Abstract
Native mass spectrometry, particularly in conjunction with gas-phase ion mobility spectrometry measurements, has proven useful as a structural biology tool for evaluating the stoichiometry, conformation, and topology of protein complexes. Here, we demonstrate the combination of trapped ion mobility spectrometry (TIMS) and surface-induced dissociation (SID) on a Bruker SolariX XR 15 T FT-ICR mass spectrometer for structural analysis of protein complexes. We successfully performed SID on mobility-selected protein complexes, including streptavidin tetramer and cholera toxin B with bound ligand. Additionally, TIMS-SID was employed on a mixture of peptides bradykinin desR1 and desR9 to mobility separate and identify the individual peptides. Importantly, results show that native-like conformations can be maintained throughout the TIMS analysis. The TIMS-SID spectra are analogous to SID spectra acquired using quadrupole mass selection, indicating little measurable, if any, structural rearrangement during mobility selection. Mobility parking was used on the ion or mobility of interest and 50 to 200 SID mass spectra were averaged. High quality TIMS-SID spectra were acquired over a period of 2–10 minutes, comparable to or slightly longer than SID coupled with ion mobility on various instrument platforms in our laboratory. The ultrahigh resolving power of the 15 T FT-ICR allowed for the identification and relative quantification of overlapping SID fragments with the same nominal m/z based on isotope patterns and shows promise as a platform to probe small mass differences, such as protein-ligand binding or post-translational modifications. These results represent the potential of TIMS-SID-MS for the analysis of both protein complexes and peptides.
Graphical Abstract
Introduction
Proteins often carry out their functions as complexes of multiple proteins. The understanding of their assembly is integral to determining how architecture affects function and possibly malfunction. Traditional biophysical methods, such as NMR, Cryo-EM, or x-ray crystallography, have long been useful tools for probing the structures and dynamics of biological molecules.1–4 Native mass spectrometry (nMS), particularly in conjunction with gas-phase ion mobility measurements, offers complementary information for evaluating the stoichiometry, conformation, and topology of protein complexes. The information provided as well as its sensitivity and ease of use have made nMS a rapidly growing technique for analyzing proteins and protein complexes prepared in solutions that mimic the cellular environment, thus maintaining their native-like structure.1,2 These biological systems are efficiently ionized, under low voltage and low activation instrument conditions, and transferred to the gas phase using nanoelectrospray ionization (nESI) in kinetically-trapped, native-like structures, in which structural disruption is limited and noncovalent interfaces are preserved.3 Importantly, nMS is capable of probing multiple conformations and proteoforms of protein complexes with low sample amounts, less extensive sample preparation, and relatively uncomplicated data processing.5–7 The combination of ion mobility spectrometry (IMS) and surface-induced dissociation (SID) in a single native mass spectrometry experiment has the potential to provide a wealth of structural information for protein complexes, including conformational landscape information and subunit stoichiometry, connectivity, and topology.8–11
IMS provides an extra dimension of structural characterization when coupled with mass spectrometry by separating ions not only by mass, but also by their rotationally averaged collision cross sections.12–21 There are several different ion mobility methods and instrument types available, and these have been compared in depth previously.21 Both drift tube ion mobility spectrometry (DTIMS) and traveling wave ion mobility spectrometry (TWIMS) have been utilized regularly by the nMS community for the analysis of protein complexes. In linear cell drift tube IMS, a potential is applied along the ion mobility cell while ions are accelerated into a quasi-stationary bath gas, providing a drag force that opposes the ion motion along the potential gradient.22–24 As a result, larger or more expanded ions undergo a greater number of collisions, therefore taking longer to traverse the IMS cell compared to more rigid or compact conformations. For ions of the same m/z, more compact ions, described as having higher mobility, will elute from the IMS cell sooner compared to expanded ions.
Trapped ion mobility spectrometry (TIMS), initially optimized and implemented for small molecules, has been gaining traction for the analysis of native proteins and protein complexes.25, 26 TIMS differs from the previously mentioned techniques in that an electric field that opposes the ion motion is used to hold ions stationary against a bath gas which pushes ions along the TIMS analyzer (an ion funnel). Ions of different mobilities are trapped at different points (potentials) along the ion optical axis using the electric field gradient and are then eluted from the device overtime, based on their mobility, as the TIMS potential gradient is reduced.27–30 Because the bath gas flows in the same direction as the ions, larger or more expanded ions are trapped farther along the TIMS analyzer and elute sooner than smaller or compact ions. Because ions are trapped and separated in the TIMS funnel, resolving power and time of analysis can be user-defined and optimized for a given application. For example, if high resolving power is necessary to separate two ions of similar mobilities, the step size of the voltage profile (or slope) is decreased, and overall time of analysis increased. For species requiring less resolving power the voltage profile is stepped at larger increments and overall analysis time is much shorter, allowing for rapid analysis. Because the resolving power of TIMS is not limited to IMS analyzer length, as traditional drift tube or travelling wave ion mobility are, higher mobility resolution has been achieved with TIMS. Park and co-workers observed a resolving power up to ~300 for separation of ubiquitin conformations, allowing for the identification of several additional conformations previously undetected using drift tube ion mobility spectrometry (R=20–50).31–35 The use of quadrupolar RF confinement in the TIMS funnel also decreases the radial spread and loss of ions compared to a traditional dc-only drift cell analyzer, therefore increasing total ion signal and allowing for ion activation of mobility-separated ions.36 This is important in the results described here because having a confined and well defined ion beam after mobility analysis facilitates surface collision experiments.
SID is a gas-phase activation technique that can probe protein complex stoichiometry, connectivity, subunit interactions, and structural rearrangements.37–42 To perform SID, ions are intentionally accelerated toward a rigid surface in the mass spectrometer. The result of the collision is a rapid increase in internal energy that often results in compact fragments indicative of the native topology of the complex, i.e., non-covalent bonds are cleaved to separate subcomplexes at the weaker of multiple interfaces. Most commercially available tandem mass spectrometers are equipped with gas-phase collision-induced dissociation (CID) capabilities, in which ions undergo numerous, low-energy collisions with neutral gas molecules.42 CID can restructure protein complexes in a variety of ways that can include collapse or expansion/unfolding, often yielding highly charged monomer and the corresponding (n-1)mer, therefore limiting the amount of structural information gained from these experiments.33 SID can access higher energy pathways with minimal restructuring, a pathway often unattainable by multi-step activation in CID, with SID often yielding lower-charge, compact subunits with symmetric charge partitioning. The implementation of SID capabilities on a Bruker SolariX XR 15T FT-ICR44–47, capable of high mass resolving power, has proven extremely beneficial for identifying overlapping oligomers produced from SID (i.e., dimer 4+ and monomer 2+) as well as probing small mass/charge differences, such as ligand binding or post translational modifications.38
TIMS combined with SID can be used to probe conformational dynamics, such as whether specific conformations can be identified as native-like, collapsed or expanded (including unfolding). With the typical symmetric charge partitioning of SID (and occasional/atypical symmetric charge partitioning of CID)48, product ions often overlap in m/z space, requiring another stage of separation for identification. The ultrahigh resolving power of the FT-ICR, however, allows for the identification of overlapping oligomers based on their isotope patterns. Here, we present the implementation and evaluation of TIMS combined with SID for the protein complexes streptavidin and cholera toxin B with GM1s ligand (sugar head group of GM1) and a peptide mixture containing bradykinin desArg1 and desArg9 on a 15 T FT-ICR mass spectrometer.
Methods and Materials
Sample Preparation and Ionization
Ammonium acetate, ethylenediamine diacetate (EDDA), cholera toxin B (CTB), the sodium salt of the pentasaccharide Galβ1–3GalNAϲβ1–4(Neu5Aϲ-α2–3)-Galβ1–4Glc (GM1s), and bradykinin desArg1 (PPGFSPFR) and desArg9 (RPPGFSPF) were purchased from Sigma-Aldrich (St. Louis, MO). Streptavidin from Streptomyces avidinii was purchased from Thermo Scientific Pierce Biotechnology (Rockford, IL) and has the known issue that each protein monomer has 0 or 1 methionine at the N-terminus. Protein complexes were buffer exchanged twice into 200 mM ammonium acetate (pH near neutral) with size exclusion chromatography spin columns (Micro Bio-Spin 6, Bio-Rad, Hercules, CA). EDDA was used as a charge-reducing agent and added to protein solutions at a final concentration of 40 mM EDDA and 160 mM ammonium acetate. Final protein concentrations ranged from 5–10 μM. For the bradykinin desArg1 and desArg9 mixture, the peptides were mixed at equal concentrations in 1:1 water: acetonitrile, at a final concentration of 5 μM each.
Instrumentation
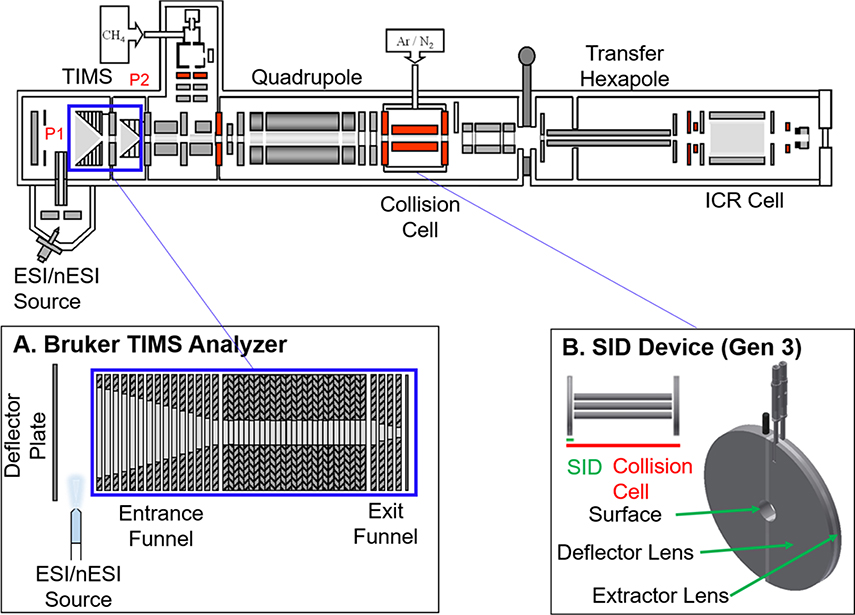
All experiments were performed on a Bruker SolariX XR 15 T FT-ICR mass spectrometer (depicted in Figure 1, Bruker Daltonics, Bremen, Germany). Ions for mass spectrometry experiments were generated using nanoelectrospray (nESI) ionization. Static nESI capillaries were prepared in-house using Sutter Instrument thin wall borosilicate capillaries with filament (outside diameter = 1.00 mm and inside diameter = 0.78 mm) and a Sutter Instrument P-97 Flaming/Brown type micropipette puller (Novato, CA). Capillary voltages of 0.6–1.1 kV were used for nESI via a 0.25mm diameter platinum wire (Sigma Aldrich, St. Louis, MO) inserted into the back of the capillary with 180 C, 5 L/min drying gas flow rate.
Figure 1.
Instrument design and modifications, including a TIMS analyzer30 (A) and SID devic44 (B). The TIMS analyzer replaced the entrance funnel (original entrance funnel region shown in blue box was replaced with the TIMS analyzer as shown in bottom blue box) and the SID device replaced the front-end cap of the commercial collision cell. The previously described SID device consists of three lenses: a semicircular surface onto which ions collide and two lenses with a non-conducting space in between: a deflector lens to push ions into the surface and an extractor lens element to extract and guide ions into the collision cell for accumulation prior to being pulsed into the ICR cell for detection.44
Once in the vacuum system ions may be analyzed and manipulated in a variety of ways. A prototype TIMS analyzer (Figure 1A) is incorporated in the first pumping region – serving the dual purpose of collecting the ions for transmission to downstream pumping stages and mobility analyzing the ions. The ions may also be mass filtered in an analytical quadrupole before arriving at the SID optic (Figure 1B). The ions may either be SID activated or allowed to pass unactivated into the collision cell. Typically, ions are collected in the collision cell before being transmitted to the ICR cell for mass analysis.
Surface-Induced Dissociation
The front endcap of the standard Bruker collision cell was replaced with a split lens SID device, termed “Gen 3”, and is described in a recent manuscript.44 Gen 3 consists of three electrodes: a surface, deflector, and extractor lens (Figure 1B).44 The device can be operated in both SID and transmission modes. The surface consists of a metal electrode (stainless steel, presumably coated with adventitious organics in the vacuum system). No significant neutralization/ion loss for large multiply-charged protein complexes was observed using a stainless steel surface compared to a gold surface coated with fluorocarbon self-assembled monolayers, as previously described by the Wysocki research group.44, 49 In SID mode, positive ions are accelerated toward the surface by applying an attractive voltage (more negative than the exit of the quadrupole mass analyzer) to the surface, while applying a more positive voltage to the deflector (Figure S1). SID energy can be increased by applying an even more negative voltage to the surface electrode, thus increasing the ion acceleration into the surface. After surface collision, the activated ions are extracted directly into the collision cell by applying a voltage to the extractor lens that is equivalent to, or slightly more positive, than the surface voltage while simultaneously holding the collision cell rods at a DC bias lower than the surface. SID lab frame collision energies (in eV) are defined as the acceleration voltage (difference between quadrupole exit lens and surface electrode) multiplied by the charge state of the precursor ion. In transmission mode, the three lenses are set to equivalent potentials and ions simply traverse the front endcap without any surface collision. Transmission mode also allows for CID experiments in the collision cell without removal or modification of the Gen 3 SID device.
Implementation of TIMS with SID
The ion funnel region was replaced with a custom TIMS analyzer (Bruker, Billerica, MA) equipped with an external power supply (Figure 1A).50, 30 In-house software, written in C++ by Danielson Software Consulting (Richland, WA) utilizing National Instruments LabVIEW resources (Austin, TX), was used to control the TIMS analyzer and was synchronized with the FT-ICR MS data acquisition program using a special pulse program. For TIMS analysis of large protein complex ions (greater than 3,000 m/z), the standard 900 kHz RF driver in the source (ion funnel) region was replaced with a 450 kHz RF driver capable of providing up to 300 Vpp. Care was taken when setting the RF amplitude as high values can unintentionally activate protein and protein complex ions. Mobility separation was performed using nitrogen as a bath gas at approximately 300 K with P1 = 3.2 mbar (entrance of TIMS) and P2 = 1.4 mbar (exit of TIMS) (Figure 1). The TIMS deflector plate (Vdef) voltage was set to 150 V for TIMS accumulation (open) and 0 V for the trapping and elution steps (closed) to prevent additional ions from entering the TIMS analyzer. The commercial quadrupole on this instrument has a theoretical upper selection limit of 6000 m/z, however, to perform successful Q-SID experiments, we have observed an experimental upper limit of 4000 m/z due to the lack of a well-defined ion beam and signal beyond this limit. The application of TIMS-SID here represents the ability for selection-activation without modification to the commercial quadrupole. The quadrupole was set to transmission mode for all experiments reported here, however if a second mode of separation is required (i.e., overlapping mobilities with different m/z), the quadrupole can be set to selection mode.
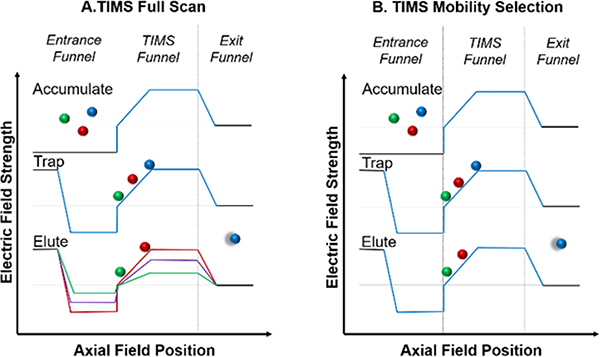
Additionally, the experimental setup includes an ion gate downstream of the TIMS funnel, as previously described elsewhere.21 Briefly, the ion gate is used to select or eliminate ions of different mobilities, allowing for the coupling of TIMS with the slower FT-ICR mass analyzer30 The TIMS analyzer was operated using an accumulate/trap/elute (with gating)/quench sequence over 34 ms per mobility scan. Ions selected with the ion gate are accumulated in the Bruker collision cell before being pulsed into the ICR cell, with an average of 25 to 100 ion mobility scans per mass spectrum. For the filling step of one TIMS scan, ions were accumulated for 15 ms of the total 34 ms TIMS profile. The TIMS funnel was quenched at the conclusion of each scan before beginning the next. An example of the voltages and timings used in the TIMS ion funnel region can be found in the supplemental information (Figures S2 and S3). For streptavidin tetramer, the starting and ending voltages of the TIMS ramp were set to −275 V and −50 V, respectively. After completion of a mobility of interest, voltage profile was lowered to the next mobility of interest.30 A pictorial representation of the accumulate/trap/elute/quench sequence can be found in Figure 2A. When performing TIMS-SID experiments the trapping voltage was held constant at the elution voltage for the mobility corresponding to the precursor of interest, resulting in isolation of a selected mobility (referred to as “mobility parking”, Figure 2B). Approximately 50–200 mass spectra were averaged to yield the multidimensional IM-MS spectrum, resulting in an average analysis time ranging from 2–10 minutes (TIMS-SID required longer averaging time compared to performing SID without mobility selection due to the lower duty cycle of combining TIMS with SID, however the total experiment time is comparable to SID-IMS experiments performed on our Waters Snyapt Q-SID-TWIMS-TOF mass spectrometers.43, 47 When calibrating TIMS mobility for proteins and protein complexes, two calibration methods were compared to assess the accuracy of Tune Mix for large, globular ions. Elution voltage values of ions were converted to reduced mobilities using both an Agilent Tune Mix calibration curve and a curve constructed using standard proteins and protein complexes (Figure S4). Collision cross sections were calculated from reduced mobilities (Ko) using the Mason-Schamp Equation (Equation S1) and compared for both calibration methods (Table S1).
Figure 2.
A visual representation of the TIMS voltage profiles used for full scans (A) or mobility selection for SID experiments (B). For the full scan, the voltage is lowered in a stepwise fashion to elute ions of different mobilities as a function of time. For mobility selection, the voltage is set for only the ion of interest to elute. The TIMS funnel is quenched after each mobility scan to remove any remaining ions before beginning the next accumulation step.
For each sample, an initial full TIMS scan was performed to determine the elution voltage for ions of particular interest, with the Gen 3 SID device held in transmission mode. To perform TIMS-SID experiments, the TIMS voltage ramp was held constant (or “parked”) at the elution voltage of interest and the Gen 3 SID device was tuned for SID mode. The collision cell accumulation time was set to a value in the range of 0.6 to 3.0 seconds (depending on number of TIMS accumulations and need for increased ion throughput, similar to Bruker’s typical CASI, continuous accumulation of selected ions, mode for CID). Post-accumulation, ion packets were pulsed into the ICR cell for detection. The process of pulsing an ion packet into the ICR was performed multiple times and the resulting mass spectra were averaged for a total analysis of time of 2–10 minutes. The time-of-flight setting, which controls the time ions have to travel between the collision cell into the ICR cell was optimized to ensure a balanced detection of low and high m/z ions (typically 2.2 to 3.0 milliseconds for protein complexes). A low time-of-flight setting results in a bias toward increased detection of low m/z ions, as high m/z ions require a longer time-of-flight. An example of the ICR tune settings used for nMS experiments are provided in Figure S5.
Results
Application of TIMS-SID for Biological Samples
TIMS-SID of Streptavidin Tetramer
To validate the performance of TIMS coupled with SID for native MS and optimization of TIMS tuning parameters for high mass trapping and transmission, a series of standard protein complexes were chosen. These systems have been well-characterized with different ion mobility spectrometry methods and SID previously.
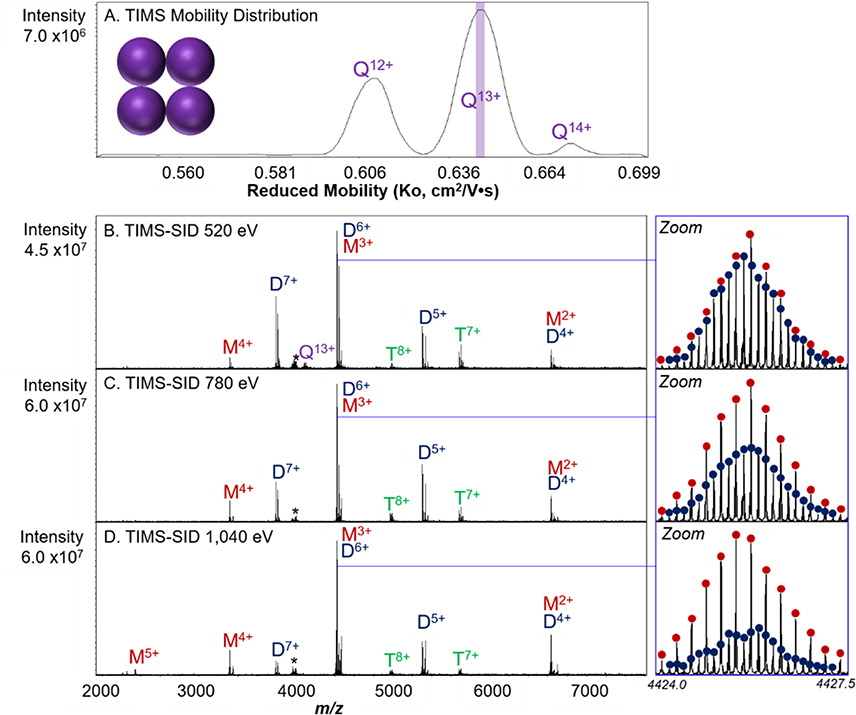
Streptavidin, a 53 kDa homotetramer, is a dimer of dimers; therefore, SID yields predominately compact dimers at low energies. At higher SID laboratory collision energies, charge-symmetric, and therefore assumed to be, compact monomers and trimers form in competition with the formation of dimers.51 As provided by Sigma, the streptavidin tetramer contains variable numbers of leading methionines, with zero or one on each monomer; these can be seen in the spectra for both the main series of peaks and for salt adducts. The results of the TIMS-SID-MS analysis of streptavidin, is shown in Figure 3. The full mass spectrum for charge-reduced streptavidin can be found in Figure S5. The full TIMS distribution for streptavidin yielded distinct mobility profiles for the 12+, 13+, and 14+ charge states (Figure 3A). In Figures 3B–D, TIMS was used to mobility select the 13+ charge state (Velution = −203 V, Ko – 0.64 cm2V−1S−1) and then activate at three different SID energies.
Figure 3.
TIMS mobility distribution of streptavidin tetramer with the mobility selection used for SID experiments highlighted by the purple slice (A). Reduced mobilities were calculated using the protein/protein complex calibration. TIMS-SID of mobility-selected 13+ streptavidin tetramer at different activation energies. At low SID energy (520 eV (B)), SID yields dimer and monomer products. As SID energy is increased to 780 eV (C) and 1,040 eV (D), trimers are also observed in the mass spectra. Even relatively low mass resolution (Rcalculated = 138,000) for the 15 T FT-ICR allows for identification of overlapping monomer and dimer based on isotopic distributions, as shown in the zoom-in panels. The ratio of total dimers to [monomer plus trimer] for each energy varies from 85:15 at 540 eV to 16:84 at 1040 eV. The asterisk denotes a noise peak.
TIMS-SID of the mobility-selected Q13+ tetramer yields mostly symmetrically charged dimer (on average, half the charge of the tetramer precursor) and symmetrically charged monomer, which can be indicative of compact subunit fragments.42 As previously mentioned, streptavidin is considered a dimer of dimers, therefore dissociation based on its native interfaces should occur between the two dimers. As SID energy is increased, however, the complex dissociates into compact, low charge (5+, 4+, 3+, and 2+) monomers and their complementary trimers (Figure 3). Because of the high resolving power of the 15 T FT-ICR, overlapping oligomers were also identified based on their isotopic distributions. For example, TIMS-SID of streptavidin shown in Figure 3B resulted in overlapping 3+ monomer and 6+ dimer. The differing amounts of monomer and dimer can be observed by zooming into the spectra at this region. An increase in SID energy resulted in the production of more monomer products, presumably from dimers that were observed to decrease at higher energies (trimers were of very low abundance at these energies). A comparison between TIMS-SID and TIMS-CID can be found in Figure SI7. Here, we obtained high signal-to-noise ratios (S/Nm/z 4424 = 120.3 for streptavidin) for protein complexes, evidenced by the accurate isotopic distributions of even species of low abundance.
TIMS-CID and TIMS-SID of Cholera Toxin B Pentamer (with GM1 Ligand)
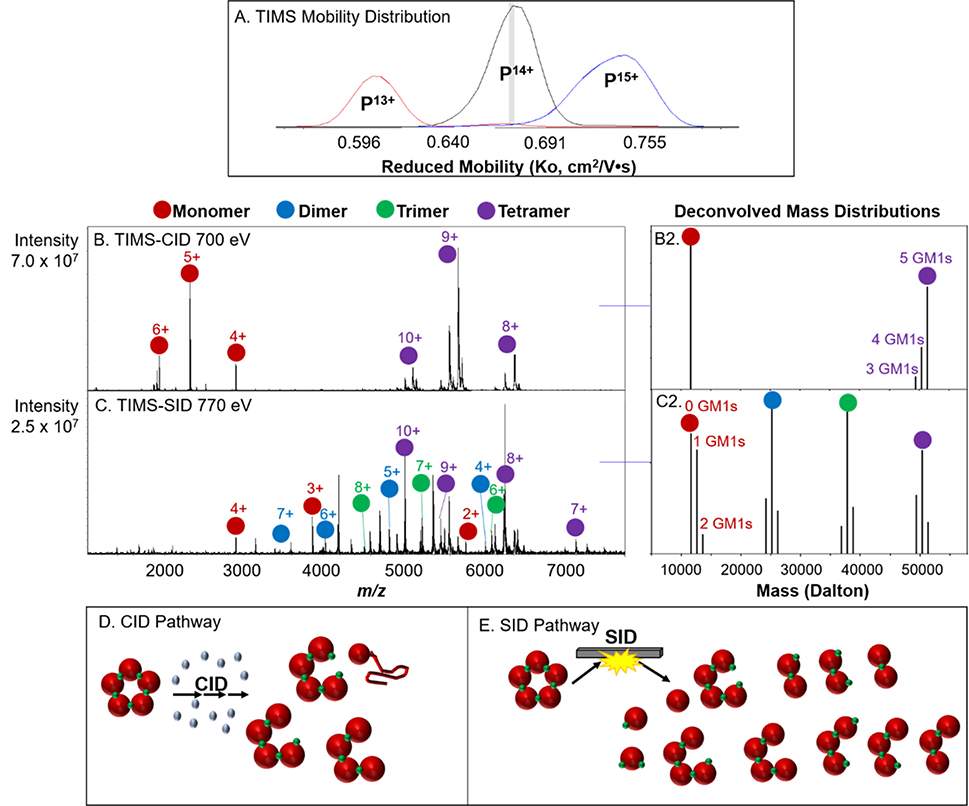
TIMS ion mobility instruments operate by holding ions stationary in an electric field for user-defined periods of time, with typically longer trapping times for higher resolution applications. It has previously been suggested that the extended trapping times and applied electric fields may increase the internal energy of ions, resulting in restructuring or even fragmentation of smaller ions.50 This raises concern when performing nMS experiments, in which maintaining the native-like structure and avoiding unintentional activation is of critical importance.30, 52, 53 It has since been clarified that that ion heating likely occurs due to non-intrinsic properties of TIMS (space charge effects and RF power absorption) and that careful tuning prevents structural rearrangement, therefore making TIMS a suitable technology for nMS.53 For the experiments performed in this manuscript, the amplitude of the RF-confining voltage in the TIMS funnel was set as low as possible, while still trapping a sufficient amount of ions, and trapping times in the TIMS analyzer were kept low to limit structural perturbation.30 To confirm the results from Bleiholder and co-workers, TIMS-SID was performed using cholera toxin B (CTB), a 58 kDa pentamer, with bound GM1s ligands and compared to TIMS-CID of the same complex (TIMS parameters were identical for both TIMS-SID and TIMS-CID experiments). The CTB complex binds five GM1s molecules, located between two neighboring subunits. The results for CTB with GM1 are shown in Figure 4 and the results agree well with those obtained previously on an Orbitrap with SID installed (monomers retain 0, 1, and 2 ligands, dimers retain 1, 2, and 3 ligands, etc, consistent with the fact that the ligands bind at the interface of 2 protein monomers). Deconvolution of the CTB + GM1s mass spectra were performed by integrating the area under each peak and normalizing against total intensity in OriginPro 9.54 The full mass spectrum for charge reduced CTB with GM1s is shown in Figure SI8.
Figure 4.
TIMS mobility distribution of CTB pentamer with GM1s ligand bound with mobility selection for CID and SID experiments highlighted with gray slice (A). Reduced mobilities were calculated using the protein/protein complex calibration. TIMS-CID and TIMS-SID of mobility-selected 14+ (B, C) with corresponding deconvolution to show variations of GM1s retention on subunits (B2, C2). CID and SID pathways of CTB with GM1s and the observed products (D, E).
Pre-activation of a protein complex can often be probed by SID. Loss of native-like structure typically results in a SID spectrum that is more consistent with CID products, that is disproportionately high-charged monomer and the corresponding (n-1) mer. Additionally, pre-activation can lead to ligand loss from the complex. TIMS-CID of the 14+ CTB with GM1s complex yields the expected highly charged monomer and corresponding tetramer, which further undergoes ligand loss (Figure 4B, 4B2, 4D). Conversely, TIMS-SID of CTB with GM1s yields monomer, dimer, trimer, and tetramer, indicative of the equal interfaces between each subunit.45 For CTB with GM1s, the GM1s ligand binds between individual subunits, therefore SID results in monomer with 0, 1, or 2 GM1s bound and dimer with 1, 2, or 3 GM1s bound, and so forth (Figure 4C, 4C2, 4E).55 The observation of charge-symmetric products, as well as retention of the ligand further suggests that carefully tuned TIMS parameters do not result in significant, measurable restructuring of protein complexes. Additionally, ICR instrument parameters downstream from the TIMS funnel were all tuned using low voltages to prevent any inadvertent activation. We likely see less measurable rearrangement for protein complexes due to their higher barrier for intramolecular rearrangement to occur compared to thermometer ions or monomeric proteins, at least within a 15 ms time scale.56
It is important to note that a current limitation of this technology is performing ion mobility spectrometry directly after ionization without the capabilities of performing an in-source activation step to “clean up” biological samples that contain several non-specific adducts. Heavily adducted samples result in a broad mobility distribution with lower total ion intensity spread across that distribution, making mobility parking on a specific point to couple with SID difficult due to low signal intensity. Solution-phase clean up (size-exclusion buffer exchange columns) and smaller diameter openings of nano electrospray capillaries were used to limit sample contamination in the work presented above. In the future, this technology will be most beneficial when investigating dynamic protein complexes, such as those that exhibit different conformations upon ligand binding or post-translational modifications, and that require ultrahigh mass resolution to decipher such small mass differences between those systems.
TIMS-SID of Bradykinin desArg1 and desArg9
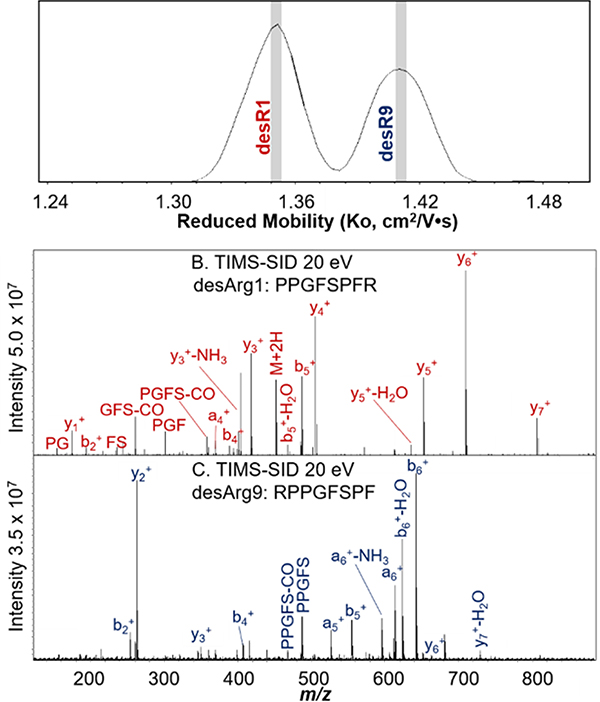
Bradykinin is a peptide comprised of nine amino acids (RPPGFSPFR). Removal of the C-terminal arginine (desArg9) or the N-terminal arginine (desArg1) results in two distinct peptides with the same exact mass. As a result, the two peptides cannot be separated using an analytical quadrupole. TIMS was performed on a 1:1 mixture of desArg1 and desArg9 bradykinin (Figure 8A) and two distinct distributions were observed for the doubly charged peptides. Each distribution was mobility-selected, and SID was performed (Figure 5B and C).
Figure 5.
TIMS of a 1:1 mixture of desArg1 and desArg9 yielded baseline resolved distributions of the isobaric peptides (A). Reduced mobilities were calculated using Tune Mix as calibrant. Each distribution was selected (as indicated by gray slices), and the corresponding peptide was assigned based on the SID fragmentation spectra (B and C).
Using TIMS, we observed baseline IMS resolution for desR1 and desR9 bradykinin, previously unachieved on a TWIMS platform.57 Despite having the same nominal mass, desArg1 and desArg9 yield significantly different SID spectra, as we have previously noted. CID and SID spectra of each of these small peptides are similar (desArg1 CID similar to desArg1 SID) with either type of spectra differing for the two isomers. Most notable is the preferential formation of y-type ions from desArg1 because the charge is sequestered on the C-terminal arginine. Comparatively, desArg9 preferentially forms more b-type fragment ions due to charge retention on the N-terminal arginine. Here, TIMS-SID is used for both the separation and identification of isobaric mixtures. The identification of the peptides was also confirmed by performing TIMS on the individual species (Figure S9). It was observed that the peptides, with lower masses than the protein complexes, were more sensitive to RF confining amplitude, as evidenced by intact peptide dissociating in the TIMS funnel at very high amplitudes. Low RF amplitude (70 Vpp for bradykinin compared to 250 Vpp for protein complexes) that did not yield any unintentional fragmentation was used for the results presented here. TIMS-SID shows promising potential application to the analysis of complex mixtures that consist of isobaric species that can be identified based on their fragmentation patterns, not just for nMS applications. TIMS also offers the advantages of being a rapid separation technique that does not require excessive amounts of sample or solvent, compared to traditional liquid chromatography methods used for mixture analysis.
Conclusions
Various protein complexes and a peptide mixture were successfully trapped and mobility-selected using trapped ion mobility spectrometry (TIMS) on an ultrahigh resolution 15T FT-ICR MS platform, further equipped with SID capabilities. The combination of TIMS and SID provided complementary structural information regarding the topology and conformation (relative drift times) of the protein complexes studied. Additionally, TIMS-SID allowed for the separation, fragmentation, and identification of peptide structural isomers present in a mixture. The SID spectra of streptavidin and CTB with GM1s ligand following IM parking were nearly identical to those from quadrupole isolation without any TIMS analysis, suggesting that little to no measurable restructuring or unfolding of protein complexes was observed during the TIMS analysis, supporting the conclusion that TIMS is applicable to nMS experiments in agreement with Bleiholder and co-workers. When analyzing peptides, however, RF Vpp must be optimized to avoid unintentional ion activation (and possibly fragmentation) due to RF heating. Additionally, the implementation of TIMS-SID on an ultrahigh resolution platform allows for identification and relative quantification of overlapping oligomeric SID products which would otherwise require a second stage of ion mobility for deconvolution.
Supplementary Material
Acknowledgments
The authors would like to thank Jacob Meier (Bruker Daltonics) for help installing the TIMS device and Desmond Kaplan (KapScience) for thoughtful discussion and advice when coupling TIMS with SID. The FT-ICR was purchased using funds from NIH award S10 OD018507. This work was supported by the NIH Resource for Native Mass Spectrometry Guided Structural Biology, P41GM128577 (VHW).
Footnotes
Supporting Information:
SID device tune settings, TIMS voltage settings, TIMS ramp voltage profile, comparison of mobility calibration curves, CCS determination, CCS values of streptavidin and CTB, FT-ICR tune settings, streptavidin full mass spectrum, streptavidin TIMS-SID and TIMS-CID comparison, Cholera toxin-B with GM1s full mass spectrum, desArg bradykinin individual mobility distributions.
References
- 1.Likeo I; Allison TM; Hopper JTS; Robinson CV. Mass spectrometry guided structural biology. Curr. Opin. Struc. Biol, 2016, 40: 136–144. [DOI] [PubMed] [Google Scholar]
- 2.Lössl P; van de Waterbeemd M; Heck AJR. The diverse and expanding role of mass spectrometry in structural and molecular biology. The EMBO Journal, 2016, 35: 2634–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rostom AA. and Robinson CV Detection of the Intact GroEL Chaperonin Assembly by Mass Spectrometry. J. Am. Chem. Soc, 1999, 121, 19, 4718–4719. [Google Scholar]
- 4.Shoemaker SC and Ando N X-rays in the Cryo-EM Era: Structural Biology’s Dynamic Future. Biochemistry, 2018. 57 (3), 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilag LL: Videler H; McKay AR; Sobott F; Fucini P; Nierhaus KH; Robinson CV. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. PNAS, 2005, 102, 23, 8192–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernández H; Makarova OV; Makarov EM; Morgner N; Muto Y; Krummel DP; Robinson CV. Isoforms of U1–70k Control Subunit Dynamics in the Human Spliceosomal U1 snRNP. PLoS ONE, 2009, 4, 9, e7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang J; Chan H; Cash DD; Miracco EJ; Ogorzalek Loo RR; Upton HE; Casicio D; Johnson RO; Collins K; Loo JA; Zhou H; Feigon J Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science, 2015, 350, 6260, aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenzen K; Olia AS; Uetrecht C; Cingolani G; Heck AJ. Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J. Mol. Biol, 2008, 379, 2, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanucara F; Holman SW; Gray CJ; Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem, 2014, 6, 4, 281–294. [DOI] [PubMed] [Google Scholar]
- 10.Song Y; Nelp MT; Bandarian V; Wysocki VH. Refining the Structural Model of a Heterohexameric Protein Complex: Surface Induced Dissociation and Ion Mobility Provide Key Connectivity and Topology Information. ACS Cent. Sci, 2015, 1,9, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurneczko E; Barran PE. How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst, 2011, 136, 1, 20–28. [DOI] [PubMed] [Google Scholar]
- 12.Revercomb HE and Mason EA Theory of plasma chromatography/gaseous electrophoresis. Anal. Chem. 1975, 47, 987–983. [Google Scholar]
- 13.Mason EA and McDaniel EW Transport properties of ions in gases. New York: Wiley; 1988. [Google Scholar]
- 14.Viehland LA and Mason EA Transport Properties of Gaseous Ions over a Wide Energy Range, IV. Atomic Data and Nuclear Data Tables, 1995, 60, 37–95. [Google Scholar]
- 15.McDaniel EW; Martin DW; Barnes WS Drift-tube Mass Spectrometer for Studies of Low-Energy Ion-Molecule Reactions. Rev. Sci. Instrum 1962, 33, 2. [Google Scholar]
- 16.Mesleh MF; Hunter JM; Shvartsburg AA; Schatz GC; Jarrold MJ Structural Information from Ion Mobility Measurements: Effects of the Long-Range Potential. Anal. Chem 1996, 100, 40, 16082–16086. [Google Scholar]
- 17.Wyttenbach T; Bleiholder C; Bowers MT Factors Contributing to the Collision Cross Section of Polyatomic Ions in the Kilodalton to Gigadalton Range: Application to Ion Mobility Measurements. Anal. Chem 2013, 85, 4, 2191, 2199. [DOI] [PubMed] [Google Scholar]
- 18.Bleiholder C; Johnson NR; Contreras A; Wyttenbach T; Bowers MT Molecular Structures and Ion Mobility Cross Sections: Analysis of the Effects of He and N2 Buffer Gas. Anal. Chem 2015, 87, 14, 7196–7203. [DOI] [PubMed] [Google Scholar]
- 19.Jurneczko E and Barran PE How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst. 2011, 136, 1, 20–28. [DOI] [PubMed] [Google Scholar]
- 20.Clemmer DE and Jarrold MF Ion Mobility Measurements and their Applications to Clusters and Biomolecules. J. Mass Spectrom 1997, 32, 577–592. [Google Scholar]
- 21.Gabelica V; Marklund E Fundamentals of ion mobility spectrometry. Curr. Opin. Chem. Biol, 2018, 42, 51–59. [DOI] [PubMed] [Google Scholar]
- 22.Stiving AQ; Jones BJ; Ujma J; Giles K; Wysocki VH. Collision Cross Sections of Charge-Reduced Proteins and Protein Complexes: A Database for CCS Calibration. Anal. Chem, 2020, 92, 6, 4475–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campuzano IDG; Giles K Historical, Current and Future Developments of Traveling Wave Ion Mobility Mass Spectrometry: A Personal Perspective. TrAC Trends Anal. Chem, 2019, 115620. [Google Scholar]
- 24.Allen SJ; Giles K; Gilbert T; Bush MF. Ion mobility mass spectrometry of peptide, protein, and protein complex ions using a radio-frequency confining drift cell. Analyst, 2016, 141, 884–891. [DOI] [PubMed] [Google Scholar]
- 25.Benigni P; Marin R; Molano-Arevalo JC; Garabedian A; Wolff JJ; Ridgeway ME; Park MA; Fernandez-Lima F Towards the Analysis of High Molecular Weight Proteins and Protein complexes using TIMS-MS. Int. J. Ion Mobil. Spectrom, 2016, 19, 2, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu FC; Cropley TC; Ridgeway ME; Park MA; Bleiholder C Structural analysis of the glycoprotein complex avidin by tandem-trapped ion mobility spectrometry-mass spectrometry (tandem TIMS/MS). Anal. Chem, 2020, 92, 6, 4459–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michelmann K; Silveria JA; Ridgeway ME; Park MA. Fundamentals of Trapped Ion Mobility Spectrometry. J. Am. Soc. Mass Spectrom, 2014, 26,1, 14–24. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Lima F; Kaplan DA; Suetering J; Park MA. Gas-phase separation using a trapped ion mobility spectrometer. Int. J. Ion Mobil. Spectrom, 2011, 14, 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridgeway ME; Lubeck M; Jordens J; Mann M; Park MA. Trapped ion mobility spectrometry: A short review. Int. J. Mass Spectrom, 2018, 425, 22–35. [Google Scholar]
- 30.Ridgeway ME; Wolff JJ; Silveira JA; Lin C; Costello CE; Park MA. Gated trapped ion mobility spectrometry coupled to fourier transform ion cyclotron resonance mass spectrometry. Int. J. Ion Mobil. Spectrom, 2016, 19, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridgeway ME.; Silveria JA; Meier JE; Park MA Microheterogeneity within conformational states of ubiquitin revealed by high resolution trapped ion mobility spectrometry. Analyst, 2015, 140, 6964. [DOI] [PubMed] [Google Scholar]
- 32.Ujma J; Giles K; Morris M; Barran PE New High Resolution Ion Mobility Mass Spectrometer Capable of Measurements of Collision Cross Sections from 150 to 520 K. Anal. Chem, 2016, 88, 9469–9478. [DOI] [PubMed] [Google Scholar]
- 33.Clemmer DE. and Jarrold MF. Ion Mobility Measurements and their Applications to Clusters and Biomolecules. J. Mass Spectrom, 1997, 32, 577–592. [Google Scholar]
- 34.Koeniger SL, Merenbloom SI, Clemmer DE: Evidence for Many Resolvable Structures within Conformation Types of Electrosprayed Ubiquitin Ions. J. Phys. Chem. B, 2006, 110, 7017–7021. [DOI] [PubMed] [Google Scholar]
- 35.Koeniger SL. and Clemmer DE. Resolution and Structural Transitions of Elongated States of Ubiquitin. J. Am. Soc. Mass Spectrom, 2006, 18, 322–331. [DOI] [PubMed] [Google Scholar]
- 36.Hernandez DR; DeBord JD; Ridgeway ME; Kaplan DA; Park MA; Fernandez-Lima F Ion dynamics in a trapped ion mobility spectrometer. Analyst, 2014, 139, 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stiving AQ; VanAernum ZL; Busch F; Harvey SR; Sarni SH; Wysocki VH Surface-Induced Dissociation: An Effective Method for Characterization of Protein Quaternary Structure. Anal. Chem, 2019, 91, 1, 190–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou M; Yan J; Romano CA; Tebo BM; Wysocki VH; Pasa-Tolič L Surface Induced Dissociation Coupled with High Resolution Mass Spectrometry Unveils Heterogenity of a 211 kDa Multicopper Oxidase Protein Complex. J. Am. Soc. Mass Spectrom, 2018, 29, 4, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harvey SR; Liu Y; Wysocki VH; Laganowsky A Surface induced dissociation as a tool to study membrane protein complexes. Chem. Commun, 2017, 53, 3106–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X; Loo JA; Wysocki VH Surface induced dissociation yields substructure of Methanosarcina thermophile 20S proteasome complexes. