Abstract
The engagement of human angiotensin-converting enzyme 2 (hACE2) and SARS-CoV-2 spike protein facilitate virus spread. Thus far, ACE2 and TMPRSS2 expression is correlated with the epithelial-mesenchymal transition (EMT) gene signature in lung cancer. However, the mechanism for SARS-CoV-2-induced EMT has not been thoroughly explored. Here, we showed that SARS-CoV-2 induces EMT phenotypic change and stemness in breast cancer cell model and subsequently identified Snail as a modulator for this regulation. The in-depth analysis identifies the spike protein (S), but not envelope (E), nucleocapsid (N), or membrane protein (M), of SARS-CoV-2 induces EMT marker changes. Suppression of Snail expression in these cells abrogates S protein-induced invasion, migration, stemness, and lung metastasis, suggesting that Snail is required for SARS-CoV-2-mediated aggressive phenotype in cancer. This study reveals an important oncogenic role of SARS-CoV-2 in triggering breast cancer metastasis through Snail upregulation.
Keywords: SARS-CoV-2, ACE2, TMPRSS2, epithelial-mesenchymal transition, EMT, spike, Snail
Introduction
Coronaviruses have been linked to several human infectious disease outbreaks, including severe acute respiratory syndrome (SARS) in 2002-2003, Middle East respiratory syndrome (MERS) in 2012, and SARS-CoV-2 in December 2019 [1-3]. The coronavirus disease 2019 (COVID-19) pandemic has resulted in a total of 89,048,345 confirmed cases and 1,930,265 deaths globally (WHO situation report, 01/12/2021). Coronavirus is a positive-sense, single-stranded RNA virus that encodes four structural proteins [envelope (E), nucleocapsid (N), spike (S), and membrane protein (M)], as well as auxiliary proteins that can be used for virus replication [4]. The interaction of viral spike (S) glycoprotein and its cell receptor, angiotensin-converting enzyme 2 (ACE2), initiates SARS-CoV-2 entering into host cells [5]. Serine protease TMPRSS2 further cleaves the spike protein allowing efficient virus entry and genome replication in host cells [6-9].
The cellular reprogramming of epithelial-mesenchymal transition (EMT) facilitates cancer metastasis, allowing primary tumor cells to intravasate blood capillaries [10]. Loss of epithelial markers (E-cadherin and ZO-1), gain of mesenchymal markers (N-cadherin, vimentin, and fibronectin), and the increase of migratory and invasive ability are the hallmark of EMT [11]. Transcription repressors such as Zeb-1/2, Twist1, Snail, and Slug induce EMT via EGF, NF-κB, or TGFβ signaling transduction [12]. Upon activation, these transcription factors recruit histone deacetylases to restrict the E-cadherin promoter susceptibility [13]. Tumor cells that undergo EMT are often associated with distant metastasis, leading to drug resistance and poor prognosis in patients [14]. Chemotherapy, targeted therapy, kinase inhibitors, or natural food compounds that revert EMT can be developed as therapeutic agents to reduce metastasis [15]. In this regard, a better understanding of EMT regulation is urgently needed for the design of effective anti-cancer strategies.
A spectrum of viruses can initiate cancer incidence, including Epstein-Barr virus (EBV), hepatitis B virus (HBV), hepatitis C virus (HCV), human Papillomavirus (HPV) and human T-lymphotropic virus 1 (HTLV-1), etc. [16]. In addition, the persistence of virus infection bona fide promotes cancer metastasis [17]. Using virus-induced hepatoma as examples, NS5A of HCV promotes EMT via activating Twist1 [18], and HBx of HBV indirectly activates the Twist1 via STAT3 [19]. Moreover, stabilization of the Snail protein through glycogen synthase kinase-3β (GSK-3β) signaling is an alternative mechanism for HBx facilitating tumor invasion and metastasis [20].
The link between SARS-CoV-2 and cancer progression remains largely unknown. Compared with normal individuals, cancer patients are three times more vulnerable to die from COVID-19 because cancer and its treatment may impair the patient’s immune system, increasing the SARS-CoV-2 infection rate [21]. COVID-19 patients with hematological cancers have the highest mortality rate, followed by lung cancer patients and esophageal cancer. In addition, COVID-19 patients with stage IV metastatic cancer are at high risks of severe illness and death [21,22]. Despite the correlation between ACE2 expression and EMT signature in lung cancer [23], the exact regulatory mechanism of SARS-CoV-2 in EMT regulation has not been unraveled. The current study aimed to dissect the underlying mechanism of SARS-CoV-2 in the EMT progression. We discovered that the spike protein of SARS-CoV-2 increases breast cancer metastatic potentials through Snail but not other EMT modulators. In-depth analysis of the relation between SARS-CoV-2 and cancer metastasis may help reduce COVID-19-mediated cancer aggressiveness.
