Abstract
In this study the emergence of rotavirus A genotype G12 in children <5 years of age is reported from Cameroon during 2010/2011. A total of 135 human stool samples were P and G genotyped by reverse transcriptase PCR. Six different rotavirus VP7 genotypes were detected, including G1, G2, G3, G8, G9, and G12 in combinations with P[4], P[6] and P[8] VP4 genotypes. Genotype G12 predominated in combination with P[8] (54.1%) and P[6] (10.4%) genotypes followed by G1P[6] (8.2%), G3P[6] (6.7%), G2P[4] (5.9%), G8P[6] (3.7%), G2P[6] (0.7%), G3P[8] (0.7%), and G9P[8] (0.7%). Genotype P[6] strains in combination with various G-types represented a substantial proportion (N = 44, 32.6%) of the genotyped strains. Partially typed strains included G12P[NT] (2.2%); G3P[NT] (0.7%); G(NT)P[6] (1.5%); and G(NT)P [8] (0.7%). Mixed infections were found in five specimens (3.7%) in several combinations including G1 + G12P[6], G2 + G3P[6] + P[8], G3 + G8P[6], G3 + G12P[6] + P[8], and G12P [6] + P[8]. The approximately 10% relative frequency of G12P[6] strains detected in this study suggests that this strain is emerging in Cameroon and should be monitored carefully as rotavirus vaccine is implemented in this country, as it shares neither G- nor P-type specificity with strains in the RotaTeq and Rotarix vaccines. These findings are consistent with other recent reports of the global spread and increasing epidemiologic importance of G12 and P[6] strains.
Keywords: genotype, surveillance, RT-PCR
INTRODUCTION
Group A rotaviruses are the most important etiological agents of acute gastroenteritis in infants and young children worldwide [Estes and Kapikian, 2007]. Current mortality estimates demonstrate that about 453,000 children <5 years of age die worldwide each year due to rotavirus [Tate et al., 2012]. Rotaviruses (family Reoviridae, genus Rotavirus) are icosahedral viruses with genome consisting of 11 segments of dsRNA [Attoui et al., 2012], encoding six structural viral proteins (VP1–VP4, VP6, and VP7) and six non-structural proteins (NSP1–NSP6) [Estes and Kapikian, 2007]. Due to the segmented nature of its genome, novel rotavirus strains can be produced in vivo by exchange of genome segments between two parental strains infecting the same cell, a process called reassortment [Ramig, 1997].
Traditionally, rotaviruses have been classified using a binomial nomenclature GxP[x] based upon serotype and genotype specificities of the outer capsid antigens, VP7 (G-type) and VP4 (P-type) [Estes and Kapikian, 2007]. Thus far, at least 27 G genotypes and 35 P genotypes have been detected from humans and animals [Abe et al., 2009; Esona et al., 2009; Schumann et al., 2009; Solberg et al., 2009; Ursu et al., 2009; Matthijnssens et al., 2011]. Five G genotypes (G1, G2, G3, G4, and G9), and two P genotypes (P[8] and P[4]) predominate worldwide, although genotype P[6] has been recognized as a common cause of diarrhea on several continents and has sometimes been described as a regionally common strain [Steele and Ivanoff, 2003; Gentsch et al., 2005; Santos and Hoshino, 2005; Bányai et al., 2012]. A large number of rare or regionally common strains have been identified during surveillance in anticipation of vaccines introduction including G5, G6, G8, G9, G10, and G12 genotypes and P[1], P[3], P[6], P[9], P[11], P[14], P[19], and P[25] genotypes [Esona et al., 2004, 2009; Gentsch et al., 2005; Rahman et al., 2005; Bányai et al., 2007; Bányai et al., 2012; Castello et al., 2009; Cunliffe et al., 2009; Payne et al., 2009; Martella et al., 2010; Matthijnssens et al., 2010]. Studies from Africa reported high prevalence of genotypes G8 and P[6] in various combinations suggesting that both of these genotypes should be considered common in Africa. [Cunliffe et al., 1999; Armah et al., 2010]. Steele and Ivanoff [2003] concluded that the predominant strains circulating across Africa during 1996–1999 were G1P[6] and G3P[6] strains.
