Abstract
The molecular characterization of genotype P[6] rotavirus strains collected from children admitted to hospital with acute dehydrating diarrhoea during a 6-year surveillance period in Taiwan is described in this study. In total, three G4P[6] strains, one G5P[6] and one G12P[6] were characterized by sequencing and phylogenetic analysis of the VP4, VP7, VP6 and NSP4 genes. Whilst all four genes of the single Taiwanese G12P[6] strain clustered with the respective genes of globally common human rotavirus strains, the G4 and G5 strains showed remarkable similarities to porcine rotavirus strains and putative porcine-origin human P[19] strains reported previously from Taiwan. The overall proportion of porcine rotavirus-like strains in Taiwan remains around 1% among hospitalized children; however, the circulation and sporadic transmission of these heterotypic strains from pigs to humans could pose a public-health concern. Therefore, continuation of strain monitoring is needed in the vaccine era to detect any possible vaccine breakthrough events associated with the introduction of such heterologous rotavirus strains.
INTRODUCTION
Group A rotaviruses are the main cause of acute dehydrating gastroenteritis in infants and young children worldwide (Estes & Kapikian, 2007). Rotavirus is characterized by a triple-layered, non-enveloped virion and a dsRNA genome consisting of 11 separate segments. The two outer-capsid proteins, VP7 and VP4, induce neutralizing antibodies in vivo, segregate independently and have served as antigens of the dual-typing classification system, the G and P serotypes, respectively (Estes & Kapikian, 2007). Additional typing schemes have been proposed for the VP6 and NSP4 genes in line with available molecular sequence data and, more recently, the molecular classification system has been extended to all 11 rotavirus gene segments, using gene-specific nucleotide similarity cut-off values (Matthijnssens et al., 2008b, 2011).
Globally, the most common human rotavirus strains are G1P[8], G3P[8], G4P[8] and G9P[8] on the Wa-like and G2P[4] on the DS1-like genomic configuration (Matthijnssens et al., 2009; Bányai et al., 2012). In general, the neutralization antigen combinations of epidemiologically important animal strains are typically different from those identified in humans. For example, in swine, the G3–G5 and G11 VP7 types and the P[6], P[7] and P[13] VP4 types are the most common (Martella et al., 2010). However, some particular antigen combinations, such as G3P[6] and G4P[6], are shared between human and porcine strains (Matthijnssens et al., 2009, 2011). In addition, other gene variants, including some of the VP6 and NSP4 genotypes, are also shared, a finding based on the postulation of common evolutionary roots of porcine and Wa-like human rotavirus strains (Matthijnssens et al., 2008a).
Genotype P[6] strains were first described in human neonates in hospital nurseries who were shedding rotavirus predominantly in the absence of gastroenteritis symptoms (Flores et al., 1986; Gentsch et al., 2005). Later, genotype P[6] strains were detected sporadically in older children with gastroenteritis worldwide, and investigations revealed that P[6] strains are epidemiologically important in parts of Africa (Bányai et al., 2012). Whilst the majority of virulent human P[6] strains clustered into a single genetic lineage together with strains isolated from neonates, subsequent studies from Japan and Hungary identified genetically highly divergent P[6] strains in children with gastroenteritis (Nakagomi et al., 1999; Bányai et al., 2004), and P[6] genotype diversity in diarrhoeic children has been observed over time (Ahmed et al., 2007; Mascarenhas et al., 2007; Nguyen et al., 2007; Li et al., 2008; Martella et al., 2008; Bányai et al., 2009a, b; Mukherjee et al., 2009, 2011; Stupka et al., 2009; Wang et al., 2010). The P[6] VP4 gene can be categorized into at least five lineages, some of which are unique to either pigs or humans, whilst others are shared between humans and swine (Bányai et al., 2004; Martella et al., 2006).
In this study, we describe the molecular characterization of five genotype P[6] strains detected during a 6-year hospital-based surveillance conducted from 2005 to 2010 in Taiwan (Table 1).
Table 1. Prevalence of rotavirus strains detected between 2005 and 2010 in Taiwan ROC.
Because genotyping was carried out by direct sequencing of the PCR products, the category non-typable (NT) should include strains whose corresponding gene sequence chromatograms were of low quality. Theoretically, this category also includes potential samples with mixed genotypes.
| P type | G type | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | G8 | G9 | G12 | GNT | Total | |
| P[4] | 111 | 1 | 27 | 139 | ||||||
| P[6] | 3 | 1 | 1 | 5 | ||||||
| P[8] | 891 | 3 | 347 | 214 | 92 | 1547 | ||||
| P[9] | 1 | 1 | 2 | |||||||
| P[14] | 1 | 1 | ||||||||
| P[19] | 3 | 1 | 1 | 5 | ||||||
| P[25] | 1 | 1 | ||||||||
| PNT | 32 | 19 | 22 | 5 | 53 | 131 | ||||
| Total | 924 | 133 | 375 | 3 | 2 | 1 | 220 | 1 | 172 | 1831 |
METHODS
Case definition and data collection.
