Rook et al. assess how mutations in acid-sensing ion channel 1 (ASIC1) affect channel desensitization. They establish that, contrary to previous reports, a glutamine to glycine substitution does not abolish desensitization, but causes complex pH-dependent effects on channel desensitization.
Abstract
Desensitization is a common feature of ligand-gated ion channels, although the molecular cause varies widely between channel types. Mutations that greatly reduce or nearly abolish desensitization have been described for many ligand-gated ion channels, including glutamate, GABA, glycine, and nicotinic receptors, but not for acid-sensing ion channels (ASICs) until recently. Mutating Gln276 to a glycine (Q276G) in human ASIC1a was reported to mostly abolish desensitization at both the macroscopic and the single channel levels, potentially providing a valuable tool for subsequent studies. However, we find that in both human and chicken ASIC1, the effect of Q276G is modest. In chicken ASIC1, the equivalent Q277G slightly reduces desensitization when using pH 6.5 as a stimulus but desensitizes, essentially like wild-type, when using more acidic pH values. In addition, steady-state desensitization is intact, albeit right-shifted, and recovery from desensitization is accelerated. Molecular dynamics simulations indicate that the Gln277 side chain participates in a hydrogen bond network that might stabilize the desensitized conformation. Consistent with this, destabilizing this network with the Q277N or Q277L mutations largely mimics the Q277G phenotype. In human ASIC1a, the Q276G mutation also reduces desensitization, but not to the extent reported previously. Interestingly, the kinetic consequences of Q276G depend on the human variant used. In the common G212 variant, Q276G slows desensitization, while in the rare D212 variant desensitization accelerates. Our data reveal that while the Q/G mutation does not abolish or substantially impair desensitization as previously reported, it does point to unexpected differences between chicken and human ASICs and the need for careful scrutiny before using this mutation in future studies.
Introduction
Desensitization is a near-ubiquitous feature of ligand-gated ion channels (LGICs), which was first described >60 yr ago (Katz and Thesleff, 1957). In general, desensitization is thought to act as a protective mechanism, terminating aberrant signaling, although other roles are possible (Gielen et al., 2020; Jones and Westbrook, 1995; Papke et al., 2011). As such, the molecular basis of desensitization has been a subject of inquiry for every type of LGIC. Mutations that essentially abolish or greatly reduce desensitization have been reported for glutamate, GABA, glycine, and nicotinic receptors (Bertrand et al., 1992; Gielen and Corringer, 2018; Gielen et al., 2015; Nayeem et al., 2009; Revah et al., 1991; Stern-Bach et al., 1998). While there have been controversies surrounding the microscopic mechanisms of particular cases (Daniels et al., 2013), these mutations have been enormously helpful in driving structure–function investigations of desensitization as well as providing insight into the physiological role (Christie et al., 2010). There are mutations that somewhat attenuate desensitization of acid-sensing ion channels (ASICs) to varying degrees (Li et al., 2010; Passero et al., 2009; Rook et al., 2020b; Roy et al., 2013; Vullo et al., 2017; Yoder et al., 2018), but no mutations purported to nearly block or abolish desensitization have been described until recently (Chen et al., 2021; Wu et al., 2019).
ASICs are sodium-selective pH-activated trimeric ion channels. They are expressed widely in the central and peripheral nervous systems, as well as other tissues (Kellenberger and Schild, 2015). Given the ubiquity of the ligand, it is unsurprising that ASICs are implicated in a host of physiological processes and disease states including ischemic cell death, fear and anxiety, learning and memory, pain, muscle fatigue, migraine, bone morphogenesis, inflammation, and cancer (Sluka and Gregory, 2015; Wemmie et al., 2013; Zhu et al., 2017). In mammals, the ASIC family includes four subunits capable of forming proton-sensitive homomers: ASIC1a, ASIC1b, ASIC2a, and ASIC3. The individual subunits all have the same topology with intracellular amino- and carboxy-terminal tails of ∼20–80 amino acid residues, separated by a large extracellular domain, two transmembrane helices, and a small amino-terminal reentrant loop (Rook et al., 2020a; Yoder and Gouaux, 2020). The extracellular domain is divided into distinct thumb, finger, knuckle, palm, and β-ball domains (Fig. 1 A). ASIC activation by acidic conditions is believed to occur through protonation of distinct residues in the interface between the thumb and finger as well as a cluster of titratable side chains in the palm domain (Jasti et al., 2007; Krauson et al., 2013; Rook et al., 2020a; Vullo et al., 2017). Protonation also triggers desensitization, either with mild acidic stimuli (pH in the 7.4–6.9 range), which leads to steady-state desensitization (SSD) in the absence of channel activation, or with strong stimuli (i.e., pH 6.8–4), which also opens the channel. Desensitization depends on the isomerization or swivel of a critical linker in the palm domain, which connects the 11th and 12th β strands (Rook et al., 2020b; Yoder et al., 2018). This linker is composed of Leu414 and Asn415 (Fig. 1 B), and in the resting and open states the Leu residue points outward, away from the central axis of the channel. However, in the desensitized state, these amino acid residues essentially switch positions, with Leu414 swiveling downward and in toward the central axis. It has been suggested that in human ASIC1a, Gln276 (Gln277 in chicken ASIC1) acts as a valve to prevent linker rotation, stabilizing the desensitized state, and, furthermore, that eliminating the 276 side chain using the Q276G mutation in effect produces a “leaky” valve that enables channels to readily escape desensitization and remain open (Wu et al., 2019).
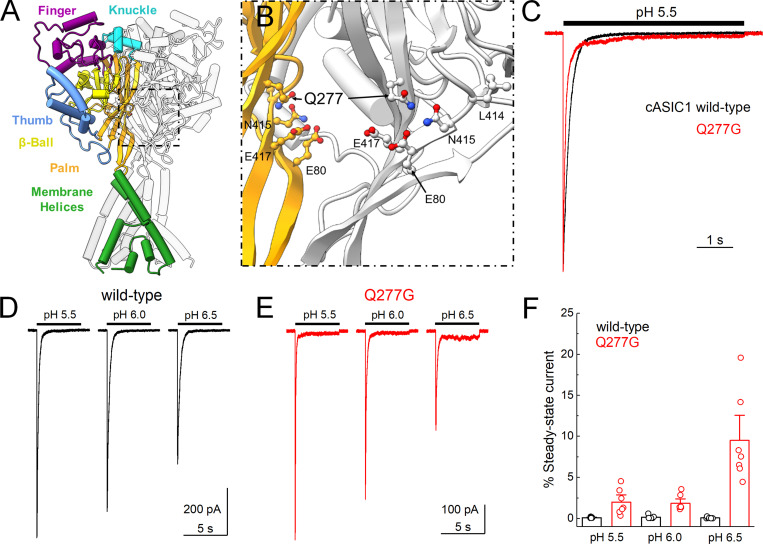
Figure 1.
cASIC1 Q277G exhibits strong desensitization over several pH values.(A) Structure of the cASIC1 resting state (PDB accession no. 6VTL). Domains are identified by color in one subunit while the remaining two subunits are colored light or darker gray. (B) Close in view of the boxed region in A showing Q277 position in two subunits as well as functionally relevant amino acids. The “front” subunit has been removed, leaving only the colored and “rear” subunits for clarity. (C) Peak normalized outside-out patch responses from cASIC1 WT (black trace) or Q277G (red trace) during a jump from pH 8.0 to pH 5.5. (D and E) Responses from single outside-out cASIC1 WT (D) or Q277G (E) patches to the indicated pH stimuli. (F) Summary of the percent steady-state current, normalized to the peak response within a pH, over several patches. The holding potential was −60 mV. Circles denote individual patches, and error bars show SEM.
To further study this intriguing Q276G mutant and relate it to the majority of structural data, we tested the Q276G equivalent in cASIC1 (Q277G) using piezo-driven fast perfusion in excised patches. We found that when using pH 6.5 to open the channels, cASIC1 Q277G does have slightly reduced desensitization; however, when using more acidic stimuli, Q276G behaves essentially like WT with desensitization principally intact. Moreover, we found that Q277G accelerates recovery from desensitization by orders of magnitude and reduces the apparent stability of the desensitized state. Based on molecular dynamics simulations, we hypothesize that Gln277 coordinates a series of hydrogen bonds within the palm domain, thereby stabilizing the desensitized conformation. Consistent with this electrostatic mechanism, a slight mutation to Q277N or Q277L also accelerates recovery from desensitization. Finally, we find that hASIC1a Q276G exhibits robust pH-dependent desensitization, in contrast to prior work.
