Abstract
Disruption of the mannose 6‐phosphate (M‐6‐P) pathway in HeLa cells by inactivation of the GNPTAB gene, which encodes the α/β subunits of GlcNAc‐1‐phosphotransferase, results in missorting of newly synthesized lysosomal acid hydrolases to the cell culture media instead of transport to the endolysosomal system. We previously demonstrated that the majority of the lysosomal aspartyl protease, cathepsin D, is secreted in these GNPTAB −/− HeLa cells. However, the intracellular content of cathepsin D in these cells was still greater than that of WT HeLa cells which retained most of the protease, indicating a marked elevation of cathepsin D expression in response to abrogation of the M‐6‐P pathway. Here, we demonstrate that HeLa cells lacking GlcNAc‐1‐phosphotransferase show a fivefold increase in cathepsin D mRNA expression over control cells, accounting for the increase in cathepsin D at the protein level. Further, we show that this increase at the mRNA level occurs independent of the transcription factors TFEB and TFE3. The intracellular cathepsin D can still be trafficked to lysosomes in the absence of the M‐6‐P pathway, but fails to undergo proteolytic processing into the fully mature heavy and light chains. Uptake experiments performed by feeding GNPTAB −/− HeLa cells with various phosphorylated cathepsins reveal that only cathepsin B is capable of partially restoring cleavage, providing evidence for a role for cathepsin B in the proteolytic processing of cathepsin D.
Keywords: cathepsin B, cathepsin D, cathepsin L, GlcNAc‐1‐phosphotransferase, lysosomes, mannose 6‐phosphate pathway
Cathepsin D mRNA expression is increased fivefold in HeLa cells when the Man‐6‐P pathway is inactivated. This increase is independent of both TFEB and TFE3. These cells also lack cathepsin B, which is partly responsible for the failure of cathepsin D processing to occur. The effect is reversed by feeding the knockout HeLa cells with cathepsin B.
Abbreviations
- GlcNAc‐1‐phosphotransferase
UDP‐GlcNAc:lysosomal enzyme N‐acetylglucosamine‐1‐phosphotransferase
- M‐6‐P
mannose 6‐phosphate
- TFEB
Transcription Factor EB
- TFE3
Transcription Factor E3
The proper delivery of the 60 or so newly synthesized acid hydrolases to the lysosome of a cell is essential for the degradation of intracellular and endocytosed materials within this organelle [1]. Key among these enzymes are the lysosomal proteases known as cathepsins, all of which are initially made as inactive pre‐proenzymes in the endoplasmic reticulum (ER) [2]. Following removal of the signal peptide within the lumen of the ER, the proenzyme, also called a zymogen, acquires N‐linked glycans as it transits from the ER through the compartments of the Golgi. Transport of the pro‐cathepsins from the trans‐Golgi network (TGN) to the endolysosomal system via the mannose 6‐phosphate (M‐6‐P)‐dependent pathway requires tagging of these acid hydrolases with the M‐6‐P moiety on their high‐mannose glycans in the cis‐Golgi, a process mediated by the enzyme UDP‐GlcNAc:lysosomal enzyme N‐acetylglucosamine‐1‐phosphotransferase (GlcNAc‐1‐phosphotransferase) [3]. Upon reaching the lysosome, the pro‐cathepsins undergo specific proteolytic cleavage events for catalytic activation.
Cathepsin D is the most ubiquitously expressed lysosomal aspartyl protease with especially high levels in the human brain [4]. In addition to the metabolic breakdown of proteins in the lysosome, cathepsin D is involved in a diverse array of physiologic processes, including activation of growth factors, hormones and enzyme precursors, processing of antigens, and regulation of apoptosis [5]. Mutations of the human gene encoding cathepsin D, CTSD, that severely impair enzyme activity gives rise to the lysosomal storage disorder neuronal ceroid lipofuscinosis [6]. Cathepsin D has also been implicated in a number of other pathological conditions including atherosclerosis, cancer, cardiovascular disease, and neurodegenerative disorders such as Alzheimer’s, Huntington’s, and Parkinson’s diseases [7].
During the course of studying a HeLa cell line (GNPTAB −/− cells) with inactivation of the M‐6‐P pathway, we found that although 80% of the newly synthesized cathepsin D was secreted by these cells, the remaining 20% of intracellular cathepsin D exceeded the level observed with the parental HeLa cells, wherein upward of 90% of the newly synthesized cathepsin D was retained [8]. This indicated that cathepsin D protein expression was markedly elevated in the GNPTAB −/− HeLa cells. Moreover, we and others noted that these cells accumulate only pro‐cathepsin D while failing to convert this single‐chain molecule to the two‐chain form [8, 9, 10]. Here, we demonstrate that in the absence of the M‐6‐P pathway, cathepsin D gene expression in HeLa cells is elevated fivefold at the mRNA level but this upregulation occurs independent of either Transcription Factor EB (TFEB) or Transcription Factor E3 (TFE3), two transcription factors that function as master regulators of lysosomal homeostasis [11]. We further determine that it is the absence of cathepsin B that is responsible for the failure to detect cathepsin D processing in the GNPTAB −/− HeLa cells.
