6-diamidino-2-phenylindole (DAPI; Beyotime, Beijing, China) was used to stain the nucleus. The Jarid2 positive cells in each section were counted and analyzed.
Flow cytometry
For flow cytometry analysis, the tumors were prepared as previous study indicated [34]. Briefly speaking, tumors were collected, washed with ice-cold PBS, and digested in RPMI-1640 medium containing collagenase IV (0.1%; Gibco), nuclease, and 1% FBS for 40-60 min at 37°C. Then the cells were collected and filtered with 40 μm sieve. Dead and live cells were discriminated by Fixable Viability Stain 620 staining (BD Biosciences, CA, USA). After washing with ice-cold PBS for three times, the cells were fixed and incubated with primary antibodies (FITC-CD4, Cy7-CD8, PE-FoxP3 and APC-IL-17). Then the cells were measured on a NovoCyte flow cytometer.
Western blotting
The tissues and T cells were collected for total protein extraction by lysing with RIPA Lysis Buffer (Beyotime, Beijing, China), containing 1% protease inhibitor cocktail (Roche, MA, USA) for 30 min on ice. Total 20 μg protein samples were used for SDS-PAGE electrophoresis. After transferring to PVDF membranes (Merck Millipore, MA, USA), TBST buffer containing 5% nonfat milk was used to block the membranes for 2 h and immunoblotted with anti-Jarid2 antibody (abcam, CA, USA) for overnight at 4°C. After incubating with peroxidase conjugatedsecondary antibodies (Zs bio, Beijing, China), the bands were detected by an ECL kit (Millipore, Bedford, MA) with iBright 2000 (Thermo Fisher, MA, USA). GAPDH was detected as an internal control.
Luciferase assay
The pMIR-REPORT Luciferase vector based plasmid of Jarid2 3’-UTR and 3’-UTR mutant were supplied by Fulengen (Guangzhou, China). The plasmids (2 μg) were transfected into 293T cells by Lipofectamine 3000 (Invitrogen, MA, USA), accompanied with 1 μg of Renilla luciferase reporter plasmid pRL-SV40. After transfection with these plasmids, lenti-miR-22 and miR-control were used to infect 293T cells separately. At 24 h later, the cells were collected for luciferase activity determination by Dual-Luciferase Reporter Assay System (Promega). Results were normalized to Renilla activity.
Statistical analysis
GraphPad Prism 5 was used for statistical analysis and all of the data were presented as the mean ± standard deviation. The data were analyzed using Students’ t test between two groups and one-way analysis of variance among three or more groups with Dunnett’s multiple comparisons. The survival rate was analyzed by the Kaplan-Meier method. P<0.05 was considered as a statistically significant difference.
Results
Downregulation of miR-22 in T cells from HCC patients
To determine the expression level of miR-22 in HCC, 30 pairs of liver malignant tissues and normal tissues were collected for quantitative PCR analysis. The result demonstrated the significant downregulation of miR-22 expression in malignant tissues, compared with that in normal tissues (Figure 1A). Further analysis indicated the lower levels of miR-22 in the malignant tissues from patients with higher HCC stage (Figure 1B). Based on miR-22 expression, 80 patients were divided into two groups as follows: one group with high miR-22 expression and other with low miR-22 expression. Prognosis analysis demonstrated that those with low miR-22 expression showed poor prognosis and overall survival time (Figure 1C). Similar results were observed in the data from The Cancer Genome Atlas (TCGA) database (Figure 1D). Furthermore, circulating T cells were collected from patients with HCC and healthy controls and the expression of miR-22 was found to be lower in the circulating T cells from patients than in those from the healthy controls (Figure 1E). The expression of miR-22 in circulating T cells was dramatically downregulated in patients with metastasis (Figure 1F). Thus, low expression of miR-22 in circulating T cells and malignant tissues correlated with poor prognosis in HCC patients.
Figure 1.
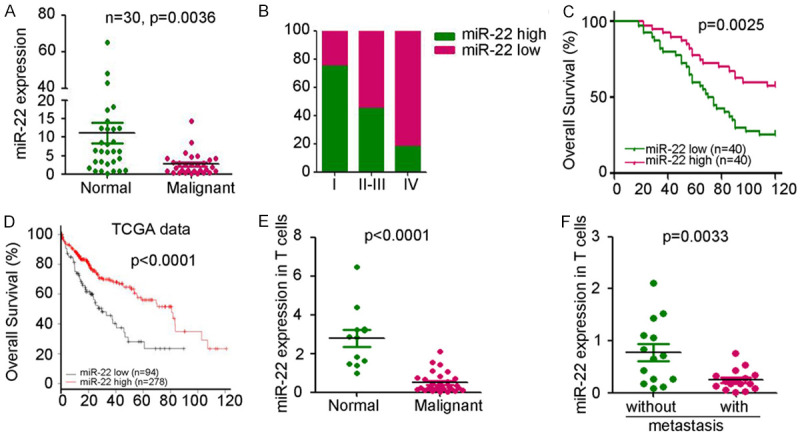
Downregulation of miR-22 in T cells of HCC patients. A. qPCR analysis of miR-22 expression in 30 pairs of liver normal and malignant tissues. U6 was used as a loading control, P=0.0036. B. Differential expression of miR-22 in different stage of patients with HCC. C. The correlation analysis between miR-22 expression and prognosis of HCC patients. The patients were divided based on the median expression of miR-22 in malignant tissues. D. The correlation analysis between miR-22 expression and prognosis of HCC patients from TCGA data base. The patients were divided based on the average expression of miR-22 in malignant tissues. E. qPCR analysis of miR-22 expression in circulating T cells from HCC patients (n=29) and healthy controls (n=11). U6 was used as a loading control, P<0.0001. F. Differential expression of miR-22 in circulating T cells from HCC patients with (n=14) or without metastasis (n=15). (P=0.0033).