Materials and methods
Cell cultures and treatments
MCF10A, MCF12A, A549, and HEK293 cells were obtained from American Type Culture Collection. MCF10A and MCF12A cells were cultured in DMEM/F12 medium supplemented with 5% horse serum, 10 μg/ml insulin, 20 ng/ml EGF, and 500 ng/ml hydrocortisone. For transient transfection, cells were transfected with DNA by lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Plasmids
The gene encoding amino acids 1-1273 of the SARS-CoV-2 spike glycoprotein (Gene ID: MN908947) was cloned into the pMD vector (RNAi core, Academia Sinica, Taiwan) to generate a pMD-spike construct for transient transfection. The gene encoding human angiotensin-converting enzyme 2 (ACE2, Gene ID: NM_021804) was cloned into the pLAS2w-pPuro lentiviral vector (RNAi core, Academia Sinica, Taiwan) to generate pLAS-ACE2-Flag construct for establishing MCF10A-ACE2 and MCF12A-ACE2. N, M, E proteins were directly cloned from SARS-CoV-2 (TCDC#4). All constructs were confirmed by enzyme digestion and DNA sequencing.
Antibodies
The following antibodies were used: β-Tubulin (66240-1-Ig; Proteintech, Chicago, IL, USA), N protein (a gift from Dr. An-Suei Yang’s lab, Genomics Research Center, Academia Sinica, Taiwan), IκBα (ab32518; Abcam, Cambridge, MA, USA), Rabbit IgG-HRP (SC-2004; Santa Cruz Biotechnology, Dallas, TX, USA), and Mouse IgG-HRP (SC-2005; Santa Cruz Biotechnology, Dallas, TX, USA). Antibodies for EMT markers were used as previously described [25].
Virus isolation and infection
The SARS-CoV-2 strain used in this study was isolated from a COVID-19 patient in Taiwan (TCDC#4) and passaged on Vero E6 cells grown in MEM supplemented with 2% FBS and incubated at 37°C with 5 % CO2. For the in vitro infection study, target cells were infected at an MOI of 0.1 (2000 pfu/well) with SARS-CoV-2 for 24-48 h. The cells were then fixed with 10% formaldehyde and permeabilized with 0.5% Triton X-100. All procedures followed the Taiwan Centers for Disease Control’s laboratory biosafety guidelines and were conducted in a biosafety level-3 facility in the IBMS, Academia Sinica.
Gene knockdown by recombinant lentivirus-expressing shRNA
The lentiviral-based shRNA (pLKO plasmid) for the knockdown of Snail was purchased from the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan). Transient transfection was performed by adding 2 μg/well of shRNA plasmids and 5 μl/well of Lipofectamine 2000 into cell suspensions. Stable clones expressing the shRNA plasmids were selected in 2 μg/ml puromycin for ten days, and cell clones were stocked for further analysis. Target sequence of shSnail are shSnail-1: 5’-TACAGCTGCTTTGAGCTACAG-3’ and shSnail-2: 5’-GCAAATACTGCAACAAGGAAT-3’.
Human phospho-kinase array analysis
The human phospho-kinase antibody array contains 39 targets, 37 antibodies recognizing phosphorylation form, and two antibodies recognizing the total form (ARY003C, R&D System). The Caco2 were infected with SARS-CoV-2 for 24 hrs, lysed with lysis buffer 6 (#895561, R&D System), and then incubated with the array overnight according to manufacturer protocols. The protein signal was detected by chemifluorescence detection (Bio-Rad, Hercules, CA, USA). The relative intensity of specific protein expression was quantified by Image J software.
Real-time quantitative PCR (RT-qPCR)
Total RNA of MCF10A-ACE2 and HEK293 cells were isolated using Quick-RNA Miniprep Kit (R1055; ZYMO Research, Irvine, CA, USA). The cDNA was prepared using ToolsQuant II Fast RT Kit (KRT-BA06-2; Biotools, Taipei, Taiwan) according to the manufacture’s protocol with 1 μg total RNA. All RT-qPCR reactions were performed in a 10 μL mixture containing 1X iQ™ SYBR® Green supermix (1708880; Bio-Rad, Hercules, CA, USA), 0.5 μmol/L of each primer, and 100 ng of cDNA template. Primers used are as follows: E-cadherin_F: 5’-TGCCCAGAAAATGAAAAAG-3’, E-cadherin_R: 5’-GTGTATGTGGCAATGCGTT-3’, N-cadherin_F: 5’-ACAGTGGCCACCTACAAAG-3’, N-cadherin_R: 5’-CCGAGATGGGGTTGATAAT-3’, Vimentin_F: 5’-GAGAACTTTGCCGTTGAAGC-3’, Vimentin_R: 5’-GCTTCCTGTAGGTGGCAATC-3’, Fibronectin_F: 5’-CAGTGGGAGACCTCGAGAAG-3’, Fibronectin_R: 5’-TCCCTCGGAACATCAGAAAC-3’, Twist1_F: 5’-GGAGTCCGCAGTCTTACG-3’, Twist1_R: 5’-TCTGGAGGACCTGGTAGA-3’, Snail_F: 5’-CCTCCCTGTCAGATGAGG-3’, Snail_R: 5’-CCAGGCTGAGGTATTCCT-3’, ZEB-1_F: 5’-GCACAACCAAGTGCAGAAGA-3’, ZEB-1_R: 5’-TGCACTGAAATCTGTCCAGC-3’, ZO-1_F: 5’-CGAAGGAGTTGAGCAGGAAATCT-3’, ZO-1_R: 5’-TCCACAGGCTTCAGGAACTTG-3’, E protein_F: 5’-ACAGGTACGTTAATAGTTAATAGCGT-3’, E protein_R: 5’-ATATTGCAGCAGTACGCACACA-3’.