Materials and methods
Cell lines
Generation of the GNPTAB −/− and GNPTG −/− HeLa cell line has been described in detail previously [12]. Mutant and parental HeLa cell lines were maintained in Dulbecco’s Modified Eagle Medium (Life Technologies, Carlsbad, CA, USA), while Expi293 cells (Life Technologies) were grown in suspension in Expi293 expression medium (Life Technologies). All media contained 0.11 g·L−1 sodium pyruvate and 4.5 g·L−1 glucose, supplemented with 10% (vol/vol) FBS (Atlanta Biologicals, Flowery Branch, GA, USA), 100 000 U·L−1 penicillin, 100 mg·L−1 streptomycin (Life Technologies), and 2 mM l‐glutamine (Life Technologies).
Antibodies
The following antibodies were used in this study: anti‐V5 mouse monoclonal antibody (Thermo Fisher Scientific, Waltham, MA, USA Cat #R960‐25), anti‐cathepsin D rabbit polyclonal antibody (generated in our lab), anti‐Lamp1 mouse monoclonal antibody (H4A3) (Development Studies Hybridoma Bank, Iowa City, IA, USA), anti‐cathepsin B 3E4 mouse monoclonal antibody (a generous gift from Bonnie Sloane, Martin Luther University, Halle‐Wittenberg, Germany), and anti‐cathepsin L goat polyclonal antibody (R&D Systems, Inc. Minneapolis, MN, USA Cat #AF1515).
DNA constructs
The construction of the full‐length GNPTAB cDNA [12] and the truncated S1‐S3 cDNA [13], both in the vector pcDNA6/V5‐His, has been described. The cDNAs for expression of cathepsins B, C, L, and Z in Expi 293 cells were all purchased from OriGene (Rockville, MD, USA).
Immunofluorescence microscopy
GNPTAB −/−, GNPTG −/−, and parental Hela cells were fixed with 4% formaldehyde (Sigma‐Aldrich, St. Louis, MO, USA) and permeabilized in 0.1% (vol/vol) Triton X‐100 in PBS. Cells were blocked for 1 h with 2% IgG‐free BSA (Jackson Immuno‐Research, West Grove, PA, USA) and probed with the indicated combinations of antibodies as described in the figure legend. The images were acquired with an LSM880 confocal microscope (Carl Zeiss, Inc., Peabody, MA, USA) in the Molecular Microbiology Imaging Facility at Washington University School of Medicine in St. Louis. Images were analyzed by imagej software (NIH, Bathesda, MD, USA).
Reverse transcriptase quantitative PCR (RT‐qPCR)
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific) from parental HeLa, GNPTAB −/−, and GNPTG −/− cells according to the manufacturer’s instructions. RNA concentration was determined by measuring the OD260 value. Two hundred nanogram of RNA from each cell line was first reverse‐transcribed using Omniscript RT Kit (Qiagen, Hilden, Germany) in a total volume of 20 µL to produce first‐strand cDNA. qPCR was then carried out using a StepOnePlus thermocycler (Applied Biosystems, Foster City, CA, USA). The reaction was performed in triplicate using SYBR Green Master Mix (Bio‐Rad, Hercules, CA, USA) following the manufacturer’s protocol, and gene expression was calculated by either the 2‐ΔCT or the 2–∆∆CT method, using the human ACTB gene as the reference gene.
The following primers were used in this study for RT‐qPCR:
CTSB fwd 5′GAATGGCACACCCTACTGG3′
rev 5′TGATCGGTGCGTGGAATTC3′
CTSC fwd 5′AACTGCACCTATCTTGACCTG3′
rev 5′CTGTATCCAGCTTCTGAAGGTAC3′
CTSD fwd 5′GGACTACACGCTCAAGGTG3′
rev 5′GTTGTCACGGTCAAACACAG3′
CTSF fwd 5′CCCTTGGCTCATTGACCATG3′
rev 5′GGACCCACGATGCAAGTAG3′
CTSH fwd 5′AACTGTGCGTGAACTCCTTAG3′
rev 5′TGTGGGCGTTTATCTTCCTC3′
CTSK fwd 5′GAAGACCCACAGGAAGCAATA3′
rev 5′TGTATGGACACCAAGAGAAGC3′
CTSL fwd 5′TTTGAGCCAGACTGTAGCAG3′
rev 5′ATCTTTACGTAGCCACCCATG3′
CTSO fwd 5′GTGGAGAAGCAAATCATGCAG3′
rev 5′GCAATACCACAAACATTACTTCCC3′
CTSS fwd 5′TGTGGTTGGCTATGGTGATC3′
rev 5′TTTCTGGGTAAGAGGGAAAGC3′
CTSV fwd 5′TGGATCATGGTGTTCTGGTG3′
rev 5′CAGTGGTTGTTCTTGTCTTTGG3′
CTSW fwd 5′AGTACCTTTCAGCTGTGACTG3′
rev 5′CGTCCACAAAATCCCAGAAAC3′
CTSZ fwd 5′TGGATGGTGTCAACTATGCC3′
rev 5′GCTCCCTTCCTCTTGATGTTG3′
ACTB fwd 5′CCCAGCACAATGAAGATCAAG3′
rev 5′GACTCGTCATACTCCTGCTTG3′
TFEB fwd 5′CTGACCCAGAAGCGAGAG3′
rev 5′TCAGCATTCCCAACTCCTTG3′
TFE3 fwd 5′CCTGCAGCTCCGAATTCAG3′
rev 5′CTGTCAGAAGCCGAAGTCG3′
Enzyme expression in Expi293 cells and enzyme uptake assays
Expi293 cells in suspension were cotransfected with the cDNAs of cathepsins B, C, D, L, and Z, along with the GNPTAB S1‐S3 mutant construct, as described [13]. Media was collected aseptically 2–3 days post‐transfection for use in uptake assays. For cell uptake experiments, GNPTAB −/− HeLa cells were plated on a 12‐well plate at ~ 80% confluence 1 day prior to the uptake experiment. The total protein concentration of the media containing the various secreted cathepsins was measured using the Bradford Assay (Bio‐Rad) and determined to be roughly equivalent. Fifty microlitre aliquots of this media containing each enzyme from the producing cells were then added to the GNPTAB −/− HeLa cells in a final volume of 500 μL. The cells were incubated with the media for 24 h before being collected and processed for immunoblotting.
