Abstract
Blood culture (BC) processes are critical to the utility of diagnostic testing, bloodstream infection (BSI) management, and antimicrobial resistance (AMR) surveillance. While Uganda has established BC guidelines, often laboratory practice does not meet the desired standards. This compromises pathogen recovery, reliability of antimicrobial susceptibility testing, and diagnostic test utility. This study assessed laboratory BC process outcomes among non-malarial febrile children below five years of age at five AMR surveillance sites in Uganda between 2017 and 2018. Secondary BC testing data was reviewed against established standards. Overall, 959 BC specimens were processed. Of these, 91% were from female patients, neonates, infants, and young children (1–48 months). A total of 37 AMR priority pathogens were identified; Staphylococcus aureus was predominant (54%), followed by Escherichia coli (19%). The diagnostic yield was low (4.9%). Only 6.3% of isolates were identified. AST was performed on 70% (18/26) of identified AMR priority isolates, and only 40% of these tests adhered to recommended standards. Interventions are needed to improve laboratory BC practices for effective patient management through targeted antimicrobial therapy and AMR surveillance in Uganda. Further research on process documentation, diagnostic yield, and a review of patient outcomes for all hospitalized febrile patients is needed.
Keywords: blood culture, bloodstream infections, febrile illness, antimicrobial resistance, antimicrobial susceptibility testing, operational research, SORT IT
1. Introduction
Due to multidrug resistance, bloodstream infections (BSI) are a growing public health concern and a common cause of morbidity and mortality globally [1,2,3,4]. Blood culture (BC) is a widely used laboratory procedure to diagnose BSI, guide antimicrobial therapy, and monitor AMR patterns [5]. BSIs are a major cause of morbidity and mortality among non-malarial febrile (NMF) children, especially in low and middle-income countries (LMICs) [6,7,8]. Common BC isolates reported in Africa include Salmonella enterica (58.4%), Streptococcus pneumoniae (18.3%), Staphylococcus aureus (9.5%), and Escherichia coli (7.3%) [8].
At the minimum, BC testing processes include specimen collection and transportation, culture, pathogen identification, AST, and timely report of results. Outcomes of these processes play a critical role in ensuring early and appropriate patient management. As part of a national AMR surveillance program, Uganda uses routine antibiotic sensitivity testing (AST) to inform treatment guidelines and AMR control strategies. It is intended that AMR surveillance sites adequately identify and perform AST on priority AMR pathogens (Table A1) [9]. However, there is limited laboratory capacity to perform BC and AST in accordance with recommended Clinical and Laboratory Standards Institute (CLSI) guidelines [10]. Good microbiological testing practices consist of standardized procedures for each process with a system for quality indicator monitoring to determine areas for improvements [11]. Microbiological testing and quality capacity gaps affect pathogen recovery and reliability reported for patient management and containment of AMR in Uganda. Monitoring BC processes and performance indicators such as the diagnostic yield, the proportion of specimens undergoing Gram staining, species identification, AST, and reporting of results in line with established standards plays a critical role in ensuring effective patient management and reliable AMR surveillance data [9,12,13,14,15,16,17,18].
Uganda’s Clinical Guidelines (2016) and National Action Plan for AMR (2018–2023) envision timely and reliable microbiology laboratory services, including BC, as an essential component for guiding targeted antimicrobial therapy and AMR containment. However, AMR surveillance capacity-building programs in Uganda suggest recurrent BC laboratory shortfalls [12,13]. This study assessed laboratory processes for specimen collection and transport, culture, pathogen identification, AST, reporting of results, and quality indicator monitoring of BC among NMF children under five years at five AMR surveillance sites in Uganda from October 2017 to September 2018. The study also documented the demographic and clinical characteristics of the affected under-five children, and identified the AMR pathogens and their resistance patterns.
2. Materials and Methods
2.1. Study Design
A cross-sectional study was conducted at the five national AMR surveillance sites using secondary data from routine laboratory BCs from October 2017 to September 2018.
2.2. Setting
2.2.1. General Setting
Uganda is a low-income country in East Africa, with approximately 41.6 million people [19]. In 2020, the population of children below five years was estimated at 6.99 million with the under-five mortality rate at 64 deaths (per 1000 live births) [20]. The national health system is comprised of both private and public sectors. The country’s public health system includes a four-tiered network of hospitals located in 14 health regions managed by the Ministry of Health (MoH).
Five regional referral public hospitals with onsite microbiology laboratories were supported to initiate AMR surveillance in 2016, through provision of technical assistance to ensure the necessary infrastructure and resources were put in place. At a minimum, each AMR surveillance site has access to a network of microbiology laboratories that offer a limited range of diagnostic services including bacterial culture, pathogen identification, and AST using disk diffusion techniques. Peripheral laboratories linked to national reference laboratories (NRLs) provide specialized testing, including advanced/confirmatory pathogen identification and quantitative AST. NRLs also provide technical support to lower-level laboratories through external quality assessments, in-service training, mentorship, and laboratory supervision by subject matter experts and implementing partners.
2.2.2. Specific Setting
The study was conducted at the five regional referral hospitals (RRHs) in Arua, Mbale, Kabale, Jinja, and Mbarara that have participated in a national AMR surveillance program since 2016 (Figure 1). These hospitals were prioritized for diagnostic microbiology capacity building by the MoH in the early efforts to implement a structured AMR surveillance system in line with the AMR National Action Plan. These sites are supported by health sector partners such as the CDC Global Health Security Agenda program to enhance microbiology laboratory capacity for specimen collection, culture, pathogen identification, and AST [12].
Figure 1.
Map showing the distribution of the five AMR surveillance sites and population density for each location in Uganda. Note: The different color coding indicates separate location demonstration.
The AMR surveillance program involves the use of standardized guidelines. Aggregate AST data is reported to the national AMR coordination office to inform AMR containment strategies, and subsequently to WHO via the GLASS portal. Each AMR surveillance site serves a population of at least 500,000 people in its catchment area, with approximately 50 pediatric beds and an annual caseload of 5000 febrile children, under five years of age, admitted. Each surveillance site has access to onsite BC and AST services. All surveillance site laboratories receive technical support from government and health partners to ensure AMR priority pathogens are identified, and AST is performed following CLSI guidelines [10]. Technical support includes the provision and maintenance of BC laboratory equipment, in-service training, mentorship of laboratory and clinical personnel on BC processes, and procurement of reagents and consumables. Pathogens prioritized for AMR surveillance from blood in Uganda include Staphylococcus aureus, Escherichia coli, Klebsiella spp., Acinetobacter spp., Pseudomonas Aeruginosa, and Salmonella spp.
At each surveillance site, specimens from hospitalized NMF children less than five years old were collected in BACTEC™ Plus Aerobic/F bottles and sent to the laboratory for testing. Blood culture bottles were processed onsite using the BD BACTEC™ 9050 automated system as per the manufacturer’s instructions. When bacterial growth was detected, a preliminary Gram stain was performed to determine and report a Gram-stain reaction to guide initial antimicrobial therapy. Consecutively, an inoculum was sub-cultured for species identification and AST, following CLSI guidelines. For quality assurance purposes, each positive blood culture isolate was referred to the national AMR reference laboratory for parallel testing. In addition, the designated AMR reference laboratory performed confirmation of any resistance patterns.
Laboratories were required to report preliminary results (Gram stain) and deliver all culture and AST results (pathogen identification and resistance patterns) to the clinic care unit within a turnaround time of one day from completion of testing. Samples that showed no sign of any growth after seven days were declared negative. All laboratories had a quality control system and were enrolled in an external quality assessment program administered by the reference laboratory.
Laboratory AMR surveillance data were captured in microbiology test request forms, specimen reception logbooks, and WHONET, a free Windows-based database software. Routine AST data generated at the facility level was subsequently submitted to the national AMR surveillance coordination center for validation, use, and reporting to GLASS, in support of the global actions for AMR containment.
2.3. Study Population
The study reviewed and analyzed secondary data from anonymized laboratory blood cultures of hospitalized, NMF children less than five years old collected from October 2017 to September 2018 at the five selected AMR surveillance sites.
2.4. Study Variables and Data Collection
At each AMR surveillance sentinel site, aggregate AST data were retrieved from standard WHONET files and submitted monthly to the national AMR coordination center. This data, by the BC testing process is summarized in Table 1 below:
Table 1.
Summary of study data variables.
| Category of Data | Variable Description | |
|---|---|---|
| 1. | Specimen collection and transportation |
|
| 2. | Blood culture parameters |
|
| 3. | Pathogen identification |
|
| 4. | AST result parameters |
|
| 5. | Reporting of results and transmission parameters |
|
2.5. Data Analysis
Routine AST data captured by surveillance sites in the WHONET software were exported and analyzed using STATA™ version 15. Categorical data were presented as frequencies and proportions in tables and figures.
3. Results
3.1. Demographic and Clinical Characteristics of Participants
A total of 959 blood culture specimens were collected from hospitalized, NMF children less than five years old at five public RRHs in the national AMR surveillance program. Jinja had the highest number of BC specimens collected, followed by Arua, Mbarara, Kabale, and Mbale, respectively (Table 2). During the study period, the majority of processed BC specimens were collected from female children (60% vs. 40% from male children, p-value < 0.001). Neonates, infants, and young children (1–48 months) were the main contributors of processed specimens, accounting for about 91% compared to 9% from those aged 49–59 months, p-value < 0.001). Patient age was not documented for three of the 959 (0.3%) specimens processed. Initial antibiotic exposure before specimen collection was recorded for only 53% of the specimens. Data on dates of fever presentation were not available.
Table 2.
Demographic and clinical characteristics of hospitalized non-malarial febrile children undergoing blood culture at five antimicrobial resistance surveillance sites in Uganda, October 2017–September 2018.