Stool specimens positive for rotavirus were collected from January 2005 to December 2010, mainly from children <5 years of age who were hospitalized for treatment of acute gastroenteritis in three sentinel hospitals located in northern, central and southern Taiwan. Acute gastroenteritis was defined as three or more episodes of watery diarrhoea or looser-than-normal stools in the 24 h before presentation. Information on basic epidemiological data (patient gender, age, contact persons and family members) and clinical data (fever, duration of vomiting and diarrhoea, underlying diseases, vaccination history and extra-intestinal symptoms) were gathered where available.
Laboratory testing.
Stool specimens were screened by antigen test (RIDASCREEN Rotavirus; R-Biopharm AG) at the participating hospitals. Rotavirus-positive specimens were transported to the Rotavirus Reference Laboratory at Taiwan Centers for Disease Control for G and P genotyping.
Viral RNA was extracted from 10% (w/v) faecal supernatants using a MagNA Pure LC DNA isolation kit (Roche Diagnostics) according to the manufacturer’s instructions. The extracted RNAs were used as template for RT-PCR with random primers (Wu et al., 2009). The viral VP4, VP6, VP7 and NSP4 genes were amplified with primer sets Con3/Con2, JRG7/JRG8 or GEN-VP6F/GEN-VP6R, Beg9-End9 and JRG30/JRG31, respectively (Gouvea et al., 1990; Gentsch et al., 1992; Matthijnssens et al., 2006; Esona et al.; 2009; Mijatovic-Rustempasic et al., 2011), and subjected to direct sequencing using the same PCR primers. Dye-labelled products were run on an ABI 3130 sequence analyser (Applied Biosystems).
Sequencing and phylogenetic analysis.
Rotavirus genotypes were determined using RotaC software (Maes et al., 2009). Multiple nucleotide sequences were aligned manually with GeneDoc software (Nicholas et al., 1997), whilst phylogenetic analysis was performed using MEGA 5.0 software using maximum-likelihood and neighbour-joining algorithms (Tamura et al., 2011).
Strain designation.
Between 2005 and 2010, a total of 1831 rotavirus strains were genotyped. Of these, five strains with genotype P[6] VP4 genes were identified (0.27%). The three G4P[6] strains were detected in 2005, 2006 and 2009, whilst the single G5P[6] and G12P[6] strains were identified in 2009 and 2006, respectively. For the nomenclature designations of these strains, we used the recently proposed scheme: RVA/human-wt/TWN/04-94s74/2005/G4P[6], RVA/human-wt/TWN/03-95s3492/2006/G4P[6], RVA/human-wt/TWN/03-98s140/2009/G4P[6], RVA/human-wt/TWN/03-98sP50/2009/G5P[6] and RVA/human-wt/TWN/03-95s1461/2006/G12P[6].
RESULTS
The finding of several highly unusual P[6] variants in recent years suggests that genetic diversity in this genotype may be underestimated and prompted us to investigate the molecular characteristics of the five P[6] strains identified among 1831 strains genotyped during a 6-year surveillance conducted in Taiwan (Table 1). Hence, we characterized their VP4, VP7, VP6 and NSP4 genes by sequencing and phylogenetic analysis.
Molecular characterization of the VP4 gene
An 831 bp fragment of the VP4 genes of all five Taiwanese P[6] strains was amplified and sequenced. We used a shorter stretch (500 bp) in subsequent sequence alignments to correspond to the length of partial VP4 gene sequences available for some reference strains. The three Taiwanese G4P[6] strains shared 93.3–94.8% nucleotide similarity among each other and 78.8–98.4% nucleotide similarity with 53 representative P[6] strains detected in humans and pigs worldwide. Comparable patterns of similarity were found when the G5P[6] strain was compared with the Taiwanese G4P[6] strains (95.0–95.8%) and international strains of the same genotype (80.0–95.8%). In contrast, the VP4 gene fragment of the G12P[6] strain had a lower nucleotide similarity to Taiwanese G4 and G5 strains of genotype P[6] (86.9–88.3%) and shared up to 98.4% nucleotide similarity with representative P[6] strains detected worldwide. In our phylogenetic analysis, the nucleotide sequence-based trees demonstrated that the Taiwanese P[6] strains fell into two discrete genetic sublineages, four strains clustering with both porcine and porcine-derived human strains and one strain clustering with the globally spread and medically important variant of human P[6] strains (Fig. 1).
Fig. 1.
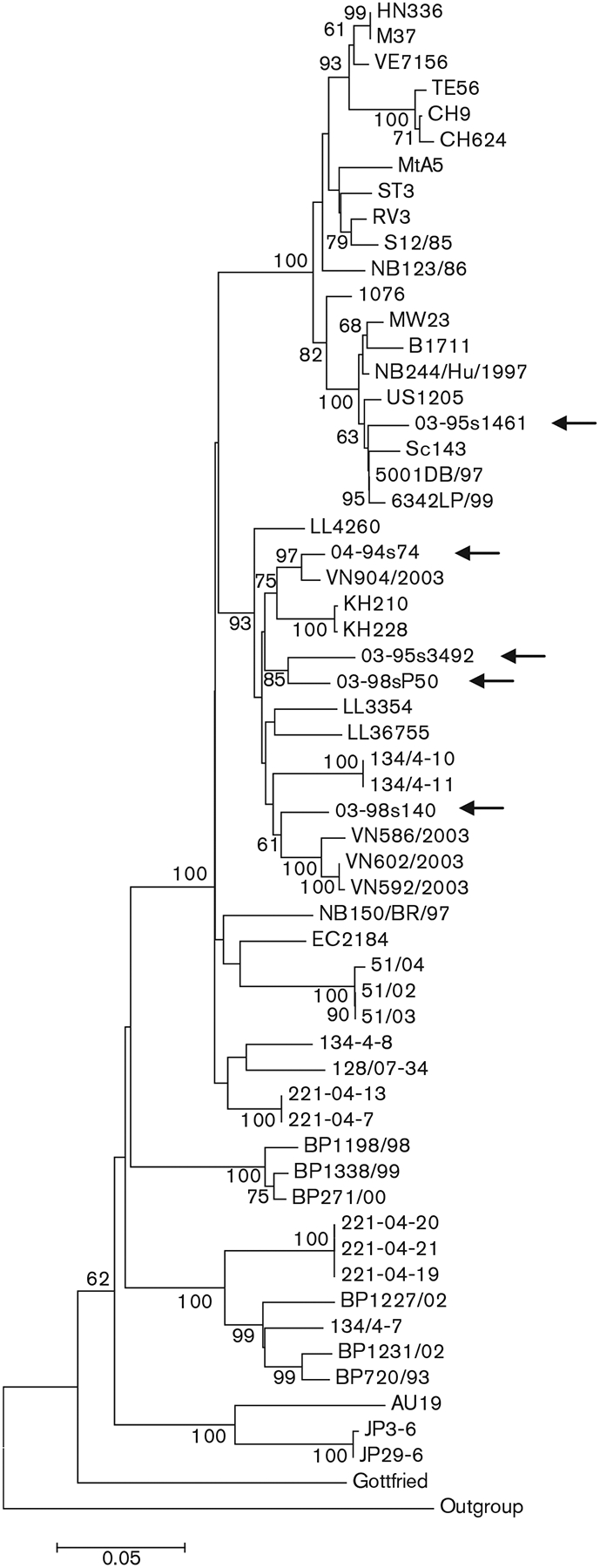
Nucleotide sequence-based neighbour-joining phylogenetic tree of the P[6] VP4 gene. Arrows indicate the Taiwanese G4P[6], G5P[6] and G12P[6] strains. Bootstrap values (500 replicates) >60% are indicated. Bar, 0.05 nucleotide substitutions per site.
Molecular characterization of the VP7 gene
As determined by nucleotide sequence-based genotyping (Wu et al., 2009), the five Taiwanese P[6] strains exhibited three different VP7 specificities: G4, G5 and G12 (see Methods).
The (nearly) full-length coding region was determined for the VP7 gene of the G4P[6] strains. These three strains shared 81.3–96.2% nucleotide similarity with each other along a 753 bp fragment. One strain had a lower VP7 gene similarity to G4 sequences (<83.2%) available in GenBank, whilst the two other G4 strains shared up to 97.5 and 98.3% nucleotide similarities, respectively, with sequences in GenBank. Phylogenetic analysis (Fig. 2a) demonstrated that two strains clustered on a major branch shared by human strains from China (E931) and Vietnam (e.g. VN592/2003), and porcine (including wild boar) strains from Japan (e.g. FGP65 and GUB88). A third Taiwanese human G4 strain, 03-95s3492, formed a new branch in the G4 VP7 gene tree, suggesting that it represents a new lineage of the G4 VP7 gene (Fig. 2a). This putative new lineage was most closely related to a Slovenian porcine strain (O-1) and an uncommon Australian human strain (M3014) (Fig. 2a).
Fig. 2.