Immunoblotting
Proteins resolved by using SDS/PAGE under reducing conditions were transferred to nitrocellulose membrane and detected with antibodies as described in the figure legends. Equal amounts of whole‐cell extract were loaded on the gels. For experiments with E‐64 and pepstatin A, parental HeLa or GNPTAB −/− HeLa cells were transfected with the human GNPTAB‐V5/His cDNA in pcDNA6 using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s protocol. Cells were treated with the indicated concentrations of the protease inhibitors for 24 h. Control cells were treated with only vehicle DMSO.
Results
CTSD gene expression is highly elevated in GNPTAB −/− HeLa cells with partial retention of the synthesized cathepsin D
The GNPTAB −/− HeLa cell line lacks the α/β subunits GlcNAc‐1‐phosphotransferase, the enzyme that generates the M‐6‐P moiety on lysosomal enzymes [3]. Consequently, the bulk of the acid hydrolases synthesized by these cells are secreted rather than sorted to lysosomes [12]. Thus, it was surprising when our laboratory determined that the cathepsin D content of these cells was actually higher than either WT parental or GNPTG −/− HeLa cells [8] (Fig. 1A). The GNPTG gene encodes the γ subunit of GlcNAc‐1‐phosphotransferase, which enhances the phosphorylation of a subset of acid hydrolases, but is dispensable for the catalytic activity of the enzyme [12, 14].
Fig. 1.
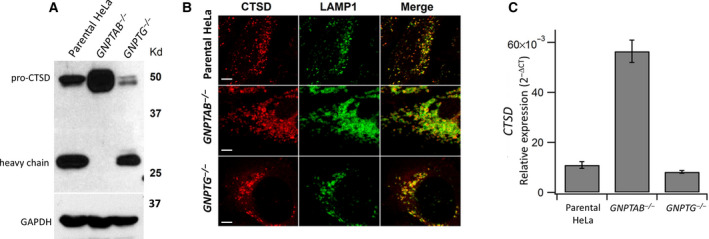
Analysis of cathepsin D in WT, GNPTAB −/−, and GNPTG −/− HeLa cells. (A) Immunoblot of endogenous cathepsin D expression in whole‐cell lysates of the indicated cell lines, as determined by probing with an anti‐cathepsin D antibody. (B) Confocal immunofluorescence images of WT, GNPTAB −/−, and GNPTG −/− HeLa cells, stained for cathepsin D and colocalized with the late endosomal/lysosomal marker LAMP1. Scale bars correspond to 5 μm. (C) Total RNA was isolated from the three cell lines, and cathepsin D mRNA levels were measured by RT‐qPCR using the ACTB gene (encoding β‐actin) as the reference gene for normalization, as described under Materials and Methods. Error bars represent the mean ± SEM of three individual determinations for the various cell lines. P = 0.00007 (Student’s t‐test) for change in CTSD expression in GNPTAB −/− cells relative parental HeLa.