MiR-22 inhibits liver tumor growth in mice
To study the functional role of miR-22 in HCC, a diethylnitrosamine (DEN)/phenobarbital (PB)-induced primary liver cancer mouse model was established. Lentivirus-based miR-22 overexpression system and lentivirus-based control were injected at 6 weeks post DEN administration (Figure 2A). After 52 weeks, the liver tissues and T cells from tumors were collected for further analysis. Quantitative PCR (qPCR) analysis confirmed the significant upregulation in the expression of miR-22 in the liver tumor tissues (Figure 2B) and T cells (Figure 2C). Lower tumor weight (Figure 2D) and tumor mass (Figure 2E) were observed in miR-22 treatment group. Hematoxylin and eosin (H&E) staining of the liver tissue also confirmed the lower tumor area for miR-22 treatment group (Figure 2F). To investigate the functional role of miR-22 in HCC, lenti-miR-22 was injected into Hepa1-6 subcutaneous tumors for five times (Figure 2G). The tumors and T cells from tumors were collected for further analysis after sacrificing the mice. qPCR analysis confirmed the significant upregulation of miR-22 expression both in tumors (Figure 2H) and T cells from tumors (Figure 2I) derived from miR-22-injected mice. As shown in Figure 2J-L, the ectopic expression of miR-22 resulted in a significant inhibition in the tumor volume by 70.5% and tumor weight by 70.2%. These results show that the ectopic expression of miR-22 could inhibit HCC growth.
Figure 2.
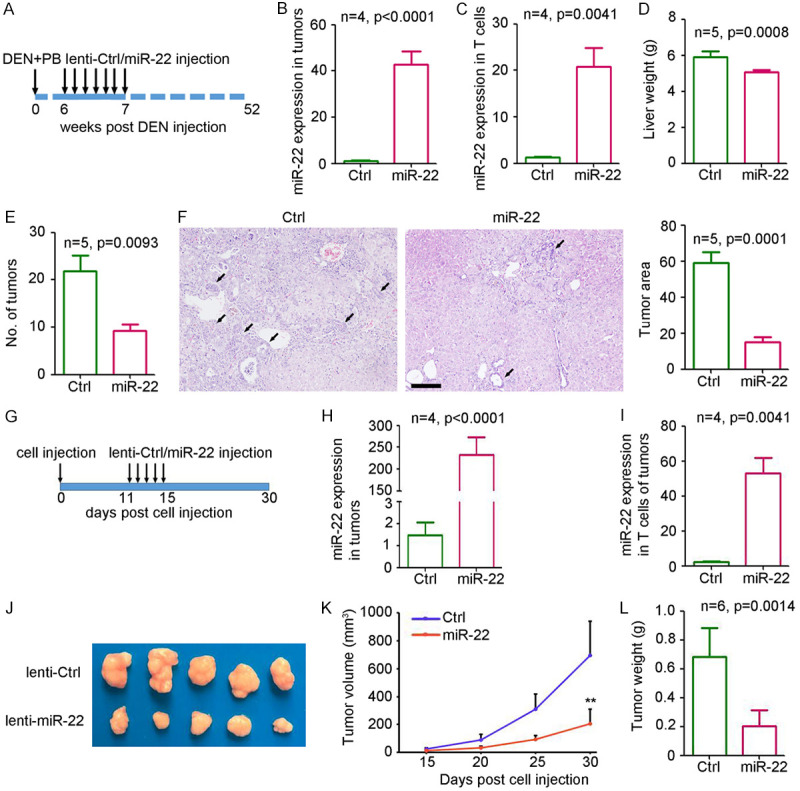
Overexpression of miR-22 inhibits liver tumor growth in mice. A. Process of miR-22 treatment in DEN/PB-mediated primary HCC model. B. qPCR analysis of miR-22 expression in tumor tissues from mice after lenti-ctrl or lenti-miR-22 treatment (n=4, P<0.0001). C. qPCR analysis of miR-22 expression in circulating T cells from mice after lenti-ctrl or lenti-miR-22 treatment (n=4, P<0.0001). D. Weight of liver from mice after lenti-ctrl or lenti-miR-22 treatment (n=5, P=0.0008). E. Number of tumors in mice after lenti-ctrl or lenti-miR-22 treatment (n=5, P=0.0093). F, H. E. staining of livers from mice after lenti-ctrl or lenti-miR-22 treatment. Scale bar =200 μm. Black arrows indicated the tumor area. The percent of tumor area was analyzed in each slides (n=5, P=0.0001). G. Process of miR-22 treatment in Hepa1-6 subcutaneous tumor model. H. miR-22 expression in subcutaneous tumors from lenti-ctrl or lenti-miR-22 treatment group (n=4, P<0.0001). I. miR-22 expression in circulating T cells from lenti-ctrl or lenti-miR-22 treatment group (n=4, P<0.0001). J. Image of tumors from lenti-ctrl or lenti-miR-22 treatment group. K. The curve of tumor volume from lenti-ctrl or lenti-miR-22 treatment group (n=5, **, P<0.01). L. The weight of tumors from lenti-ctrl or lenti-miR-22 treatment group (n=5, **, P=0.0014).
