Abstract
The microbiota of female reproductive tract have attracted considerable attention in recent years due to their effects on host fitness. However, the microbiota throughout the chicken oviduct and its symbiotic relationships with the host have not been well characterized. Here, we characterized the microbial composition of six segments of the reproductive tract, including the infundibulum, magnum, isthmus, uterus, vagina and cloaca, in pedigreed laying hens with phenotypes of egg quality and quantity. We found that the microbial diversity gradually increased along the reproductive tract from the infundibulum to the cloaca, and the microbial communities were distinct among the cloaca, vagina and four other oviductal segments. The magnum exhibited the lowest diversity, given that the lysozyme and other antimicrobial proteins are secreted at this location. The results of correlation estimated showed that the relationship between host genetic kinship and microbial distance was negligible. Additionally, the genetically related pairwise individuals did not exhibit a more similar microbial community than unrelated pairs. Although the egg might be directly contaminated with potential pathogenic bacteria during egg formation and oviposition, some microorganisms provide long-term benefits to the host. Among these, we observed that increased abundance of vaginal Staphylococcus and Ralstonia was significantly associated with darker eggshells. Meanwhile, vaginal Romboutsia could be used as a predictor for egg number. These findings provide insight into the nature of the chicken reproductive tract microbiota and highlight the effect of oviductal bacteria on the process of egg formation.
Key words: chicken reproductive tract, microbiota, host genetics, microbial contamination, egg quality
INTRODUCTION
All animals host a diversity of microbial communities in and on their bodies (Ursell et al., 2012), and these microbiota can significantly influence host biology (Gill et al., 2006; Nicholson et al., 2012). Numerous studies have focused on the gut microbiota (Ussar et al., 2015; Gensollen et al., 2016), whereas relatively little is known about the microbiota throughout the reproductive tract. As the crucial component of reproductive system, the genital tract of female also harbours a mass of microorganisms in the endosomatic lumen surfaces, representing a finely balanced mutualistic association.
In humans, the vaginal microbiota is dominated by taxa of Lactobacillus or other lactic acid bacteria that maintain a low pH and prevent pathogen infection (Ravel et al., 2011). This characterization is largely different from other mammals including nonhuman primates (Yildirim et al., 2014), cows, pigs, and sheep, in which the relative abundance of Lactobacillus accounts for less than 1% and the pH of the vagina is near neutral (Miller et al., 2016). Although the specific roles of the microbiota in the reproductive tract have not been precisely illustrated, microbiota dysbiosis is related to bacterial vaginosis (Ma et al., 2012) and uterine-related diseases (Chen et al., 2017). Several studies have indicated that sex steroid hormones (Larsen et al., 1977; Bezirtzoglou et al., 2008) and mucosal immunity (Mehta et al., 2020) might play crucial roles in driving the composition and abundance of the genital microbiota.
The reproductive tract of female poultry is not the site for embryo development but the formation of each compartment of eggs, including egg yolk (ovary), albumen (magnum), eggshell membrane (isthmus) and eggshell (uterus) (Sah and Mishra, 2018). Hence, the avian oviduct exhibits a close relationship with egg internal and external qualities. A detailed understanding of the oviductal microbiota is important for the improvement of egg production. However, limited studies have been performed on the reproductive tract microbiota in avian species. Previous studies indicate that uterine proteins or metabolites can affect the process of eggshell calcification (Gautron et al., 1997) and determine eggshell quality represented by its mechanical properties (eggshell strength and thickness) (Sun et al., 2013) and appearance (eggshell colour and gross) (Nys et al., 1991). In addition, the microbiota colonizing the reproductive tract also importantly impacts the process of egg formation. The overgrowth of Gallibacterium anatis has a causal role in salpingitis in egg-laying chickens, resulting in reduced egg production (Zhang et al., 2017).
Recently, Lee et al. (2019) characterized the microbial communities of the chicken oviduct and found a relatively high abundance of Pseudomonas and high similarity between the embryo and egg white, suggesting the transfer of maternal oviduct microbiota to the embryo through the egg white, which is consistent with one view in mammals that the foetus initially colonizes the microbiota from the placenta in utero (Collado et al., 2016). Depending on the location within the female reproductive tract, the microbiota could be directly transmitted into the yolk, egg white, eggshell membrane and eggshell during egg formation. Poultry egg are one of the major nutrient sources for humans worldwide (Réhault-Godbert et al., 2019), and their contamination with microorganisms can lead to spoilage and pathogen transmission, inducing food-borne infection in consumers (Salihu et al., 2015). For instance, microorganisms of Escherichia coli, Salmonella and Campylobacter have been isolated from egg surfaces and contents (Schoeni and Doyle, 1994; Hope et al., 2002). In addition to horizontal transmission by penetration through the eggshell, bacterial contamination can also occur through the trans-oviduct route (Gantois et al., 2009).
Hence, characterizing the microbiota along the avian oviduct is crucial for the assessment of the production of safe and good-quality eggs. In the current study, we performed 16S rRNA sequencing on the chicken reproductive tract from six segments (including the cloaca, vagina, uterus, isthmus, magnum and infundibulum) to comprehensively characterize the microbial composition of each segment, examine the host genetic contribution to the microbial community, and further investigate the contribution of the oviductal microbiota to chicken egg quality traits.
MATERIALS AND METHODS
Ethics Statement
The protocol was approved by the Animal Care and Use Committee of China Agricultural University.
Animals
A pure line derived from Rhode Island Red chicken from Beijing Huadu Yukou Poultry Breeding Co., Ltd. was used as the experimental animal. The breed had been previously selected based on laying intensity and persistency. This population comprised 705 individuals, including 183 pairs of full siblings. All hens were housed in individual cages and properly identified by their cage number. All hens were kept in the same environment under the 16L:8D lighting schedule and had access to feed and water ad libitum. The process of artificial insemination was conducted by using a sterile injector. Artificial insemination was conducted on all of the hens during the pedigree in chicken purebred reproduction.
Phenotype Measurements
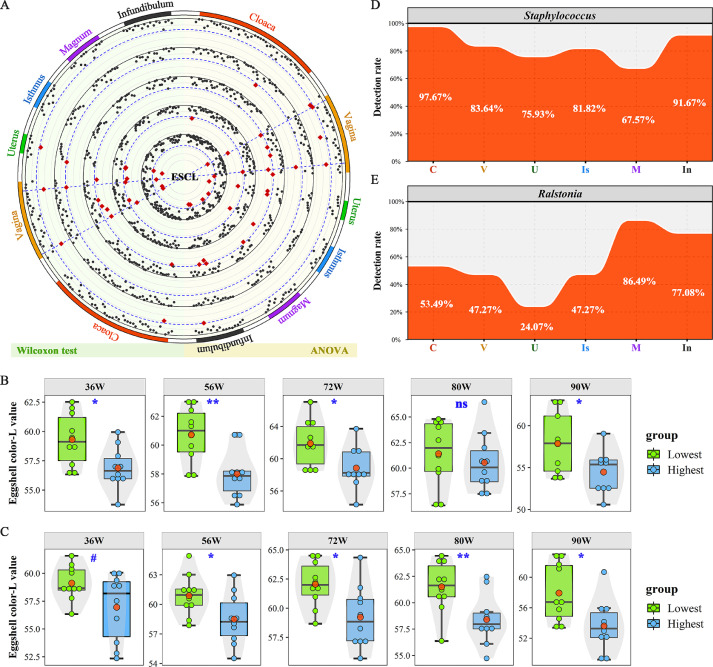
The number of eggs produced from the age at the first egg to 90 weeks of age was recorded daily. The egg qualities were detected at 36, 56, 72, 80 and 90 weeks of age, which was followed the breeding schedule. All eggs were identified by the QR code. At each point of time, a total of three eggs per hen were used to measure egg weight (EW), eggshell colour (ESC), eggshell gloss (ESG) and eggshell strength (ESS). All egg characteristics were measured within 24 h after laying. In brief, EW was measured with an electronic scale accurate to 0.01 g. ESC was measured with a CM-2600D reflectometer (Konica Minolta, Tokyo, Japan) using the three following parameters: L* represents lightness, a* measures the balance of red and green, and b* describes hue of blue-yellow scale. A commercial glossmeter (Konica Minolta CM600, Tokyo, Japan) was used to measure the ESG of fresh eggs. The gloss of each egg was evaluated by the mean of three points: the blunt end, equator, and sharp end. Then, ESS (i.e., breaking strength) of each egg was measured vertically with the Eggshell Force Gauge (Model-II, Robotmation, Tokyo, Japan).