Materials and methods
Human embryonic kidney (HEK) 293T ASIC knockout (KO) cell creation
A guide RNA sequence (5′-GGCTAAAGCGGAACTCGTTG-PAM-3′) targeting the coding region of ASIC1 was cloned into Bbsl-linearized pSpCas9(BB)-2A-GFP vector (a kind gift from Feng Zhang, Massachusetts Institute of Technology, Boston, MA; Addgene plasmid #48138) as previously described (Ran et al., 2013). Transfected HEK cells (HEK293T; American Type Culture Collection no. CRL-3216) were verified for GFP expression and clonally expanded following serial dilution. Clonal lines were screened for on-target genome editing by Sanger sequencing of PCR products (Fwd: 5′-TTGGAGGAACCCTGGATGTGTC-3′, and Rev: 5′-TAACTCCTCTGCTGTGAGTGGC-3′). KO was confirmed by Western blotting. Briefly, 105 cell equivalents of radioimmunoprecipitation assay buffer lysate from the parental cell line, KO clone, and clone transiently transfected with human ASIC1a cDNA were resolved on an acrylamide gel and transferred to nitrocellulose membranes. Blots were blocked with bovine serum albumin and probed with an ASIC1-specific antibody (NeuroMab clone N271/44) overnight at a 1:1,000 dilution. Blots were washed with Tris-buffered saline supplemented with 0.1% Tween-20 and probed with goat anti-mouse IgG horseradish peroxidase (HRP)–conjugated secondary antibody. Blots were imaged with an Azure 300 Imaging System. After inactivation of HRP with sodium azide, the blot was probed again with Direct-Blot HRP anti-GAPDH (Biolegend) antibody as a loading control.
Cell culture, mutagenesis, and transfection
HEK293T ASIC KO cells were maintained in Dulbecco’s modified Eagle’s medium with 4.5 g/liter glucose, L-glutamine, and sodium pyruvate (Corning/Mediatech, Inc.) or Eagle’s minimum essential medium with Glutamax & Earle’s Salts (Gibco), supplemented with 10% FBS (Atlas Biologicals) and penicillin/streptomycin (Invitrogen). Cells were passaged every 2–3 d when ∼90% confluence was achieved. HEK293T KO cells were plated on tissue culture–treated 35-mm dishes, transfected 24–48 h later, and recorded from 12–48 h after transfection. Cells were transiently transfected with the indicated ASIC construct and enhanced GFP (eGFP) using an ASIC:eGFP ratio of between 2.5 and 10:1 µg of cDNA per 10 ml of medium, depending on the construct. For hASIC1a whole cell experiments (Fig. 6), a ratio of 0.25:0.25:1 μg of hASCI1a, eGFP, and pUC empty vector was used. We used commercial gene synthesis (IDT) to create human codon optimized cDNA of hASIC1a with the G212 variant that was then inserted into pcDNA 3 at the EcoRV site. Transfections were performed using polyethylenimine 25k (Polysciences, Inc.) following the manufacturer’s instructions, with media change at 1–8 h after transfection. In general, WT recordings were obtained sooner after transfection using short transfection durations (i.e., 24 h and 1–2 h incubation), while Q276G recordings were obtained later and with longer transfection durations (i.e., 48 h and 8 h incubation). Mutations were introduced using site-directed mutagenesis PCR and confirmed by sequencing (Thermo Fisher Scientific/Eurofins Genomics).
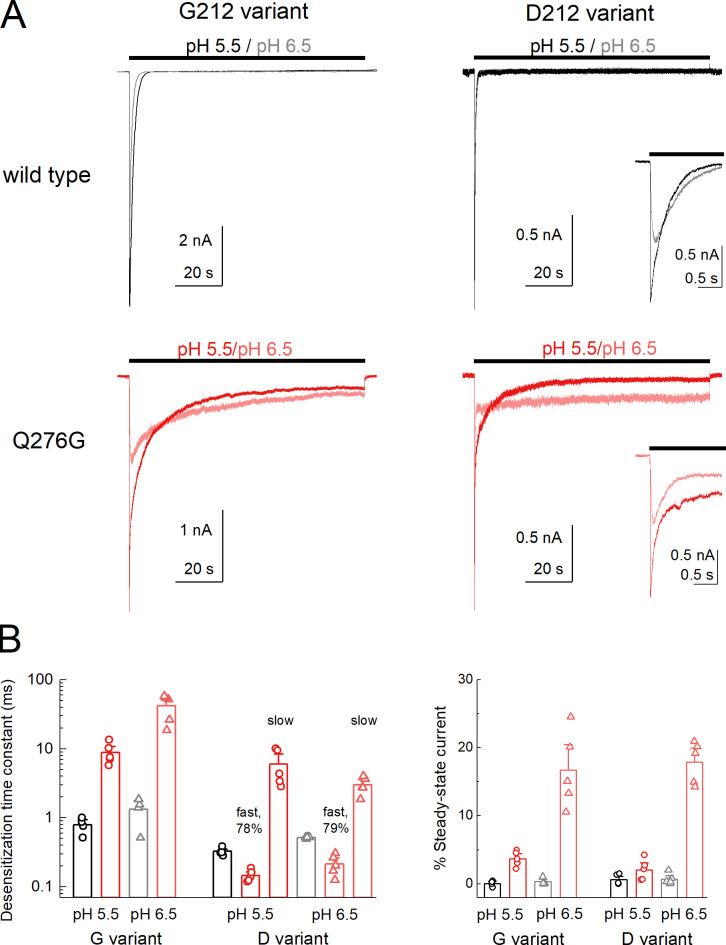
Figure 6.
Human ASIC1a Q276G also does not abolish desensitization.(A) Whole-cell recordings of hASIC1a WT or Q276G, on either the G212 (left) or D212 (right) background during pH 5.5 (red trace) or 6.5 (light red trace) application from baseline pH 8.0. The inset shows the D212 variant traces on expanded time base. (B) Summary of desensitization time constants (left) and percent of steady-state current (right) at pH 5.5 (circles) and 6.5 (triangles) for WT (black) and Q277G (red) in the indicated background. The decay of Q276G in D212 background required double exponential fits. The fast component (78–79% of area) and slow components are reported. The percent of steady state is relative to the peak current of the test pH. Symbols denote single cells, and error bars are SEM. Holding potential is −60 mV.
Electrophysiology
Culture dishes were visualized with phase contrast on a Nikon Ti2 microscope using a 20× objective. GFP was excited using a 455-nm or 470-nm LED (Thorlabs) and dichroic filter cube for emission detection. Outside-out patches were excised using heat-polished, thick-walled borosilicate glass pipettes of 3–15 MΩ resistance. The pipette internal solution contained (in mM) 135 CsF, 33 CsOH, 11 EGTA, 10 HEPES, 2 MgCl2, and 1 CaCl2, pH 7.4. External solutions with pH values >7 were composed of (in mM) 150 NaCl, 10 HEPES, 1 CaCl2, and 1 MgCl2, with pH values adjusted to their respective values using NaOH. For solutions with a pH value <7, HEPES was replaced with Mes. All recordings were performed at room temperature with a holding potential of −60 mV using an Axopatch 200B amplifier (Molecular Devices). Data were acquired using AxoGraph software (Axograph) at 20 kHz, filtered at 10 kHz, and digitized using a USB-6343 DAQ (National Instruments). Series resistance was routinely compensated by 90–95% where the peak amplitude exceeded 100 pA. Rapid perfusion was performed using home-built, double- or triple-barrel application pipettes (Vitrocom), manufactured according to a prior method (MacLean, 2015). Application pipettes were translated using piezo actuators driven by voltage power supplies. The command voltages were generally low-pass–filtered (50–100 Hz, eight-pole Bessel). Whole-cell recording (Fig. S1 and Fig. 6) used identical conditions except patch pipette and application pipette diameters tended to be larger.
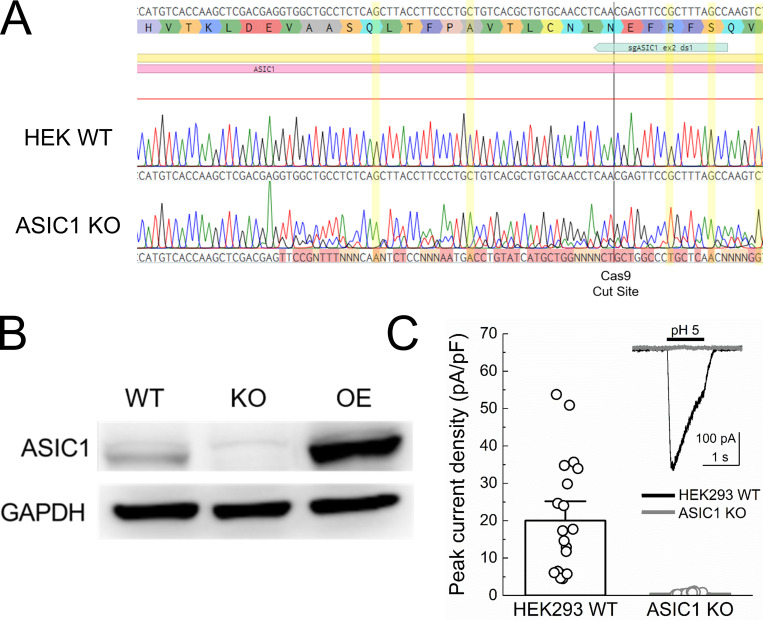
Figure S1.
Validation of human ASIC1 KO cells. (A) Sequencing of genomic DNA from HEK293T WT cells or ASIC1 KO cells of human ASIC1 gene’s second exon. Positions highlighted in yellow show that all alleles of the KO line have been frame-shifted. (B) Western blot of WT HEK293T cells, ASIC1 KO cells, and KO cells overexpressing human ASIC1a (OE). (C) pH 5–evoked whole-cell current densities from HEK293T WT (black) and KO (gray) cells. Raw traces are inset. pH 5–evoked peak current density at −60 mV holding potential: WT, 20 ± 3 pA/pF, n = 20 cells; KO pH 5.0, 0.51 ± 0.05 pA/pF, n = 20 cells; P < 1e−5, Mann–Whitney U test. Circles represent individual cells, and error bars depict SEM.