Promoter assay
X-tremeGENE HP DNA Transfection Reagent (XTGHP-RO, Roche, Mannheim, Germany) was used to co-transfect the E-cadherin promoter construct [24] with various structural proteins in MCF7 cells. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System kit (E1980, Promega, Madison, WI, USA).
Confocal microscopy
Immunofluorescence was performed as previously described [24]. Briefly, MCF10A-ACE2 transfectants were seeded on Lab-Tek chamber slides one day before the experiment and stained with the indicated antibodies. Slides were examined using a Zeiss Axiovert 200 inverted microscope equipped with a cooled charge-coupled device camera (Carl Zeiss, Thornwood, NY).
Sphere formation assay
For mammary tumor spheroid formation, cells were grown in serum-free conditions with growth factors in an ultra-low adherent plate as described earlier [25]. Briefly, 70~80% confluent cells were trypsinized and washed with PBS. Cells were then suspended into a single cell suspension with complete MammoCultTM Medium (Stemcell Technologies, Vancouver, Canada) at the concentration of 1,000 cells/ml. 2 ml of suspension was added to each well in a 6-well ultra-low adherent plate. Suspension cultures were incubated for five days. Mammospheres with a size of more than 100 mm were counted and imaged.
In vitro cell migration and invasion assays
Invasion assays were performed using Transwell permeable supports (Corning-Costar, Cambridge, MA) with uncoated porous filters (8-μm pore size) as earlier described [25]. Briefly, the filters were precoated with Matrigel matrix (BD Biosciences, Bedford, MA) and air-dried for 2 h. The cells were serum-starved overnight before the experiment. Approximately 5,000 MCF7 cells were placed onto the upper chamber in 0.25 ml serum-free DMEM. After incubation for 2 days, cells that had migrated to the lower surface of the filters were fixed in 4% paraformaldehyde, visualized with 1% crystal violet, and counted.
A mouse model for lung metastasis
Six weeks old female BALB/c mice were purchased from the National Laboratory Animal Center (Taipei, Taiwan) and were housed at the Institute of Biomedical Sciences Animal Care Facility. The tumor metastasis assays were performed using a breast cancer mouse model through intravenous injection. The mouse breast tumor cell line 4T1 was infected with lentivirus-based shRNA clones. 1×105 cells were injected into the lateral tail vein of BALB/c mice (5 mice per group). Ten days later, lung nodules were stained with India ink to determine the ability of metastasis. All animals were handled in compliance with the protocols approved by the Academia Sinica Institutional Animal Care and Utilization Committee.
Statistical analysis
The data from individual experiments are assessed by one-way or two-way ANOVA with Tukey’s post hoc test for multiple comparisons (GraphPad Prism Software Inc, San Diego, CA, USA) and presented as mean ± SD (standard deviation). A p-value < 0.05 was considered statistically significant. The relationships between Ace2/Tmprss2 and Snail in the TCGA breast cancer dataset retrieved from the TCGA database were analyzed by Pearson correlation analysis.
Results
EMT gene signature upregulated upon SARS-CoV-2 infection
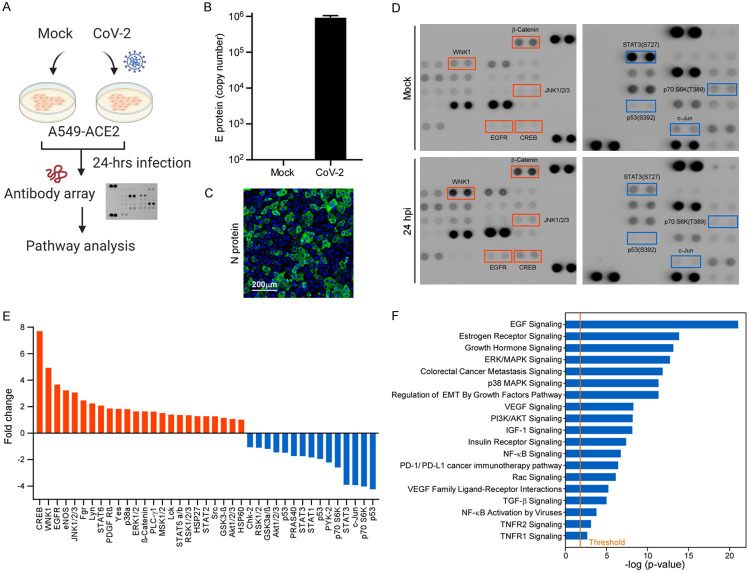
To identify SARS-CoV-2-mediated cancer cell signaling regulation, we performed a non-biased screening on human lung cancer cell line A549 overexpressing the ACE2 receptor (Figure 1A). A549-ACE2 cells were infected with SARS-CoV-2 (MOI of 0.1) for 48 h. The copy number of virus E protein was firstly quantified by qPCR and converted with a DNA standard (Figure 1B). Immunofluorescence microscopy analysis further indicated that over 60% of the infected cells expressed the nucleocapsid of SARS-CoV-2 in the A549-ACE2 (Figure 1C, the anti-N protein signals in the cytoplasm). To identify the downstream signaling, SARS-CoV-2 infected A549-ACE2 cells were harvested and subjected to examine the differential expression of 39 phospho-kinases using an antibody array (Figure 1D). Oncogenic signaling like pEGFR or pErK was upregulated concomitantly with the downregulation of tumor suppressor P53 signaling (Figure 1D). To further examine which oncogenic pathway affected by SARS-CoV-2, we performed an IPA analysis based on our antibody array data. Nineteen canonical pathways related to cancer signaling were upregulated, including EGF signaling, regulation of EMT by growth factors pathway, and TGF-β signaling (Figure 1F). Among these pathways, EMT particularly caught our attention as virus-induced metastasis has been found in many cancers [17].