Many of the newly described genotypes are thought to be of animal origin, including G9 that emerged subsequently as globally common G9P[8] strains [Gentsch et al., 2005; Santos and Hoshino, 2005; Bányai et al., 2012]. While G9 strains were reported from the late 1990s in Africa, G12 strains were found spreading only from mid-2000s onward, in Cameroon, Ethiopia, Ghana, Malawi, South Africa, and Zimbabwe [Bányai et al., 2012] at low-to-moderate frequencies (<20%). However, reports from the majority of African countries is scanty and only one third of all countries in the WHO’s African region reported any data during 1990–2006 [Todd et al., 2010; Bányai et al., 2012]. Although rotaviruses have been reported as an important etiological agent of diarrhea in Cameroon [Koulla-Shiro et al., 1995; Esona et al., 2003; Ndze et al., 2012], there is a need to obtain comprehensive baseline data on rotavirus genetic diversity for Cameroon in anticipation of the introduction of rotavirus vaccine [Esona et al., 2004, 2009, 2010; Armah et al., 2010].
Two rotavirus vaccines have been developed as effective interventions for severe diarrhea and mortality associated with rotavirus infection. The two live oral vaccines from Merck (RotaTeq, Whitehouse Station, NJ) and GlaxoSmithKline (Rotarix, Rixensart, Belgium) have been licensed in more than 100 countries and have been introduced into routine immunization programs in the United States and some countries in Latin America, Europe, Africa and Asia [Dennehy, 2008]. RotaTeq possesses genes encoding human rotavirus serotypes G1–G4 and P1A[8] on a bovine rotavirus background. This mixture of the five reassortant vaccine strains was designed to stimulate serotype-specific protection to these common rotavirus serotypes. In contrast, a G1P[8] vaccine, Rotarix, was constructed to induce both homotypic responses to G1P[8] strains and heterotypic protective immune responses against different serotypes.
It is considered important for countries considering vaccine introduction into their routine immunization programs to assess the need for the vaccine through disease burden studies. These studies provide baseline data on disease burden and strain prevalence to assess better the impact of the vaccination program on rotavirus disease and circulating strains, and to identify the possible emergence of serotypes that escape vaccine-induced immunity. In this study data on circulating genotypes are presented in the Far Northern (two sites) and North Western (two sites) regions of Cameroon during 2010–2011.
MATERIALS AND METHODS
Study Population and Specimens
In this study, rotavirus strains detected during 2010–2011 in two regions of Northern Cameroon [Ndze et al., 2012] were characterized. In brief, samples were collected from children <5 years of age who presented with acute diarrhea at the Regional Hospital Maroua and at the Domayo Djama integrated health center in the Far North region; and at the Regional hospital Bamenda and at the Esu integrated health centre in the North West region. The population is urban (Maroua and Bamenda) and rural (Esu) and the major occupation in the study area is subsistence agriculture supplemented with fishing and dairy farming in the Far North region.
The Far North is located at latitude 11°00′N and longitude 14°30′E characterized by tropical and Sahelian climates with rainfall ranging from 400 to 900 mm per year. The region is generally dry and hot with rains falling relatively more frequently in the Mandara region. The Far North has two seasons: one dry, and one wet. The climate in Maroua is of the Sahel type. Temperatures reach their highest levels from January to May. The North West region of Cameroon is located between latitude 5°20′ and 7°10′N and longitudes 9°4′ and 11°15′E with altitude range of 300–3,000 m above sea level. Temperatures vary within the year between 10 and 28°C and mean annual rainfall is 2,000 mm, while relative humidity ranges from 49% to 87% [Bayemi et al., 2005].