In a recent study using pulse‐chase experiments, we found that 93% of the newly synthesized cathepsin D of WT HeLa cells remained intracellular within a 5‐h time period. This is in stark contrast to the GNPTAB −/− HeLa cells where only 22.5% of the newly synthesized cathepsin D remained intracellular, with 77.5% being secreted into the cell culture medium within the same time period [8]. Still, the amount of the partially retained cathepsin D within the latter cells exceeded the level seen in the WT cells, indicating that in the absence of the M‐6‐P pathway, HeLa cells either upregulate expression of cathepsin D and/or decrease the turnover rate of the protein within lysosomes. Moreover, only pro‐cathepsin D was detected in the immunoblot of whole‐cell lysates from GNPTAB −/− cells, whereas the fully processed heavy chain, in addition to the proform, was seen with both WT and GNPTG −/− HeLa cells [8] (Fig. 1A). A similar observation has been reported by Boonen et al. [9] and Miller et al. [10] with our GNPTAB −/− cells. Despite the absence of GlcNAc‐1‐phosphotransferase, confocal immunofluorescence microscopy showed that the pro‐cathepsin D in the GNPTAB −/− cells colocalized with the lysosomal membrane protein, LAMP1, in enlarged late endosomes/lysosomes, indicating transport of the aspartyl protease to this organelle (Fig. 1B). The morphology of the late endosomes/lysosomes of the GNPTG −/− cells was intermediate between WT and GNPTAB −/− cells, as previously reported [12].
Reverse transcriptase quantitative PCR (RT‐qPCR) was next performed to assess the level of cathepsin D mRNA in the three cell lines. Using total RNA isolated from the individual cell lines, RT‐qPCR analysis determined that the cathepsin D mRNA level was markedly elevated (between fivefold to sixfold) in the GNPTAB −/− cells compared with the WT and GNPTG −/− cells (Fig. 1C).
Elevation of cathepsin D and cathepsin L gene expression in GNPTAB −/− HeLa cells occurs independent of TFEB and TFE3
Since the mRNA level of cathepsin D was markedly elevated when the GNPTAB gene was inactivated in HeLa cells, we next asked if expression of other cathepsins at the mRNA level was also altered in the GNPTAB −/− cells. Of the 11 cathepsins tested, RT‐qPCR analysis showed that only five had detectable mRNA levels in WT HeLa cells (CTSB, CTSC, CTSF, CTSL, and CTSZ), and of these, only the mRNA of CTSL was also increased in the GNPTAB −/− cells, relative to either WT or GNPTG −/− cells (Fig. 2A). The slightly higher mRNA levels of CTSB, CTSC, and CTSZ under these conditions were not statistically significant.
Fig. 2.
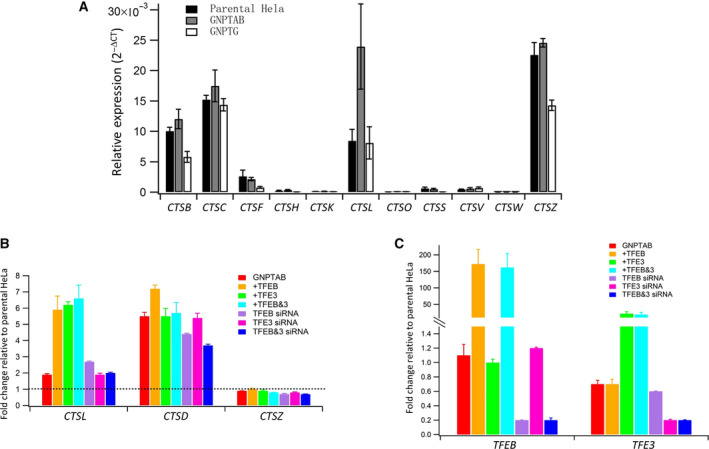
Analysis of the expression of CTS genes in WT, GNPTAB −/−, and GNPTG −/− HeLa cells. (A) Total RNA was isolated from the three cell lines, and the mRNA levels of the indicated cathepsins were measured by RT‐qPCR using the ACTB gene as the reference gene for normalization. Delta Ct was calculated using the ACTB qPCR Ct value minus the qPCR Ct values of the various cathepsins. Error bars represent the mean ± SEM of three individual determinations for the various cell lines. The following P values (Student’s t‐test) were obtained for the change in gene expression between GNPTAB −/− cells and parental HeLa; CTSB P = 0.11286, CTSC P = 0.21899, CTSL P = 0.02059, and CTSZ P = 0.18135. (B) Total RNA was isolated from either untransfected GNPTAB −/− cells (red), or cells transfected with plasmids expressing TFEB and TFE3, individually or in combination, or cells transfected with oligonucleotides to silence TFEB and TFE3, individually or in combination. The level of the indicated cathepsins under the various conditions was then measured by RT‐qPCR using the ACTB gene as the reference gene for normalization. (C) The same cells used in (B) were also analyzed for the message levels of either TFEB or TFE3 using the ACTB gene as the reference gene for normalization. Error bars represent the mean ± SEM of three individual determinations for the different conditions (B and C).