Figure 1.
SARS-CoV-2 induces cancer cells to undergo EMT. A. Flowchart of the non-biased screening using a phospho-antibody array. B. Virus loading of SARS-CoV-2 in A549-ACE2 cells. Copy number of the viral genome was determined using a single strand DNA standard. C. Immunofluorescence microscopy of N protein expression in SARS-CoV-2 infected A549-ACE2 cells. D. Representative image of antibody array result. E. Quantification results of the human phospho-kinase array. After infected by SARS-CoV-2 for 24 hours, some kinases were either up-regulated or down-regulated. Candidates down-regulated for 2-fold, including p53, STAT3, and c-Jun, were selected. Candidates up-regulated for 3-fold, including STAT2, PLC-γ1, PDGF Rβ, p38α, Lyn, Lck, JNK1/2/3, HSP27, EGFR, and CREB, were chosen. F. IPA analysis of several oncogenic pathways upon SARS-CoV-2 infection in A549-ACE2 cells.
SARS-CoV-2 induces EMT reprogramming in breast epithelial cells
To validate whether SARS-CoV-2 induces EMT, we stably transduced human ACE2 receptor in immortal human normal mammary epithelial cell lines (MCF10A and MCF12A). Upon SARS-CoV-2 infection, the viral genome was effectively replicated in the cells (data not shown). Western blot analysis indicated that the typical EMT markers, N-cadherin, fibronectin, and vimentin, are upregulated concomitantly with downregulation of E-cadherin (Figure 2A). Prolong the virus infection to 48 hours post injection (hpi), EMT markers exhibited enhanced expression, suggesting a positive correlation between virus load and EMT. Next, we compared the mRNA expression of the EMT markers N-cadherin, Vimentin, Fibronectin, and several EMT transcription regulators between SARS-CoV-2 infected and uninfected MCF10A-ACE2 cells (Figure 2B) and confirmed that mRNA levels of these markers were increased in the infected cells. Immunofluorescence microscopy analysis of morphologic changes showed that E-cadherin and vimentin expression are significantly changed upon SARS-CoV-2 infection (Figure 2C). SARS-CoV-2 induces NF-κB activation for cytokine release, and NF-κB is also known to enhance EMT by inducing Twist1 [25] and Snail [26]. We next overexpressed the dominant-negative form of IκBα (IκBα-DN) and found that inhibition of NF-κB reverts SARS-CoV-2-mediated EMT (Figure 2D). Taken together, these results indicate that SARS-CoV-2 contributes to cytoskeleton rearrangement and mesenchymal changes.
Figure 2.
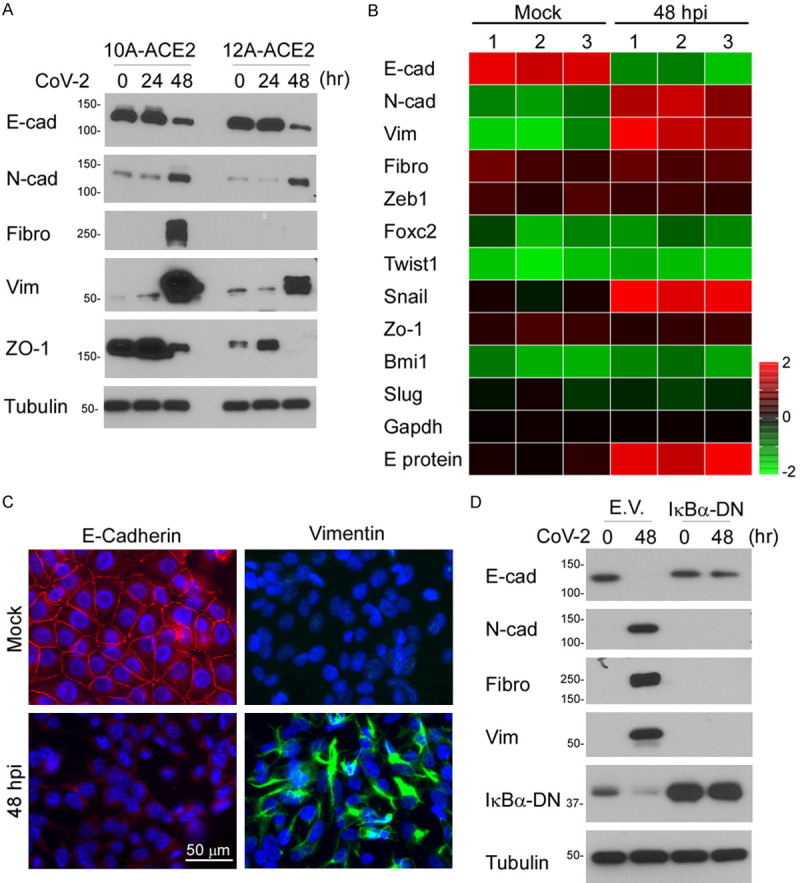
SARS-CoV-2 induces EMT phenotypic changes in breast cancer cells. A. Western blot analysis of EMT markers in MCF10A-ACE2 and MCF12A-ACE2. B. Gene expression heatmap of E-cadherin, N-cadherin, Vimentin, Fibronectin, and EMT regulators in MCF10A-ACE2 cells. C. Morphological changes of MCF10A-ACE2 upon SARS-CoV-2 infection. Expression of EMT markers was detected using anti-E-cadherin and anti-vimentin antibodies by confocal microscopy. Cell nuclei were stained with DAPI. D. Western blot analysis of several EMT markers. A dominant-negative form of IκBα (IκBα-DN) was ectopically expressed in MCF10A-ACE2 cells. E.V. indicates an empty vector.