MiR-22 promoted Th17 differentiation
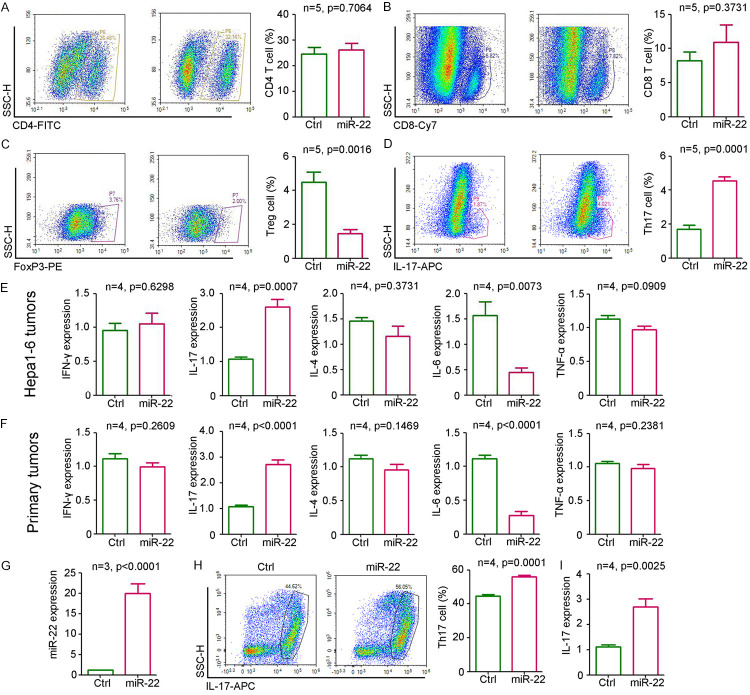
To investigate the function of miR-22 in the tumor microenvironment, T cells from Hepa1-6 subcutaneous tumors were analyzed by flowy cytometry. Our results indicated that the ectopic expression of miR-22 had no significant effect on the infiltration of CD4-positive T cells but promoted the infiltration of CD8-positive T cells in tumors (Figure 3A, 3B). Furthermore, miR-22 inhibited Treg differentiation (Figure 3C) but promoted Th17 differentiation (Figure 3D). Enzyme-linked immunosorbent assay (ELISA) results showed that miR-22 enhanced IL-17 and IFN-γ expression but inhibited IL-6 expression in Hepa1-6 subcutaneous tumors (Figure 3E). No significant change was observed in IL-4 and tumor necrosis factor (TNF)-α levels (Figure 3E). Furthermore, similar expression of cytokines was observed in DEN/PB-induced primary liver tumors (Figure 3F). To confirm the regulatory role of miR-22 on T cell differentiation, the T cells from the spleen were extracted for in vitro experiments. qPCR results confirmed the significant upregulation of miR-22 expression in the T cells infected with lenti-miR22 (Figure 3G). After stimulation with several cytokines (IL-6, IL-1β, IL-23, and TGF-β) and antibodies (anti-IFN-γ and anti-IL-4) for Th17 differentiation induction, more IL-17-positive cells were detected in miR-22-overexpressing T cells (Figure 3H). This was accompanied with a significant upregulation in IL-17 expression (Figure 3I). Thus, miR-22 promoted Th17 differentiation.
Figure 3.
miR-22 regulated T cell differentiation. A-D. Flow cytometry analysis of CD4 T cells, CD8 T cells, FoxP3 positive T cells and IL-17 positive T cells in tumors from lenti-ctrl or lenti-miR-22 treatment group (n=4). E. Expression of IFN-γ, IL-17, IL-4, IL-6 and TNF-α in Hepa1-6 tumors from lenti-ctrl or lenti-miR-22 treatment group (n=4). F. Expression of IFN-γ, IL-17, IL-4, IL-6 and TNF-α in primary liver tumors from lenti-ctrl or lenti-miR-22 treatment group (n=4). G. qPCR analysis of miR-22 expression in T cells that were infected with lenti-miR-22 or control group (n=3, P<0.0001). H. Flow cytometry analysis of IL-17 T positive cells that were infected with lenti-miR-22 or control group (n=4, P=0.0001). I. Determination of IL-17 expression in T cells that were infected with lenti-miR-22 or control group (n=3, P<0.0001).
Injection of miR-22-overexpressing T cells impairs the growth of the liver tumor in mice
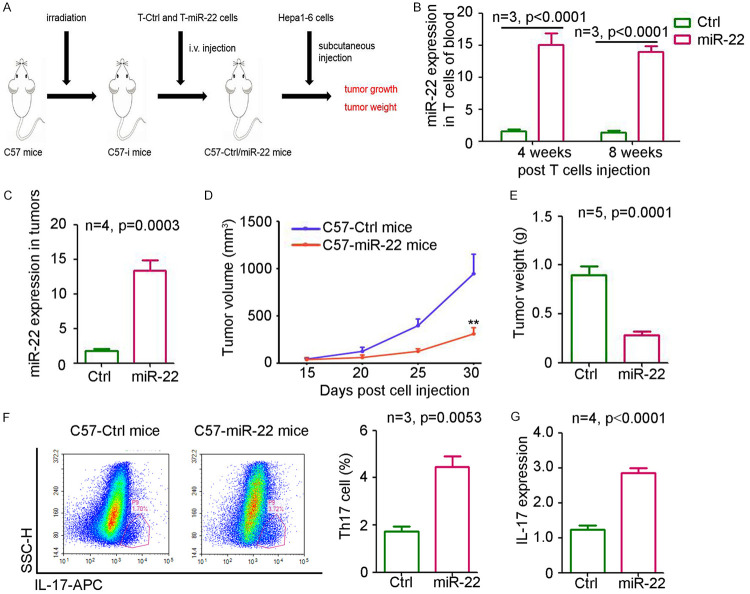
To exclude the antitumor effect of miR-22 on tumor cells, miR-22-overexpressing T cells were used for constructing an immunoreconstituted mouse model. As shown in Figure 4A, miR-22-overexpressing T cells and control cells were injected into irradiated mice, which were then injected with Hepa1-6 cells to generate subcutaneous tumors. The results demonstrated that miR-22 expression in circulating T cells was significant upregulated at 4 and 8 weeks post-injection of miR-22-overexpressing T cells (Figure 4B). Furthermore, a dramatic increase in miR-22 level was also detected in the tumors from mice injected with miR-22-overexpressing T cells (Figure 4C). As shown in Figure 4D, 4E, the injection of miR-22-overexpressing T cells significantly inhibited the tumor volume by 67.5% and tumor weight by 73.3%. More Th17 cells were detected in the tumors from mice injected with miR-22-T cells than in those from the control group (Figure 4F), consistent with the higher levels of IL-17 (Figure 4G). Thus, the injection of miR-22-overexpressing T cells could impair the growth of liver tumors in mice.