Sample Collection
To evaluate the microbial composition throughout the reproductive tract and its relationship with host genetics and egg formation, 28 full-sib pairs of laying hens from 21 paternal half-sib families were selected for microbiome analysis. These hens were euthanized by cervical dislocation followed by decapitation at 90 weeks of age. The mucosal surface of each compartment of the oviduct, including the infundibulum, magnum, isthmus, uterus and vagina was scraped with a small steel spoon. The cloaca was sampled with sterile cotton swabs. All sample collection operations were performed on a clean bench that was sterilized using 75% ethanol as an extra precaution to prevent sample contamination. The mucosa and swabs were collected in 2-mL swab tubes (CY-98000, Shenzhen, China). All samples were stored at 4 °C and then transported to the laboratory for DNA extraction.
DNA Extraction and 16S rRNA Gene Sequencing
Microbial DNA was extracted using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA) according to the manufacturer's instructions. The DNA extract was checked on a 1% agarose gel, and DNA concentration and purity were determined with a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA). 40 samples were excluded due to poor DNA quality. Finally, a total of 296 samples (including 43 cloaca, 55 vagina, 55 uterus, 55 isthmus, 38 magnum, 50 infundibulum) were used for subsequent sequencing.
The hypervariable region V4 of the bacterial 16S rRNA gene was amplified with the primer pair 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Shterzer et al., 2020) using an ABI GeneAmp® 9700 PCR thermocycler (ABI, Foster City, CA, USA). PCR amplification of the 16S rRNA gene was performed as follows: 95°C for 3 min; 27 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 45 s; followed by a single extension at 72°C for 10 min. Purified amplicons were pooled in equimolar amounts and sequenced on an Illumina MiSeq PE300 platform to generate 300-bp paired-end reads (Illumina, San Diego, CA, USA).
Sequencing Processing
If the average quality score at the tail of the read with 50-bp window was less than 20, we cut off the back-end bases from the window. Then, the low-quality reads that met the following criteria were discarded: (1) reads containing ambiguous characters and (2) read lengths shorter than 200 bp. We also removed the sample with fewer than 30,000 sequence reads. Four samples (one for the uterus, one for the magnum and two for the infundibulum) were excluded from the subsequent analysis. The paired-end reads of the remaining samples were assembled with FLASH (Magoc and Salzberg, 2011). Sequences were processed and taxonomy assigned using QIIME2 (Bolyen et al., 2019). amplicon sequence variants (ASVs) were determined with DADA2 using the denoise-paired method. Afterward, singletons were filtered from the dataset. The SILVA 138 release was used as the reference database for the taxonomic assignment (Quast et al., 2012). The taxonomic classification of phyla, classes, orders, families, genera and species was then obtained.
Statistical Analysis
Alpha and Beta diversity were calculated using QIIME2 software. Bray-Curtis distance metrics were obtained to generate principal coordinate analysis (PCoA). To determine the relationships among each microorganism in the six segments of the reproductive tract, Spearman's correlation for microbial genera that were detected in greater than 40% of individuals was calculated using the psych package in the R program, and P-values were adjusted for FDRs using the BH method. Correlations between detected genera in specific segments were also inferred. The correlation patterns were further filtered to select only adjusted P-value <0.05. Co-occurrence networks were then constructed by using Cytoscape (Shannon et al., 2003).
The host genetic relationship was calculated based on pedigree information. To explore the effects of host genetics on the reproductive tract microbiota. We calculated the correlation between host genetic relationships and microbial dissimilarity based on the weighted and unweighted UniFrac distances in each segment. Pairs of hens with an estimated genetic relationship less than 0.015 were considered genetically unrelated. We further compared the difference in the weighted UniFrac distance among full sibs, half sibs, first cousins, and genetically unrelated individuals for each segment.
For the detection of taxa that were significantly associated with egg qualities and quantitative analysis, ANOVA was used to test the difference in phenotypic traits between chickens with the highest (N=10) and lowest (N=10) abundances of specific taxa. Meanwhile, the Wilcoxon rank-sum test was performed to determine the relative abundance of each taxon between the highest (N=10) and lowest (N=10) trait-ranked chickens. Adjusted P-values < 0.05 were considered significant.
Data Availability
The raw data are available from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with accession number SRP292828.
RESULTS
Descriptive Statistics for Host Phenotypes and Sequencing Outputs
The descriptive statistics of phenotypic observations, including EW, ESC, ESG, ESS and egg number (EN), at 5 age points from 36 to 90 wks of age are presented in Table 1. The chicken reproductive tract from external to internal can be divided into the cloaca, vagina, uterus, isthmus, magnum, and infundibulum (Figure 1A). To characterize the microbial composition of the chicken reproductive tract, we performed 16S rRNA sequencing on the abovementioned six segments. After quality control, up to 17,700,993 high-quality reads were generated from 292 samples with an average of 59,200 reads per sample (Supplementary Table S1). The resulting sequences were assigned by the DADA2 analysis pipelines in QIIME2. A total of 892 ASVs were identified. These ASVs were subsequently classified into 22 phyla, 39 classes, 91 orders, 155 families, 313 genera and 473 species. It should be noted that only 64.29% of the magnum samples (36/56) could successfully detect microbiota, which is consistent with hypotheses that the magnum may be resistant to bacteria due to the secretion of antimicrobial proteins such as lysozyme.
Table 1.
Descriptive statistics of egg quality traits.
| Age (weeks) | ESCL* | ESCa* | ESCb* | EN | EW (g) | ESS (kg/cm2) | ESG (GU) |
|---|---|---|---|---|---|---|---|
| 36 | 58.22 ± 2.56 | 18.04 ± 1.17 | 25.61 ± 0.97 | 113.14 ± 6.60 | 56.08 ± 3.09 | 3.72 ± 0.46 | 2.91 ± 0.69 |
| 56 | 59.46 ± 2.33 | 18.45 ± 0.97 | 29.14 ± 1.68 | 244.95 ± 8.33 | 59.71 ± 3.63 | 3.89 ± 0.55 | 2.70 ± 054 |
| 72 | 60.83 ± 3.12 | 17.99 ± 1.36 | 28.35 ± 1.85 | 348.76 ± 12.52 | 58.93 ± 3.55 | 3.45 ± 0.50 | 2.61 ± 0.55 |
| 80 | 60.87 ± 2.69 | 17.36 ± 1.25 | 28.12 ± 2.21 | 398.60 ± 15.11 | 59.04 ± 3.87 | 3.02 ± 0.61 | 2.32 ± 0.48 |
| 90 | 63.01 ± 3.67 | 18.33 ± 1.21 | 25.58 ± 1.58 | 456.63 ± 17.84 | 61.26 ± 4.55 | 2.90 ± 0.55 | 2.49 ± 0.46 |
Abbreviations: ESC, eggshell colour (L*, a*, b*); EN, egg numbers; EW, egg weight; ESS, eggshell strength; ESG, eggshell gloss.