| Characteristic | Jinja n (%) | Arua n (%) | Mbarara n (%) | Kabale n (%) | Mbale n (%) | Total n (%) |
|---|---|---|---|---|---|---|
| Age in months ^ | ||||||
| <1 ¥ | 66 (8.7) | 32 (27.6) | 8 (25.0) | 6 (20.0) | 7 (36.8) | 119 (12.4) |
| 1 to 48 | 622 (81.6) | 81 (69.8) | 24 (75.0) | 14 (46.7) | 12 (63.2) | 753 (78.5) |
| 49–59 | 74 (9.7) | 1 (0.9) | 0 (0.0) | 9 (30.0) | 0 (0.0) | 84 (8.8) |
| Not recorded | 0 (0.0) | 2 (1.7) | 0 (0.0) | 1 (3.3) | 0 (0.0) | 3 (0.3) |
| Sex ^ | ||||||
| Male | 307 (40.3) | 52 (44.8) | 21 (65.6) | 18 (60.0) | 9 (47.4) | 407 (42.4) |
| Female | 455 (59.7) | 64 (55.2) | 11 (34.4) | 12 (40.0) | 10 (52.6) | 552 (57.6) |
| Initial antibiotic exposure | ||||||
| Yes | 0 (0.0) | 44 (37.9) | 0 (0.0) | 0 (0.0) | 9 (47.4) | 53 (5.5) |
| No * | 0 (0.0) | 59 (50.9) | 1 (3.1) | 0 (0.0) | 8 (42.1) | 68 (0.7) |
| Not recorded Ω | 762 (100.0) | 13 (11.2) | 31 (96.9) | 30 (100.0) | 2 (10.5) | 838 (87.3) |
| Total N (%) | 762 (79.5) | 116 (12.1) | 32 (3.3) | 30 (3.1) | 19 (2.0) | 959 |
^ p-value < 0.001; ¥ Neonates; * Hospitalized non-malaria febrile children undergoing blood culture with a record of antibiotic exposure during the current infection at specimen collection; Ω Initial antibiotic exposure during the current infection was not recorded.
3.2. Specimen Collection and Transportation
A total of 959 BC specimens were routinely collected from hospitalized, NMF children less than five years old during the study period. All of the 959 specimens did not have data on specimen volume, time of collection, and culture contamination. All laboratories did not track all prescribed quality indicators related to specimen collection and transportation.
3.3. Blood Culture
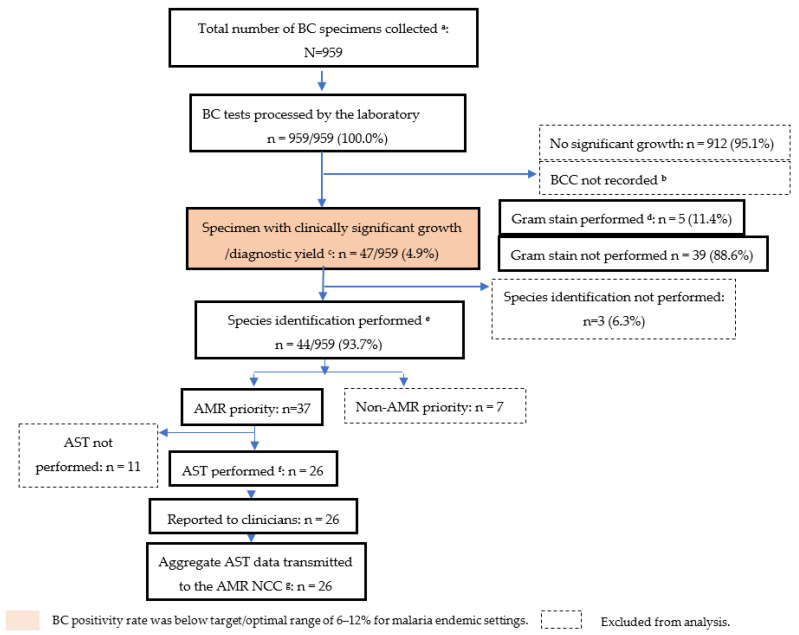
All of the 959 collected specimens underwent a complete laboratory workup to identify organisms. Of these, only 4.9% showed clinically significant growth (diagnostic yield) (Figure 2). BC contamination (BCC) was not monitored at any of the five AMR surveillance sites.
Figure 2.
Cascade of blood culture positivity, isolate identification, and AST among hospitalized, non-malarial febrile patients undergoing blood culture at the five AMR surveillance sites in Uganda, October 2017–September 2018. a One BC specimen bottle collected from each child; b Performance target for BCC rates range from 2–3%; c Performance target for BC diagnsotic yield range from 6–12%. Only one isolate per specimen was included in our analysis; d Performance target for Gram-staining and reporting for all specimens showing clinically significant growth is 100%; e Performance target for pathogen identification and reporting is 100%; f Performance target for AST and reporting for all identified pathogens is 100%; g AST data for all AMR priority pathogens should be reported to clinicians and transmitted to the AMR NCC.
3.4. Pathogen Identification
Table 3 shows BC specimens collected, and the AMR priority pathogens recovered at each AMR surveillance site. Overall, 44 pathogens were recovered and identified, and no polymicrobial infections were reported (i.e., >1 pathogen recovered from one blood specimen). Of these, 37 (84%) were AMR priority pathogens. Staphylococcus aureus was the most predominant isolate (54%), followed by Escherichia coli (19%), Salmonella spp. (11%), Streptococcus pneumoniae (11%), and Klebsiella spp. (5%).
Table 3.