Figure 4.
Injection of miR-22 overexpressed T cells impairs liver tumor growth in mice. A. Process of Hepa1-6 subcutaneous cancer model based on immunological reconstitution mice. B. qPCR analysis of miR-22 expression in blood from mice at 4 and 8 weeks post T cells injection (n=3). C. miR-22 expression in subcutaneous tumors from lenti-ctrl or lenti-miR-22 treatment group (n=4, P<0.0001). D. The curve of tumor volume from lenti-ctrl or lenti-miR-22 treatment group (n=5, **, P<0.01). E. The weight of tumors from lenti-ctrl or lenti-miR-22 treatment group (n=5, **, P=0.0014). F. Flow cytometry analysis of IL-17 T positive cells in subcutaneous tumors from lenti-ctrl or lenti-miR-22 treatment group (n=3, P<0.0001). G. Determination of IL-17 expression in subcutaneous tumors from lenti-ctrl or lenti-miR-22 treatment group (n=4, P<0.0001).
MiR-22 directly targets Jarid2
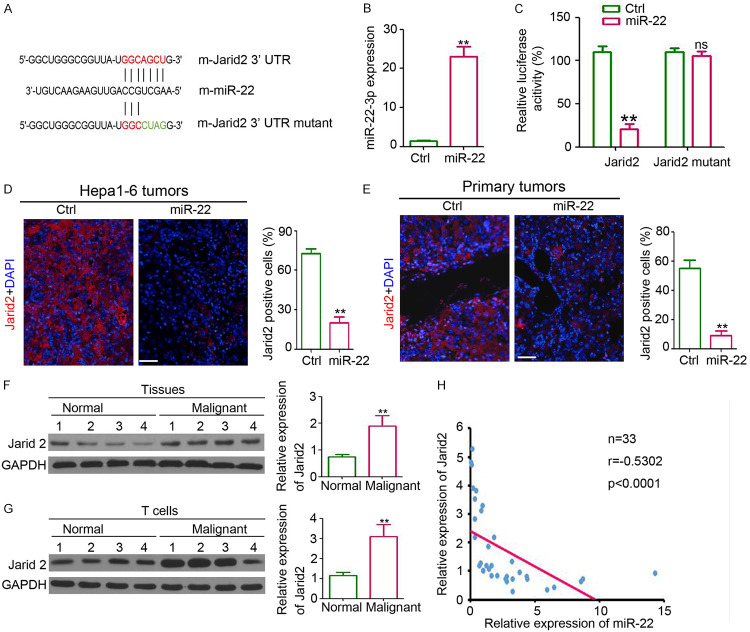
To determine the direct effect of miR-22 on T cell differentiation, the potential direct target of miR-22 was predicted with bioinformatic tools (miRbase and TargetScan). The results revealed a potential binding site of miR-22 on the 3’-untranslated region (UTR) of Jarid2. To determine this analysis, the Jarid2 3’-UTR carrying the miR-22-binding site and Jarid2 mutant 3’-UTR carrying a mutated miR-22 binding site were cloned into the luciferase-report plasmid (Figure 5A). qPCR results showed that the expression of miR-22 in lenti-miR-22-infected cells was higher than that in the control cells (Figure 5B). After luciferase-Jarid2-3’-UTR transfection, the expression of luciferase in miR-22-transfected 293T cells was significantly downregulated (Figure 5C), whereas no dramatic reduction in luciferase expression was observed in luciferase-Jarid2-mutant 3’-UTR-transfected cells (Figure 5C). Immunofluorescence staining showed fewer Jarid2-positive cells in Hepa1-6 tumors (Figure 5D) and primary liver tumors (Figure 5E). We also detected upregulation in Jarid2 expression in malignant tissues (Figure 5F) and circulating T cells (Figure 5G) from patients with HCC. Pearson’s analysis revealed the inverse correlation between miR-22 and Jarid2 in the malignant tissues from patients with HCC (Figure 5H). These results suggest that miR-22 could directly target Jarid2 and inhibit the expression of Jarid2.