Molecular dynamics simulations
The systems for molecular dynamics simulations were constructed using the cASIC1 structure proposed to illustrate the desensitized state (Protein Data Bank [PDB] accession no. 4NYK; resolution: 3.00 Å; Gonzales et al., 2009). Residues 42–455 were resolved in the crystal structure; of these, 23 residues had missing atoms, which were added using Modeller v9.21 (Sali and Blundell, 1993). The model was oriented for placement in a lipid bilayer by aligning the complete structure with the corresponding structure from the Orientations of Proteins in Membranes database (Lomize et al., 2012). Protonation states of specific residues were set using pdb2gmx during the system setup in GROMACS (Abraham et al., 2015). In all cases, residues Glu98, His111, Glu239, His328, Glu354, Glu374, Asp408, and Asp433 were protonated, leaving the acidic residues neutral and histidine residues with a positive charge. This was considered the “background” protonation setup, and the purpose was to maintain an overall stable protein structure. However, the importance of the presence of these individual protons was not tested in this study as they are relatively far away from our region of interest. On the given background, Glu80, Glu412, and Glu417 were protonated or deprotonated in accordance with Table 1 to test the importance of protonation of these specific residues.
Table 1. Protonation state setup for molecular dynamics simulations.
| H/H/H | H/H/− | H/−/H | −/H/H | H/−/− | −/H/− | −/−/H | −/−/− | |
|---|---|---|---|---|---|---|---|---|
| E80 | H | H | H | − | H | − | − | − |
| E412 | H | H | − | H | − | H | − | − |
| E417 | H | − | H | H | − | − | H | − |
All systems were prepared from scratch from the original protein structure and with the given protonation scheme applied (“background” plus a specific combination for E80, E412, and E417) before insertion into the membrane and equilibration of each system. The Charmm36m force field was applied (Huang et al., 2017). The initial POPC lipid bilayer (120 Å × 120 Å) was generated using the membrane builder of the CHARMM-GUI with PDB accession no. 4NYK inserted using the replacement method (Jo et al., 2008). The protein structures with different protonation states were then inserted into this original lipid bilayer using the InflateGro method (Kandt et al., 2007). The crystallographic water molecules and chloride ions were retained. Water molecules (TIP3P model; Jorgensen et al., 1983) were further generated to fill the box (120 Å × 120 Å × 161 Å) with solvent, and sodium and chloride ions were added to neutralize the system at a concentration of 0.15 M NaCl.
The simulations were performed using GROMACS 2019.4 (Abraham et al., 2015). All systems were minimized until convergence or to a maximum of 5,000 steps. The systems were then equilibrated in six steps totaling 375 ps, using the standard method from the CHARMM-GUI. The first three equilibration runs used a time step of 1 fs, while the last three and the production run used a time step of 2 fs. The first three equilibration runs were each 25 ps long, and the final three were each 100 ps long. The position restraints were gradually lifted during the equilibration steps as suggested in the default CHARMM-GUI protocol. Periodic boundary conditions were applied. The Verlet cutoff scheme was used throughout with a force-switch modifier starting at 10 Å and a cutoff of 12 Å. A cutoff of 12 Å was used for short-range electrostatics, and the particle mesh Ewald method was used for long-range electrostatics (Darden et al., 1993; Essmann et al., 1995). A Berendsen thermostat was used for all steps of the equilibration, and a Nosé–Hoover thermostat (Hoover, 1985; Nosé, 1984) was used in the production run to maintain the temperature at 310.15 K for all steps. Using semi-isotropic pressure coupling, the pressure was maintained at 1 bar in the last four steps of equilibration and in the production run using the Berendsen barostat (Berendsen et al., 1984) and the Parrinello–Rahman barostat (Parrinello and Rahman, 1981), respectively. The LINCS algorithm was used to constrain covalent bonds to hydrogen atoms (Hess, 2008). The production runs were 100 ns long with a total of three repeats for each system. Each repeat had different initial velocities. The system for the Q277N mutant was prepared as above, with the exception that the Gln277 side chain was manually mutated to Asn before system setup. The same background protonation scheme was used, and additionally, Glu412 and Glu417 were protonated. The system was simulated as described with three repeats of 100 ns each.
Potential hydrogen bonds between the residues Glu80, Gln277, Glu412, Leu414, and Glu417 were sampled every 10 ps. The donor atoms include Q277NH1, Q277NH2, E80HE2 (if protonated), E412HE2 (if protonated), and E417HE2 (if protonated). The acceptor atoms include Q277OE1, E80OE1, E80OE2, E412OE1, E412OE2, L414O, E417OE1, and E417OE2. The 4.3.1 Hydrogen Bond Analysis module (Smith et al., 2019) of MDAnalysis (Michaud-Agrawal et al., 2011; Gowers, R.J., et al. 2016. SciPy2016. https://doi.org/10.25080/majora-629e541a-00e) was used for the analysis, using an updated and adapted version of M. Chavent’s Jupyter Notebook available on GitHub (https://github.com/MChavent/Hbond-analysis; Del Toro et al., 2020). Default cutoffs were used for the donor-acceptor distance (3.0 Å) and the donor-hydrogen-acceptor angle (150°). The presence of each unique hydrogen bond was calculated over the trajectory and expressed as a percentage of the total trajectory; the presence of equivalent hydrogen bonds (e.g., from 417OE1 and 417OE2 in the deprotonated state) was added to give one overall percentage for the given interaction. Plots were prepared using the Matplotlib package in Python. Figures were prepared using visual molecular dynamics (Humphrey et al., 1996).
Statistics and data analysis
Current desensitization decays were fitted using exponential decay functions in Clampfit (Molecular Devices). The percent of steady-state current was the current at the end of a pH application that had reached equilibrium divided by the peak current. For recovery from desensitization experiments, the test peak (i.e., the second response) was normalized to the conditioning peak (i.e., the first response). OriginLab (OriginLab Corp.) was used to fit the normalized responses to
| (1) |
where It is the fraction of the test peak at an interpulse interval of t compared with the conditioning peak, τ is the time constant of recovery, and m is the slope of the recovery curve. The slope, m, is not intended to convey any specific mechanistic information but is needed to properly fit the recovery time course of faster (or slower) recovering ASIC mutations (Rook et al., 2020b). Each protocol was performed between one and three times on a single patch, with the resulting test peak/conditioning peak ratios averaged together. Patches were individually fit, and averages for the fits were reported in the text. N was taken to be a single patch. For activation and SSD curves, peak currents within a patch were normalized to the peak response evoked by pH 5.5 and fit to
| (2) |
where Ix is the current at a given pH value, pHx, pH50 is the pH yielding half-maximal response, and n is the Hill slope. Patches were individually fit, and averages for the fits were reported in the text. N was taken to be a single patch.
Unless otherwise noted, to avoid assumptions about the underlying distribution of the data, statistical testing was done using nonparametric permutation tests with at least 100,000 iterations implemented using the mlxtend package in Python (http://rasbt.github.io/mlxtend/). Statistical comparisons of recovery from desensitization were based and reported on differences in recovery time constant.
Online supplemental material
Fig. S1 shows the validation of hASIC1 KO cells using sequencing, Western blot, and patch clamp. Fig. S2 depicts the hydrogen bond stability of all possible protonation schemes for residues Glu80, Glu412, and Glu417. Fig. S3 shows a similar hydrogen bond analysis of the Glu80/Glu412H/Glu417H scheme for both WT and Q277N simulations. Video 1 and Video 2 show trajectories of Gln277 or Asn277, respectively, upon deprotonation using the E80-/E412H/E417H scheme.
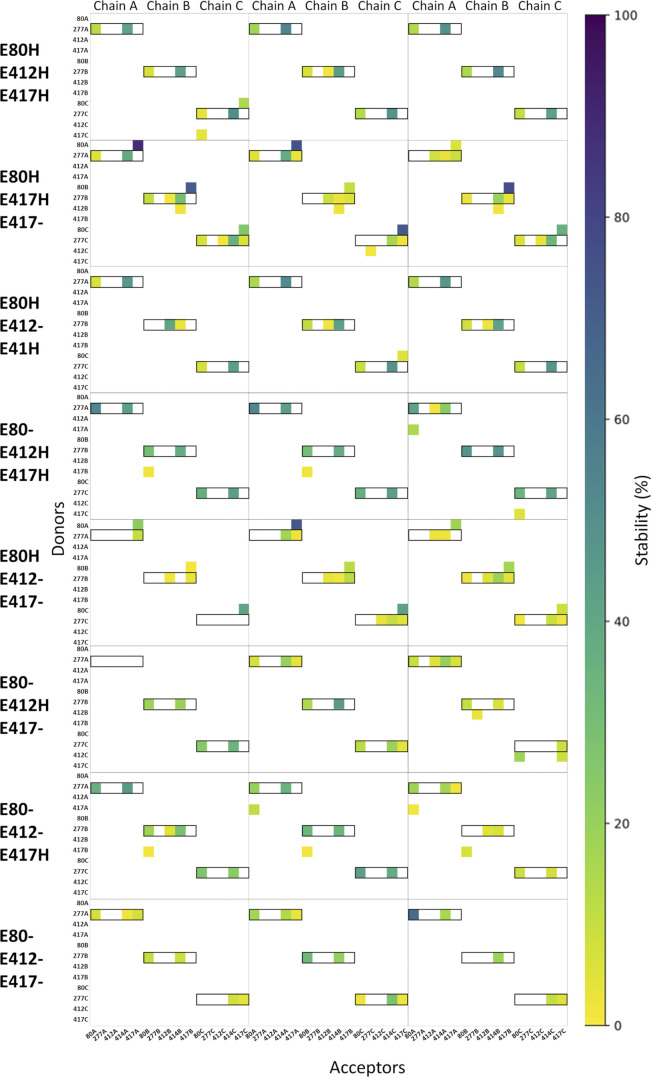
Figure S2.