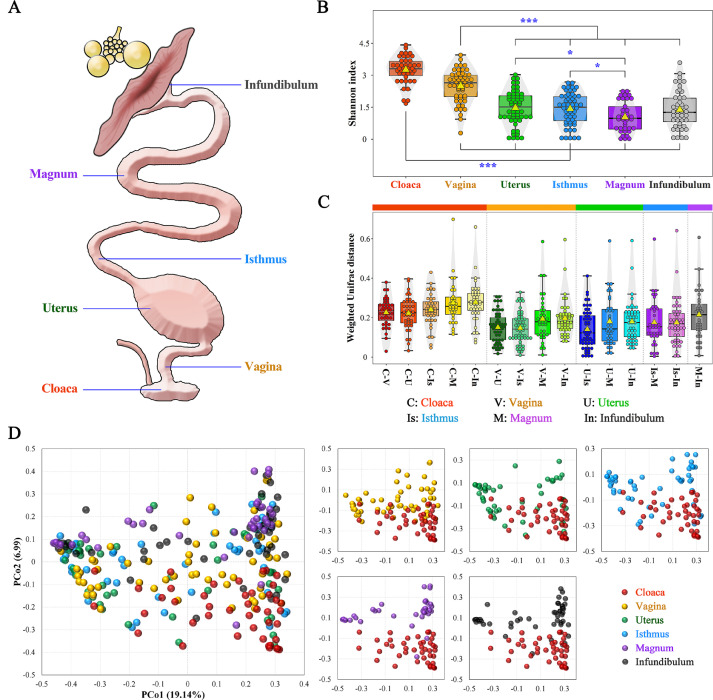
Figure 1.
Spatial variation in microbial diversity in the chicken reproductive tract. (A) Sampling sites in the reproductive tract of chickens. (B) Alpha-diversity comparison based on the Shannon diversity index. ⁎⁎⁎ and * indicate adjusted P-value less than 0.001 and 0.05, respectively. (C) Beta-diversity comparison based on the weighted UniFrac distance among six segments. The centre yellow triangle indicates the mean value in the corresponding group. (D) Principal coordinate analysis based on Bray-Curtis distance. Each point represents a sample.
Diversity of Microbiota in Various Segments of the Chicken Reproductive Tract
As the shared site for the digestive, urinary and reproductive tracts, the cloaca exhibits a more complicated environment. Consistent with this notion, the cloaca exhibited the highest Shannon index, which is commonly used to measure microbial diversity (Figure 1B). Interestingly, the alpha diversity of the microbiota gradually decreased from the cloaca to the infundibulum (Figure 1B). The microbial diversity observed in the magnum and infundibulum was significantly reduced compared with that in other segments. In addition, the beta diversity based on the weighted UniFrac distance between two segments increased with physical distance (Figure 1C). We then performed principal coordinate analysis to visualize the differences in microbial communities among the various segments. Although no significant differences in the uterus, isthmus, magnum or infundibulum were noted, obvious separation was observed among the cloaca, vagina and other segments, especially between the cloaca and the other oviductal segments (Figure 1D).
Variations in Microbial Composition Along the Reproductive Tract
The phyla Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria, and Bacteroidetes were the dominant microbiota in the chicken reproductive tract, among which Firmicutes and Proteobacteria were the two most abundant. However, appreciable differences in microbial compositions were noted among these segments (Figure 2A and Supplementary Table S2). For instance, the Firmicutes and Actinobacteria phyla reached 57.35% and 17.11% in the cloaca, and less than 39% and 3% in other segments, respectively. In contrast, Proteobacteria accounted for 60∼80% of the total in the segment from the vagina to the infundibulum but only reached 22.70% in the cloaca. In addition, the abundance of Bacteroidetes which is typically highly present in the digestive tract, was less than 4% in the chicken reproductive tract.
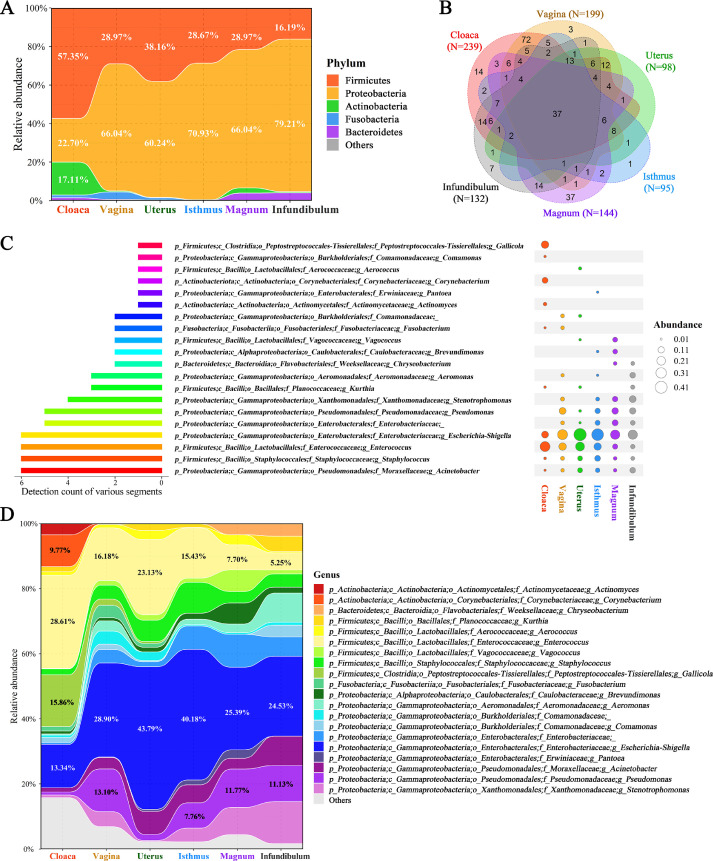
Figure 2.
Community composition in diverse segments. (A) Relative abundance of the dominant microbial phyla in various segments. (B) The number of microbial genera at the six anatomical segments and their overlaps. (C) The top 10 most abundant genera of each segment and their overlaps. After omitting the overlapping genera, a list of 20 genera was obtained. The size of bubbles indicates the relative abundance. (D) Relative abundance of the 20 genera in the six segments.
At the genus level, a total of 239, 199, 98, 95, 144 and132 genera were identified in the cloaca, vagina, uterus, isthmus, magnum, and infundibulum with a set of 37 genera present in all segments (Figure 2B). The top 10 abundant genera of each segment were then extracted, and after omitting the overlapping genera, a list of 20 genera was obtained (Figure 2C). Among them, four genera, Escherichia-Shigella, Enterococcus, Staphylococcus and Acinetobacter were simultaneously present in six segments but with varied abundances. Enterococcus, Gallicola, Corynebacterium and Escherichia-Shigella were the four most abundant genera in the cloaca, whereas Escherichia-Shigella, Enterococcus, Pseudomonas, Acinetobacter and Staphylococcus represented the majority of the genera in the other five oviductal sections (Figure 2D and Supplementary Table S3). Specifically, the potential pathogenic bacteria of Escherichia-Shigella exhibited the highest abundance in the segments from the vagina to the infundibulum, spanning from 24.53% to 43.79%, but the abundance was relatively low in the cloaca at 13.34%. A similar pattern was also observed in the genus Pseudomonas. However, the genus Enterococcus displayed the opposite pattern with relatively higher abundance in the cloaca (28.61%) compared to the others.