RNA Extraction and Genotyping
This study included a subset of the rotavirus-positive stool specimens analyzed previously by a one-step reverse transcription polymerase chain reaction (RT-PCR) for VP6 protein [Ndze et al., 2012]. Rotavirus dsRNA was extracted from 100 μl stool suspension with the Promega DNA & RNA Purification kit (Madison, WI) using an automated extractor [Gyuranecz et al., 2011]. The extracted dsRNA of each strain was denatured at 97°C for 5 min and then reverse transcription-PCR (RT-PCR) was carried out using a One-Step RT-PCR kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. Previously published consensus primers [Gentsch et al., 1992; Das et al., 1994; Iturriza-Gomara et al., 2001] were used for the amplification of the VP4 and VP7 gene segments. Genotyping of the VP4 and VP7 genes was done by semi-nested RT-PCR assay [Gouvea et al., 1990; Gentsch et al., 1992; Das et al., 1994] using first round PCR products generated by consensus primers. For G- and P-type determination, a series of type-specific primers were utilized (including G1–G4, G8, G9, G10, and G12 primers and P[4], P[6], P[8], P[9], P[10], and P[11] primers [Gouvea et al., 1990; Gentsch et al., 1992; Iturriza-Gomara et al., 2000, 2001, 2004]. For a randomly selected subset of strains representing distinct genotypes sequencing was carried out to confirm G- and P-typing data obtained by the multiplex genotyping PCR assay. Sequencing was also utilized occasionally when samples remained non-typeable.
RESULTS
Prevalence of Individual G Types
A total of 144 samples were subjected to G genotyping by a multiplex RT-PCR method. After the first amplification step, PCR products for the VP7 gene were obtained for 135 (94%) of the specimens, and the remaining nine specimens yielded no visible first-round product. Only the 135 specimens with visible first-round amplification products were subjected to further analysis (Table I).
TABLE I.
Rotavirus G- and P-Types Circulating in the North West and Far North Regions of Cameroon (2010–2011)
| No. of strains of the following G-type | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-type | G1 | G2 | G3 | G8 | G9 | G12 | G1G12 | G2G3 | G3G8 | G3G12 | GNT | Total |
| P[4] | — | 8 | — | — | — | — | — | — | — | — | — | 8 |
| P[6] | 11 | 1 | 9 | 5 | — | 14 | 1 | — | 1 | — | 2 | 44 |
| P[8] | — | — | 1 | — | 1 | 73 | — | — | — | — | 1 | 76 |
| P[6] + P[8] | — | — | — | — | — | 1 | — | 1 | — | 1 | — | 3 |
| P[NT] | — | — | 1 | — | — | 3 | — | — | — | — | — | 4 |
| Total | 11 | 9 | 11 | 5 | 1 | 91 | 1 | 1 | 1 | 1 | 3 | 135 |
NT, non-typeable.
In all, a total of six different rotavirus VP7 genotypes were observed, including G1, G2, G3, G8, G9, and G12. The predominant genotype was G12 (67.4%), followed by G1 and G3 (8.1% each), G2 (6.6%), G8 (3.7%), and G9 (0.7%). Mixed infections (G1 + G12, G2 + G3, G3 + G8, and G3 + G12) were observed in four specimens (2.9%; Table I).
Prevalence of Individual P-Types
First round PCR products for the VP4 gene were obtained for 135 specimens. A total of three different VP4 genotypes (P[4], P[6], and P[8]) were observed. Genotype P[8] (56.3%) predominated followed by P[6] (32.6%) and P[4] (5.9%). Mixed infections P[6] + P[8] were found in three specimens (2.2%).
G–P Combinations
When G- and P-type data were combined the following genotype prevalence was observed: G12P[8] (54.1%), G12P[6] (10.4%), G1P[6] (8.2%), G3P[6] (6.7%), G2P[4] (5.9%), G8P[6] (3.7%), and G2P[6], G3P[8] plus G9P[8] (0.7% each). The following partial G–P combinations were identified; G12P[NT] (2.2%); G3P[NT] (0.7%); GNTP[6] (1.5%); and GNTP[8] (0.7%). Mixed infections were found in five specimens (3.7%) and presented with these combinations: G1 + G12P[6], G2 + G3P[6] + P[8], G3 + G8P[6], G3 + G12P[6] + P[8], and G12P[6] + P[8].
Regional Distribution of Most Common Rotavirus Strains
The G–P combination analysis by site showed that G12P[8] (54%) dominated in Esu followed by G8P[6] (31%), while in Bamenda G2P[4] (38%) dominated, followed by G12P[6] (33%) and G12P[8] (29%), and in Maroua G12P[8] (59%) dominated, followed by G1P [6] (11%) and G3P[6] (9%; Fig. 1). Mixed infections were detected only in Maroua.
Fig. 1.
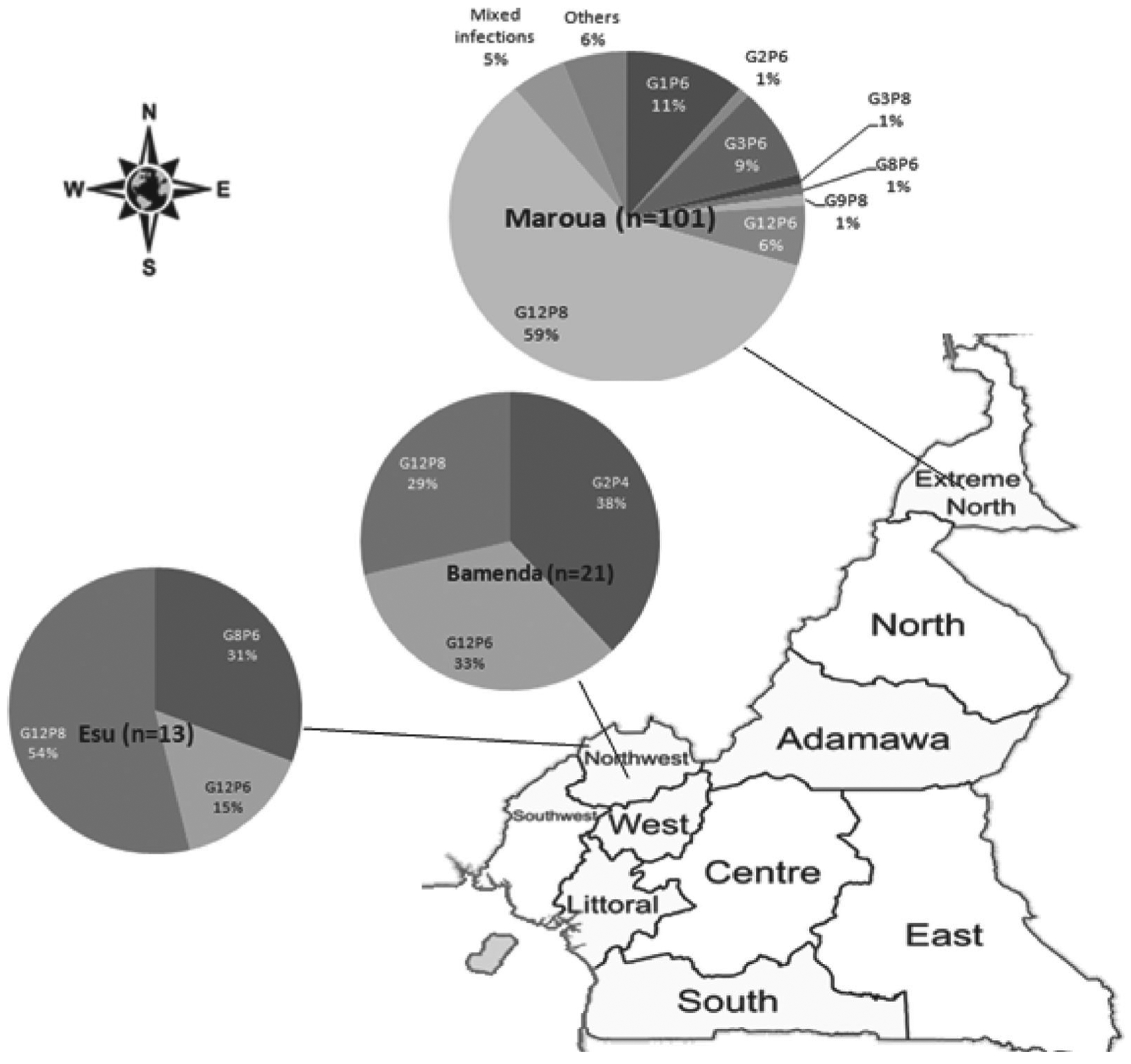
Rotavirus G and P genotypes combinations circulating in Northern Cameroon (North West: Esu and Bamenda; Far North: Maroua).
DISCUSSION
The objective of this study was to identify the circulating rotavirus genotypes from infected hospitalized and outpatient children <5 years of age in Cameroon. Previous studies from Cameroon reported the circulation of G1–G5, G8, G9, and G12 VP7 types as well as P[4] and P[6]–P[10] VP4 types during the 1996–2000 and the 2007–2008 surveillance period [Esona et al., 2004, 2009, 2010; Armah et al., 2010; Mwenda et al., 2010], although G12 strains were found only in 11% of samples during 2007–2008 [Mwenda et al., 2010]. Despite the limited length period and the relatively small sample size some novel findings relevant to strain diversity were made in the present study as well.