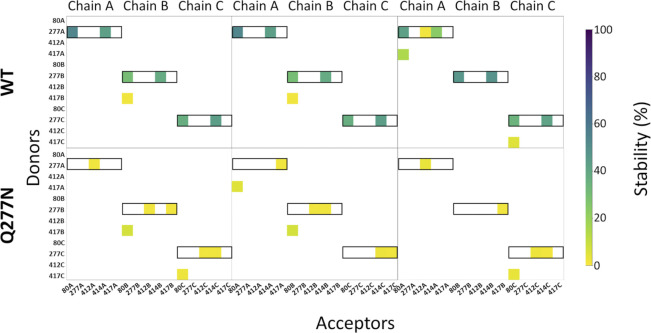
Hydrogen bond analysis for all eight possible protonation setups concerning E80 (H/−), E412 (H/−), and E417 (H/−). All hydrogen bonds formed between donors and acceptors of the side chains of E80, Q277, E412, and E417 are considered, as well as hydrogen bonds in which the backbone oxygen atom of L414 participates as an acceptor. Hydrogen bond stability, following the color scale, is illustrated for each chain in each of three runs (100 ns in duration). Hydrogen bonds in which Q277 participates as a donor are highlighted by black boxes.
Figure S3.
Q277N reduces hydrogen bond stability. Same analysis as Fig. S2, looking at potential hydrogen bonds around residue 277. In both sets of simulations (WT and Q277N), E80 is deprotonated, while E412 and E417 are both protonated. The hydrogen bond stability for each chain in each of three runs (100 ns in duration) is illustrated. Q277N shows heavily reduced presence of hydrogen bonds between 80, 277, and 414 relative to WT for every chain in all repeats.
Video 1.
Representative trajectory illustrating Q277 from a single chain (cyan carbon atoms) and the surrounding residues (from top right and clockwise: E412, L414, E80, and E417; gray carbon atoms) in the WT cASIC1. Q277 is generally kept in a stable conformation to hydrogen bond to E80Oε and L414O. E80 is deprotonated, while E412 and E417 are protonated. The animation shows 100 ns of simulation time. Non-polar hydrogen atoms are omitted. Playback speed, 3 ns/s.
Video 2.
Representative trajectory illustrating N277 from a single chain (cyan carbon atoms) and the surrounding residues (from top right and clockwise: E412, L414, E80, and E417; gray carbon atoms) in the Q277N mutant cASIC1. N277 is highly dynamic and rarely forms hydrogen bonds to E80Oε and L414O. E80 is deprotonated, while E412 and E417 are protonated. The animation shows 100 ns of simulation time. Non-polar hydrogen atoms are omitted. Playback speed, 3 ns/s.
Results
Our goal was to investigate the functional properties of the Q276G mutation in a cASIC1 background, to permit easy comparison with structural data and molecular dynamic simulations. HEK293T cells are an ideal system for this as they are easily cultured and transfected, and amenable to patch clamp. However, HEK293T cells express endogenous human ASIC1, which may complicate interpretation. Therefore, we removed the endogenous human ASIC1 using CRISPR. To do this, exon 2 of the human ASIC1 gene was targeted with a guide RNA cloned into a Cas9-GFP expressing vector (see Materials and methods). Single GFP-positive HEK293T cells were clonally expanded and screened using PCR followed by sequencing (Fig. S1 A). One such clonal population was selected for further characterization. As seen in Fig. S1, this cell line had negligible ASIC1 immunoreactivity compared with either WT HEK293T cells or HEK293T cells transfected with human ASIC1a. Furthermore, whole-cell patch clamp recordings from these presumptive KO cells found no significant currents in response to pH 5.0 application. All other experiments in this study used this HEK293T ASIC1 KO cell line where endogenous ASIC1 has been removed.
To investigate the kinetic consequences of Q277G in cASIC1, we excised outside-out patches from HEK293T KO cells transfected with either WT cASIC1 or cASIC1 Q277G, along with eGFP. Patches were jumped from pH 8, to populate the resting state, into pH 6.5, 6, or 5.5, to activate and desensitize the channels. We were surprised to find that desensitization is completely intact in Q277G (Fig. 1 C). Indeed, the rate of desensitization was accelerated by about twofold (pH 5 desensitization time constant: WT, 180 ± 7 ms, n = 6; Q277G, 78 ± 8 ms, n = 7; P < 1e−6; Fig. 1 C). We also noted that there was a slight elevation of the steady-state or equilibrium current with pH 5.5 stimuli (Fig. 1, C and F; percent steady-state current: WT, 0.09 ± 0.03%, n = 6; Q277G, 2.0 ± 0.6%, n = 7; P = 0.005). To better compare with past work on hASIC1a (Wu et al., 2019), we used the same pH 6.5 stimulus in our cASIC1 experiments. Interestingly, the elevated steady-state current was more prominent with less acidic stimuli, increasing to ∼10% of the peak response using pH 6.5 (Fig. 1, D–F; percent steady-state current: pH 6.0, 1.8 ± 0.4%; pH 6.5, 10 ± 2%, n = 7, P = 0.0001). Such a pH-dependent increase in steady-state current was not detectable in WT channels, although the amplitudes of these steady-state currents are exceedingly small and hence difficult to measure (Fig. 1, D and F; percent steady-state current: pH 6.0, 0.15 ± 0.09%; pH 6.5, 0.06 ± 0.04%, n = 6, P = 0.32 versus pH 5.5 steady-state).
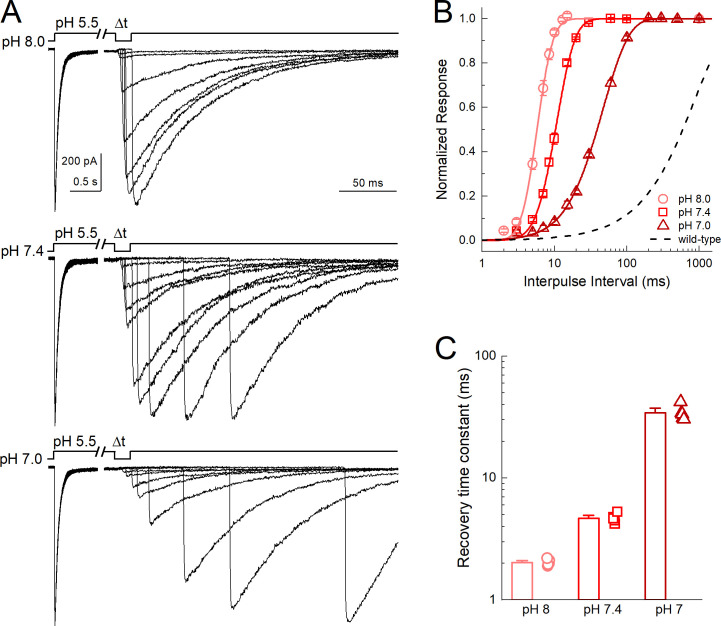
The robust desensitization of Q277G was unexpected given prior work. However, we did observe a small yet significant increase in the current at steady-state, particularly at more alkaline stimulating values (Fig. 1, E and F). We hypothesize that this phenotype arises from a weaker pH dependence of recovery from desensitization. ASIC recovery from desensitization is strongly dependent on the pH between the conditioning and test stimuli. Relatively alkaline inter-stimuli pH values accelerate recovery, while more acidic inter-stimuli pH values slow recovery (Alijevic et al., 2020; Gwiazda et al., 2015; MacLean and Jayaraman, 2016; Rook et al., 2020b). If one extrapolates this trend, then at more acidic values (i.e., pH 5.5) recovery is very slow, and transitions from the desensitized state to the open or resting states are very unfavorable. Consequently, there is minimal steady-state current. The elevated steady-state current of Q277G suggests that Q277G recovery may be faster than WT and/or less influenced by inter-stimuli pH values. To test this, we examined Q277G recovery from desensitization using several inter-stimuli pH values in the same patch. Consistent with our hypothesis, Q277G recovery from desensitization is substantially faster than WT cASIC1 (Fig. 2). Specifically, Q277G recovery had a time constant (and slope m) of 2.03 ± 0.05 ms (m = 12 ± 1, n = 5) at pH 8.0, which is roughly 400-fold faster than WT cASIC1 (840 ± 90 ms, m = 0.96 ± 0.05, n = 5, P < 1e−5; Rook et al., 2020b). Furthermore, the recovery time constants remained very fast at pH 7.4 and 7.0 (pH 7.4 τrec = 4.7 ± 0.2 ms, m = 6.1 ± 0.4; pH 7.0 τrec = 34 ± 2 ms, 1.8 ± 0.1, n = 5). Thus, these data support the notion that the elevated steady-state current in Q277G arises from faster recovery from desensitization in general.