Cathepsin D gene expression has been shown to be regulated by TFEB, a member of the MiT/TFE family, which binds to a common E‐box response element termed CLEAR (Coordinated Lysosomal Expression and Regulation), found in the promoter region of many lysosomal genes [15]. In addition, TFE3, a closely related paralog of TFEB, was also shown to upregulate expression of cathepsin D, and it was proposed that TFE3 controls lysosomal biogenesis by regulating an overlapping gene network [16]. We, therefore, asked if either TFEB or TFE3, or both could be responsible for the highly increased gene expression of CTSD observed in GNPTAB −/− HeLa cells. Total RNA isolated from these cells was subject to RT‐qPCR to assess the mRNA levels of cathepsins D, L, and Z under basal, TFEB/TFE3 overexpression, or TFEB/TFE3 siRNA conditions. Under basal conditions, CTSL gene expression was increased twofold in GNPTAB −/− HeLa cells relative to WT parental HeLa cells, which is set at 1 (Fig. 2A and 2B, CTSL). The mRNA level of the CTSL gene was increased a further threefold upon overexpression of TFEB and TFE3 in GNPTAB −/− cells, either individually or in combination, relative to the untransfected GNPTAB −/− cells (Fig. 2B, CTSL). In contrast to CTSL (twofold increase in gene expression under basal conditions), CTSD gene expression was elevated fivefold to sixfold in GNPTAB −/− cells relative to WT cells (Figs 1C and 2B, CTSD). However, any further increase in CTSD mRNA expression when TFEB was overexpressed was only marginal (Fig. 2B, CTSD). The overexpression of TFE3, in this case, did not elicit an increase, as was the case when TFEB and TFE3 were coexpressed together. It should be noted that the mRNA level of TFEB is not significantly different between the parental WT and GNPTAB −/− HeLa cells (Fig. 2C, TFEB), while the mRNA level of TFE3 is ~ 30% lower in GNPTAB −/− HeLa cells relative to WT cells, which is set at 1 (Fig. 2C, TFE3).
We next performed RNA interference (RNA‐i) of TFEB and TFE3. Knocking down TFEB or TFE3, either alone or in combination, did not significantly decrease CTSD or CTSL gene expression in GNPTAB −/− HeLa cells (Fig. 2B, CTSL and CTSD). The expression of CTSZ remained unchanged under any circumstances in these experiments (Fig. 2B, CTSZ). This result was somewhat surprising, especially when TFEB and TFE3 were overexpressed, since the promoter region of the CTSZ gene has been shown to contain a CLEAR element [15]. However, it is worth noting that the CTSZ gene was not identified among the list of lysosomal hydrolase genes that represent the most likely direct targets of TFEB in HeLa cells [17].
The specific effects of either overexpression or RNA‐i of TFEB and TFE3, on their respective mRNA levels, were confirmed by the RT‐qPCR data shown in Fig. 2C. Taken together, these data indicate that the markedly increased expression of the CTSD gene in GNPTAB −/− cells is likely occurring through a mechanism that is independent of TFEB and TFE3.
The retained cathepsin D in GNPTAB −/− cells does not undergo maturation to the two‐chain species
While the level of the retained cathepsin D in the GNPTAB −/− cells exceeded that of the WT cells, there was a complete absence of the 30 kDa form, indicating that the proform of the protease was not being converted to the mature two‐chain form of the enzyme. Transfection of these cells with the α/β cDNA construct, which restores phosphorylation to endogenous acid hydrolases for transport to the lysosome [12], gave rise to the 48 kDa intermediate and 30 kDa mature heavy chain of cathepsin D (Fig. 3A, compare lanes 1 and 2). The WT parental HeLa cells in this experiment mainly contained the mature/heavy chain form of cathepsin D (Fig. 3A, lane 7). The lack of quantitative conversion of the pro‐cathepsin D to the mature 2‐chain form is most likely due to the low transfection efficiency of HeLa cells. Treatment of the transfected GNPTAB −/− cells with the selective cysteine protease inhibitor E‐64 completely abrogated cathepsin D processing at a concentration of 100 μm (Fig. 3A, lane 5). This concentration of E‐64 also mostly, but not completely, inhibited the formation of mature cathepsin D in the WT cells whereas 10 µm was without effect (Fig. 3A, compare lanes 9 and 10), in agreement with the previously published study using E‐64 to inhibit processing of cathepsin D [18]. This finding confirms the reports of others implicating cysteine proteases in the processing of cathepsin D [18, 19].
Fig. 3.
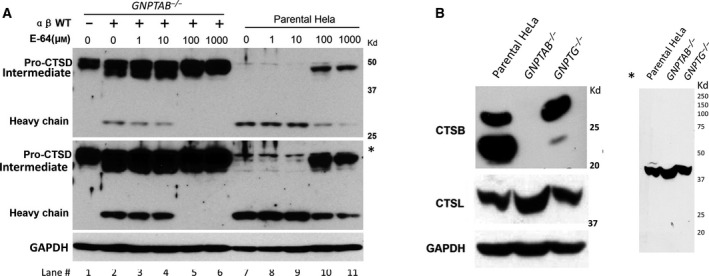
Effect of the cysteine protease inhibitor E‐64 on cathepsin D processing. (A) GNPTAB −/− cells transfected with the human GNPTAB‐V5/His cDNA construct, and untransfected WT parental HeLa cells were treated with the indicated concentrations of E‐64 for 24 h. Equivalent amount of whole‐cell lysates were loaded for immunoblotting and probed for endogenous cathepsin D. * Indicates a longer exposure of the cathepsin D blot. (B) Immunoblot of endogenous cathepsin B and cathepsin L in whole‐cell lysates of the indicated cell lines, as determined by probing with their respective antibodies. * Indicates a longer exposure of the whole cathepsin L blot.