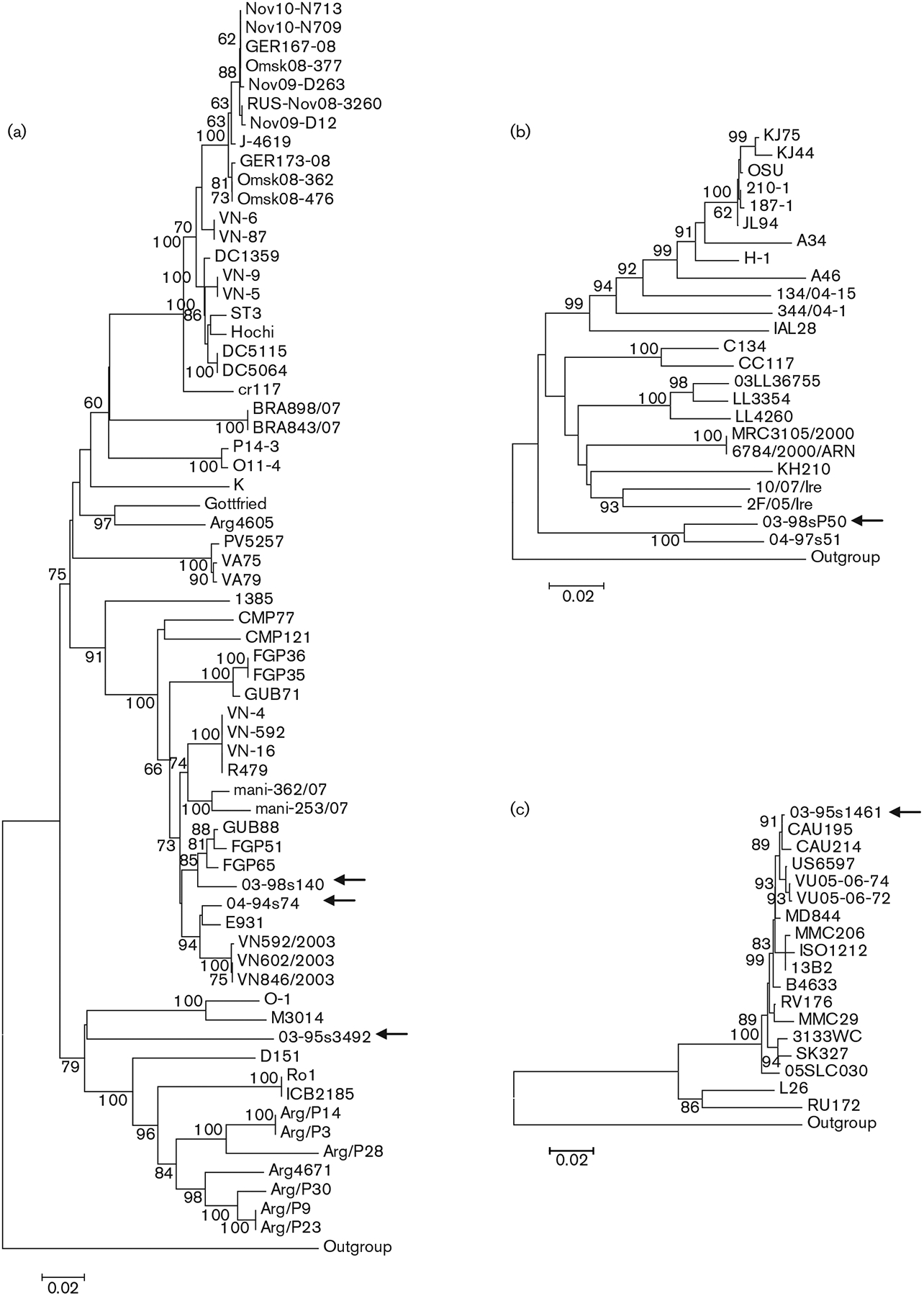
Nucleotide sequence-based neighbour-joining phylogenetic trees of the G4 (a), G5 (b) and G12 (c) VP7 genes. Arrows indicate the Taiwanese genotype P[6] strains. Bootstrap values (500 replicates) >60% are indicated. Bars, 0.02 nucleotide substitutions per site.
Sequence analysis of an 826 bp stretch of the VP7 gene of the Taiwanese G5P[6] strain revealed moderate nucleotide sequence similarity to a variety of reference G5 strains of both animal and human origin (range 82.6–86.4%) from several countries. The most closely related G5 strain was another Taiwanese G5 strain, which carried the P[19] VP4 gene (nucleotide similarity 94.4%). This result reaffirmed that the VP7 genes of Taiwanese human G5 rotaviruses form an individual lineage within this specificity (Wu et al., 2011) (Fig. 2b).
The fifth Taiwanese P[6] strain belonged to genotype G12. Sequencing and phylogenetic analysis of its VP7 gene classified this strain into a geographically widespread lineage that emerged during the late 1990s (Fig. 2c). This strain shared a nucleotide similarity of ⩾99% with strains belonging to the globally common lineage and <91% nucleotide similarity with an early human G12 isolate (L26) and a porcine G12 rotavirus (RU172), representing a unique lineage of the G12 VP7 gene.
Molecular characterization of the VP6 and NSP4 genes
The full or partial coding regions of the VP6 and NSP4 genes were determined to further characterize the five Taiwanese P[6] strains.
For the VP6 gene, a 1000 bp fragment was used in the analysis. The rotavirus genotyping tool, RotaC (Maes et al., 2009), identified two VP6 genotypes among the Taiwanese P[6] strains (Fig. 3a). Two G4P[6] strains and the G12P[6] strain belonged to the I1 VP6 genotype, whilst a third G4P[6] strain and the G5P[6] strain had the I5 VP6 genotype. Phylogenetic analysis clustered the two G4 strains with I1 VP6 specificity into a common lineage with Wa and two human–porcine reassortant strains, identified in Ecuador and India, respectively. The G12 strain shared high nucleotide similarity (up to 100%) with modern I1 strains from a global collection. Of interest, the G4 and G5 strains that shared the I5 genotype clustered on the same branch of the phylogenetic tree and were more closely related to Taiwanese P[19] strains (Wu et al., 2011).