Figure 2.
Q277G rapidly recovers from desensitization over a wide pH range.(A) Outside-out patch recordings of cASIC1 Q277G recovery from desensitization with interpulse pH values of 8.0, 7.4, and 7.0 (upper, middle, and lower traces, respectively). All data from the same patch. Note the break and change in time base between conditioning and test pulses. (B and C) Summary recovery curves (B) and time constants (C) for Q277G recovery at different interpulse pH values. All pH values tested in the same patch. Symbols denote individual patches, and error bars show SEM. Dotted line is the recovery time course of WT cASIC1 with pH 8.0 drawn from Rook et al., 2020a.
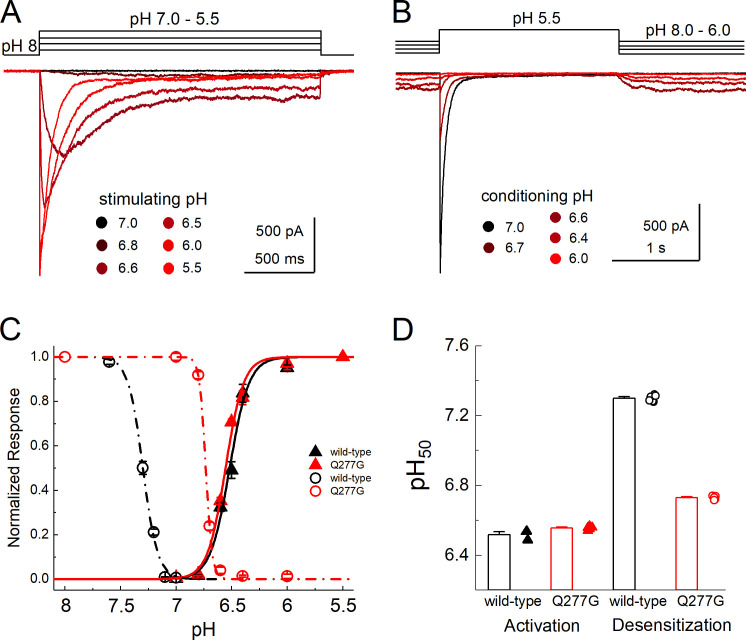
SSD at any given pH value reflects a balance between channels entering and exiting the desensitized state. We hypothesized that the 400-fold faster exit from desensitization is a consequence of a less stable desensitized state. Another consequence of a less stable desensitized state is that a stronger pH stimulus is needed to induce desensitization. This could be observed as a right shift in the SSD curve. To test this, we constructed both activation and SSD curves of Q277G and WT cASIC1 in excised patches (Fig. 3). We found that the pH dependence of activation of Q277G was very slightly more alkaline compared with WT (WT pH50 = 6.51 ± 0.01, n = 5; Q277G pH50 = 6.55 ± 0.01, n = 6, P = 0.006; Fig. 3). However, the SSD of Q277G was considerably right-shifted (Fig. 3, B–D). Specifically, the pH50 of SSD shifted from 7.30 ± 0.01 in WT to 6.73 ± 0.01 in Q277G (n = 6 for both, P < 1e−5). The magnitude of the right shift was sufficiently large to induce overlap with the activation curve, leading to distinct standing currents with baseline pH values such as pH 6.8 or 6.4 (Fig. 3 B). Taken together, we have found that Q277G produces only a small reduction in desensitization (or enhanced steady-state current) while dramatically accelerating recovery from desensitization and right-shifting SSD curves with minimal effect on activation curves. We hypothesize that the recovery and SSD phenotypes all result from reducing the stability of the desensitized state. It has previously been suggested that the conformation of Gln276 (human ASIC numbering) controls the stability of the desensitized state by acting as a valve or steric barrier to regulate isomerization of the β11–12 linker (Wu et al., 2019). To gain insight into the structural mechanism, we turned to molecular dynamics simulations.
Figure 3.
Q277G right-shifts SSD without altering activation.(A) Outside-out patch recording of cASIC1 Q277G responses to increasingly acidic solutions. (B) Responses of Q277G to pH 5.5 application when preincubated with solutions ranging from pH 8.0 to 6. Note solutions of intermediate acidity (pH 6.8–6.4) produce persistent currents at equilibrium. (C) Response curves to activation (solid triangles) or SSD (open circles) for WT (black) or Q277G (red). (D) Mean ± SEM pH50s of activation (left) and SSD (right) for WT (black) or Q277G (red). Fits from individual patches are shown as symbols following the legend for C. All recordings done at −60 mV holding potential. Error bars depict SEM.
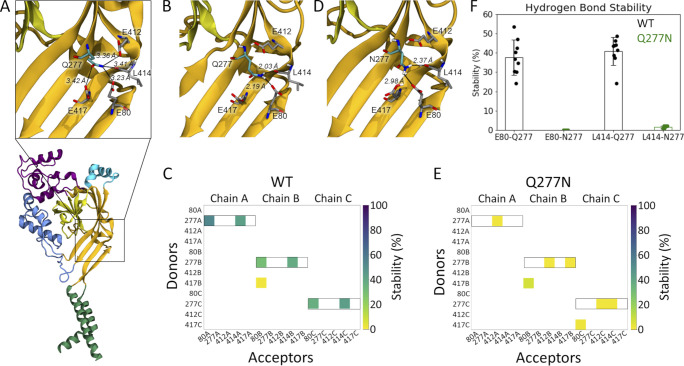
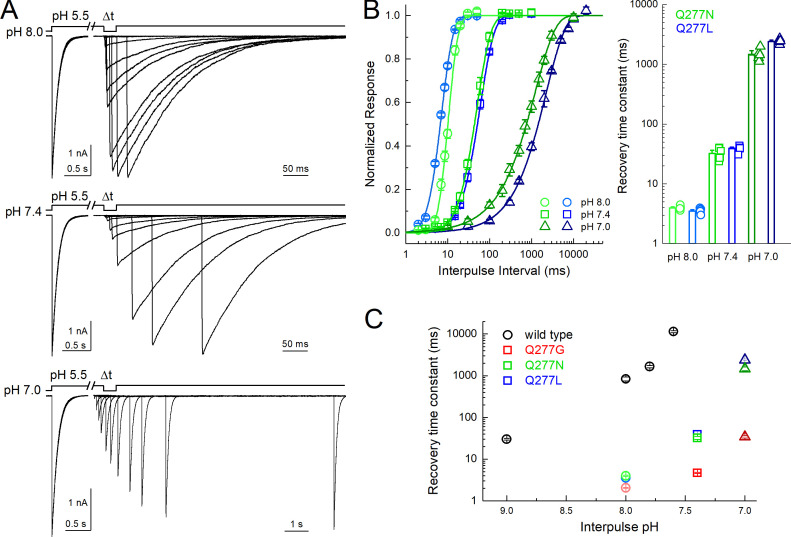
Examining the proposed desensitized state structure of cASIC1 suggested that the Gln277 side chain might form a hydrogen bond to the backbone oxygen atom of Leu414 when the linker is in the desensitized conformation (Fig. 4 A). Rather than acting as a valve, Gln277 could potentially stabilize the desensitized conformation through this hydrogen bond, as we proposed from previous simulations (Rook et al., 2020b). Additionally, three acidic residues, Glu80 and Glu417 in the lower palm domain and Glu412 in the upper palm domain, are within potential hydrogen bond distance of Gln277, partly depending on the protonation states of the acidic residues. Thus, the structure suggests that Glu277 could play a role in a larger hydrogen bond network (Fig. 4 A). Because protonation states of the acidic side chains cannot be observed but must be inferred, we first tested the relative stability of the potential hydrogen bonds in the presence of different protonation states of Glu80, Glu412, and Glu417. Each residue can be protonated or not, giving rise to eight possible protonation combinations. Using the desensitized state structure (PDB accession no. 4NYK; Gonzales et al., 2009) as a starting point, we simulated each protonation scheme for three repeats of 100 ns each. To quantify the stability of potential hydrogen bond interactions between the residues of interest, we measured the fraction of time that each potential hydrogen bond was present over the course of the simulations. Potential hydrogen bond donors considered were the side chains of Glu80, Gln277, Glu412, and Glu417, while potential hydrogen bond acceptors were the same side chains along with the backbone oxygen atom of Leu414. An interaction was considered as a hydrogen bond when the donor-acceptor distance was within 3.0 Å and the donor-hydrogen-acceptor angle >150°. The overall hydrogen bond analysis (Fig. S2) illustrated that no matter the protonation states, Gln277 very rarely acted as a hydrogen bond acceptor. On the contrary, Gln277 often participated as a hydrogen bond donor in fairly stable hydrogen bonds. Looking at all 72 chains analyzed (8 setups × 3 repeats × 3 chains), Gln277 formed hydrogen bonds of varying stability to Glu80 in 75% of cases, to Glu412 in 25% of cases, to L414 in 90% of cases, and to Glu417 in 35% of cases. Therefore, we deemed the hydrogen bonds to Glu80 and to Leu414 to be most important. These two hydrogen bonds showed the highest stability in the setup in which Glu412 and Glu417 were protonated while Glu80 was deprotonated (E80-/E412H/E417H in Fig. S2). Thus, we chose this protonation setup to be the most stable for the desensitized state. Under these conditions, the side chain conformation of Gln277 was generally stable and positioned to hydrogen bond with the side chain of Glu80 and the backbone carbonyl oxygen of Leu414 (Fig. 4 B and Video 1). These interactions are noteworthy as mutations of either Glu80 or Leu414 can profoundly alter desensitization kinetics (Cushman et al., 2007; Della Vecchia et al., 2013; MacLean and Jayaraman, 2017; Rook et al., 2020b; Roy et al., 2013). In particular, motion of Leu414 is a critical regulator of ASIC desensitization, underscoring the potential significance of these contacts. Fig. 4 and Fig. S3 illustrate this analysis, showing that Q277 spends considerable time in putative hydrogen bond interactions with both Glu80 and Leu414. We hypothesized that such a network stabilizes the desensitized state with Gln277 acting as a critical hub. This role of Gln277 as an electrostatic hub is in contrast to the purely steric “valve” model of Gln277 proposed previously (Wu et al., 2019). We reasoned that a Q277N mutation may delineate between these hypotheses. If the “steric” hypothesis is true, then shortening the side chain (Q277N) should produce only a small effect on desensitization kinetics. However, if the electrostatic hub model is more accurate, then the sub-optimal bonding distances of Q277N should result in much faster recovery from desensitization. To confirm that Q277N does attenuate hydrogen bond interactions, we repeated simulations using the Q277N mutation and observed that Q277N showed a greatly reduced capacity to participate in hydrogen bonds with Glu80 and Leu414 (Fig. 4, C–F; Fig. S3; and Video 2). Therefore, we measured the recovery from desensitization of Q277N in excised patches. Consistent with the electrostatic hub hypothesis, Q277N recovers from desensitization much faster than WT at all pH values tested. Specifically, at pH 8.0, the recovery time constant (and slope m) for Q277N is 4.0 ± 0.1 ms (m = 8.7 ± 0.6, n = 7, P < 1e−5 versus WT; Fig. 5). This is slowed to 32 ± 3 ms (m = 2.4 ± 0.1) and 1,500 ± 150 ms (m = 0.79 ± 0.07) at pH 7.4 and 7 (n = 6 and 5, respectively; Fig. 5). Next, we eliminated any residual capacity of the 277 position to participate in hydrogen bonds by using the Q277L mutation, which has identical steric factors as Q277N but no capacity for electrostatic interactions with nearby side chains. Consistent with the electrostatic hub hypothesis, Q277L has comparable recovery kinetics to Q277N (pH 8: τrec = 3.5 ± 0.1 ms, m = 4.3 ± 0.3; pH 7.4: τrec = 39 ± 2 ms, m = 2.23 ± 0.07; pH 7: τrec = 2,400 ± 90 ms, m = 0.88 ± 0.03, n = 5–7; Fig. 5, B–D). While these time constants are slower than Q277G, they are orders of magnitude faster than WT, suggesting the essential feature of Gln277’s function is as a hydrogen bond hub or coordinator and not a steric valve.