The finding that E‐64 prevented maturation of pro‐cathepsin D in the WT HeLa cells prompted us to ask whether the two cysteine proteases previously shown to mediate cathepsin D processing, namely cathepsin B and cathepsin L, are also present in GNPTAB −/− HeLa cells [18, 19]. Immunoblot analysis of whole‐cell lysates showed no detectable cathepsin B in the GNPTAB −/− cells whereas the WT and GNPTG −/− cells had readily detectable levels of this enzyme (Fig. 3B). It should be noted that while the parental HeLa cells show both the active 2‐chain form of cathepsin B (around 25 kDa) and the active single‐chain form (around 30 kDa) [20], only the latter form is detected in the GNPTG −/− cells. The reason for this discrepancy is not clear. The fact that GNPTAB −/− cells have the same level of CTSB mRNA as WT cells (Fig. 2A), but no detectable cathepsin B protein, suggests that all of the synthesized enzyme is likely being secreted in the absence of the M‐6‐P pathway. On the other hand, only pro‐cathepsin L was detected in all 3 cell lines with the anti‐cathepsin L antibody used in this study, even with the long exposure (Fig. 3B*).
Cell uptake of cathepsin B facilitates processing of cathepsin D
We next asked whether delivery of exogenous cathepsin B to the lysosomes of the GNPTAB −/− cells could partially restore processing of the endogenous pro‐cathepsin D. To this end, cathepsins B, C, L, and Z were coexpressed with a truncated form of GlcNAc‐1‐phosphotransferase (S1‐S3) that exhibits high activity toward lysosomal enzymes in Expi293 cells [13]. These four cathepsins were selected on the basis of their expression in HeLa cells (Fig. 2A). We have previously shown that coexpression of a number of lysosomal enzymes with S1‐S3 in Expi293 cells resulted in the secretion of highly phosphorylated enzymes into the media [13]. Since the uptake of the secreted enzymes via the cell‐surface cation‐independent M‐6‐P receptor is dependent upon the extent of phosphorylation, we surmised that coexpression with S1‐S3 would result in the production of enzymes with a higher M‐6‐P content for use in the uptake experiments.
The media collected from these cultures contained high levels of each protease (Fig. 4A, arrows). The conditioned media were then added to cultures of GNPTAB −/− cells for 24 h. Immunoblot analysis of whole‐cell lysates probed for cathepsin D showed that only cathepsin B‐containing conditioned media facilitated processing of pro‐cathepsin D to the mature species (Fig. 4B, lane 4). Differential processing of the cathepsin D N‐linked glycans due to the lack of phosphorylation of the high mannose‐type sugar chains results in the slower migration of the cathepsin D heavy chain in GNPTAB −/− cells compared with WT HeLa cells (Fig. 4B, compare lanes 1 and 4) [21, 22].
Fig. 4.
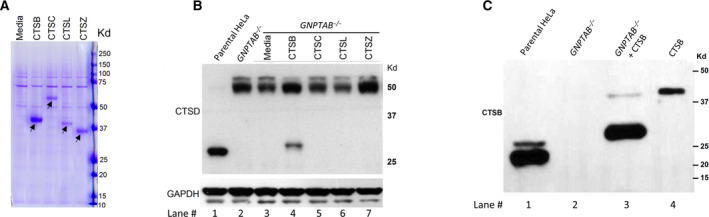
Cell uptake of exogenously added cathepsins. (A) Enzyme produced by Expi293 cells was collected from the media of cells transfected with the cDNAs of the respective cathepsins and cotransfected with the α/β precursor or the S1‐S3 mutant. Ten microlitre media were resolved by SDS/PAGE and the gel stained with Coomassie Brilliant Blue‐250. The position of the secreted cathepsins are indicated by black arrows. (B) Immunoblot of GNPTAB −/− cells following uptake of enzyme from the media, and probed for endogenous cathepsin D. (C) Immunoblot analysis of parental HeLa cells (lane 1), and untreated GNPTAB −/− cells (lane 2), or treated with media containing secreted cathepsin B (lane 3), and probed for cathepsin B. Ten microlitre of the Expi293 cell media containing the secreted cathepsin B was loaded in lane 4.
The cathepsin B that was fed to the cells (Fig. 4C, lane 4) and taken up from the conditioned media was also processed to a mature species that migrated somewhat slower than the endogenous, mature cathepsin B in WT Hela cells (Fig. 4C, compare lanes 1 and 3). The basis for the discrepancy is not clear at this time.