A total of six different rotavirus VP7 genotypes were identified, including G1, G2, G3, G8, G9, and G12. The predominant genotype was G12 (67.4%; 91/135) in combination with P[8] (54.1%) and P[6] (10.4%) genotypes. Although G12 strains were in circulation in 2007/2008 in sub-Saharan regions [Mwenda et al., 2010], to the best of our knowledge, no study in Africa has demonstrated such a high prevalence of G12 strains, suggesting a major out-break of this strain in the study area during 2010/2011. In neighboring and other sub-Saharan countries different strains were more prevalent in this period as documented in the annual report of the WHO’s Global Rotavirus Surveillance Network information bulletin [WHO, 2012]. For example, G3P[6] and G12P[8] strains were prevalent in Togo, G2P[4] and G2P[6] predominated in Guinea Bissau and in Democratic Republic of the Congo, G3P[6] was common in Nigeria, and G9P[8] was abundant in Cote d’Ivoir. Genotype G12 strains (mainly G12P[8]) were found to be in circulation at relatively low detection rates in Nigeria, Ghana, and Democratic Republic of the Congo, but were also detected in East African countries, including Uganda and Tanzania, and were very prevalent in Ethiopia (with >30% relative frequency). Data on strain prevalence from this period have been not specified for Cameroon in the WHO’s Global Rotavirus Information and Surveillance Bulletin.
Data on mixed infections and non-typeable strains in this study showed lower frequency than reported usually from developing countries [Bányai et al., 2012]. The utilization of an improved genotyping protocol implemented by the European rotavirus strain monitoring network (EuroRotaNet [Iturriza-Gomara et al., 2009, 2011]) combined with nucleotide sequencing could be one reason that the proportion of non-typeable strains could be minimized. In fact, Esona et al. [2010] demonstrated that the majority of non-typeable strains collected across Africa could be genotyped and most of them belonged to one of the major genotypes when improved typing reagents (i.e., modified typing primers) or methods (i.e., sequencing) were utilized. Nonetheless, in this study a small portion (~6%) of strains that did not produce a first round product were omitted from routine genotyping and it is possible that some of the rare human rotavirus genotypes were present among these few strains.
Some studies have shown that both vaccines currently in use worldwide are protective against a variety of circulating strains. Examples include G1 to G4, G8, and G9. Vaccine effectiveness has been demonstrated against G12 strains in the United States with an estimated 85–100% protection [Payne et al., 2011], which was somewhat reduced in parts of Africa (20–60% effectiveness) [Madhi et al., 2010; Cunliffe et al., 2012; Steele et al., 2012]. Current theories suggest that vaccine failure may not be a direct consequence of the circulation of uncommon rotavirus strains in African countries [Cunliffe et al., 2012]. Nonetheless it will be important to monitor the spatiotemporal dynamics of emerging strains, which might become predominant after rotavirus vaccines are used in massive immunization programs. Those strains that share no G- and P-type specificities with strains in RotaTeq and Rotarix may be challenging to existing rotavirus vaccines. This hypothesis was raised during mass vaccination conducted with the Rotarix vaccine in parts of Latin America, Australia, or Europe, where G2P[4] strains dominated other strains over several consecutive rotavirus seasons. However, rotavirus experts agree that the possibility that this re-emergence of G2P[4] strains was the result of natural fluctuation of rotavirus strains could not be excluded either [Matthijnssens et al., 2009].
The findings from this study document the rotavirus genotypes circulating in Cameroon and the emergence of genotype G12. The predominance of genotype G12 strengthens the evidence indicating that G12 is emergent worldwide. Any vaccine used in Cameroon will need to be effective against unusual rotavirus strains, such as G8, G12 and P[6]. The detection of emerging new strains re-enforces the need for enhanced rotavirus surveillance in humans and animals. Complete genome studies are needed to understand better the full picture of the circulating rotavirus strains in this geographic region.