Fig. 3.
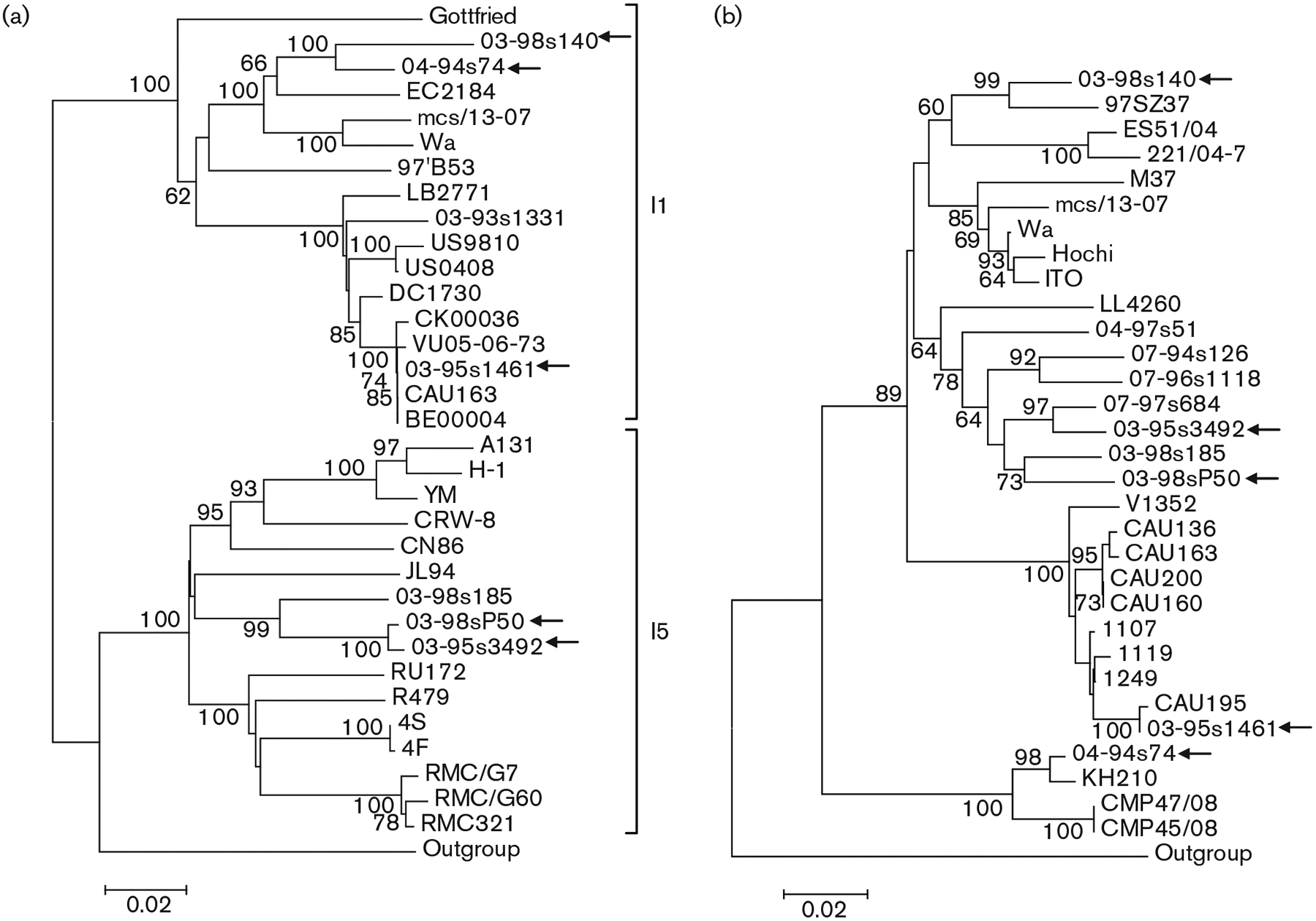
Nucleotide sequence-based neighbour-joining phylogenetic trees of the VP6 (a) and NSP4 (b) genes of Taiwanese P[6] strains. Arrows indicate the Taiwanese P[6] strains. Bootstrap values (500 replicates) >60% are indicated. In (a), the genotypes are indicated. Bars, 0.02 nucleotide substitutions per site.
RotaC classified all five Taiwanese P[6] strains into a single NSP4 genotype, E1, with >85% nucleotide similarity to the reference strains. The Taiwanese P[6] strains clustered into four distantly related phylogenetic lineages. The single G12P[6] strain formed a common branch with recent Indian, South Korean and US G12 strains. One G4 strain, 04-94s74, clustered with a rare human G5P[6] strain identified in Vietnam and with porcine strains from Thailand. Another G4 strain, 03-98s140, clustered with an unusual Chinese human G9 strain, and was also related to European porcine strains. The third Taiwanese G4P[6] strain together with the Taiwanese G5P[6] strain clustered with previously characterized uncommon P[19] strains identified in Taiwanese children with diarrhoea (Fig. 3b).