Discussion
Numerous studies published over the last several decades have implicated cathepsin D in a variety of physiological processes as well as diseased states [23]. Hence, a clear understanding of the gene regulatory networks underlying control of CTSD gene expression, as well as the mechanisms involved in the proteolytic processing of cathepsin D under normal and pathological conditions, is important. Here, we present evidence for the sustained elevation of CTSD gene expression at the transcriptional level when the M‐6‐P pathway is abrogated in HeLa cells. This lysosomal stress results in an approximately sixfold increase in the steady‐state message level of the CTSD gene that is still maintained when the transcription factors TFEB and TFE3 are simultaneously silenced by RNA‐i. In addition, GFP‐tagged TFEB or TFE3 did not show any discernible difference in subcellular localization between the WT parental and GNPTAB −/− HeLa cells (data not shown), in contrast to the enhanced nuclear translocation of TFEB that has been observed with renal glomerular cells isolated from Gnptab −/− mice [24]. Taken together, these results suggest that an alternate mechanism/s must be present in HeLa cells that contribute to the upregulation of gene expression due to inactivation of the α/β subunits of GlcNAc‐1‐phosphotransferase. The GNPTAB −/− HeLa cells have been subject to long‐term adaptation to lysosomal stress due to many passages of the cell line over time. The roles of TFEB and TFE3 in lysosomal regulation for cellular adaptation under a variety of acute stress conditions such as nutrient deprivation and pathogen infection are well documented [11]. However, this does not rule out the existence of additional pathways that may be involved in lysosomal homeostasis, especially when lysosome function is impaired in the long run.
Previously, we showed that the cathepsin D synthesized by GNPTAB −/− cells is not phosphorylated and is hyper‐secreted [12]. In view of the substantial increase in cathepsin D expression at both the mRNA and protein levels in these cells, it is possible that a fraction of the newly synthesized cathepsin D is nonspecifically incorporated into clathrin‐coated vesicles that bud off the TGN and are trafficked to the endosome/lysosome compartment. In addition, some of the cathepsin D may be targeted to the lysosome by a M‐6‐P‐independent mechanism [9, 25, 26]. Our finding that GNPTAB −/− cells are unable to process this retained pro‐cathepsin D to the mature heavy and light chains provided a useful system for searching for a protease that acts on this intermediate. The first clue that implicated cathepsin B came from immunoblot analysis of whole‐cell lysates which showed good expression of the cysteine protease cathepsin L, but no detectable cathepsin B, in the mutant cells. Since the mutant cells had similar levels of CTSB mRNA as WT cells, the lack of cathepsin B is presumably due to hypersecretion of the enzyme. Evidence for the role of cathepsin B in this process was then obtained by the cellular uptake experiment wherein conditioned media containing each of the five expressed cathepsins, which we show are synthesized by the GNPTAB −/− cells (Fig. 2A), were individually fed to this cell type. Our results show that only cathepsin B was capable of cleaving the endogenous pro‐cathepsin D. Consistent with a role for cathepsin B in this processing is the finding that the cysteine protease inhibitor, E‐64, blocked pro‐cathepsin D conversion in the WT HeLa cells, as well as in the GNPTAB −/− cells transfected with a cDNA construct encoding the α/β subunits of GlcNAc‐1‐phosphotransferase. Cathepsin L, in addition to cathepsin B, has been reported to be involved in pro‐cathepsin D maturation [18]. While our cell uptake data favor cathepsin B acting alone in this process, at least in HeLa cells, we cannot exclude a role for cathepsin L since our anti‐cathepsin L antibody detected only the proform of cathepsin L in both parental and GNPTAB −/− HeLa cells, and not the mature active form.
In conclusion, we present our data that inactivation of the M‐6‐P pathway in HeLa cells results in an upregulation of CTSD gene expression at the transcriptional level that occurs independent of TFEB and TFE3. In addition, we determine that it is the lack of cathepsin B in these GNPTAB −/− cells that is responsible for the failure of the retained cathepsin D to undergo proteolytic processing and maturation.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
LL conceived and designed the project. LL acquired the data. LL and BD analyzed and interpreted the data. BD wrote the paper.
Acknowledgements
We thank Stuart Kornfeld (Washington University in St. Louis) and Marco Sardiello (Washington University in St. Louis) for their valuable input and insight in the preparation of this manuscript. This study was supported by National Institutes of Health Grant 2R01CA008759‐53 to Stuart Kornfeld.
Data Accessibility
Data will be available from the corresponding author upon reasonable request.