Figure 5.
miR-22 regulates Jarid2 expression in T cells. A. Direct binding between miR-22 and Jarid2. B. qPCR analysis of miR-22 expression in HEK293 cells that infected with miR-22 or group (n=4, **, P<0.01). C. Relative luciferase activity in HEK293 cells that infected with miR-22 or group (n=4, **, P<0.01). D. Immunofluorescent staining of Jarid2 expression in Hepa1-6 tumors. Scale bar = 100 μm. Analysis of Jarid2 positive cells in Hepa1-6 tumors (n=4, **, P<0.01). E. Immunofluorescent staining of Jarid2 expression in primary liver tumors. Scale bar = 100 μm. Analysis of Jarid2 positive cells in primary tumors (n=4, **, P<0.01). F. Western blotting analysis of Jarid2 expression in liver normal and malignant tissues. GAPDH was used a loading control. Relative expression analysis of Jarid2 (n=4, **, P<0.01). G. Western blotting analysis of Jarid2 expression in circulating T cells from healthy controls and patients with HCC. GAPDH was used a loading control. Relative expression analysis of Jarid2 (n=4, **, P<0.01). H. Correlation analysis between Jarid2 expression and miR-22 expression in malignant tissues from HCC patients (n=33, r=-0.5302, P<0.0001).
Jarid2 plays an important role during miR-22-mediated regulation of T cell differentiation
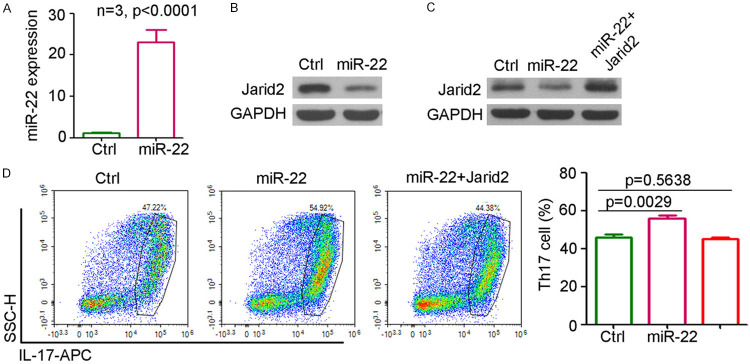
To determine the role of Jarid2 during the miR-22-mediated regulation of T cell differentiation, T cells from the spleen were extracted for in vitro experiments. qPCR analysis confirmed the significant upregulation in miR-22 expression in the T cells infected with lenti-miR-22 (Figure 6A). Western blotting results also suggested that the ectopic expression of miR-22 could inhibit Jarid2 expression in T cells (Figure 6B). Infection of T cells with lenti-Jarid2 overexpression system rescued Jarid2 expression in miR-22-infected T cells (Figure 6C). Th17 differentiation analysis indicated that miR-22 promoted the differentiation of T cells into Th17 cells (IL-17 positive), whereas the ectopic expression of Jarid2 attenuated this effect on Th17 differentiation by miR-22 (Figure 6D). Taken together, Jarid2 plays an important role during the miR-22-mediated regulation of T cell differentiation.
Figure 6.
Jarid2 plays a necessary role during miR-22 regulating T cell differentiation. A. qPCR analysis of miR-22 expression in T cells that infected with miR-22 or control group (n=4, **, P<0.01). B. Western blotting of Jarid2 expression in T cells that infected with miR-22 or control group. C. Western blotting of Jarid2 expression in T cells that infected with miR-22, miR-22+Jarid2 or control group. D. Flow cytometry analysis of IL-17 positive cells in T cells that infected with miR-22, miR-22+Jarid2 or control group (n=4).
Discussion
Our study clarfied the decrease of miR-22 expression in malignant tissues and circulating T cells from patients with HCC and an inverse correlation between miR-22 expression and poor prognosis of patients. Ectopic expression of miR-22 impaired the DEN/PB-induced primary HCC formation and the growth of Hepa1-6 subcutaneous tumors by promoting Th17 differentiation. Injection of miR-22-overexpressing T cells in irradiated mice resulted in the inhibition of Hepa1-6 subcutaneous tumor growth via Th17 differentiation promotion. MiR-22 could directly bind to Jarid2, which played an important role during the miR-22-mediated regulation of Th17 differentiation. Together the results of our study expand the understanding of miR-22 function and provide a therapeutic target for HCC.
Several microRNAs have been employed as prognostic factors in various cancers. Low expression of miR-22 correlates with unfavorable prognosis in osteosarcoma, epithelial ovarian cancer, breast cancer, and gastric cancer [16,35-37]. Furthermore, serum miR-22 level was found to be significantly associated with a short survival and poor prognosis in all patients with breast cancer and different subgroups of B-cell lymphoma [17,38]. In advanced non-small cell lung cancer, circulating miR-22 level could act as a predictive biomarker of pemetrexed-based treatment [39]. Our study also demonstrated that the downregulation of miR-22 expression in malignant tissues correlated with poor prognosis of patients with HCC, consistent with the results of a previous study by Zhang et al [24]. Furthermore, we clarified the downregulation of miR-22 expression in T cells from patients with HCC and the inverse correlation between miR-22 expression in T cells and cancer metastasis. Thus, the downregulated expression of miR-22 in malignant tissues, serum, and T cells acts as a prognostic factor for HCC.