Identified pathogens and yield among hospitalized/non-malarial febrile children undergoing blood culture at five antimicrobial resistance surveillance sites in Uganda, October 2017–September 2018.
| Pathogen | Jinja n | Arua n | Mbarara n | Kabaale n | Mbale n | Total n |
|---|---|---|---|---|---|---|
| AMR Priority Pathogens | ||||||
| Gram-Negative | ||||||
| Escherichia coli | 2 | 1 | 2 | 1 | 1 | 7 |
| Klebsiella spp. | 2 | 0 | 0 | 0 | 0 | 2 |
| Salmonella spp. | 0 | 4 | 0 | 0 | 0 | 4 |
| Gram-positive | ||||||
| Staphylococcus aureus | 7 | 0 | 6 | 4 | 3 | 20 |
| Streptococcus pneumoniae | 2 | 0 | 1 | 1 | 0 | 4 |
| Non-AMR priority pathogens | ||||||
| Staphylococcus epidermidis | 1 | 0 | 0 | 0 | 0 | 1 |
| Pseudomonas spp. | 1 | 0 | 0 | 0 | 0 | 1 |
| Candida spp. | 2 | 0 | 0 | 0 | 0 | 2 |
| Coagulase-Negative Staphylococcus | 3 | 0 | 0 | 0 | 0 | 3 |
| Diagnosic yield π (%) | 20/762 (2.6) | 5/116 (4.3) | 9/32 (28.1) | 6/30 (20.0) | 4/19 (21.0) | 44/959 (4.6%) |
π Diagnostic yield = total number of isolates recovered at each site/total number of blood specimens processed × 100.
3.5. Antimicrobial Susceptibility Testing
AST was performed on only 70% of the identified AMR priority pathogens; non-AMR priority pathogens were excluded from our AST analysis. Table 4 displays resistance patterns of the five AMR priority isolates to the 14 recommended antimicrobial agents under surveillance and laboratory adherence to established AST guidelines (CLSI). Resistance to commonly used antimicrobials was observed among AMR priority pathogens. Staphylococcus aureus, Escherichia coli, Klebsiella spp., and Salmonella spp. were resistant to two or more antimicrobials. Streptococcus pneumoniae isolates were susceptible to all tested antimicrobial agents. Overall, only 40% of these tests adhered to recommended CLSI guidelines. Areas of non-conformity included the following:
-
(1)
none of the recommended antimicrobial agents were tested (28%),
-
(2)
some of the recommended antimicrobial agents were tested (22%), and
-
(3)
incorrect pathogen–antimicrobial agent combination was used (10%).
Table 4.
Compliance with antimicrobial susceptibility testing guidelines and resistance patterns of identified AMR surveillance priority pathogens among hospitalized, non-malarial febrile children undergoing blood culture at five AMR surveillance sites in Uganda, October 2017–September 2018.
| Antibiotic | Gram-Negative Bacteria | Gram-Positive Bacteria | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
E coli, n = 3 |
Klebsiella spp., n = 1 |
Salmonella spp., n = 4 |
S aureus, n = 15 |
S pneumoniae, n = 3 |
||||||
| Tested | Resistant | Tested | Resistant | Tested | Resistant | Tested | Resistant | Tested | Resistant | |
| AMP | 3 | 3 | 1 | 1 | 0 | NR | 0 | NR | 0 | NR |
| FEP | 1 | 0 | 0 | NR | 0 | NR | 1 | 0 | 0 | NR |
| FOX | 0 | NR | 0 | NR | 0 | NR | 0 | NR | 0 | NR |
| CRO | 2 | 1 | 0 | NR | 4 | 0 | 0 | NR | 0 | NR |
| CTX | 1 | 1 | 0 | NR | 0 | NR | 1 | 0 | 1 | 0 |
| CAZ | 1 | 1 | 0 | NR | 0 | NR | 1 | 1 | 0 | NR |
| CIP | 3 | 1 | 1 | 1 | 4 | 4 | 8 | 1 | 0 | NR |
| COL | 0 | NR | 0 | NR | 0 | NR | 1 | 0 | 0 | NR |
| GEN | 1 | 1 | 1 | 1 | 4 | 0 | 3 | 0 | 0 | NR |
| AMK | 2 | 2 | 0 | NR | 0 | NR | 6 | 0 | 1 | 0 |
| MEM | 0 | NR | 0 | NR | 4 | 0 | 2 | 2 | 1 | 0 |
| OXA | 0 | NR | 0 | NR | 0 | NR | 3 | 0 | 0 | NR |
| PEN | 0 | NR | 0 | NR | 0 | NR | 2 | 2 | 0 | NR |
| SXT | 0 | NR | 1 | 1 | 4 | 1 | 5 | 3 | 0 | NR |
| Not recommended and not tested | ||||||||||
| Not recommended but tested in at least one | ||||||||||
| Recommended and all the isolates were tested | ||||||||||
| Recommended but not all the isolates were tested | ||||||||||
| Recommended but none were tested | ||||||||||
Abbreviations: AMR = antimicrobial resistance; AMP = ampicillin; FEP = cefepime; FOX = cefoxitin; CRO = ceftriaxone; CTX = cefotaxime; CAZ = ceftazidime; CIP = ciprofloxacin; COL = colistin; GEN = gentamicin; AMK = amikacin; MEM = -meropenem; OXA = oxacillin; PEN = penicillin G; SXT = co-trimoxazole; NR = not reported.
3.6. Results Delivery and AMR Data Submission
During the study period, all the surveillance site laboratories did not have patient result delivery data (from the laboratory to patient care points/wards).
4. Discussion
This study reported findings of BC testing process outcomes, including sampled patient profiles, AMR priority pathogen etiology, quality management, and compliance to recommended AST guidelines at five RRHs, after the initiation of AMR surveillance. We identified areas for further strengthening and research. There were substantial deficits in BC laboratory process outcomes, potentially limiting blood culture test utilization in routine patient care and introducing bias regarding AMR surveillance data reported nationally.