Snail is positively associated with the expressions of SARS-CoV-2 receptors in breast cancer cells
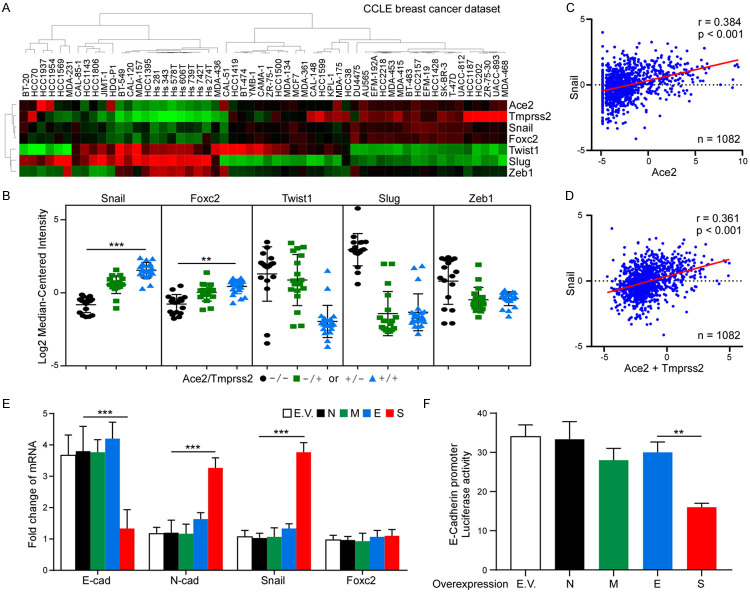
To elucidate the clinical relevance of SARS-CoV-2 infection and EMT regulation, the mRNA expression of EMT regulators was examined by reanalyzing a public dataset generated from 917 cancer cell lines (CCLE) [27]. Unsupervised hierarchical clustering analysis was performed based on the SARS-CoV-2 receptors and EMT regulators, including Snail, Foxc2, Twist1, Slug, and Zeb1 (Figure 3A). A strong correlation was found between Snail, Ace2, and Tmprss2 expression. We next grouped 56 breast cancer cell lines into Ace2-/Tmprss2-, Ace2+/Tmprss2- and Ace2-/Tmprss2+, and Ace2+/Tmprss2+ subtypes based on the gene expression of Ace2 and Tmprss2. Snail and Foxc2 expression were positively correlated with Ace2 and Tmprss2 (Figure 3B). To further confirm our findings in human breast tumors, we investigated the expression of Snail, Twist1, Foxc2, Zeb1, and Slug in 1,082 human primary breast tumor specimens using the TCGA dataset. Among these candidates, only Snail showed a positive association with Ace2 (Figure 3C) and Ace2+Tmprss2 (Figure 3D). To investigate which virus product has the potential to induce EMT progression in breast cancer cells, we transiently expressed the structural proteins of SARS-CoV-2, such as N, M, E, or S, in MCF7 cells. Results from qPCR revealed that only S protein induces downregulation of E-cadherin and upregulation of N-cadherin and Snail (Figure 3E). Similarly, the expression of S in MCF7 cells repressed the E-cadherin promoter activity (Figure 3F), supporting the role of spike protein in facilitating breast cancer cell EMT.
Figure 3.
Snail expression is correlated with ACE2 in breast cancer cells. A. Nonsupervised clustering of 56 breast cancer cell lines based on listed seven genes. Gene expression heatmap showing distinct expression patterns. Heatmap was generated using TreeView. B. Expression plots showing the average expression level of Snail, Foxc2, Twist1, Slug or Zeb1 in Ace2-/Tmprss2-, Ace2+/Tmprss2- and Ace2-/Tmprss2+, and Ace2+/Tmprss2+ subtypes. C. Correlation between Snail and Ace2 expression in TCGA breast cancer patient samples. D. Correlation between Snail and Ace2+Tmprss2 expression in TCGA breast cancer patient samples. E. qPCR analysis of E-cadherin, N-cadherin, Snail, and Foxc2 in MCF7 cells expressing N, M, E, or S. Two-way ANOVA with Tukey’s post hoc test. ***P < 0.001. F. Luciferase reporter assay of E-cadherin. MCF7 cells were transfected with the E-cadherin reporter together with SARS-CoV-2 structural protein N, M, E, or S. One-way ANOVA with Tukey’s post hoc test. **P < 0.01.