ACKNOWLEDGMENTS
We acknowledge the support from the Ministry of Women Empowerment and the Family Cameroon for financial support during sample collection. Further supports were given by the Hungarian Scientific Research Fund (OTKA, K100727). We also thank the technical assistance of Anett at the pathogen discovery unit, Veterinary Medical research institute, Budapest Hungary and Ngeng Marcel Bong at Regional Hospital Maroua.
Glossary
- NT
non-typeable
Footnotes
Publisher's Disclaimer: Disclaimer: The findings and conclusions in this report are those of the authors and do not represent necessarily the views of the CDC.
REFERENCES
- Abe M, Ito N, Morikawa S, Takasu M, Murase T, Kawashima T, Kawai Y, Kohara J, Sugiyama M. 2009. Molecular epidemiology of rotaviruses among healthy calves in Japan: Isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Res 144:250–257. [DOI] [PubMed] [Google Scholar]
- Armah GE, Steele AD, Esona MD, Akran VA, Nimzing L, Pennap G. 2010. Diversity of rotavirus strains circulating in West Africa from 1996 to 2000. J Infect Dis 202:S64–S71. [DOI] [PubMed] [Google Scholar]
- Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, Chappell JD, Ciarlet M, del Vas M, Dermody TS, Dormitzer PR, Duncan R, Fcang Q, Graham R, Guglielmi KM, Harding RM, Hillman B, Makkay A, Marzachì C, Matthijnssens J, Milne RG, Mohd Jaafar F, Mori H, Noordeloos AA, Omura T, Patton JT, Rao S, Maan M, Stoltz D, Suzuki N, Upadhyaya NM, Wei C, Zhou H. 2012. Reoviridae. In: King A, Adams M, Carstens E, Lefkwitz E, editors. Virus taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. Waltham, MA: Elsevier Academic Press, pp 497–650. [Google Scholar]
- Bányai K, Bogdan A, Kisfali P, Molnar P, Mihaly I, Melegh B, Martella V, Gentsch JR, Szucs G. 2007. Emergence of serotype G12 rotaviruses, Hungary. Emerg Infect Dis 13:916–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bányai K, Laszlo B, Vojdani J, Steel AD, Nelson EAS, Gentch JR, Parashar UD. 2012. Systematic review of regional and temporal trends in the global rotavirus strain diversity in the pre rotavirus vaccine era: Insights for understanding the impact of rotavirus vaccination programs. Vaccines 30:A122–A130. [DOI] [PubMed] [Google Scholar]
- Bayemi PH, Bryant MJ, Perera BMAO, Mbanya JN, Cavestany D, Webb EC. 2005. Milk production in Cameroon: A review. Livestock Res Rural Dev 17:60. [Google Scholar]
- Castello AA, Nakagomi T, Nakagomi O, Jiang B, Kang JO, Glass RI, Glikmann G, Gentsch JR. 2009. Characterization of genotype P[9]G12 rotavirus strains from Argentina: High similarity with Japanese and Korean G12 strains. J Med Virol 81:371–381. [DOI] [PubMed] [Google Scholar]
- Cunliffe NA, Gondwe JS, Broadhead RL, Molyneux ME, Woods PA, Bresee JS, Glass RI, Gentsch JR, Hart CA. 1999. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: Predominance of novel P[6]G8 strains. J Med Virol 57:308–312. [PubMed] [Google Scholar]
- Cunliffe NA, Ngwira BM, Dove W, Nakagomi O, Nakagomi T, Perez A, Hart CA, Kazembe PN, Mwansambo CCV. 2009. Serotype G12 rotaviruses, Lilongwe, Malawi. Emerg Infect Dis 15:87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe NA, Witte D, Ngwira BM, Todd S, Bostock NJ, Turner AN, Chimpeni P, Victor JC, Steele AD, Bouckenooghe A, Neuzil KM. 2012. Efficacy of human rotavirus vaccine against severe gastroenteritis in Malawian children in the first two years of life: A randomized, double-blind, placebo controlled trial. Vaccine 30S:A36–A43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, Kumar R, Bhan MK, Glass RI. 1994. Characterization of rotavirus strains from newborns in New-Delhi, India. J Clin Microbiol 32:1820–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy PH. 2008. Rotavirus vaccines: An overview. Clin Microbiol Rev 21:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona MD, Armah GE, Steele AD. 2003. Molecular epidemiology of rotavirus infection in Western Cameroon. J Trop Pediatr 49:160–163. [DOI] [PubMed] [Google Scholar]