Figure 4.
Gln277 links Leu414 and Glu80 via hydrogen bond network. (A) A single subunit of cASIC1 in the desensitized state, illustrating residues within potential hydrogen bonding distance of Gln277 (inset). Colors as in Fig. 1 A. (B) Snapshot from a WT simulation illustrating the hydrogen bond network with Gln277 in the center, hydrogen bonding to L414 and E80. The snapshot was taken at 8.6 ns. (C) Hydrogen bond analysis for a representative repeat (100 ns) of WT with E80 deprotonated and E412 and E417 protonated. All hydrogen bonds formed between donors and acceptors of the side chains of E80, Q277, E412, and E417 are considered, as well as hydrogen bonds in which the backbone oxygen atom of L414 participates as an acceptor. Acceptors are listed horizontally, donors vertically. The colored squares illustrate that a given hydrogen bond is present for part of the 100 ns of simulation, following the color bar given to the right. Hydrogen bonds in which Q277 participates as a donor are highlighted by black boxes. (D) Snapshot from a Q277N simulation illustrating that the inserted Asn residue is too short to form the same hydrogen bond network as Gln277. The snapshot was taken at 19.2 ns. (E) Hydrogen bond analysis as in D, but for the Q277N mutant. (F) Average stability (bars) of the E80-Q277 and the L414-Q277 hydrogen bonds in the WT (black) and Q277N simulations (green; WT: E80-Q277: 38 ± 9%; L414-Q277: 41 ± 7%; Q277N: E80-N277: 0.02 ± 0.01%; L414-N277: 1.6 ± 0.6%. The nine data points (three chains × three repeats) are illustrated as points, and the error bar depicts SD.
Figure 5.
Q277N recovers nearly as fast as Q277G.(A) Outside-out patch recordings of cASIC1 Q277N recovery from desensitization with interpulse pH values of 8.0, 7.4, and 7.0 (upper, middle, and lower traces, respectively). All data are from the same patch. Note the break and change in time base between conditioning and test pulses. (B) Summary recovery curves (left) and time constants (right) for Q277N and Q277L recovery at different interpulse pH values. All pH values tested in the same patch. Symbols denote individual patches and error bars show SEM. (C) Summary of recovery time constants at various pH values for WT, Q277G, Q277N, and Q277L. WT data drawn from Rook et al., 2020a.
These data demonstrate that in cASIC1, Q277G does not block desensitization. Rather, Q277G induces a slight increase in steady-state current that is pH-dependent. Given this, we reexamined the Q276G mutation in human ASIC1a as was previously published (Wu et al., 2019), as well as mouse ASIC1a. In both cases, the Q276G mutation gave small, barely detectable currents in excised patches, necessitating whole-cell recording. In the case of mouse ASIC1a Q276G, even whole-cell currents were too small to resolve and examine convincingly (43 ± 13 pA, n = 10). Therefore, we confined ourselves to hASIC1a Q276G. WT hASIC1a contains a Gly at position 212, as does cASIC1, but many laboratories have been inadvertently using a rare human variant containing an Asp at that position (Vaithia et al., 2019). We measured the decay kinetics of both WT G212 and the rare variant D212. As expected (Vaithia et al., 2019), the D212 variant had faster desensitization kinetics (pH 5.5 desensitization time constant: G212, 790 ± 100 ms, n = 4; D212, 330 ± 20 ms, n = 5; P < 1e−6; Fig. 6), while both had comparable percent steady-state currents (pH 5.5 percent steady-state current: G212, 0.1 ± 0.2%, n = 4; D212, 0.6 ± 0.3%, n = 5; P = 0.27; Fig. 6). Interestingly, combining the Q276G mutation with either variant produced different results. In the G212 background, we observed rather fast rundown or inhibition when using a 5-s stimulus and 20-s intervals. This stimulus and interval duration have proven adequate for WT hASIC1a in our hands. To properly measure the desensitization time course and allow for complete recovery, we progressively extended both the stimulus and interval times. Ultimately, using a 100-s pH application spaced by 120 s, we found that hASIC1a Q276G channels desensitized very slowly (Q276G: 8,800 ± 1,400 ms, n = 5; WT: 790 ± 100 ms, n = 4; P < 1e−5). Q276G in the D212 background also required long intervals for recovery. It was impractical to fully map the recovery time course, but we estimate the pH 8.0 recovery time constant to be ∼60 s for G212 and 35 s for D212. Interestingly, and in contrast to the G212, D212/Q276G showed an acceleration of decay kinetics. Fitting the D212/Q276G decays required double exponentials where ∼80% of the current was fit by a rather fast component (150 ± 10 ms for pH 5.5, 517 ± 5 ms for pH 6.5, n = 5) and the rest with a slower component (6.0 ± 1.5 s for pH 5.5, 3.0 ± 0.4 ms for pH 6.5, n = 5). However, in both backgrounds, Q276G increased the steady-state current to ∼20% with pH 6.5 but only a few percent with pH 5.5 (Fig. 6), as was observed with cASIC1 Q277G. Taken together, we demonstrate that the Q/G mutation does not abolish desensitization as previously reported. Rather, in cASIC1 this mutation elevates the steady-state current, accelerates recovery from desensitization, and reduces the stability of the desensitized state. Molecular dynamics simulations and subsequent mutagenesis suggest that these phenotypes arise by destabilizing a critical hydrogen bond network, which in the WT stabilizes the desensitized state. In hASIC1a, this mutation also does not abolish desensitization and either slows or accelerates decay kinetics, depending on the variant.
Discussion
We explored the properties of the recently described Q276G hASIC1a mutation (Wu et al., 2019; human numbering) using a combination of fast perfusion electrophysiology and molecular dynamics simulations. In contrast to prior work on hASIC1a, we find that this mutation does not abolish cASIC1 desensitization. Rather, this mutation leads to a slight elevation in steady-state current that is more pronounced with weaker pH stimuli (Fig. 1). In cASIC1, Q277G also markedly accelerates recovery from desensitization over a wide pH range (Fig. 2) and right-shifts the pH dependence of SSD without substantially altering activation (Fig. 3). All-atom simulations of the cASIC1 desensitized state indicate that this conformation is stabilized by a network of hydrogen bonds linking the lower palm residue Glu80, through Gln277, with the β11–12 linker (Fig. 5). It is worth noting that our analysis focused on interactions centered around Gln277. Thus, we cannot exclude that effects stemming from longer-range electrostatic interactions within or between subunits also affect this hydrogen bond network and thus the stability of the desensitized conformation. Consistent with the hydrogen bond analysis, compromising the hydrogen bond network by slightly shortening the Q277 side chain (Q277N) or eliminating the capacity for hydrogen bonding (Q277G and Q277L) has a profound impact on the stability of the desensitized state as measured by recovery from desensitization (Fig. 5). Finally, we found that in hASIC1a, Q276G also desensitizes but enters the desensitized state slower or faster depending on the hASIC1a variant (Fig. 6).