Snail is required for spike-induced EMT progression
Upregulation of Snail by S protein prompts us to investigate if Snail is required for S protein-induced EMT. To this end, we knocked down Snail expression in MCF10A-ACE2 cells and then examined the alternation of EMT markers by Western blot. Downregulation of Snail indeed impaired SARS-CoV-2-induced EMT (Figure 4A). To explore the functional significance, we expressed spike protein in MCF7 cells. We found that downregulation of Snail inhibits cell migration (Figure 4B), and invasion (Figure 4C), suggesting that Snail is required for spike-induced EMT phenotype. Because EMT usually accompanies cancer cell stemness, we further investigate the tumorsphere formation ability of MCF7-spike cells (Figure 4D). While spike increases tumorsphere by two-fold, downregulation of Snail compromised spike-mediated cancer stemness (Figure 4D). We next used a xenograft metastasis model to validate the earlier results by overexpressing spike protein in 4T1 cells (4T1-spike). Consistent with the in vitro results, knockdown of Snail antagonized spike-induced metastasis by measuring the number of lung nodules in mice (Figure 4E) (45% lower in shSnail versus shCTRL). Together with the data from cell migration, invasion assay, and stemness, a prerequisite role of Snail in SARS-CoV-2-mediated metastasis was identified (Figure 4F).
Figure 4.

Snail is required for SARS-CoV-2 and spike-induced EMT. A. Western blot analysis of MCF10A-ACE2 cells upon SARS-CoV-2 infection. B. Quantification of migration activity of MCF7-spike cells. C. Quantification of invasion activity of MCF7-spike cells. D. Tumor initiation ability measured by mammosphere formation. MCF7 cells were transiently transfected with indicated plasmids. E. 4T1-spike cells with shSnail stable clones were injected into female BALB/c mice via the tail vein. Lung nodules were stained by India ink at the experimental endpoint. Error bars represent the mean ± SD of five mice. F. Proposed working model of this study. Statistic method: One-way ANOVA with Tukey’s Post Hoc Test. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
With the ongoing pandemic of COVID-19, SARS-CoV-2 not only causes severe pneumonia but exacerbates many pre-existing diseases, such as diabetes, stroke, and cancers. COVID-19 patients with cancer have a higher mortality rate compared to those who do not have cancer [28]. A few observational studies have evaluated the positive association with adverse prognosis in cancer patients compared with those without cancer [29,30]. At the same time, the inflammatory factors triggered by the virus in the tumor microenvironment might implicate reactivating dormant breast cancer cells [31]. Thus, our present study could provide a strong scientific base to support a potential risk of cancer metastasis at the early stage of COVID-19 infection. Several lines of evidence suggest that SARS-CoV-2-mediated Snail expression in breast cancer cells contributes to aggressive phenotype. First, we demonstrated that SARS-CoV-2 upregulates many oncogenic pathways, including EMT. Secondly, Ace2 expression is positively correlated with Snail expression in breast cancer cells. Thirdly, many canonical modules in NF-κB signaling were known to induce Snail expression [26]. Lastly, Snail expression is required for the SARS-CoV-2 or spike-mediated EMT changes. Although Snail contains a functional p65 binding motif on the promoter, we propose that SARS-CoV-2 induced breast cancer EMT is coordinately through canonical NF-κB signaling involved in Snail.
We found several phosphorylated proteins related to EMT by performing the human phospho-kinase array analysis. p38α (Mitogen-activated Protein Kinase 14, MAPK14) signaling pathway plays important roles in the stress response. Moreover, inflammatory cytokines and variant non-stress stimuli can also trigger p38α signaling, leading to numerous critical cell processes [32]. Interruption of the p38α signaling pathway contributes to many human pathologies, and cancer is one of them. Previous studies showed that p38α is related to EMT, which is the initial stage of cancer metastasis [33-35]. In our result, we noticed both p38α and HSP27 are up-regulated after SARS-CoV-2 infection. A previous study confirmed that p38α phosphorylates HSP27 leading to actin polymerization and reorganization into stress fibers [36], an important EMT process. Accumulation of evidence also suggests that p38 MAPK induces EMT in different types of cancers, including melanoma [37], breast cancer [38,39], colon cancer [40], ovarian cancer [41], etc.
In addition to growth factors, microbes can also induce EMT. The patients with gastric cancer, which is caused by Helicobacter pylori, had been reported expression of Snail and Slug, two of the important EMT-related protein, in gastric epithelial cells [42]. The viruses also known to induce EMT. For example, the latent membrane protein 1 and 2A (LMPs) produced by the Epstein-Barr virus (EBV) hijack the host cell signaling and induce EMT [43]. SARS-CoV-2, which belongs to the respiratory infectious virus, was reported to induce EMT-like change through zinc finger E-box-binding homeobox 1 (ZEB1) in lung cancer [23]. However, this kind of change suppresses the viral receptor ACE2 expression, losing the ACE2 protective effect of the acute respiratory distress syndrome (ARDS), the high-mortality complication of COVID-19 [44]. As mentioned previously, p38α plays a key role in EMT. Mizutani et al. discovered SARS-CoV infection triggers the p38 MAPK phosphorylation, leading the downstream targets MAPKAPK-2, HSP27, CREB, and eIF4E phosphorylation in virus-infected cells [45]. Though the study suggested it may be related to cell apoptosis, it may also induce the infected cell to undergo EMT. This finding correlated SARS-CoV-2 infected cells with upregulated phosphorylation of p38α, HSP27, and CREB. The virus infection may also trigger the p38 MAPK pathway to affect the cytoskeleton rearrangement and make EMT-like changes. Therefore, SARS-CoV-2 may lead to respiratory disease and cancer metastasis through p38 MAPK activation.