The antitumor effect of miR-22 has been demonstrated in several cancers, including HCC. miR-22 inhibits the proliferation, metastasis, and EMT of HCC [26,27,40]. In the present study, we confirmed the tumor suppressor role of miR-22 in DEN/PB-induced primary liver cancer and Hepa1-6 subcutaneous tumor models. The results observed in DEN/PB-induced primary liver cancer mice demonstrate for the first time the antitumor effects of miR-22 in a spontaneous tumor model. Tumor microenvironment analysis indicated that miR-22 exerted no significant effect on CD4+ T cell infiltration but promoted their differentiation into Th17 cells. Th17 cells play opposite roles under different conditions and exert antitumor effects by activating effector CD8+ T cells [41]. Thus, the upregulation in the expression of CD8+ T cells in miR-22-overexpressing tumors would be attributed to the enhanced population of Th17 cells. IFN-γ overexpression provides further evidences of the antitumor effects of Th17 in miR-22-overexpressing tumors [15,42]. To exclude the direct antitumor effect of miR-22 on tumor cells [43], including inhibition of proliferation, miR-22-overexpressing T cells were injected into irradiated mice to construct an immunoreconstituted mouse model. As a result we found that the injection of miR-22-overexpressing T cells could impair the growth of the liver tumor, suggestive of the miR-22-mediated inhibition of the liver tumor growth through the regulation of the differentiation of T cells into Th17 cells. Thus, miR-22-overpressing T cells impaired liver tumor growth by promoting Th17 cell differentiation, indicative of the important role of miR-22 in cancers.
Jarid2 is a non-catalytic member of the polycomb-repressive complex 2 and functions as an oncogene in different cancers [44-46]. Jarid2 plays an important role in embryonic development [47], vascular patterning [48], and differentiation of lineage-committed cells [49]. A recent study by Renata et al suggested that Jarid2 controls the maturation of invariant natural killer T cells and influences Th17 differentiation through recruiting Polycomb Repressive Complex 2 to epigenetically silence the IL-22 locus. Our study shows that miR-22 directly binds to and inhibits the expression of Jarid2 in T cells. As a result, it promotes the differentiation of Th17 cells in miR-22-overexpressing T cells, consistent with the results of miR-155 [51]. Furthermore, rescuing Jarid2 expression in T cells resulted in the attenuation of the effect on Th17 differentiation mediated by miR-22, suggesting that Jarid2 plays an important role during the miR-22-mediated regulation of Th17 differentiation.
Thus, the present study provides solid evidences on the function of miR-22. These results suggest that miR-22 would be a prognostic factor and a potential therapeutic target for HCC.
Acknowledgements
This work was supported by the Shanghai Xuhui District Medical Peak Subject Project (SHXH201710), The Xuhui District Medical Science and Technology Project (SHXH201709) and the Xuhui District Medical Science and Technology Project (SHXH201736).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Yuen MF, Lai CL. Serological markers of liver cancer. Best Pract Res Clin Gastroenterol. 2005;19:91–99. doi: 10.1016/j.bpg.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Tian Z. HBV-induced immune imbalance in the development of HCC. Front Immunol. 2019;10:2048. doi: 10.3389/fimmu.2019.02048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Couri T, Pillai A. Goals and targets for personalized therapy for HCC. Hepatol Int. 2019;13:125–137. doi: 10.1007/s12072-018-9919-1. [DOI] [PubMed] [Google Scholar]
- 5.Singhal A, Agrawal A, Ling J. Regulation of insulin resistance and type II diabetes by hepatitis C virus infection: a driver function of circulating miRNAs. J Cell Mol Med. 2018;22:2071–2085. doi: 10.1111/jcmm.13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Guo P, Xie X, Wang Y, Chen G. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J Cell Mol Med. 2017;21:648–657. doi: 10.1111/jcmm.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding JJ, El Dika I, Abou-Alfa GK. Immunotherapy in hepatocellular carcinoma: primed to make a difference? Cancer. 2016;122:367–377. doi: 10.1002/cncr.29769. [DOI] [PubMed] [Google Scholar]
- 8.Cheng HS, Lee JXT, Wahli W, Tan NS. Exploiting vulnerabilities of cancer by targeting nuclear receptors of stromal cells in tumor microenvironment. Mol Cancer. 2019;18:51. doi: 10.1186/s12943-019-0971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senthebane DA, Rowe A, Thomford NE, Shipanga H, Munro D, Mazeedi M, Almazyadi HAM, Kallmeyer K, Dandara C, Pepper MS, Parker MI, Dzobo K. The Role of tumor microenvironment in chemoresistance: to survive, keep your enemies closer. Int J Mol Sci. 2017;18:1586. doi: 10.3390/ijms18071586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wacleche VS, Landay A, Routy JP, Ancuta P. The Th17 lineage: from barrier surfaces homeostasis to autoimmunity, cancer, and HIV-1 pathogenesis. Viruses. 2017;9:303. doi: 10.3390/v9100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asadzadeh Z, Mohammadi H, Safarzadeh E, Hemmatzadeh M, Mahdian-Shakib A, Jadidi-Niaragh F, Azizi G, Baradaran B. The paradox of Th17 cell functions in tumor immunity. Cell Immunol. 2017;322:15–25. doi: 10.1016/j.cellimm.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Chang SH. T helper 17 (Th17) cells and interleukin-17 (IL-17) in cancer. Arch Pharm Res. 2019;42:549–559. doi: 10.1007/s12272-019-01146-9. [DOI] [PubMed] [Google Scholar]