Comparison with previous studies
The original report that Q276G blocks desensitization used human ASIC1a in a Xenopus laevis oocyte expression system primarily using bath perfusion, pH 6.5 as a stimulus with pH 7.4 as a baseline pH (Wu et al., 2019). Oocyte experiments have an inherently slower perfusion speed. Moreover, if recordings were obtained from the faster desensitizing phenotype of hASIC1a D212 Q276G, the peak response might have been missed or at least insufficiently resolved. Thus, the experimenter would predominantly observe the larger steady-state current produced by pH 6.5, leading to the conclusion that desensitization has been blocked.
Rather than Gln277 controlling desensitization and recovery by the proposed valve mechanism (Wu et al., 2019), we provide evidence that Gln277 is central to an important hydrogen bond network linking the influential Glu80 residue in the lower palm with the critical β11–12 linker that governs desensitization. How might such a network function in the ASIC gating cycle? In our simulations, both Glu412 and Glu417 are protonated, leaving the Gln277 amide to act as a hydrogen bond donor to the deprotonated Glu80 and the backbone carbonyl of Leu414. The interaction with the carbonyl is the most commonly observed (Fig. 4, Fig. S2, and Fig. S3). We propose that in the desensitized state, Gln277 partly contributes to the stability of Leu414 by this hydrogen bond, with Gln277 being held in this advantageous position by Glu80. Upon alkalization, Glu412, Glu417, or both tend to become deprotonated, acting as alternative hydrogen bond acceptors and thereby helping to pull the amide group of Q277 away from the backbone carbonyl of Leu414, releasing Leu414. This would enable the swivel of the β11–12 linker and thus recovery from desensitization. However, it is difficult to reconcile this hypothesis with the hASIC1a Q276G data that show an apparent slowing of recovery from desensitization.
Human versus chicken data
In our hands, the Q/G mutant gives distinct effects in cASIC1 versus hASIC1a. In every case, the channels still desensitize quite strongly when stimulated with pH 5.5. However, in the G212 (or equivalent) background, cASIC1 and hASIC1a show opposing effects of the Q/G mutation on desensitization kinetics, accelerating decays in cASIC1 but slowing them in hASIC1a (Fig. 1 versus Fig. 6). This is reminiscent of the effects of psalmotoxin, which inhibits mammalian ASICs by stabilizing a desensitized state (Chen et al., 2005) yet activates cASIC1, promoting an unusual nonselective open state (Baconguis and Gouaux, 2012; Samways et al., 2009). Another recent example is the blunted effect of mambalgin in cASIC1 compared with hASIC1a, which can largely be reversed by several point mutations (Sun et al., 2020). Presently it is unclear what the source of these differences is. Human and chicken ASIC1 contain 56 amino acid differences, 31 of which are in the extracellular domain. A number of these are concentrated in the wrist region, including a 2–amino acid insertion. Given the wrist region’s involvement in gating (Li et al., 2009), it is possible that many species-specific differences arise from here. Further differences relevant for our kinetic experiments include the TRL versus SQL substitutions around amino acids 84–86 (Coric et al., 2003) as well as Ser275Ala, Val368Leu, and Ala413Val, which are all relatively proximal to Gln277 (chicken to human differences). We hypothesize that one or more of these changes subtly alter the structure of hASIC1a, potentially imparting distinct pKa values on critical palm residues and thus changing the phenotype of Q276G. As more hASIC1a structures become available in distinct functional states (Sun et al., 2020), we hope to explore the source of these differences and the conservation of mechanisms in more detail. A similar examination may uncover why the equivalent Q269G mutation in ASIC3 does appear to inhibit desensitization even with pH 5.0 (Klipp et al., 2020). Regardless of phenotypic differences, our data clearly indicate that both cASIC1 and hASIC1a Q277G mutants desensitize to a large extent. Therefore, using these mutations to explore either biophysical mechanisms of desensitization, or its physiological consequences, may be problematic.
Acknowledgments
Christopher J. Lingle served as editor.
We thank Dr. Matthieu Chavent for initial discussions concerning the hydrogen bond analysis.
Funding for this work was provided by National Institutes of Health grant T32GM068411-15 and a Joan Wright Goodman Fellowship to M.L. Rook; National Science Foundation Graduate Reseach Fellowship Program to T. Couch; an National Science and Engineering Research Council Discovery Grant (RGPIN 2019-06864) and Canada Research Chairs grant (950-232154) to M. Musgaard; and National Institutes of Health grants R00NS094761 and R35GM137951 and a National Alliance for Research on Schizophrenia and Depression Young Investigator Award (27518) to D.M. MacLean.
The authors declare no competing financial interests.
Author contributions: M.L. Rook, M. Miaro, D.L. Kneisley, T. Couch, and D.M. MacLean conducted experiments and analyzed data. M.L. Rook, M. Miaro, M. Musgaard, and D.M. MacLean interpreted results and edited the manuscript.
References
- Abraham, M.J., Murtola T., Schulz R., Pall S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1-2:19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Alijevic, O., Bignucolo O., Hichri E., Peng Z., Kucera J.P., and Kellenberger S.. 2020. Slowing of the Time Course of Acidification Decreases the Acid-Sensing Ion Channel 1a Current Amplitude and Modulates Action Potential Firing in Neurons. Front. Cell. Neurosci. 14:41. 10.3389/fncel.2020.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baconguis, I., and Gouaux E.. 2012. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature. 489:400–405. 10.1038/nature11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., and Haak J.R.. 1984. Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 81:3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Bertrand, D., Devillers-Thiéry A., Revah F., Galzi J.L., Hussy N., Mulle C., Bertrand S., Ballivet M., and Changeux J.P.. 1992. Unconventional pharmacology of a neuronal nicotinic receptor mutated in the channel domain. Proc. Natl. Acad. Sci. USA. 89:1261–1265. 10.1073/pnas.89.4.1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Kalbacher H., and Gründer S.. 2005. The tarantula toxin psalmotoxin 1 inhibits acid-sensing ion channel (ASIC) 1a by increasing its apparent H+ affinity. J. Gen. Physiol. 126:71–79. 10.1085/jgp.200509303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Kuenze G., Meiler J., and Canessa C.M.. 2021. An arginine residue in the outer segment of hASIC1a TM1 affects both proton affinity and channel desensitization. J. Gen. Physiol. 153:e202012802. 10.1085/jgp.202012802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, L.A., Russell T.A., Xu J., Wood L., Shepherd G.M., and Contractor A.. 2010. AMPA receptor desensitization mutation results in severe developmental phenotypes and early postnatal lethality. Proc. Natl. Acad. Sci. USA. 107:9412–9417. 10.1073/pnas.0908206107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric, T., Zhang P., Todorovic N., and Canessa C.M.. 2003. The extracellular domain determines the kinetics of desensitization in acid-sensitive ion channel 1. J. Biol. Chem. 278:45240–45247. 10.1074/jbc.M304441200 [DOI] [PubMed] [Google Scholar]
- Cushman, K.A., Marsh-Haffner J., Adelman J.P., and McCleskey E.W.. 2007. A conformation change in the extracellular domain that accompanies desensitization of acid-sensing ion channel (ASIC) 3. J. Gen. Physiol. 129:345–350. 10.1085/jgp.200709757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, B.A., Andrews E.D., Aurousseau M.R., Accardi M.V., and Bowie D.. 2013. Crosslinking the ligand-binding domain dimer interface locks kainate receptors out of the main open state. J. Physiol. 591:3873–3885. 10.1113/jphysiol.2013.253666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden, T., York D., and Pedersen L.. 1993. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Del Toro, D., Carrasquero-Ordaz M.A., Chu A., Ruff T., Shahin M., Jackson V.A., Chavent M., Berbeira-Santana M., Seyit-Bremer G., Brignani S., et al. 2020. Structural Basis of Teneurin-Latrophilin Interaction in Repulsive Guidance of Migrating Neurons. Cell. 180:323–339.e19. 10.1016/j.cell.2019.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Vecchia, M.C., Rued A.C., and Carattino M.D.. 2013. Gating transitions in the palm domain of ASIC1a. J. Biol. Chem. 288:5487–5495. 10.1074/jbc.M112.441964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera L., Berkowitz M.L., Darden T., Lee H., and Pedersen L.G.. 1995. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 103:8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Gielen, M., and Corringer P.J.. 2018. The dual-gate model for pentameric ligand-gated ion channels activation and desensitization. J. Physiol. 596:1873–1902. 10.1113/JP275100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen, M., Thomas P., and Smart T.G.. 2015. The desensitization gate of inhibitory Cys-loop receptors. Nat. Commun. 6:6829. 10.1038/ncomms7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen, M., Barilone N., and Corringer P.J.. 2020. The desensitization pathway of GABAA receptors, one subunit at a time. Nat. Commun. 11:5369. 10.1038/s41467-020-19218-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, E.B., Kawate T., and Gouaux E.. 2009. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 460:599–604. 10.1038/nature08218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwiazda, K., Bonifacio G., Vullo S., and Kellenberger S.. 2015. Extracellular Subunit Interactions Control Transitions between Functional States of Acid-sensing Ion Channel 1a. J. Biol. Chem. 290:17956–17966. 10.1074/jbc.M115.641688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B.2008. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4:116–122. 10.1021/ct700200b [DOI] [PubMed] [Google Scholar]