- Esona MD, Armah GE, Geyer A, Steele AD. 2004. Detection of an unusual human rotavirus strain with G5P[8] specificity in a Cameroonian child with diarrhea. J Clin Microbiol 42:441–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona MD, Geyer A, Banyai K, Page N, Aminu M, Armah GE, Hull J, Steele AD, Glass RI, Gentsch JR. 2009. Novel human rotavirus genotype G5P[7] from child with diarrhea, Cameroon. Emerg Infect Dis 15:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona MD, Armah GE, Steele AD. 2010. Rotavirus VP4 and VP7 genotypes circulating in Cameroon: Identification of unusual types. J Infect Dis Suppl 202:S205–S211. [DOI] [PubMed] [Google Scholar]
- Estes MK, Kapikian AZ. 2007. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields virology. vol. 2, 5th edition. Philadelphia, PA: Kluwer/Lippincott, Williams and Wilkins, pp 1917–1974. [Google Scholar]
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol 30:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. 2005. Serotype diversity and reassortment between human and animal rotavirus strains: Implications for rotavirus vaccine programs. J Infect Dis 192:S146–S159. [DOI] [PubMed] [Google Scholar]
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. 1990. Polymerase chain-reaction amplification and typing of rotavirus nucleic-acid from stool specimens. J Clin Microbiol 28:276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuranecz M, Rigo K, Dan A, Foldvari G, Makrai L, Denes B, Fodor L, Majoros G, Tirjak L, Erdelyi K. 2011. Investigation of the ecology of Francisella tularensis during an inter-epizootic period. Vector Borne Zoonot Dis 8:1031–1035. [DOI] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Green J, Brown DWG, Desselberger U, Gray J. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: Implications for reverse transcription-PCR genotyping. J Clin Microbiol 38:898–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Cubitt D, Desselberger U, Gray J. 2001. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J Clin Microbiol 39:3796–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Kang G, Mammen A, Jana AK, Abraham M, Desselberger U, Brown D, Gray J. 2004. Characterization of G10P[11] rotaviruses causing acute gastroenteritis in neonates and infants in Vellore, India. J Clin Microbiol 42:2541–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Dallman T, Bányai K, Bottiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Korsun N, Kroneman A, Lappalainen M, László B, Maunula L, Matthijnssens J, Midgley S, Mladenova Z, Nawaz S, Poljsak-Prijatelj M, Pothier P, Ruggeri FM, Sanchez-Fauquier A, Schreier E, Steyer A, Sidaraviciute I, Tran AN, Usonis V, Van Ranst M, De Rougemont A, Gray J. 2009. Rotavirus surveillance in Europe: Web-enabled reporting and real-time analysis of genotyping and epidemiological data. J Infect Dis 200:S215–S221. [DOI] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Dallman T, Bányai K, Bottiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Koopmans M, Korsun N, Koukou D, Kroneman A, László B, Lappalainen M, Maunula L, Mas Marques A, Matthijnssens J, Midgley S, Mladenova Z, Nawaz S, Poljsak-Prijatelj M, Pothier P, Ruggeri FM, Sanchez-Fauquier A, Steyer A, Sidaraviciute-Ivaskeviciene I, Syriopoulou V, Tran AN, Usonis V, Van Ranst M, De Rougemont A, Gray J. 2011. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol Infect 139:895–909. [DOI] [PubMed] [Google Scholar]
- Koulla-Shiro S, Loe C, Ekoe T. 1995. Prevalence of Campylobacter enteritis in children from Yaounde (Cameroon). Cent Afr J Med 41:91–94. [PubMed] [Google Scholar]
- Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C, Ngwira C, Victor JC, Gillard PH, Cheuvart BB, Han HH, Neuzil KM. 2010. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med 362:289–298. [DOI] [PubMed] [Google Scholar]