- 13.Ye J, Livergood RS, Peng G. The role and regulation of human Th17 cells in tumor immunity. Am J Pathol. 2013;182:10–20. doi: 10.1016/j.ajpath.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey SR, Nelson MH, Himes RA, Li Z, Mehrotra S, Paulos CM. Th17 cells in cancer: the ultimate identity crisis. Front Immunol. 2014;5:276. doi: 10.3389/fimmu.2014.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guery L, Hugues S. Th17 cell plasticity and functions in cancer immunity. Biomed Res Int. 2015;2015:314620. doi: 10.1155/2015/314620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B, Tang H, Liu X, Liu P, Yang L, Xie X, Ye F, Song C, Xie X, Wei W. miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett. 2015;356:410–417. doi: 10.1016/j.canlet.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Shao Y, Yao Y, Xiao P, Yang X, Zhang D. Serum miR-22 could be a potential biomarker for the prognosis of breast cancer. Clin Lab. 2019;65 doi: 10.7754/Clin.Lab.2018.180825. [DOI] [PubMed] [Google Scholar]
- 18.Hussein NA, Kholy ZA, Anwar MM, Ahmad MA, Ahmad SM. Plasma miR-22-3p, miR-642b-3p and miR-885-5p as diagnostic biomarkers for pancreatic cancer. J Cancer Res Clin Oncol. 2017;143:83–93. doi: 10.1007/s00432-016-2248-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Li Y, Wang D, Wei X. miR-22 suppresses tumorigenesis and improves radiosensitivity of breast cancer cells by targeting Sirt1. Biol Res. 2017;50:27. doi: 10.1186/s40659-017-0133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zong W, Feng W, Jiang Y, Cao Y, Ke Y, Shi X, Ju S, Cong H, Wang X, Cui M, Jing R. LncRNA CTC-497E21.4 promotes the progression of gastric cancer via modulating miR-22/NET1 axis through RhoA signaling pathway. Gastric Cancer. 2019;23:228–240. doi: 10.1007/s10120-019-00998-w. [DOI] [PubMed] [Google Scholar]
- 21.Xu M, Li J, Wang X, Meng S, Shen J, Wang S, Xu X, Xie B, Liu B, Xie L. MiR-22 suppresses epithelial-mesenchymal transition in bladder cancer by inhibiting Snail and MAPK1/Slug/vimentin feedback loop. Cell Death Dis. 2018;9:209. doi: 10.1038/s41419-017-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang K, Li XY, Wang ZM, Han ZF, Zhao YH. MiR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharmacol Sci. 2017;21:3598–3604. [PubMed] [Google Scholar]
- 23.Jiang W, Han X, Wang J, Wang L, Xu Z, Wei Q, Zhang W, Wang H. miR-22 enhances the radiosensitivity of small-cell lung cancer by targeting the WRNIP1. J Cell Biochem. 2019;120:17650–17661. doi: 10.1002/jcb.29032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, Wu M, Pan Z, Zhou W. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215–1220. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zekri AN, Youssef AS, El-Desouky ED, Ahmed OS, Lotfy MM, Nassar AA, Bahnassey AA. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol. 2016;37:12273–12286. doi: 10.1007/s13277-016-5097-8. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Wu FX, Luo HL, Liu JJ, Luo T, Bai T, Li LQ, Fan XH. Berberine upregulates miR-22-3p to suppress hepatocellular carcinoma cell proliferation by targeting Sp1. Am J Transl Res. 2016;8:4932–4941. [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M, Hu W, Xiong CL, Qu Z, Yin CQ, Wang YH, Luo CL, Guan Q, Yuan CH, Wang FB. miR-22 targets YWHAZ to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Oncotarget. 2016;7:80751–80764. doi: 10.18632/oncotarget.13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo LJ, Zhang LP, Duan CY, Wang B, He NN, Abulimiti P, Lin Y. The inhibition role of miR-22 in hepatocellular carcinoma cell migration and invasion via targeting CD147. Cancer Cell Int. 2017;17:17. doi: 10.1186/s12935-016-0380-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang F, Gong J, Wang G, Chen P, Yang L, Wang Z. Waltonitone inhibits proliferation of hepatoma cells and tumorigenesis via FXR-miR-22-CCNA2 signaling pathway. Oncotarget. 2016;7:75165–75175. doi: 10.18632/oncotarget.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You Y, Tan JX, Dai HS, Chen HW, Xu XJ, Yang AG, Zhang YJ, Bai LH, Bie P. MiRNA-22 inhibits oncogene galectin-1 in hepatocellular carcinoma. Oncotarget. 2016;7:57099–57116. doi: 10.18632/oncotarget.10981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez Cetina Biefer H, Heinbokel T, Uehara H, Camacho V, Minami K, Nian Y, Koduru S, El Fatimy R, Ghiran I, Trachtenberg AJ, de la Fuente MA, Azuma H, Akbari O, Tullius SG, Vasudevan A, Elkhal A. Mast cells regulate CD4(+) T-cell differentiation in the absence of antigen presentation. J Allergy Clin Immunol. 2018;142:1894–1908. e1897. doi: 10.1016/j.jaci.2018.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai L, Liu Y, Cheng L, Wang H, Lin Y, Shi G, Dong Z, Li J, Fan P, Wang Q, Su X, Zhang S, Yang Y, Hu X, Huang W, Zhou Z, Yu D, Probert C, Wei Y, Deng H. SARI attenuates colon inflammation by promoting STAT1 degradation in intestinal epithelial cells. Mucosal Immunol. 