The high number of hospitalized febrile neonates, infants, and young children (1–48 months) whose BC were processed were consistent with studies in Uganda, Switzerland, and the United States of America [10,12,21,22]. In contrast, the Switzerland study found a majority of bacterial BSI occurring in male children. These observations could be attributed to differences in the study population (children up to 15 years of age).
The low diagnostic yield/pathogen recovery (below the recommended benchmark of 6–12% considered for malaria-endemic settings). These findings are consistent with previous reports in Uganda [12]. Studies beyond Uganda reported a higher diagnostic yield [23,24]. This observation could be due to multiple factors: variations in patient sampling, inadequate specimen volumes, the number of blood specimens drawn per patient, inadequate laboratory quality management, and considerable exposure to antibiotics before sampling, as noted at the surveillance sites of Arua and Mbale that documented clinical information on antibiotic exposure before hospitalization. Antimicrobial administration before blood sample collection at the community and health facilities is common in many resource-limited settings, including Uganda [25,26,27]. Our observation of Staphylococcus aureus and Escherichia coli as the predominant AMR priority pathogens identified, and their resistance patterns were not surprising. These results were similar to prior blood culture studies [4,12,13,21,28,29]. The exception was the predominant Salmonella spp. isolated in Arua RRH, which likely was due to differences in underlying patient conditions, predisposing factors, specimen collection, and laboratory techniques.
Jinja RRH recovered a wider variety of AMR priority pathogens as it had the largest number of specimens processed. Arua RRH was the only surveillance site that recovered Salmonella spp. isolates from blood. Other factors that could influence the recovery of Salmonella spp. include circulating bacterial load, specimen collection timing, volumes of blood collected, and exposure to amicrobial agents before specimen collection [10,24].
Our study highlights considerable gaps in quality of laboratory performance. Most surveillance sites were unable to adhere to basic standards for culture and pathogen identification. There were variations in the identification of bacterial pathogens. The choice of antimicrobial agents for AST was not consistent with national guidelines (40% level of compliance). Laboratory performance on key quality indicators such as specimen volume, turnaround time, and specimen contamination rates was not routinely documented and monitored. Reporting of preliminary Gram-stain results for positive BC specimen was not adequately documented and tracked. Both pediatric and adult studies evaluating laboratory BC culture practices in Uganda, the Democratic Republic of Congo, Nigeria, Nepal, India, Switzerland, and the United States of America similarly reported shortfalls in microbiology laboratory quality capacity as major bottlenecks to BC execution [12,13,14,16,17,18,23,27,30]. These observations emphasize the need for targeted interventions for improving the reliability of laboratory data to guide targeted antimicrobial therapy and appropriately contribute to AMR surveillance.
Challenges related to supply chain reliability for critical laboratory commodities (including BC specimen collection supplies), personnel technical competence, and equipment functionality were potential impediments to the inadequate quality and poor compliance to standard testing guidelines. These observations are comparable to previous studies that reported laboratory supplies, personnel technical competence, and equipment functionality as major impediments to quality test utilization by clinicians [14,16,31]. Consequently, the reliability of AST reported for the national surveillance of AMR is often compromised/biased and may not sufficiently inform treatment guidelines.
To our knowledge, this study is one of the first blood culture testing cascade-analyses at AMR surveillance sites in Uganda. Strengths of our study included the inclusion of regional referral hospitals that pioneered the national AMR surveillance in 2016 and received external technical assistance and resources to enhance AST practices in support of a national AMR surveillance program. This approach minimized bias due to restrictive sampling. A systematic approach to reviewing the entire blood culture process against established technical and quality standards revealed laboratory technical challenges and their impact on patient care and AMR surveillance.
This study was limited by missing data for crucial patient demographics, clinical characteristics, and diagnostic microbiological variables. Whereas specimen collection, transportation and handling, communication of results (Gram stain, bacterial species identification, and AST), and patient outcomes are important factors in blood culture execution, this study did not assess them due to incomplete documentation and missing variables. Because of the low diagnostic yield and number of recovered pathogens (< 20 for the most predominant pathogen), this study did not perform statistical analysis comparing patient demographics and pathogens and conclude on resistance patterns to minimize potential bias. Lastly, our study focused on hospitalized non-malaria febrile children below five years of age, limiting the generalizability of pathogen etiology and their resistance patterns.
The quality of AST data generated and reported to inform treatment guidelines and AMR control strategies nationally and globally was inadequate. Site level capacity-building interventions premised on implementing laboratory procedures and practice changes could substantially improve diagnostic test utility for patient management and AMR surveillance.
5. Conclusions
This study has provided insight into BC testing process indicators, including specimen collection and transportation, bacterial culture, AST, and quality indicator monitoring at the five RRHs, after the initiation of AMR surveillance. Blood culture was mostly performed in neonates, infants, and young children (1–48 months). Staphylococcus aureus, followed by Escherichia coli, were the most predominant AMR priority pathogens. Laboratory BC testing process outcomes did not conform to established testing and quality standards. Interventions to improve BC testing processes, including specimen collection, bacterial culture, AST, and quality indicator monitoring at AMR surveillance sites are needed. If these processes are improved, routine microbiological diagnostic data can adequately guide targeted antimicrobial therapy for patient management, and reliably contribute to AMR surveillance in Uganda. Other studies with improved documentation of routine BC processes, a higher diagnostic yield, and analysis of patient outcomes for all hospitalized febrile patients will be necessary to further assess specimen collection practices and priority pathogen resistance patterns at AMR surveillance sites.