Acknowledgements
This work was funded in part by the following: Ministry of Science and Technology (MOST 109-2314-B-001-002 and MOST 109-2314-B-001-008 to C.-W.Li). Ministry of Science and Technology (109-2628-B-009-004 to C.-H.Chao). University of Massachusetts Lowell (Faculty start-up D50210000000022 to Y.-J.Lai). Additionally, we thank Taiwan CDC for providing SARS-CoV-2 TCDC#4 (hCoV-19/Taiwan/4/2020) and funding support from Academia Sinica for IBMS P3 facility (AS-CFII-108-102) and the Ministry of Science and Technology, Taiwan for COVID-19 study (MOST 109-3114-Y-001-001). We thank DNA Sequencing Core Facility for their service. The core facility is funded by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-115).
Disclosure of conflict of interest
None.
References
- 1.Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stöhr K, Osterhaus AD. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arabi YM, Balkhy HH, Hayden FG, Bouchama A, Luke T, Baillie JK, Al-Omari A, Hajeer AH, Senga M, Denison MR, Nguyen-Van-Tam JS, Shindo N, Bermingham A, Chappell JD, Van Kerkhove MD, Fowler RA. Middle east respiratory syndrome. N Engl J Med. 2017;376:584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry Depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 10.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 11.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, Weinberg RA. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 14.Hong D, Fritz AJ, Zaidi SK, van Wijnen AJ, Nickerson JA, Imbalzano AN, Lian JB, Stein JL, Stein GS. Epithelial-to-mesenchymal transition and cancer stem cells contribute to breast cancer heterogeneity. J Cell Physiol. 2018;233:9136–9144. doi: 10.1002/jcp.26847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoletov K, Beatty PH, Lewis JD. Novel therapeutic targets for cancer metastasis. Expert Rev Anticancer Ther. 2020;20:97–109. doi: 10.1080/14737140.2020.1718496. [DOI] [PubMed] [Google Scholar]
- 16.Krump NA, You J. Molecular mechanisms of viral oncogenesis in humans. Nat Rev Microbiol. 2018;16:684–698. doi: 10.1038/s41579-018-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Bode AM, Dong Z, Cao Y. The epithelial-mesenchymal transition (EMT) is regulated by oncoviruses in cancer. FASEB J. 2016;30:3001–3010. doi: 10.1096/fj.201600388R. [DOI] [PubMed] [Google Scholar]
- 18.Akkari L, Gregoire D, Floc’h N, Moreau M, Hernandez C, Simonin Y, Rosenberg AR, Lassus P, Hibner U. Hepatitis C viral protein NS5A induces EMT and participates in oncogenic transformation of primary hepatocyte precursors. J Hepatol. 2012;57:1021–1028. doi: 10.1016/j.jhep.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 19.Teng J, Wang X, Xu Z, Tang N. HBx-dependent activation of Twist mediates STAT3 control of epithelium-mesenchymal transition of liver cells. J Cell Biochem. 2013;114:1097–1104. doi: 10.1002/jcb.24450. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Xu L, He H, Zhu Y, Liu J, Wang S, Chen L, Wu Q, Xu J, Gu J. Hepatitis B virus X protein promotes hepatoma cell invasion and metastasis by stabilizing Snail protein. Cancer Sci. 2012;103:2072–2081. doi: 10.1111/cas.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z, Zhang Z, You H, Wu M, Zheng Q, Xiong Y, Xiong H, Wang C, Chen C, Xiong F, Zhang Y, Peng Y, Ge S, Zhen B, Yu T, Wang L, Wang H, Liu Y, Chen Y, Mei J, Gao X, Li Z, Gan L, He C, Shi Y, Qi Y, Yang J, Tenen DG, Chai L, Mucci LA, Santillana M, Cai H. Patients with cancer appear more vulnerable to SARS-CoV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov. 2020;10:783–791. doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathania AS, Prathipati P, Abdul BA, Chava S, Katta SS, Gupta SC, Gangula PR, Pandey MK, Durden DL, Byrareddy SN, Challagundla KB. COVID-19 and cancer comorbidity: therapeutic opportunities and challenges. Theranostics. 2021;11:731–753. doi: 10.7150/thno.51471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart CA, Gay CM, Ramkumar K, Cargill KR, Cardnell RJ, Nilsson MB, Heeke S, Park EM, Kundu ST, Diao L, Wang Q, Shen L, Xi Y, Maria Della Corte C, Fan Y, Kundu K, Pickering CR, Johnson FM, Zhang J, Kadara H, Minna JD, Gibbons DL, Wang J, Heymach JV, Byers LA. SARS-CoV-2 infection induces EMT-like molecular changes, including ZEB1-mediated repression of the viral receptor ACE2, in lung cancer models. bioRxiv. 2020 2020.05.28.122291. [Google Scholar]
- 24.Moreno-Bueno G, Peinado H, Molina P, Olmeda D, Cubillo E, Santos V, Palacios J, Portillo F, Cano A. The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat Protoc. 2009;4:1591–1613. doi: 10.1038/nprot.2009.152. [DOI] [PubMed] [Google Scholar]