- Hoover, W.G.1985. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 31:1695–1697. 10.1103/PhysRevA.31.1695 [DOI] [PubMed] [Google Scholar]
- Huang, J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., and MacKerell A.D. Jr. 2017. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 14:71–73. 10.1038/nmeth.4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38: 27–28. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jasti, J., Furukawa H., Gonzales E.B., and Gouaux E.. 2007. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 449:316–323. 10.1038/nature06163 [DOI] [PubMed] [Google Scholar]
- Jo, S., Kim T., Iyer V.G., and Im W.. 2008. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29:1859–1865. 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- Jones, M.V., and Westbrook G.L.. 1995. Desensitized states prolong GABAA channel responses to brief agonist pulses. Neuron. 15:181–191. 10.1016/0896-6273(95)90075-6 [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L., Chandrasekhar J., Madura J.D., Impey R.W., and Klein M.L.. 1983. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 79:926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kandt, C., Ash W.L., and Tieleman D.P.. 2007. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 41:475–488. 10.1016/j.ymeth.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Katz, B., and Thesleff S.. 1957. A study of the desensitization produced by acetylcholine at the motor end-plate. J. Physiol. 138:63–80. 10.1113/jphysiol.1957.sp005838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger, S., and Schild L.. 2015. International Union of Basic and Clinical Pharmacology. XCI. structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol. Rev. 67:1–35. 10.1124/pr.114.009225 [DOI] [PubMed] [Google Scholar]
- Klipp, R.C., Cullinan M.M., and Bankston J.R.. 2020. Insights into the molecular mechanisms underlying the inhibition of acid-sensing ion channel 3 gating by stomatin. J. Gen. Physiol. 152:e201912471. 10.1085/jgp.201912471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauson, A.J., Rued A.C., and Carattino M.D.. 2013. Independent contribution of extracellular proton binding sites to ASIC1a activation. J. Biol. Chem. 288:34375–34383. 10.1074/jbc.M113.504324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., Yang Y., and Canessa C.M.. 2009. Interaction of the aromatics Tyr-72/Trp-288 in the interface of the extracellular and transmembrane domains is essential for proton gating of acid-sensing ion channels. J. Biol. Chem. 284:4689–4694. 10.1074/jbc.M805302200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., Yang Y., and Canessa C.M.. 2010. Asn415 in the beta11-beta12 linker decreases proton-dependent desensitization of ASIC1. J. Biol. Chem. 285:31285–31291. 10.1074/jbc.M110.160382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize, M.A., Pogozheva I.D., Joo H., Mosberg H.I., and Lomize A.L.. 2012. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40(D1):D370–D376. 10.1093/nar/gkr703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean, D.M.2015. Constructing a Rapid Solution Exchange System. InIonotropic Glutamate Receptor Technologies. Popescu G.K., editor. Springer, New York. 165–183. [Google Scholar]
- MacLean, D.M., and Jayaraman V.. 2016. Acid-sensing ion channels are tuned to follow high-frequency stimuli. J. Physiol. 594:2629–2645. 10.1113/JP271915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean, D.M., and Jayaraman V.. 2017. Deactivation kinetics of acid-sensing ion channel 1a are strongly pH-sensitive. Proc. Natl. Acad. Sci. USA. 114:E2504–E2513. 10.1073/pnas.1620508114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud-Agrawal, N., Denning E.J., Woolf T.B., and Beckstein O.. 2011. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32:2319–2327. 10.1002/jcc.21787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayeem, N., Zhang Y., Schweppe D.K., Madden D.R., and Green T.. 2009. A nondesensitizing kainate receptor point mutant. Mol. Pharmacol. 76:534–542. 10.1124/mol.109.056598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosé, S.1984. A Molecular-Dynamics Method for Simulations in the Canonical Ensemble. Mol. Phys. 52:255–268. 10.1080/00268978400101201 [DOI] [Google Scholar]
- Papke, D., Gonzalez-Gutierrez G., and Grosman C.. 2011. Desensitization of neurotransmitter-gated ion channels during high-frequency stimulation: a comparative study of Cys-loop, AMPA and purinergic receptors. J. Physiol. 589:1571–1585. 10.1113/jphysiol.2010.203315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello, M., and Rahman A.. 1981. Polymorphic Transitions in Single-Crystals-a New Molecular-Dynamics Method. J. Appl. Phys. 52:7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Passero, C.J., Okumura S., and Carattino M.D.. 2009. Conformational changes associated with proton-dependent gating of ASIC1a. J. Biol. Chem. 284:36473–36481. 10.1074/jbc.M109.055418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revah, F., Bertrand D., Galzi J.L., Devillers-Thiéry A., Mulle C., Hussy N., Bertrand S., Ballivet M., and Changeux J.P.. 1991. Mutations in the channel domain alter desensitization of a neuronal nicotinic receptor. Nature. 353:846–849. 10.1038/353846a0 [DOI] [PubMed] [Google Scholar]
- Rook, M.L., Musgaard M., and MacLean D.M.. 2020a. Coupling structure with function in acid-sensing ion channels: challenges in pursuit of proton sensors. J. Physiol. 10.1113/JP278707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, M.L., Williamson A., Lueck J.D., Musgaard M., and Maclean D.M.. 2020b. β11-12 linker isomerization governs acid-sensing ion channel desensitization and recovery. eLife. 9:e51111. 10.7554/eLife.51111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Boiteux C., Alijevic O., Liang C., Bernèche S., and Kellenberger S.. 2013. Molecular determinants of desensitization in an ENaC/degenerin channel. FASEB J. 27:5034–5045. 10.1096/fj.13-230680 [DOI] [PubMed] [Google Scholar]
- Sali, A., and Blundell T.L.. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Samways, D.S., Harkins A.B., and Egan T.M.. 2009. Native and recombinant ASIC1a receptors conduct negligible Ca2+ entry. Cell Calcium. 45:319–325. 10.1016/j.ceca.2008.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluka, K.A., and Gregory N.S.. 2015. The dichotomized role for acid sensing ion channels in musculoskeletal pain and inflammation. Neuropharmacology. 94:58–63. 10.1016/j.neuropharm.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P., Ziolek R.M., Gazzarrini E., Owen D.M., and Lorenz C.D.. 2019. On the interaction of hyaluronic acid with synovial fluid lipid membranes. Phys. Chem. Chem. Phys. 21:9845–9857. 10.1039/C9CP01532A [DOI] [PubMed] [Google Scholar]
- Stern-Bach, Y., Russo S., Neuman M., and Rosenmund C.. 1998. A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron. 21:907–918. 10.1016/S0896-6273(00)80605-4 [DOI] [PubMed] [Google Scholar]
- Sun, D., Liu S., Li S., Zhang M., Yang F., Wen M., Shi P., Wang T., Pan M., Chang S., et al. 2020. Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1. eLife. 9:e57096. 10.7554/eLife.57096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaithia, A., Vullo S., Peng Z., Alijevic O., and Kellenberger S.. 2019. Accelerated Current Decay Kinetics of a Rare Human Acid-Sensing ion Channel 1a Variant That Is Used in Many Studies as Wild Type. Front. Mol. Neurosci. 12:133. 10.3389/fnmol.2019.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullo, S., Bonifacio G., Roy S., Johner N., Bernèche S., and Kellenberger S.. 2017. Conformational dynamics and role of the acidic pocket in ASIC pH-dependent gating. Proc. Natl. Acad. Sci. USA. 114:3768–3773. 10.1073/pnas.1620560114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie, J.A., Taugher R.J., and Kreple C.J.. 2013. Acid-sensing ion channels in pain and disease. Nat. Rev. Neurosci. 14:461–471. 10.1038/nrn3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Chen Z., and Canessa C.M.. 2019. A valve-like mechanism controls desensitization of functional mammalian isoforms of acid-sensing ion channels. eLife. 8:e45851. 10.7554/eLife.45851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, N., and Gouaux E.. 2020. The His-Gly motif of acid-sensing ion channels resides in a reentrant ‘loop’ implicated in gating and ion selectivity. eLife. 9:e56527. 10.7554/eLife.56527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, N., Yoshioka C., and Gouaux E.. 2018. Gating mechanisms of acid-sensing ion channels. Nature. 555:397–401. 10.1038/nature25782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S., Zhou H.Y., Deng S.C., Deng S.J., He C., Li X., Chen J.Y., Jin Y., Hu Z.L., Wang F., et al. 2017. ASIC1 and ASIC3 contribute to acidity-induced EMT of pancreatic cancer through activating Ca2+/RhoA pathway. Cell Death Dis. 8:e2806. 10.1038/cddis.2017.189 [DOI] [PMC free article] [PubMed] [Google Scholar]