References
- 1. Grayson M (2016) Lysosomal storage disorders. Nature 537, S145. [DOI] [PubMed] [Google Scholar]
- 2. Stoka V, Turk V and Turk B (2016) Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res Rev 32, 22–37. [DOI] [PubMed] [Google Scholar]
- 3. Lang L, Reitman M, Tang J, Roberts RM and Kornfeld S (1984) Lysosomal enzyme phosphorylation. Recognition of a protein‐dependent determinant allows specific phosphorylation of oligosaccharides present on lysosomal enzymes. J Biol Chem 259, 14663–14671. [PubMed] [Google Scholar]
- 4. Zaidi N, Maurer A, Nieke S and Kalbacher H (2008) Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun. 376, 5–9. [DOI] [PubMed] [Google Scholar]
- 5. Benes P, Vetvicka V and Fusek M (2008) Cathepsin D–many functions of one aspartic protease. Crit Rev Oncol Hematol 68, 12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mukherjee AB, Appu AP, Sadhukhan T, Casey S, Mondal A, Zhang Z and Bagh MB (2019) Emerging new roles of the lysosome and neuronal ceroid lipofuscinoses. Mol Neurodegener 14, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dubey V and Luqman S (2017) Cathepsin D as a promising target for the discovery of novel anticancer agents. Curr Cancer Drug Targets 17, 404–422. [DOI] [PubMed] [Google Scholar]
- 8. Doray B, Liu L, Lee WS, Jennings BC and Kornfeld S (2020) Inactivation of the three GGA genes in HeLa cells partially compromises lysosomal enzyme sorting. FEBS Open Bio 11, 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boonen M, Staudt C, Gilis F, Oorschot V, Klumperman J and Jadot M (2016) Cathepsin D and its newly identified transport receptor SEZ6L2 can modulate neurite outgrowth. J Cell Sci 129, 557–568. [DOI] [PubMed] [Google Scholar]
- 10. Miller HE, Hoyt FH and Heinzen RA (2019) Replication of Coxiella burnetii in a lysosome‐like vacuole does not require lysosomal hydrolases. Infect Immun 87, e00493‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martina JA and Puertollano R (2017) TFEB and TFE3: the art of multi‐tasking under stress conditions. Transcription 8, 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Meel E, Lee WS, Liu L, Qian Y, Doray B and Kornfeld S (2016) Multiple domains of GlcNAc‐1‐phosphotransferase mediate recognition of lysosomal enzymes. J Biol Chem 291, 8295–8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu L, Lee WS, Doray B and Kornfeld S (2017) Engineering of GlcNAc‐1‐phosphotransferase for production of highly phosphorylated lysosomal enzymes for enzyme replacement therapy. Mol Ther Methods Clin Dev 5, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qian Y, Lee I, Lee WS, Qian M, Kudo M, Canfield WM, Lobel P and Kornfeld S (2010) Functions of the alpha, beta, and gamma subunits of UDP‐GlcNAc:lysosomal enzyme N‐acetylglucosamine‐1‐phosphotransferase. J Biol Chem 285, 3360–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS et al. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- 16. Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N and Puertollano R (2014) The nutrient‐responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M and Ballabio A (2011) Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20, 3852–3866. [DOI] [PubMed] [Google Scholar]
- 18. Laurent‐Matha V, Derocq D, Prebois C, Katunuma N and Liaudet‐Coopman E (2006) Processing of human cathepsin D is independent of its catalytic function and auto‐activation: involvement of cathepsins L and B. J Biochem 139, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wille A, Gerber A, Heimburg A, Reisenauer A, Peters C, Saftig P, Reinheckel T, Welte T and Buhling F (2004) Cathepsin L is involved in cathepsin D processing and regulation of apoptosis in A549 human lung epithelial cells. Biol Chem 385, 665–670. [DOI] [PubMed] [Google Scholar]
- 20. Weber E, Barbulescu E, Medek R, Reinheckel T, Sameni M, Anbalagan A, Moin K and Sloane BF (2015) Cathepsin B‐deficient mice as source of monoclonal anti‐cathepsin B antibodies. Biol Chem 396, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boonen M, van Meel E, Oorschot V, Klumperman J and Kornfeld S (2011) Vacuolization of mucolipidosis type II mouse exocrine gland cells represents accumulation of autolysosomes. Mol Biol Cell 22, 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gelfman CM, Vogel P, Issa TM, Turner CA, Lee WS, Kornfeld S and Rice DS (2007) Mice lacking alpha/beta subunits of GlcNAc‐1‐phosphotransferase exhibit growth retardation, retinal degeneration, and secretory cell lesions. Invest Ophthalmol Vis Sci 48, 5221–5228. [DOI] [PubMed] [Google Scholar]
- 23. Khalkhali‐Ellis Z and Hendrix MJ (2014) Two faces of cathepsin D: physiological guardian angel and pathological demon. Biol Med 6, 1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sachs W, Sachs M, Kruger E, Zielinski S, Kretz O, Huber TB, Baranowsky A, Westermann LM, Voltolini Velho R, Ludwig NF et al. (2020) Distinct modes of balancing glomerular cell proteostasis in mucolipidosis type II and III prevent proteinuria. J Am Soc Nephrol 31, 1796–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rijnboutt S, Kal AJ, Geuze HJ, Aerts H and Strous GJ (1991) Mannose 6‐phosphate‐independent targeting of cathepsin D to lysosomes in HepG2 cells. J Biol Chem 266, 23586–23592. [PubMed] [Google Scholar]
- 26. Capony F, Braulke T, Rougeot C, Roux S, Montcourrier P and Rochefort H (1994) Specific mannose‐6‐phosphate receptor‐independent sorting of pro‐cathepsin D in breast cancer cells. Exp Cell Res 215, 154–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available from the corresponding author upon reasonable request.