Acknowledgments
This research was conducted through the Structured Operational Research and Training Initiative (SORT IT), a global partnership coordinated by TDR, the Special Programme for Research and Training in Tropical Diseases at the World Health Organization (TDR). The specific SORT IT program that led to these publications included a partnership of TDR with WHO country offices of Ghana, Sierra Leone, and Uganda and was implemented along with The Tuberculosis Research and Prevention Center Non-Governmental Organization, Armenia; The International Union Against Tuberculosis and Lung Diseases, Paris and South East Asia offices; Institute of Tropical Medicine, Antwerp, Belgium; Sustainable Health Systems, Freetown, Sierra Leone; Médecins Sans Frontières- Luxembourg (LuxOR) Centre National de Formation et de Recherche en Santé Rurale de Maferinyah, Guinea; BahirDar University BahirDar, Ethiopia, Makerere, and Lira Universities, and the University of Salford, United Kingdom. Sincere appreciation also goes to the Global Health Security Program at the Infectious Disease Institute, Makerere University, College of Health Sciences for their support in providing the program data used for this analysis. We also express our appreciation to the Uganda National Health Laboratory Services (UNHLS), MoH-Uganda, the National One Health Platform, MoH-Uganda, hospital directors, heads of microbiology laboratory sections, and focal person(s) at Arua, Mbale, Kabale, Jinja, and Mbarara regional referral hospitals (RRHs).
Appendix A
Table A1.
Pathogen–antimicrobial combinations for national AMR surveillance (GLASS).
| Pathogen | Antimicrobial Class | Antimicrobial Agents Used for AST ‡ |
|---|---|---|
| S. pneumoniae | Penicillins | Penicillin G |
| Sulfonamides and trimethoprim | Co-trimoxazole | |
| Third-generation cephalosporins | Ceftriaxone | |
| Staphylococcus aureus | Penicillinase-stable beta-lactams | Cefoxitin b and Oxacillin a |
| Escherichia coli | Sulfonamides and trimethoprim | Co-trimoxazole |
| Fluoroquinolones | Ciprofloxacin | |
| Third-generation cephalosporins | Ceftriaxone and ceftazidime | |
| Fourth-generation cephalosporins | Cefepime | |
| Carbapenems | Meropenem | |
| Polymyxins | Colistin c | |
| Penicillins | Ampicillin | |
| Klebsiella species | Sulfonamides and trimethoprim | Co-trimoxazole |
| Fluoroquinolones | Ciprofloxacin | |
| Third-generation cephalosporins | Ceftriaxone and ceftazidime | |
| Fourth-generation cephalosporins | Cefepime | |
| Carbapenems | Meropenem | |
| Polymyxins | Colistin c | |
| Acinetobacter species | Tetracyclines | Tigecycline |
| Aminoglycosides | Gentamicin and Amikacin | |
| Carbapenems * | Meropenem | |
| Polymyxins | Colistin c | |
| Pseudomonous Aeruginosa | Carbapenems | Meropenem |
| Salmonella spp. | Fluoroquinolones | Ciprofloxacin |
| Third-generation cephalosporins | Ceftriaxone and ceftazidime | |
| Carbapenems * | Meropenem |
Key:‡ The listed antimicrobials are priorities for surveillance of resistance in each pathogen, although they may not be first-line options for treatment. One or more of the drugs listed may be tested. a Oxacillin is a surrogate for testing reduced susceptibility or resistance to penicillin; the AST report to clinicians should state reduced susceptibility or resistance to penicillin. b Cefoxitin is a surrogate for testing susceptibility to oxacillin (methicillin, nafcillin); the AST report to clinicians should state susceptibility or resistance to oxacillin. c Minimum inhibitory concentration (MIC). * Imipenem or meropenem is preferred to represent the group when available.
Author Contributions
Conceptualization, R.K., R.N., J.v.G., and M.L.; methodology, R.K., R.N., J.v.G., A.D., F.E.K., K.R., and M.L.; software, R.K., K.T., T.K., M.O.; validation, R.K., R.N., J.v.G., F.E.K., K.R., K.T., and P.T.; formal analysis, R.K., R.N., J.v.G., T.K.; investigation, R.K., R.N., J.v.G., A.D., P.T., F.E.K., R.W., F.K., I.M., K.R., T.K., M.S.O., M.O., and M.L.; resources, R.K., F.K., R.W., and M.L.; data curation, R.K., M.O., and K.R.; writing—original draft preparation, R.K., R.N., J.v.G., K.R., T.K., and M.L.; writing—review and editing, R.K., R.N., J.v.G., F.K, R.W., M.S.O., and M.L.; visualization, R.K., P.T., and T.K.; supervision, R.K., F.E.K., R.W., I.M., M.S., and M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by the SORT IT AMR program funded by the National Institute of Health Research, Department of Health & Social Care of the United Kingdom, and supported by implementing partners.