Correlation and Interaction of the Microbiota
We further calculated the Spearman correlations between the same microbiota detected in any two of the segments. The results showed that the genus Escherichia-Shigella, which has a detection rate close to 100%, exhibited highly positive correlation coefficients among the oviduct segments except cloaca (Figure 3A). In addition, the genus Staphylococcus displayed significant correlations between adjacent segments from cloaca to magnum. In addition, Acinetobacter and Enterococcus also had positive correlations between certain segment pairs.
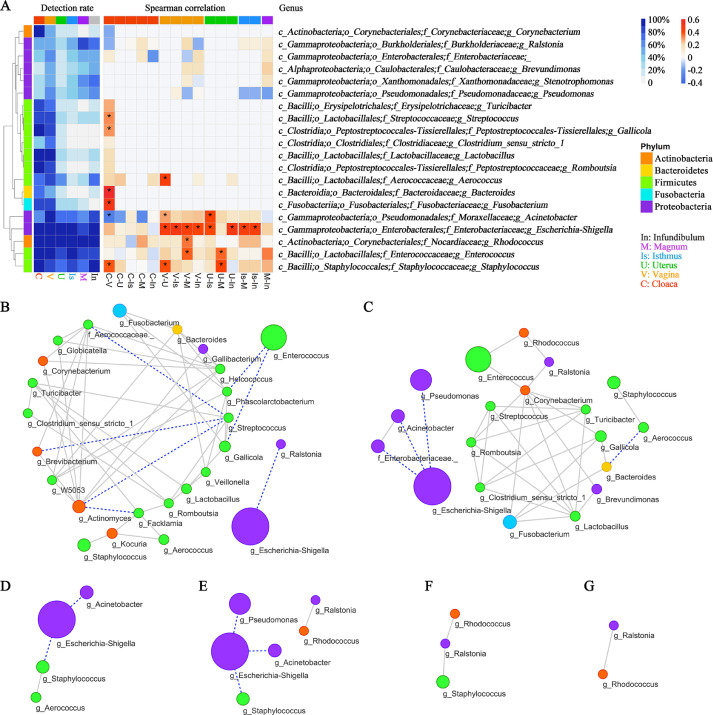
Figure 3.
Correlation and interaction of the microbiota. (A) The detection rate and Spearman's correlation for microbial genera that were detected in more than 40% of individuals. * means the correlation was significant (P < 0.05). Microbial co-occurrence network in the (B) cloaca, (C) vagina, (D) uterus, (E) isthmus (F) magnum and (G) infundibulum. The size of the nodes was proportional to the relative abundance of genera. The colour of nodes represent the phylum. The gray lines and blue dashed lines represent positive and negative correlations, respectively.
A co-occurrence network of core genera was then constructed to explore the microbial interactions along with the reproductive tract. Figure 3B-3G shows the interactions of microbiota in the cloaca, vagina, uterus, isthmus magnum and infundibulum, and the interaction complexity decreased from the external to internal parts (cloaca to infundibulum). Positive correlations were mainly observed in the core genera of the cloaca and vagina (Figure 3B-3C and Supplementary Tables S4-S5). However, the highly abundant genus Escherichia-Shigella had negative associations with limited microorganisms (Ralstonia in cloaca, Pseudomonas, Acinetobacter and Enterobacteraceae in vagina) and no interaction with most of the other microorganisms. Consistent results were found in the uterus and isthmus for Escherichia-Shigella, which was also negatively associated with the Acinetobacter genus (Figure 3D-3E). For the magnum and infundibulum, the interaction was simple, but both exhibited interactions between Ralstonia and Rhodococcus (Figure 3F- 3G).
Effect of Genetic Kinship on Microbiota Community
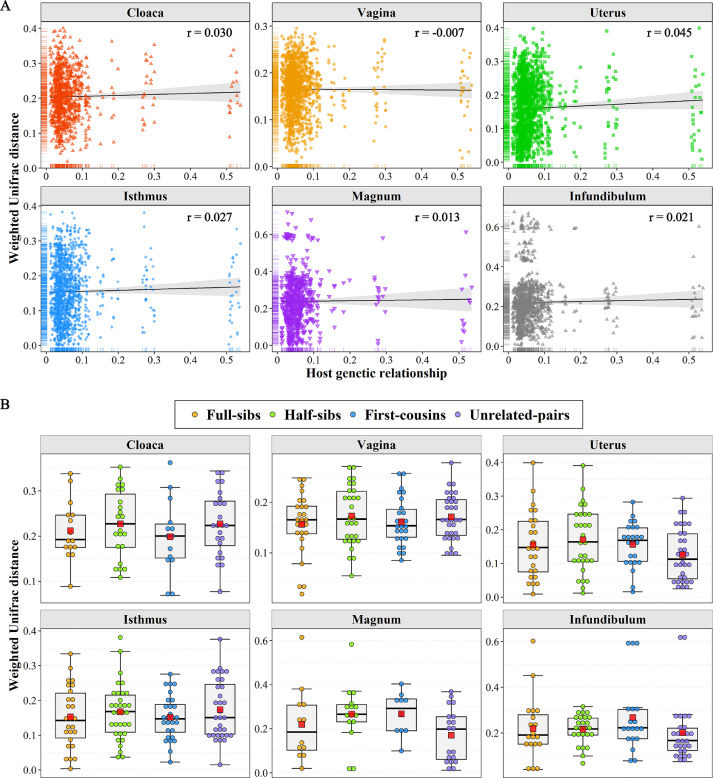
Since the abovementioned results indicated that the microbial communities were distinct among the cloaca, vagina and the other four segments, we further evaluated the effects of host genetics on the microbial composition in diverse segments. We first calculated the correlation coefficients between host genetic relationships and weighted UniFrac distances in each segment, and the correlations were very weak ranging from -0.007 to 0.045 (Figure 4A). Similar results were also observed between host genetic relationships and unweighted UniFrac distance (Supplementary Figure S1). Because most pairs of chickens exhibited no or a low degree of genetic relationship, we further compared the difference in the weighted UniFrac distance among full sibs, half sibs, first cousins, and genetically unrelated individuals for each segment. Consistently, no significant difference was observed among the four groups (Figure 4B).
Figure 4.
Effect of genetic kinship on reproductive tract microbiota. (A) The association between host genetic relationships and microbial dissimilarity based on weighted UniFrac distances. (B) Comparison of the weighted UniFrac distance among full sibs, half sibs, first cousins, and genetically unrelated individuals in six segments. The centre red square indicates the mean value in the corresponding group.