DISCUSSION
It was once thought that genotype P[6] rotaviruses were restricted to asymptomatic nosocomial infections of neonates born in hospital nurseries (Gentsch et al., 2005). Subsequently, this genotype has been implicated in acute dehydrating diarrhoea and has been found as the third most common VP4 genotype in human rotaviruses worldwide (Bányai et al., 2012). However, the various lineages of P[6] strains associated with human infections show marked geographical differences. For example, epidemiologically major P[6] strains, including the abundant African P[6] strains carrying various G types as well as the globally spread G9 and G12 strains, mainly carry a single common P[6] lineage, Ia (Freeman et al., 2009; László et al., 2009; Pietsch & Liebert, 2009; Jere et al., 2011; Le et al., 2011). Based on epidemiological evidence, it is thought that these P[6] strains represent a true human rotavirus VP4 lineage. However, a recent report of a bat G25 rotavirus carrying the same P[6] lineage has indicated that animals may also play a role in the circulation of this gene. Whether such potential animal reservoirs of P[6]-Ia could transmit the virus to humans remains to be elucidated (Esona et al., 2010).
In many regions where genotype P[6] strains are uncommon causes of gastroenteritis in humans, evolutionary analysis of the P[6] VP4 gene has usually identified lineages that are highly divergent from the original P[6] gene found in neonates. Examples include the G4P[6] strains from Hungary, a G3P[6] strain from Italy, G5P[6] strains from China and Vietnam, G4P[6] strains from Brazil and Argentina, a G9P[6] strain from India and a G11P[6] strain from Ecuador. These uncommon lineages are often shared among human and animal rotaviruses, and it is thought that their occurrence in humans might be the result of interspecies transmission and reassortment between human and porcine strains. Both partial and whole genome-based characterization studies provide convincing evidence for this hypothesis (Ahmed et al., 2007; Nguyen et al., 2007; Mascarenhas et al., 2007; Li et al., 2008; Martella et al., 2008; Bányai et al., 2009a, b; Mukherjee et al., 2009, 2011; Stupka et al., 2009; Wang et al., 2010).
This report has described the molecular characterization of P[6] strains from sporadic cases of rotavirus gastroenteritis in Taiwanese children admitted to hospital. In total, five out of 1831 strains carrying P[6] specificity were identified that belonged to VP7 genotypes G4 (n=3), G5 (n=1) and G12 (n=1). These findings demonstrated that the P[6] genotype had little epidemiological importance in Taiwan during 2005–2010. Molecular characterization of these five P[6] strains revealed that the VP4 gene of the G12P[6] strain clustered in the globally common human lineage (Ia), whilst the other four Taiwanese P[6] strains clustered with strains in a lineage shared between porcine and human strains.
The VP7, VP6 and NSP4 genes of the unusual P[6] strains were related more closely to the respective genes of porcine rotaviruses or to a distantly related strain whose origin is unclear, but is clearly different from human strains, further strengthening a possible common origin with porcine strains. In contrast, the G12P[6] strain carried corresponding genes related to common human rotaviruses. Whilst the G12P[6] strain did not seem to have the potential to cause large outbreaks in Taiwan during 2006 or later, this study is the first to report the occurrence of this strain in Taiwan, implying that G12 rotaviruses may have been introduced only recently. In some countries, G12P[6] strains have been found to be able to spread and cause local epidemics (Bányai et al., 2012). Thus, surveillance is needed to determine whether this particular genotype will emerge over time and become medically important in Taiwan.
Another conclusion from the molecular characterization of porcine rotavirus-like Taiwanese P[6] strains was that they are related more closely to local non-P[6] strains than to unusual P[6] strains detected worldwide, suggesting that they could have emerged locally. For example, these P[6] strains shared more sequence similarity with some Taiwanese P[19] strains than with other known strains from global collections (Wu et al., 2011). This finding reaffirms that locally circulating animal rotaviruses – porcine strains, in this case – may serve as a source for infection in infants and young children through interspecies transmission coupled with gene reassortment. That such putative interspecies transmission and reassortment events are independent events is quite likely, based on the finding that a porcine rotavirus-like G5 VP7 gene combined with either a P[19] or an uncommon P[6] VP4 gene, or the shared P[6] VP4 gene combined with porcine rotavirus-like G4 and G5 VP7 genes, were identified in various Taiwanese surveillance areas from different detection periods. Thus, the probability that these reassortment events occurred in the human host with the involvement of two different porcine strains seems negligible. Whilst the proportion of porcine rotavirus-like strains in Taiwan is low, the circulation and sporadic transmission of these heterotypic strains from pigs to humans could pose a public-health concern even in the vaccine era. Continuous surveillance is needed to detect such possible vaccine breakthrough events associated with the introduction of heterologous strains.