- Martella V, Banyai K, Matthijnssens J, Buonavoglia C, Ciarlet M. 2010. Zoonotic aspects of rotaviruses. Vet Microbiol 140:246–255. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Bilcke J, Ciarlet M, Martella V, Bányai K, Rahman M, Zeller M, Beutels P, Van Damme P, Van Ranst M. 2009. Rotavirus disease and vaccination: Impact on genotype diversity. Future Microbiol 4:1303–1316. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Rahman M, Ciarlet M, Zeller M, Heylen E, Nakagomi T, Uchida R, Hassan Z, Azim T, Nakagomi O, Van Ranst M. 2010. Reassortment of human rotavirus gene segments into G11 rotavirus strains. Emerg Infect Dis 16:625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PPC, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156:1397–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwenda JM, Ntoto KM, Abebe A, Enweronu-Laryea C, Amina I, Mchomvu J, Kisakye A, Mpabalwani EM, Pazvakavambwa I, Armah GE, Seheri LM, Kiulia NM, Page N, Widdowson MA, Steele AD. 2010. Burden and epidemiology of rotavirus diarrhea in selected African countries: Preliminary results from the African Rotavirus Surveillance Network. J Infect Dis 202: S5–S11. [DOI] [PubMed] [Google Scholar]
- Ndze VN, Achidi EA, Gonsu KH, Lyonga EE, Esona MD, Banyai K, Obama Abena MT. 2012. Epidemiology of rotavirus diarrhea in children <5 years in Northern Cameroon. Pan Afr Med J 11:73. [PMC free article] [PubMed] [Google Scholar]
- Payne DC, Szilagyi PG, Staat MA, Edwards KM, Gentsch JR, Weinberg GA, Hall CB, Curns AT, Clayton H, Griffin MR, Fairbrother G, Parashar UD. 2009. Secular variation in United States rotavirus disease rates and serotypes. Pediatr Infect Dis J 28:948–953. [DOI] [PubMed] [Google Scholar]
- Payne DC, Wikswo M, Parashar UD. 2011. Rotavirus. VPD surveillance manual, 5th edition, 2011. [Google Scholar]
- Rahman M, Matthijnssens J, Nahar S, Podder G, Sack DA, Azim T, Van Ranst M. 2005. Characterization of a novel P[25],G11 human group A rotavirus. J Clin Microbiol 43:3208–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig RF. 1997. Genetics of the rotaviruses. Annu Rev Microbiol 51:225–255. [DOI] [PubMed] [Google Scholar]
- Santos N, Hoshino Y. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15:29–56. [DOI] [PubMed] [Google Scholar]
- Schumann T, Hotzel H, Otto P, Johne R. 2009. Evidence of interspecies transmission and reassortment among avian group A rotaviruses. Virology 386:334–343. [DOI] [PubMed] [Google Scholar]
- Solberg OD, Hasing ME, Trueba G, Eisenberg JNS. 2009. Characterization of novel VP7, VP4, and VP6 genotypes of a previously untypeable group A rotavirus. Virology 385:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele AD, Ivanoff B. 2003. Rotavirus strains circulating in Africa during 1996–1999: Emergence of G9 strains and P[6] strains. Vaccine 21:361–367. [DOI] [PubMed] [Google Scholar]
- Steele AD, Neuzil KM, Cunliffe NA, Madhi SA, Bos P, Ngwira B, Witte D, Todd S, Louw C, Kirsten M, Aspinall S, Van Doorn LJ, Bouckenooghe A, Suryakiran PV, Han HH. 2012. Human rotavirus vaccine Rotarix provides protection against diverse circulating rotavirus strains in African infants: A randomized controlled trial. BMC Infect Dis 12:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD. 2012. 2008 Estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect Dis 12:136–141. [DOI] [PubMed] [Google Scholar]
- Todd S, Page NA, Duncan Steele A, Peenze I, Cunliffe NA. 2010. Rotavirus strain types circulating in Africa: Review of studies published during 1997–2006. J Infect Dis 202 Suppl:S34–S42. [DOI] [PubMed] [Google Scholar]
- Ursu K, Kisfali P, Rigo D, Ivanics E, Erdelyi K, Dan A, Melegh B, Martella V, Banyai K. 2009. Molecular analysis of the VP7 gene of pheasant rotaviruses identifies a new genotype, designated G23. Arch Virol 154:1365–1369. [DOI] [PubMed] [Google Scholar]
- World Health Organization. 2012. Annex: January through December 2011 rotavirus surveillance data. Global Rotavirus Inf Surveill Bull 6:12–15. [Google Scholar]