Screening of Microorganisms Related to Egg Quality and Production Traits
We further screened the crucial microorganisms related to egg quality and egg production traits, including ESC, ESS, ESG, EW and EN. ANOVA and Wilcoxon tests were used to detect the trait differences between divergent microbial groups and microbial differences between divergent trait groups, respectively. Only the microorganisms that were screened by both methods were considered candidate microorganisms. As shown in Figure 5A, two genera, Staphylococcus and Ralstonia were found associated with eggshell colour, i.e., eggshell brownness. The higher abundances of Staphylococcus (Figure 5B and Supplementary Table S6) and Ralstonia (Figure 5C and Supplementary Table S7) was related to darker brownness of the eggshell, and the results were consistent along the life cycle at 36, 56, 72, 80 and 90 weeks of age. In addition, the genus Staphylococcus exhibited a relatively high detection rate spanning from 67.57 to 97.67% in various segments (Figure 5D). However, for the genus Ralstonia, the detection rate was relatively low, ranging from 24.07 to 86.49% (Figure 5E).
Figure 5.
Screening of microorganisms related to eggshell colour (ESC). (A) Significant P-values for Wilcoxon rank-sum test (left) and ANOVA (right). Displayed from the inner to the outer circle are 36, 56, 72, 80 and 90 weeks of age. The P-values for the significance test are plotted as −log2 (P). The blue dashed line shows the significance threshold (P = 0.05). Each point represents a microorganism, and the red point indicates that the adjusted P-value passed the significance threshold. (B, C) Difference in the eggshell colour L* value between the two groups with the highest and lowest abundance of vaginal Staphylococcus and Ralstonia, respectively. **, * and ns represent adjusted P-values<0.01, <0.05 and >0.05, respectively. The centre red point indicates the mean value in the corresponding group. (D, E) The detection rate of Staphylococcus and Ralstonia in the six oviductal segments.
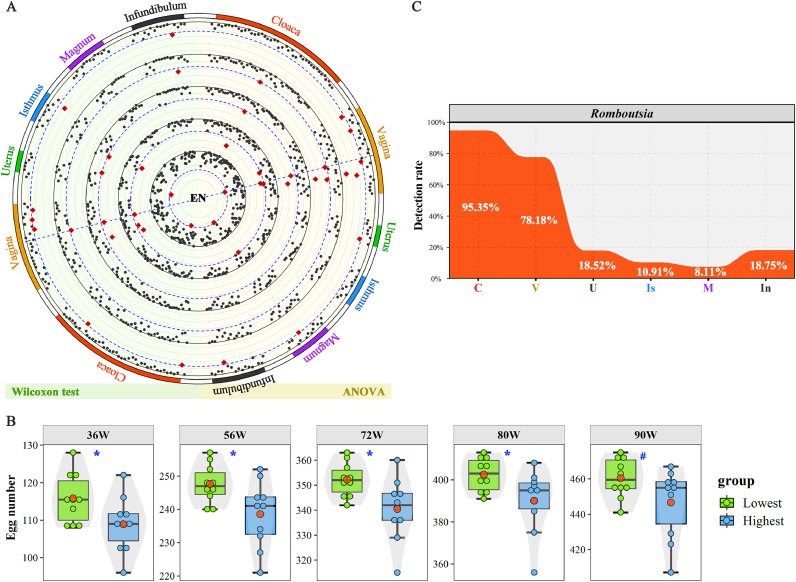
Regarding egg number, the genus Romboutsia in the vagina was screened (Figure 6A). The high abundance of Romboutsia was related to low egg number production at different age points (Figure 6B and Supplementary Table S8), and this genus exhibited a relatively high detection rate (78.1%) in the vagina (Figure 6C). For the other egg quality traits, including ESS, ESG and EW, no microorganisms were consistently related during along the laying period.
Figure 6.
Screening of microorganisms related to egg number (EN). (A) Significant P-values for Wilcoxon rank-sum test (left) and ANOVA (right). Displayed from the inner to the outer circle are 36, 56, 72, 80 and 90 weeks of age. The P-values for the significance test are plotted as −log2 (P). The blue dashed line shows the significance threshold (P = 0.05). Each point represents a microorganism and the red point indicates that the adjusted P-value passed the significance threshold. (B) Difference in egg number between the two groups with the highest and lowest abundance of vaginal Romboutsia. **, *, # and ns indicate adjusted P-values <0.01, 0.05, <0.10 and >0.1, respectively. The centre red point indicates the mean value in the corresponding group. (C) The detection rate of Romboutsia in the six oviductal segments.
DISCUSSION
Poultry eggs are valuable food source for humans due to their high quality of nutrients and relatively low cost. The quantity and quality of eggs are related to poultry production efficiency and food safety. In this study, we confirmed that chicken eggs are not formed in a sterile environment. The microbiota of the chicken reproductive tract spanned a continuum; however, the microbial communities among the cloaca, vagina and other sections of the chicken oviduct were obviously different. The reproductive tract of chickens harbours a rich microbiota, including some pathogenic bacteria. Eggs could be directly contaminated with microorganisms during egg formation and oviposition. Moreover, part of the microorganisms might be vertically transmitted to the embryo through the egg and constitute the initial gut microbiota of chick. Although the microbial diversity decreased from external to internal segments of the chicken reproductive tract due to the presence of antibacterial mechanisms, the correlation between host genetic kinship and all microbial distance were feeble. Of course, not all colonizers are pathogenic, and some bacteria may have long-term benefits to the host. Among these microorganisms, we identified several that were significantly associated with eggshell colour and egg number, highlighting that the notion that oviductal bacteria play an important role in the process of egg formation and might be useful to improve egg quality in chickens.
The microbiota of the female reproductive tract in humans has attracted considerable attention in recent years due to its impacts on reproductive health (Ravel et al., 2011; Chen et al., 2017). Indeed, the effects of the reproductive microbiome on host ecology, evolution and fitness are widespread across the animal kingdom, as previously alluded by Rowe et al. (2020). With the development of sequencing technology, demonstrating new roles of the microbial community in the reproductive tract is becoming increasingly active. To characterize the microbiota residing in the chicken reproductive tract, we systematically collected samples from six segments throughout the reproductive tract, including the cloaca, vagina, uterus, isthmus, magnum and infundibulum. Consistent with a previous report in humans, where the microbiota in the upper reproductive tract was significantly different from that in the lower reproductive tract (Chen et al., 2017), we found distinct microbial communities in the uterus, isthmus, magnum and infundibulum compared to those of the cloaca and vagina. A recent study in chickens also showed that the oviductal microbiota was significantly distinct from those in the cloaca (Lee et al., 2019). It is worthwhile to understand the origin of the microbiota in chicken reproductive tract. Anatomically, the cloaca is the end of the reproductive tract; moreover, it connects to the digestive and urinary systems of birds. Therefore, the cloacal microbiota is a mixture of bacteria from these sources (Lee et al., 2020) as well as from the external environment (Escallón et al., 2019), resulting in a particularly complex bacterial community. The vaginal microbiota is partially derived from semen and contacts during artificial vaginal insemination (Kulkarni and Heeb, 2007; Rowe et al., 2020).