- 25.Li CW, Xia W, Huo L, Lim SO, Wu Y, Hsu JL, Chao CH, Yamaguchi H, Yang NK, Ding Q, Wang Y, Lai YJ, Labaff AM, Wu TJ, Lin BR, Yang MH, Hortobagyi GN, Hung MC. Epithelial-mesenchyme transition induced by TNF-a requires NF-kB mediated transcriptional upregulation of Twist1. Cancer Res. 2012;72:1290–300. doi: 10.1158/0008-5472.CAN-11-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15:416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakouny Z, Hawley JE, Choueiri TK, Peters S, Rini BI, Warner JL, Painter CA. COVID-19 and cancer: current challenges and perspectives. Cancer Cell. 2020;38:629–646. doi: 10.1016/j.ccell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z, Zhang Z, You H, Wu M, Zheng Q, Xiong Y, Xiong H, Wang C, Chen C, Xiong F, Zhang Y, Peng Y, Ge S, Zhen B, Yu T, Wang L, Wang H, Liu Y, Chen Y, Mei J, Gao X, Li Z, Gan L, He C, Li Z, Shi Y, Qi Y, Yang J, Tenen DG, Chai L, Mucci LA, Santillana M, Cai H. Patients with cancer appear more vulnerable to SARS-CoV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov. 2020;10:783–791. doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian J, Yuan X, Xiao J, Zhong Q, Yang C, Liu B, Cai Y, Lu Z, Wang J, Wang Y, Liu S, Cheng B, Wang J, Zhang M, Wang L, Niu S, Yao Z, Deng X, Zhou F, Wei W, Li Q, Chen X, Chen W, Yang Q, Wu S, Fan J, Shu B, Hu Z, Wang S, Yang XP, Liu W, Miao X, Wang Z. Clinical characteristics and risk factors associated with COVID-19 disease severity in patients with cancer in Wuhan, China: a multicentre, retrospective, cohort study. Lancet Oncol. 2020;21:893–903. doi: 10.1016/S1470-2045(20)30309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francescangeli F, De Angelis ML, Zeuner A. COVID-19: a potential driver of immune-mediated breast cancer recurrence? Breast Cancer Res. 2020;22:117. doi: 10.1186/s13058-020-01360-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 33.del Barco Barrantes I, Nebreda AR. Roles of p38 MAPKs in invasion and metastasis. Biochem Soc Trans. 2012;40:79–84. doi: 10.1042/BST20110676. [DOI] [PubMed] [Google Scholar]
- 34.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115:3193–3206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 35.Naffa R, Vogel L, Hegedus L, Paszty K, Toth S, Kelemen K, Singh N, Remenyi A, Kallay E, Cserepes M, Tovari J, Grusch M, Enyedi A. P38 MAPK promotes migration and metastatic activity of BRAF mutant melanoma cells by inducing degradation of PMCA4b. Cells. 2020;9:1209. doi: 10.3390/cells9051209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landry J, Huot J. Regulation of actin dynamics by stress-activated protein kinase 2 (SAPK2)-dependent phosphorylation of heat-shock protein of 27 kDa (Hsp77) Biochem Soc Symp. 1999;64:79–89. [PubMed] [Google Scholar]
- 37.Wen SY, Cheng SY, Ng SC, Aneja R, Chen CJ, Huang CY, Kuo WW. Roles of p38alpha and p38beta mitogenactivated protein kinase isoforms in human malignant melanoma A375 cells. Int J Mol Med. 2019;44:2123–2132. doi: 10.3892/ijmm.2019.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parvani JG, Taylor MA, Schiemann WP. Noncanonical TGF-beta signaling during mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2011;16:127–146. doi: 10.1007/s10911-011-9207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, Zhang W, Font-Burgada J, Palmer T, Hamil AS, Biswas SK, Poidinger M, Borcherding N, Xie Q, Ellies LG. Ubiquitin-conjugating enzyme Ubc13 controls breast cancer metastasis through a TAK1-p38 MAP kinase cascade. Proc Natl Acad Sci U S A. 2014;111:13870–13875. doi: 10.1073/pnas.1414358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urosevic J, Garcia-Albéniz X, Planet E, Real S, Céspedes MV, Guiu M, Fernandez E, Bellmunt A, Gawrzak S, Pavlovic M. Colon cancer cells colonize the lung from established liver metastases through p38 MAPK signalling and PTHLH. Nat Cell Biol. 2014;16:685–694. doi: 10.1038/ncb2977. [DOI] [PubMed] [Google Scholar]
- 41.Lu L, Wang J, Wu Y, Wan P, Yang G. Rap1A promotes ovarian cancer metastasis via activation of ERK/p38 and notch signaling. Cancer Med. 2016;5:3544–3554. doi: 10.1002/cam4.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin Y, Grabowska AM, Clarke PA, Whelband E, Robinson K, Argent RH, Tobias A, Kumari R, Atherton JC, Watson SA. Helicobacter pylori potentiates epithelial: mesenchymal transition in gastric cancer: links to soluble HB-EGF, gastrin and matrix metalloproteinase-7. Gut. 2010;59:1037–1045. doi: 10.1136/gut.2009.199794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horikawa T, Yang J, Kondo S, Yoshizaki T, Joab I, Furukawa M, Pagano JS. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res. 2007;67:1970–1978. doi: 10.1158/0008-5472.CAN-06-3933. [DOI] [PubMed] [Google Scholar]
- 44.Cheng H, Wang Y, Wang GQ. Organ-protective effect of angiotensin-converting enzyme 2 and its effect on the prognosis of COVID-19. J Med Virol. 2020;92:726–730. doi: 10.1002/jmv.25785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem Biophys Res Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]