Institutional Review Board Statement
Permission to use the national AMR surveillance data was obtained from the Commissioner, National Health Laboratory Services, MoH-Uganda. Ethics approval was received from the Makerere University School of Biomedical Sciences Research and Ethics Committee (Approval No. SBS-REC-008 dated 6th Jan 2021). International ethics approval was also obtained from the Ethics Advisory Group of the International Union against Tuberculosis and Lung Disease, Paris, France (Approval No. EAG 79/19 dated 29/10/2019). The research ethics committee review was also received from the Infectious Diseases Institute (IDI) Scientific Review Committee (SRC) (review no. 24/2020 dated 25th September 2020).
Informed Consent Statement
As this study used anonymized program data without identifiers, the issue of informed consent did not apply.
Data Availability Statement
The data presented in this study is available on request (due to ethics and confidentiality restrictions) from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, data collation, analyses, data interpretation, manuscript writing, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goodman C.W., Brett A.S. Gabapentin and Pregabalin for Pain—Is Increased Prescribing a Cause for Concern? N. Engl. J. Med. 2017;377:411–414. doi: 10.1056/NEJMp1704633. [DOI] [PubMed] [Google Scholar]
- 2.Aung A.K., Skinner M.J., Lee F.J., Cheng A.C. Changing epidemiology of bloodstream infection pathogens over time in adult non-specialty patients at an Australian tertiary hospital. Commun. Dis. Intell. Q. Rep. 2012;36:E333–E341. [PubMed] [Google Scholar]
- 3.Kern W.V., Rieg S. Burden of bacterial bloodstream infection—A brief update on epidemiology and significance of multidrug-resistant pathogens. Clin. Microbiol. Infect. 2020;26:151–157. doi: 10.1016/j.cmi.2019.10.031. [DOI] [PubMed] [Google Scholar]
- 4.Tumuhamye J., Sommerfelt H., Bwanga F., Ndeezi G., Mukunya D., Napyo A., Nankabirwa V., Tumwine J.K. Neonatal sepsis at Mulago national referral hospital in Uganda: Etiology, antimicrobial resistance, associated factors and case fatality risk. PLoS ONE. 2020;15:e0237085. doi: 10.1371/journal.pone.0237085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J., Vincent J.L., Adhikari N.K.J., Machado F.R., Angus D.C., Calandra T., Jaton K., Giulieri S., Delaloye J., Opal S., et al. Sepsis: A roadmap for future research. Lancet Infect. Dis. 2015;15:581–614. doi: 10.1016/S1473-3099(15)70112-X. [DOI] [PubMed] [Google Scholar]
- 6.Shrestha P., Dahal P., Ogbonnaa-Njoku C., Das D., Stepniewska K., Thomas N.V., Hopkins H., Crump J.A., Bell D., Newton P.N., et al. Non-malarial febrile illness: A systematic review of published aetiological studies and case reports from Southern Asia and South-eastern Asia, 1980–2015. BMC Med. 2020;18:1–17. doi: 10.1186/s12916-020-01745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sears D., Mpimbaza A., Kigozi R., Sserwanga A., Chang M.A., Kapella B.K., Yoon S., Kamya M.R., Dorsey G., Ruel T. Quality of inpatient pediatric case management for four leading causes of child mortality at six government-run Ugandan hospitals. PLoS ONE. 2015;10:e0127192. doi: 10.1371/journal.pone.0127192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy E.A., Shaw A.V., Crump J.A. Community-acquired bloodstream infections in Africa: A systematic review and meta-analysis. Lancet Infect. Dis. 2010;10:417–432. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization (WHO) Global Antimicrobial Resistance Surveillance System. Volume 36 WHO; Geneva, Switzerland: 2015. [Google Scholar]
- 10.Towns M.L., Jarvis W.R., Hsueh P.R. Guidelines on Blood Cultures. J. Microbiol. Immunol. Infect. 2010;43:347–349. doi: 10.1016/S1684-1182(10)60054-0. [DOI] [PubMed] [Google Scholar]
- 11.Plebani M. Appropriateness in programs for continuous quality improvement in clinical laboratories. Clin. Chim. Acta. 2003;333:131–139. doi: 10.1016/S0009-8981(03)00177-3. [DOI] [PubMed] [Google Scholar]
- 12.Lamorde M., Mpimbaza A., Walwema R., Kamya M., Kapisi J., Kajumbula H., Sserwanga A., Namuganga J.F., Kusemererwa A., Tasimwa H., et al. A Cross-Cutting Approach to Surveillance and Laboratory Capacity as a Platform to Improve Health Security in Uganda. Health Secur. 2018;16:S76–S86. doi: 10.1089/hs.2018.0051. [DOI] [PubMed] [Google Scholar]