In fact, the oviductal ecosystem is maintained through mutualistic relationships between the host and microbiota. The host immune system can alter microbial colonization due to microbiota-host interactions (Pekmezovic et al., 2019; Al-Nasiry et al., 2020). Under the protection of efficient host immune and antibacterial system (Silphaduang et al., 2006), the diversity of microbiota incrementally decreased from the cloaca to the infundibulum, indicating a microbiota continuum along the reproductive tract. The lowest diversity was observed in the magnum since lysozyme and other proteins with antimicrobial activity are secreted and concentrated in this location (Edwards et al., 1976). Recently, Mehta et al. (2020) revealed that genetic variation in the host innate immune system and cell signallings have an important effect on the human vaginal microbiota based on a genome-wide association study. A twin study reported greater similarity of the vaginal microbiota between monozygotic twins compared with that between dizygotic twin pairs and identified Prevotella as the most heritable vaginal bacteria (Si et al., 2017). However, it has not been well illustrated whether host genetics shapes the chicken oviductal microbiota. Correlation estimation between host genetic relationships and microbial similarity, as well as microbial distance comparisons between genetically related and unrelated individuals have been used as efficient measures for determining the influence of genetics on a population (Goodrich et al., 2014; Si et al., 2017; Rothschild et al., 2018; Wen et al., 2019). By utilizing the two analytical tools, we found that the correlation between host genetic relationships and microbial distance was negligible. Meanwhile, the genetically related pairwise individuals did not exhibit a more similar microbial community than unrelated pairs. These results indicated that the chicken oviductal microbiota is predominantly shaped by factors other than host genetics. The present study only performed the correlation analysis and microbial similarity comparison. However, in the future the host genetic variants are needed to explore the association between host genetics and oviductal microbiota.
Greater than 99% of the oviductal microbiota of egg-type chickens evaluated here comprised of Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria and Bacteroidetes. Similar results were also reported by Shterzer et al. (2020) in meat-type chickens. The microbiota of the cloaca was dominated by the phylum Firmicutes, which was similar to the faecal specimens (Wen et al., 2019). At the lower phylogenetic levels, previous studies highlighted that Gallicola is a typical genus in chicken faeces (Naphtali et al., 2019), and Pseudomonas, Acinetobacter, Enterococcus, Corynebacterium and Staphylococcus are the core bacterial genera in the mature hen reproductive tract (Lee et al., 2019). These major genera were obviously different from those in humans (Yildirim et al., 2014; Chen et al., 2017), where the vagina was predominantly comprised of Lactobacillus, whereas Acinetobacter and Pseudomonas constituted a notable fraction of the microbiota in the human upper reproductive tract. In contrast to Lactobacillus in the human vagina, which is thought to inhibit pathogenic bacteria through lactic acid and hydrogen peroxide production (Anahtar et al., 2018), explanations for the prevalence of Acinetobacter and Pseudomonas in the female reproductive tract have not been well documented. Based on previous and current studies, the prevalence of these genera may be involved in protecting the oviduct against inflammation and bacterial pathogens (Ligon et al., 2000; El-Fouly et al., 2015; Bassols et al., 2016), such as inhibiting the production of Escherichia-Shigella pathogens as noted in our results.
As oviparous animals, the chicken oviduct provides the biological environment for egg formation. The bacteria might be directly deposited into the yolk, albumen, eggshell membrane and eggshell during egg formation before oviposition as a result of microbial existence in the reproductive tissue. In addition, some microorganisms of the maternal oviduct could be vertically transmitted to the embryo through the egg and constitute the initial chick gut bacterial population. A recent study detected as many as 21 shared genera in the maternal oviduct, eggshell, egg white and intestinal tract of embryo (Lee et al., 2019). Most surprisingly, egg white and embryo gut exhibited similar microbial compositions. Another study observed a moderate correlation (0.52) between the microbiota of the embryo and chick (Ding et al., 2017). In the present study, Escherichia-Shigella and Enterococcus were two dominant genera in the chicken reproductive tract and could also be detected in freshly laid eggs (Schwaiger et al., 2010; Trudeau et al., 2020). Furthermore, Jurburg et al. (2019) reported that Escherichia-Shigella accounted for 42.5% of the gut microbial community in chicks on day 1 after hatching, but decreased gradually with age. A similar result was also observed by Sekelja et al. (2012), suggesting that Escherichia-Shigella might be vertically transmitted through eggs. Additionally, the relative abundance of the genus Enterococcus was reduced in 19-day embryonic intestines but increased in chicks (Ding et al., 2017). Most Enterococcus spp. detected in our study were unclassified below the genus level, but a small fraction of these was Enterococcus cecorum, which has emerged as an emerging pathogen in the poultry industry worldwide (Jung et al., 2018). It is largely unknown whether the Enterococcus that colonizes the chicken oviduct is pathogenic. However, zoonotic Escherichia-Shigella infections acquired from contaminated chicken eggs are possible (Shi et al., 2014).
As mentioned above, not all potential pathogen exposure results in disease, and some colonized bacteria might be beneficial and provide crucial physiological functions for the host. The entire process of egg formation takes approximately 24 h, with most of the time used for shell formation (∼20 h), in which pigmentation occurs to produce colour eggshells (Samiullah et al., 2015). Previous studies have confirmed that various bacteria have pigment synthesis ability (Narsing Rao et al., 2017). Therefore, investigating the effect of oviductal microbiota on eggshell traits might represent a breakthrough to explore the functions of inhabitant bacteria. Fortunately, we observed that higher abundance of vaginal Staphylococcus and Ralstonia was significantly associated with darker brownness of eggshells (lower L value). The major eggshell pigment in brown-egg laying hens is protoporphyrin IX with traces of uroporphyrin and coproporphyrin (Samiullah et al., 2015). Samiullah and Roberts (2013) demonstrated that the majority of the pigment was located in the calcareous part of the eggshell, and a small fraction (13∼20%) was noted within the cuticle. Previous studies have demonstrated that Staphylococcus have a high biosynthesis ability of porphyrins and pyrrole pigments, which were mainly uroporphyrin and coproporphyrin (Fuente et al., 1986). Staphylococcus could inhabit various segments of the chicken reproductive tract, and correlations of Staphylococcus abundances between adjacent sections were significant and positive. A previous theory suggested that pigments are secreted from the uterine epithelium into the uterine fluid and hence onto the eggshell during eggshell formation (Sparks, 2011). Here, our results indicated that the oviductal microbiota is also involved in the synthesis and deposition of eggshell pigments, and vaginal Staphylococcus might affect the content of pigments within the shell cuticle.
The success of identifying eggshell colour-related bacteria led us to further investigate the effect of specific microbiota on egg production. We found that chickens with a higher abundance of vaginal Romboutsia exhibited increased egg production compared with chickens with a lower abundance of this genus. The genus Romboutsia was identified by Gerritsen et al. (2014) after the first isolation from a digesta sample. In our study, Romboutsia was largely detected in the cloaca and vagina, but its detection rate in the uterus, isthmus, magnum and infundibulum was less than 20%. Thus, Romboutsia might be employed as a marker to predict egg number. Although we noticed that specific microorganisms were correlated with egg formation, further investigation should be performed to verify the specific role of these microorganisms in the chicken oviduct.
In summary, our results confirmed that the microbial communities are consecutive and exhibit moderate spatial heterogeneity throughout the chicken reproductive tract, and the association between host genetic kinship and microbial similarity of each segment was weak. Egg form in nonsterile environment; thus, microbial contamination may occur during egg formation and oviposition. Meanwhile, some bacteria have a favourable effect on their host. In particular, the genera Staphylococcus and Ralstonia are significantly associated with eggshell colour. In addition, vaginal Romboutsia could be used as a predictor for egg number. These findings highlight an important contribution of oviductal bacteria in the process of egg formation and provide a useful reference for investigating the reproductive tract microbiota of avian and other livestock species.
ACKNOWLEDGMENTS
We thank Beijing Huadu Yukou Poultry Industry Co., Ltd. for providing the experimental chickens. This work was supported by the National Natural Science Foundation of China (31930105), China Agriculture Research Systems (CARS-40) and the China Postdoctoral Science Foundation (2020M680028).