ACKNOWLEDGEMENTS
This study was financially supported in part by the Centers for Disease Control, Department of Health, Execute Yuan, Taiwan (research grant DOH98-DC-1005) and by the National Research Program for Genomic Medicine (supported grants: 94-0324-19-F-01-00-00, 95-0324-19-F01-00-00-00-35, 96-0324-01-F-01). K. B. is funded by the Hungarian Academy of Sciences (OTKA, PD76364 and K100727; ‘Momentum Program’, LP2011-010/2011).
REFERENCES
- Ahmed K, Anh DD & Nakagomi O (2007). Rotavirus G5P[6] in child with diarrhea, Vietnam. Emerg Infect Dis 13, 1232–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bányai K, Martella V, Jakab F, Melegh B & Szücs G (2004). Sequencing and phylogenetic analysis of human genotype P[6] rotavirus strains detected in Hungary provides evidence for genetic heterogeneity within the P[6] VP4 gene. J Clin Microbiol 42, 4338–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bányai K, Esona MD, Kerin TK, Hull JJ, Mijatovic S, Vásconez N, Torres C, de Filippis AM, Foytich KR & Gentsch JR (2009a). Molecular characterization of a rare, human–porcine reassortant rotavirus strain, G11P[6], from Ecuador. Arch Virol 154, 1823–1829. [DOI] [PubMed] [Google Scholar]
- Bányai K, Bogdán Á, Domonkos G, Kisfali P, Molnár P, Tóth A, Melegh B, Martella V, Gentsch JR & Szucs G (2009b). Genetic diversity and zoonotic potential of human rotavirus strains, 2003– 2006, Hungary. J Med Virol 81, 362–370. [DOI] [PubMed] [Google Scholar]
- Bányai K, László B, Duque J, Steele AD, Nelson EAS, Gentsch JR & Parashar UD (2012). Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine 30 (Suppl. 1), A122–A130. [DOI] [PubMed] [Google Scholar]
- Esona MD, Geyer A, Banyai K, Page N, Aminu M, Armah GE, Hull J, Steele DA, Glass RI & Gentsch JR (2009). Novel human rotavirus genotype G5P[7] from child with diarrhea, Cameroon. Emerg Infect Dis 15, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona MD, Mijatovic-Rustempasic S, Conrardy C, Tong S, Kuzmin IV, Agwanda B, Breiman RF, Banyai K, Niezgoda M & other authors (2010). Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum). Emerg Infect Dis 16, 1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes MK & Kapikian AZ (2007). Rotaviruses. In Fields Virology, 5th edn, vol. 2, pp. 1917–1974. Edited by Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B & Straus SE. Philadelphia, PA: Lippincott Williams & Wilkins. [Google Scholar]
- Flores J, Midthun K, Hoshino Y, Green K, Gorziglia M, Kapikian AZ & Chanock RM (1986). Conservation of the fourth gene among rotaviruses recovered from asymptomatic newborn infants and its possible role in attenuation. J Virol 60, 972–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MM, Kerin T, Hull J, Teel E, Esona M, Parashar U, Glass RI & Gentsch JR (2009). Phylogenetic analysis of novel G12 rotaviruses in the United States: a molecular search for the origin of a new strain. J Med Virol 81, 736–746. [DOI] [PubMed] [Google Scholar]
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK & Bhan MK (1992). Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol 30, 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O & other authors (2005). Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis 192 (Suppl. 1), S146–S159. [DOI] [PubMed] [Google Scholar]
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B & Fang ZY (1990). Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol 28, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jere KC, Mlera L, O’Neill HG, Potgieter AC, Page NA, Seheri ML & van Dijk AA (2011). Whole genome analyses of African G2, G8, G9, and G12 rotavirus strains using sequence-independent amplification and 4541 pyrosequencing. J Med Virol 83, 2018–2042. [DOI] [PubMed] [Google Scholar]
- László B, Nyúl Z, Kisfali P, Deák J, Kovács J, Kónya J, Mészner Z, Molnár P, Pátri L & other authors (2009). First detection of P[6],G9 rotaviruses in Hungary – an imported strain from India? J Travel Med 16, 141–143. [DOI] [PubMed] [Google Scholar]
- Le VP, Kim JB, Shon DH, Chung IS, Yoon Y, Kim K, Chung S-I, Lim I & Kim W (2011). Molecular characterization of rare G12P[6] rotavirus isolates closely related to G12 strains from the United States, CAU 195 and CAU 214. Arch Virol 156, 511–516. [DOI] [PubMed] [Google Scholar]
- Li D-D, Duan Z-J, Zhang Q, Liu N, Xie Z-P, Jiang B, Steele D, Jiang X, Wang Z-S & Fang Z-Y (2008). Molecular characterization of unusual human G5P[6] rotaviruses identified in China. J Clin Virol 42, 141–148. [DOI] [PubMed] [Google Scholar]
- Maes P, Matthijnssens J, Rahman M & Van Ranst M (2009). RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol 9, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V, Bányai K, Ciarlet M, Iturriza-Gómara M, Lorusso E, De Grazia S, Arista S, Decaro N, Elia G & Cavalli A (2006). Relationships among porcine and human P[6] rotaviruses: evidence that the different human P[6] lineages have originated from multiple interspecies transmission events. Virology 344, 509–519. [DOI] [PubMed] [Google Scholar]