- 13.Kajumbula H., Fujita A.W., Mbabazi O., Najjuka C., Izale C., Akampurira A., Aisu S., Lamorde M., Walwema R., Bahr N.C., et al. Antimicrobial drug resistance in blood culture isolates at a tertiary hospital, Uganda. Emerg. Infect. Dis. 2018;24:174–175. doi: 10.3201/eid2401.171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buehler S.S., Madison B., Snyder S.R., Derzon J.H., Cornish N.E., Saubolle M.A., Weissfeld A.S., Weinstein M.P., Liebow E.B., Wolk D.M. Effectiveness of practices to increase timeliness of providing targeted therapy for inpatients with bloodstream infections: A laboratory medicine best practices systematic review and meta-analysis. Clin. Microbiol. Rev. 2015;29:59–103. doi: 10.1128/CMR.00053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemigisha E., Nanjebe D., Ii Y.B., Langendorf C., Aberrane S., Nyehangane D., Nackers F., Mueller Y., Charrel R., Murphy R.A., et al. Antimicrobial treatment practices among Ugandan children with suspicion of central nervous system infection. PLoS ONE. 2018;13:e0205316. doi: 10.1371/journal.pone.0205316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chukwuemeka I., Samuel Y. Quality assurance in blood culture: A retrospective study of blood culture contamination rate in a tertiary hospital in Nigeria. Niger. Med. J. 2014;55:201. doi: 10.4103/0300-1652.132038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bard J.D., TeKippe E.M.E. Diagnosis of bloodstream infections in children. J. Clin. Microbiol. 2016;54:1418–1424. doi: 10.1128/JCM.02919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pradhan R., Shrestha U., Gautam S.C., Thorson S., Shrestha K., Yadav B.K., Kelly D.F., Adhikari N., Pollard A.J., Murdoch D.R. Bloodstream Infection among Children Presenting to a General Hospital Outpatient Clinic in Urban Nepal. PLoS ONE. 2012;7:e47531. doi: 10.1371/journal.pone.0047531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uganda Bureau of Statistics National Mid Year Population Projections by Single Age, 2015–2050. UBOS; Kampala, Uganda: 2015. [Google Scholar]
- 20.National Population Council . State of Uganda Population Report 2019. NPC; Kampala, Uganda: 2019. [Google Scholar]
- 21.Buetti N., Atkinson A., Kottanattu L., Bielicki J., Marschall J., Kronenberg A., Auckenthaler R., Cherkaoui A., Dolina M., Dubuis O., et al. Patterns and trends of pediatric bloodstream infections: A 7-year surveillance study. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:537–544. doi: 10.1007/s10096-016-2830-6. [DOI] [PubMed] [Google Scholar]
- 22.Biondi E.A., Mischler M., Jerardi K.E., Statile A.M., French J., Evans R., Lee V., Chen C., Asche C., Ren J., et al. Blood culture time to positivity in febrile infants with bacteremia. JAMA Pediatr. 2014;168:844–849. doi: 10.1001/jamapediatrics.2014.895. [DOI] [PubMed] [Google Scholar]
- 23.Kalonji L.M., Post A., Phoba M.F., Falay D., Ngbonda D., Muyembe J.J., Bertrand S., Ceyssens P.J., Mattheus W., Verhaegen J., et al. Invasive salmonella infections at multiple surveillance sites in the Democratic Republic of the Congo, 2011–2014. Clin. Infect. Dis. 2015;61:S346–S353. doi: 10.1093/cid/civ713. [DOI] [PubMed] [Google Scholar]
- 24.Baron E.J., Weinstein M.P., Dunne W.M., Yagupsky P., Welch D.F., Wilson D.M. Cumitech 1C, Blood Cultures IV. ASM Press; Washington, DC, USA: 2005. [Google Scholar]
- 25.Scheer C.S., Fuchs C., Gründling M., Vollmer M., Bast J., Bohnert J.A., Zimmermann K., Hahnenkamp K., Rehberg S., Kuhn S.O. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: A prospective clinical cohort study. Clin. Microbiol. Infect. 2019;25:326–331. doi: 10.1016/j.cmi.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Mukonzo J.K., Namuwenge P.M., Okure G., Mwesige B., Namusisi O.K., Mukanga D. Over-the-counter suboptimal dispensing of antibiotics in Uganda. J. Multidiscip. Healthc. 2013;6:303–310. doi: 10.2147/JMDH.S49075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris A.M., Bramley A.M., Jain S., Arnold S.R., Ampofo K., Self W.H., Williams D.J., Anderson E.J., Grijalva C.G., McCullers J.A., et al. Influence of antibiotics on the detection of bacteria by culture-based and culture-independent diagnostic tests in patients hospitalized with community-acquired pneumonia. Open Forum Infect. Dis. 2017;4 doi: 10.1093/ofid/ofx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parajuli N.P., Parajuli H., Pandit R., Shakya J., Khanal P.R. Evaluating the trends of bloodstream infections among pediatric and adult patients at a teaching hospital of Kathmandu, Nepal: Role of drug resistant pathogens. Can. J. Infect. Dis. Med. Microbiol. 2017;2017 doi: 10.1155/2017/8763135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buys H., Muloiwa R., Bamford C., Eley B. Klebsiella pneumoniae bloodstream infections at a South African children’s hospital 2006-2011, a cross-sectional study. BMC Infect. Dis. 2016;16:1–10. doi: 10.1186/s12879-016-1919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dharmapalan D., Shet A., Yewale V., Sharland M. High reported rates of antimicrobial resistance in Indian neonatal and pediatric blood stream infections. J. Pediatric Infect. Dis. Soc. 2017;6:e62–e68. doi: 10.1093/jpids/piw092. [DOI] [PubMed] [Google Scholar]
- 31.Dailey P.J., Osborn J., Ashley E.A., Baron E.J., Dance D.A.B., Fusco D., Fanello C., Manabe Y.C., Mokomane M., Newton P.N., et al. Defining system requirements for simplified blood culture to enable widespread use in resource-limited settings. Diagnostics. 2019;9:10. doi: 10.3390/diagnostics9010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study is available on request (due to ethics and confidentiality restrictions) from the corresponding author.