DISCLOSURES
The authors declare that they have no conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101104.
Appendix. Supplementary materials
REFERENCES
- Al-Nasiry S., Ambrosino E., Schlaepfer M., Morre S.A., Wieten L., Voncken J.W., Spinelli M., Mueller M., Kramer B.W. The interplay between reproductive tract microbiota and immunological system in human reproduction. Front. Immunol. 2020;11:378. doi: 10.3389/fimmu.2020.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anahtar M.N., Gootenberg D.B., Mitchell C.M., Kwon D.S. Cervicovaginal microbiota and reproductive health: the virtue of simplicity. Cell Host. Microbe. 2018;23:159–168. doi: 10.1016/j.chom.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Bassols J., Serino M., Carreras-Badosa G., Burcelin R., Blasco-Baque V., Lopez-Bermejo A., Fernandez-Real J. Gestational diabetes is associated with changes in placental microbiota and microbiome. Pediatr. Res. 2016;80:777–784. doi: 10.1038/pr.2016.155. [DOI] [PubMed] [Google Scholar]
- Bezirtzoglou E., Voidarou C., Papadaki A., Tsiotsias A., Kotsovolou O., Konstandi M. Hormone therapy alters the composition of the vaginal microflora in ovariectomized rats. Microb. Ecol. 2008;55:751–759. doi: 10.1007/s00248-007-9317-z. [DOI] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodriguez A.M., Chase J., Cope E.K., Da Silva R., Diener C., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibbons S.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G.A., Janssen S., Jarmusch A.K., Jiang L., Kaehler B.D., Bin Kang K., Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G.I., Lee J., Ley R., Liu Y., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J.T., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S.I., Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., van der Hooft J.J.J., Vargas F., Vazquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Walters W., Wan Y., Wang M., Warren J., Weber K.C., Williamson C.H.D., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y., Zhu Q., Knight R., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Song X., Wei W., Zhong H., Dai J., Lan Z., Li F., Yu X., Feng Q., Wang Z., Xie H., Chen X., Zeng C., Wen B., Zeng L., Du H., Tang H., Xu C., Xia Y., Xia H., Yang H., Wang J., Wang J., Madsen L., Brix S., Kristiansen K., Xu X., Li J., Wu R., Jia H. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 2017;8:875. doi: 10.1038/s41467-017-00901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M.C., Rautava S., Aakko J., Isolauri E., Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Dai R., Yang L., He C., Xu K., Liu S., Zhao W., Xiao L., Luo L., Zhang Y., Meng H. Inheritance and establishment of gut microbiota in chickens. Front. Microbiol. 2017;8:1967. doi: 10.3389/fmicb.2017.01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards N.A., Luttrell V., Nir I. The secretion and synthesis of albumen by the magnum of the domestic fowl (Gallus domesticus) Comp. Biochem. Physiol. B. 1976;53:183–186. doi: 10.1016/0305-0491(76)90032-8. [DOI] [PubMed] [Google Scholar]
- El-Fouly M.Z., Sharaf A.M., Shahin A.A.M., El-Bialy H.A., Omara A.M.A. Biosynthesis of pyocyanin pigment by Pseudomonas aeruginosa. J. Radiat. Res. Appl. Sci. 2015;8:36–48. [Google Scholar]
- Escallón C., Belden L.K., Moore I.T. The cloacal microbiome changes with the breeding season in a wild bird. Integr. Org. Biol. 2019;1:oby009. doi: 10.1093/iob/oby009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente R., Schleifer K.H., Götz F., Köst H.P. Accumulation of porphyrins and pyrrole pigments by Staphylococcus aureus ssp. Anaerobius and its aerobic mutant. FEMS Microbiol. Lett. 1986;35:183–188. [Google Scholar]
- Gantois I., Ducatelle R., Pasmans F., Haesebrouck F., Gast R., Humphrey T.J., Van Immerseel F. Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol. Rev. 2009;33:718–738. doi: 10.1111/j.1574-6976.2008.00161.x. [DOI] [PubMed] [Google Scholar]
- Gautron J., Hincke M.T., Nys Y. Precursor matrix proteins in the uterine fluid change with stages of eggshell formation in hens. Connect. Tissue Res. 1997;36:195–210. doi: 10.3109/03008209709160220. [DOI] [PubMed] [Google Scholar]
- Gensollen T., Iyer S.S., Kasper D.L., Blumberg R.S. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritsen J., Fuentes S., Grievink W., van Niftrik L., Tindall B.J., Timmerman H.M., Rijkers G.T., Smidt H. Characterization of Romboutsia ilealis gen. Nov., Sp. Nov., Isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera Romboutsia gen. Nov., Intestinibacter gen. Nov., Terrisporobacter gen. Nov. And Asaccharospora gen. Nov. Int. J. Syst. Evol. Microbiol. 2014;64:1600–1616. doi: 10.1099/ijs.0.059543-0. [DOI] [PubMed] [Google Scholar]
- Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T., Spector T.D., Clark A.G., Ley R.E. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope B.K., Baker R., Edel E.D., Hogue A.T., Schlosser W.D., Whiting R., McDowell R.M., Morales R.A. An overview of the Salmonella enteritidis risk assessment for shell eggs and egg products. Risk Anal. 2002;22:203–218. doi: 10.1111/0272-4332.00023. [DOI] [PubMed] [Google Scholar]
- Jung A., Chen L.R., Suyemoto M.M., Barnes H.J., Borst L.B. A review of Enterococcus cecorum infection in poultry. Avian Dis. 2018;62:261–271. doi: 10.1637/11825-030618-Review.1. [DOI] [PubMed] [Google Scholar]
- Jurburg S.D., Brouwer M.S.M., Ceccarelli D., Goot J., Jansman A.J.M., Bossers A. Patterns of community assembly in the developing chicken microbiome reveal rapid primary succession. Microbiology Open. 2019;8:e00821. doi: 10.1002/mbo3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S., Heeb P. Social and sexual behaviours aid transmission of bacteria in birds. Behav. Processes. 2007;74:88–92. doi: 10.1016/j.beproc.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Larsen B., Markovetz A.J., Galask R.P. Role of estrogen in controlling the genital microflora of female rats. Appl. Environ. Microbiol. 1977;34:534–540. doi: 10.1128/aem.34.5.534-540.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Cho S., La T., Lee H., Lee J., Park S., Song C., Choi I., Lee S. Comparison of microbiota in the cloaca, colon, and magnum of layer chicken. Plos One. 2020;15 doi: 10.1371/journal.pone.0237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., La T., Lee H., Choi I., Song C., Park S., Lee J., Lee S. Characterization of microbial communities in the chicken oviduct and the origin of chicken embryo gut microbiota. Sci. Rep. 2019;9:6838. doi: 10.1038/s41598-019-43280-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon J.M., Hill D.S., Hammer P.E., Torkewitz N.R., Hofmann D., Kempf H.J., van Pee K.H. Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag. Sci. 2000;56:688–695. [Google Scholar]
- Ma B., Forney L.J., Ravel J. Vaginal microbiome: rethinking health and disease. Annu. Rev. Microbiol. 2012;66:371–389. doi: 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S.D., Nannini D.R., Otieno F., Green S.J., Agingu W., Landay A., Zheng Y., Hou L. Host genetic factors associated with vaginal microbiome composition in kenyan women. mSystems. 2020;5:e00502–e00520. doi: 10.1128/mSystems.00502-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.A., Beasley D.E., Dunn R.R., Archie E.A. Lactobacilli dominance and vaginal pH: why is the human vaginal microbiome unique? Front. Microbiol. 2016;7:1936. doi: 10.3389/fmicb.2016.01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naphtali P., Mohiuddin M.M., Paschos A., Schellhorn H.E. Application of high-throughput 16S rRNA sequencing to identify fecal contamination sources and to complement the detection of fecal indicator bacteria in rural groundwater. J. Water Health. 2019;17:393–403. doi: 10.2166/wh.2019.295. [DOI] [PubMed] [Google Scholar]
- Narsing Rao M.P., Xiao M., Li W. Fungal and bacterial pigments: secondary metabolites with wide applications. Front. Microbiol. 2017;8:1113. doi: 10.3389/fmicb.2017.01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Nys Y., Zawadzki J., Gautron J., Mills A.D. Whitening of brown-shelled eggs: mineral composition of uterine fluid and rate of protoporphyrin deposition. Poult. Sci. 1991;70:1236–1245. doi: 10.3382/ps.0701236. [DOI] [PubMed] [Google Scholar]
- Pekmezovic M., Mogavero S., Naglik J.R., Hube B. Host–pathogen interactions during female genital tract infections. Trends Microbiol. 2019;27:982–996. doi: 10.1016/j.tim.2019.07.006. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J., Gajer P., Abdo Z., Schneider G.M., Koenig S.S., McCulle S.L., Karlebach S., Gorle R., Russell J., Tacket C.O., Brotman R.M., Davis C.C., Ault K., Peralta L., Forney L.J. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réhault-Godbert S., Guyot N., Nys Y. The golden egg: nutritional value, bioactivities, and emerging benefits for human health. Nutrients. 2019;11:684. doi: 10.3390/nu11030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., Costea P.I., Godneva A., Kalka I.N., Bar N., Shilo S., Lador D., Vila A.V., Zmora N., Pevsner-Fischer M., Israeli D., Kosower N., Malka G., Wolf B.C., Avnit-Sagi T., Lotan-Pompan M., Weinberger A., Halpern Z., Carmi S., Fu J., Wijmenga C., Zhernakova A., Elinav E., Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- Rowe M., Veerus L., Trosvik P., Buckling A., Pizzari T. The reproductive microbiome: An emerging driver of sexual selection, sexual conflict, mating systems, and reproductive isolation. Trends Ecol. Evol. 2020;35:220–234. doi: 10.1016/j.tree.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Sah N., Mishra B. Regulation of egg formation in the oviduct of laying hen. World's Poult. Sci. J. 2018;74:509–522. [Google Scholar]
- Salihu M., Garba B., Isah Y. Evaluation of microbial contents of table eggs at retail outlets in Sokoto metropolis. Nigeria. Sokoto J. Vet. Sci. 2015;13:22–28. [Google Scholar]
- Samiullah S., Roberts J.R., Chousalkar K. Eggshell color in brown-egg laying hens - a review. Poult. Sci. 2015;94:2566–2575. doi: 10.3382/ps/pev202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samiullah S., Roberts J.R. The location of protoporphyrin in the eggshell of brown-shelled eggs. Poult. Sci. 2013;92:2783–2788. doi: 10.3382/ps.2013-03051. [DOI] [PubMed] [Google Scholar]
- Schoeni J.L., Doyle M.P. Variable colonization of chickens perorally inoculated with Escherichia coli O157:H7 and subsequent contamination of eggs. Appl. Environ. Microbiol. 1994;60:2958–2962. doi: 10.1128/aem.60.8.2958-2962.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger K., Schmied E.M., Bauer J. Comparative analysis on antibiotic resistance characteristics of Listeria spp. and Enterococcus spp. isolated from laying hens and eggs in conventional and organic keeping systems in Bavaria, Germany. Zoonoses Public Health. 2010;57:171–180. doi: 10.1111/j.1863-2378.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- Sekelja M., Rud I., Knutsen S.H., Denstadli V., Westereng B., Naes T., Rudi K. Abrupt temporal fluctuations in the chicken fecal microbiota are explained by its gastrointestinal origin. Appl. Environ. Microbiol. 2012;78:2941–2948. doi: 10.1128/AEM.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome. Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Yang X., Chen L., Chang H., Liu H., Zhao J., Wang X., Wang C. Pathogenicity of Shigella in chickens. Plos One. 2014;9:e100264. doi: 10.1371/journal.pone.0100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shterzer N., Rothschild N., Sbehat Y., Stern E., Nazarov A., Mills E. Large overlap between the intestinal and reproductive tract microbiomes of chickens. Front. Microbiol. 2020;11:1508. doi: 10.3389/fmicb.2020.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si J., You H.J., Yu J., Sung J., Ko G. Prevotella as a hub for vaginal microbiota under the influence of host genetics and their association with obesity. Cell Host Microbe. 2017;21:97–105. doi: 10.1016/j.chom.2016.11.010. [DOI] [PubMed] [Google Scholar]
- Silphaduang U., Hincke M.T., Nys Y., Mine Y. Antimicrobial proteins in chicken reproductive system. Biochem. Bioph. Res. Commun. 2006;340:648–655. doi: 10.1016/j.bbrc.2005.12.054. [DOI] [PubMed] [Google Scholar]
- Sparks N.H.C. Eggshell pigments–from formation to deposition. Avian Biol. Res. 2011;4:162–167. [Google Scholar]
- Sun C., Xu G., Yang N. Differential label-free quantitative proteomic analysis of avian eggshell matrix and uterine fluid proteins associated with eggshell mechanical property. Proteomics. 2013;13:3523–3536. doi: 10.1002/pmic.201300286. [DOI] [PubMed] [Google Scholar]
- Trudeau S., Thibodeau A., Côté J., Gaucher M., Fravalo P. Contribution of the broiler breeders’ fecal microbiota to the establishment of the eggshell microbiota. Front. Microbiol. 2020;11:666. doi: 10.3389/fmicb.2020.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell L.K., Clemente J.C., Rideout J.R., Gevers D., Caporaso J.G., Knight R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J. Allergy Clin. Immunol. 2012;129:1204–1208. doi: 10.1016/j.jaci.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S., Griffin N.W., Bezy O., Fujisaka S., Vienberg S., Softic S., Deng L., Bry L., Gordon J.I., Kahn C.R. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 2015;22:516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Yan W., Sun C., Ji C., Zhou Q., Zhang D., Zheng J., Yang N. The gut microbiota is largely independent of host genetics in regulating fat deposition in chickens. ISME. J. 2019;13:1422–1436. doi: 10.1038/s41396-019-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim S., Yeoman C.J., Janga S.C., Thomas S.M., Ho M., Leigh S.R., White B.A., Wilson B.A., Stumpf R.M. Primate vaginal microbiomes exhibit species specificity without universal Lactobacillus dominance. ISME. J. 2014;8:2431–2444. doi: 10.1038/ismej.2014.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lu C., Li Y., Yang X., Wang X., Chang H., Liu H., Chen L., Zhao J., Wang C., Chang Y. In vitro adherence and invasion of primary chicken oviduct epithelial cells by Gallibacterium anatis. Vet. Microbiol. 2017;203:136–142. doi: 10.1016/j.vetmic.2017.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data are available from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with accession number SRP292828.