2019;12:1130–1140. doi: 10.1038/s41385-019-0178-9. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Shi G, Yang Q, Zhang Y, Jiang Q, Lin Y, Yang S, Wang H, Cheng L, Zhang X, Li Y, Wang Q, Liu Y, Wang Q, Zhang H, Su X, Dai L, Liu L, Zhang S, Li J, Li Z, Yang Y, Yu D, Wei Y, Deng H. Modulating the tumor microenvironment via oncolytic viruses and CSF-1R inhibition synergistically enhances Anti-PD-1 immunotherapy. Mol Ther. 2019;27:244–260. doi: 10.1016/j.ymthe.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan WN, Zhang YQ, Wang XM, Liu YJ, Zhang YX, Que YH, Zhao WJ, Li P. Down-regulated miR-22 as predictive biomarkers for prognosis of epithelial ovarian cancer. Diagn Pathol. 2014;9:178. doi: 10.1186/s13000-014-0178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G, Shen N, Cheng L, Lin J, Li K. Downregulation of miR-22 acts as an unfavorable prognostic biomarker in osteosarcoma. Tumour Biol. 2015;36:7891–7895. doi: 10.1007/s13277-015-3379-1. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Li F, Zhang Y, Tu Y, Yang Q, Gao X. Reduced expression of miR-22 in gastric cancer is related to clinicopathologic characteristics or patient prognosis. Diagn Pathol. 2013;8:102. doi: 10.1186/1746-1596-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchesi F, Regazzo G, Palombi F, Terrenato I, Sacconi A, Spagnuolo M, Donzelli S, Marino M, Ercolani C, Di Benedetto A, Blandino G, Ciliberto G, Mengarelli A, Rizzo MG. Serum miR-22 as potential non-invasive predictor of poor clinical outcome in newly diagnosed, uniformly treated patients with diffuse large B-cell lymphoma: an explorative pilot study. J Exp Clin Cancer Res. 2018;37:95. doi: 10.1186/s13046-018-0768-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franchina T, Amodeo V, Bronte G, Savio G, Ricciardi GR, Picciotto M, Russo A, Giordano A, Adamo V. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J Cell Physiol. 2014;229:97–99. doi: 10.1002/jcp.24422. [DOI] [PubMed] [Google Scholar]
- 40.Su YH, Huang WC, Huang TH, Huang YJ, Sue YK, Huynh TT, Hsiao M, Liu TZ, Wu AT, Lin CM. Folate deficient tumor microenvironment promotes epithelial-to-mesenchymal transition and cancer stem-like phenotypes. Oncotarget. 2016;7:33246–33256. doi: 10.18632/oncotarget.8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ankathatti Munegowda M, Deng Y, Mulligan SJ, Xiang J. Th17 and Th17-stimulated CD8(+) T cells play a distinct role in Th17-induced preventive and therapeutic antitumor immunity. Cancer Immunol Immunother. 2011;60:1473–1484. doi: 10.1007/s00262-011-1054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilke CM, Kryczek I, Wei S, Zhao E, Wu K, Wang G, Zou W. Th17 cells in cancer: help or hindrance? Carcinogenesis. 2011;32:643–649. doi: 10.1093/carcin/bgr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong J. Emerging roles of microRNA-22 in human disease and normal physiology. Curr Mol Med. 2012;12:247–258. doi: 10.2174/156652412799218886. [DOI] [PubMed] [Google Scholar]
- 44.Cao J, Li H, Liu G, Han S, Xu P. Knockdown of JARID2 inhibits the proliferation and invasion of ovarian cancer through the PI3K/Akt signaling pathway. Mol Med Rep. 2017;16:3600–3605. doi: 10.3892/mmr.2017.7024. [DOI] [PubMed] [Google Scholar]
- 45.Lei X, Xu JF, Chang RM, Fang F, Zuo CH, Yang LY. JARID2 promotes invasion and metastasis of hepatocellular carcinoma by facilitating epithelial-mesenchymal transition through PTEN/AKT signaling. Oncotarget. 2016;7:40266–40284. doi: 10.18632/oncotarget.9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Lyu J, Ji A, Zhang Q, Liao G. Jarid2 enhances the progression of bladder cancer through regulating PTEN/AKT signaling. Life Sci. 2019;230:162–168. doi: 10.1016/j.lfs.2019.05.053. [DOI] [PubMed] [Google Scholar]
- 47.Cho E, Mysliwiec MR, Carlson CD, Ansari A, Schwartz RJ, Lee Y. Cardiac-specific developmental and epigenetic functions of Jarid2 during embryonic development. J Biol Chem. 2018;293:11659–11673. doi: 10.1074/jbc.RA118.002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh BN, Tahara N, Kawakami Y, Das S, Koyano-Nakagawa N, Gong W, Garry MG, Garry DJ. Etv2-miR-130a-Jarid2 cascade regulates vascular patterning during embryogenesis. PLoS One. 2017;12:e0189010. doi: 10.1371/journal.pone.0189010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Raawi D, Jones R, Wijesinghe S, Halsall J, Petric M, Roberts S, Hotchin NA, Kanhere A. A novel form of JARID2 is required for differentiation in lineage-committed cells. EMBO J. 2019;38:e98449. doi: 10.15252/embj.201798449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pereira RM, Martinez GJ, Engel I, Cruz-Guilloty F, Barboza BA, Tsagaratou A, Lio CW, Berg LJ, Lee Y, Kronenberg M, Bandukwala HS, Rao A. Jarid2 is induced by TCR signalling and controls iNKT cell maturation. Nat Commun. 2014;5:4540. doi: 10.1038/ncomms5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escobar TM, Kanellopoulou C, Kugler DG, Kilaru G, Nguyen CK, Nagarajan V, Bhairavabhotla RK, Northrup D, Zahr R, Burr P, Liu X, Zhao K, Sher A, Jankovic D, Zhu J, Muljo SA. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity. 2014;40:865–879. doi: 10.1016/j.immuni.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]