- Martella V, Colombrita D, Lorusso E, Draghin E, Fiorentini S, De Grazia S, Bányai K, Ciarlet M, Caruso A & Buonavoglia C (2008). Detection of a porcine-like rotavirus in a child with enteritis in Italy. J Clin Microbiol 46, 3501–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V, Bányai K, Matthijnssens J, Buonavoglia C & Ciarlet M (2010). Zoonotic aspects of rotaviruses. Vet Microbiol 140, 246–255. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JD, Leite JP, Lima JC, Heinemann MB, Oliveira DS, Araújo IT, Soares LS, Gusmão RH, Gabbay YB & Linhares AC (2007). Detection of a neonatal human rotavirus strain with VP4 and NSP4 genes of porcine origin. J Med Microbiol 56, 524–532. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gómara M, Maes P & other authors (2008a). Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 82, 3204–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Bányai K, Estes MK, Gentsch JR, Iturriza-Gómara M, Kirkwood CD & other authors (2008b). Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153, 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Bilcke J, Ciarlet M, Martella V, Bányai K, Rahman M, Zeller M, Beutels P, Van Damme P & Van Ranst M (2009). Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol 4, 1303–1316. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Bányai K, Brister JR, Buesa J, Esona MD, Estes MK & other authors (2011). Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156, 1397–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Rahman M, Martella V, Xuelei Y, De Vos S, De Leener K, Ciarlet M, Buonavoglia C & Van Ranst M (2006). Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J Virol 80, 3801–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijatovic-Rustempasic S, Bányai K, Esona MD, Foytich K, Bowen MD & Gentsch JR (2011). Genome sequence based molecular epidemiology of unusual US rotavirus A G9 strains isolated from Omaha, USA between 1997 and 2000. Infect Genet Evol 11, 522–527. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Dutta D, Ghosh S, Bagchi P, Chattopadhyay S, Nagashima S, Kobayashi N, Dutta P, Krishnan T & other authors (2009). Full genomic analysis of a human group A rotavirus G9P[6] strain from Eastern India provides evidence for porcine-to-human interspecies transmission. Arch Virol 154, 733–746. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Ghosh S, Bagchi P, Dutta D, Chattopadhyay S, Kobayashi N & Chawla-Sarkar M (2011). Full genomic analyses of human rotavirus G4P[4], G4P[6], G9P[19] and G10P[6] strains from North-eastern India: evidence for interspecies transmission and complex reassortment events. Clin Microbiol Infect 17, 1343–1346. [DOI] [PubMed] [Google Scholar]
- Nakagomi T, Horie Y, Koshimura Y, Greenberg HB & Nakagomi O (1999). Isolation of a human rotavirus strain with a super-short RNA pattern and a new P2 subtype. J Clin Microbiol 37, 1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TA, Khamrin P, Trinh QD, Phan TG, Pham D, Hoang P, Hoang KT, Yagyu F, Okitsu S & Ushijima H (2007). Sequence analysis of Vietnamese P[6] rotavirus strains suggests evidence of interspecies transmission. J Med Virol 79, 1959–1965. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, & Deerfield DW II (1997). GeneDoc: analysis and visualization of genetic variation. EMBnet News 4 (2, ), 1–4. http://www.nrbsc.org/gfx/genedoc/ebinet.htm [Google Scholar]
- Pietsch C & Liebert UG (2009). Human infection with G12 rotaviruses, Germany. Emerg Infect Dis 15, 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupka JA, Carvalho P, Amarilla AA, Massana M, Parra GI & Argentinean National Surveillance Network for Diarrheas (2009). National rotavirus surveillance in Argentina: high incidence of G9P[8] strains and detection of G4P[6] strains with porcine characteristics. Infect Genet Evol 9, 1225–1231. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M & Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H, Kobayashi N, Nagashima S, Zhou X, Ghosh S, Peng J-S, Hu Q, Zhou D-J & Yang Z-Q (2010). Full genomic analysis of a porcine–bovine reassortant G4P[6] rotavirus strain R479 isolated from an infant in China. J Med Virol 82, 1094–1102. [DOI] [PubMed] [Google Scholar]
- Wu FT, Liang SY, Tsao KC, Huang CG, Lin CY, Lin JS, Su CY, Eng HL, Yang JY & other authors (2009). Hospital-based surveillance and molecular epidemiology of rotavirus infection in Taiwan, 2005–2007. Vaccine 27 (Suppl. 5), F50–F54. [DOI] [PubMed] [Google Scholar]
- Wu F-T, Bányai K, Huang JC, Wu H-S, Chang F-Y, Yang J-Y, Hsiung CA, Huang Y-C, Lin J-S & other authors (2011). Diverse origin of P[19] rotaviruses in children with acute diarrhea in Taiwan: detection of novel lineages of the G3, G5, and G9 VP7 genes. J Med Virol 83, 1279–1287. [DOI] [PubMed] [Google Scholar]


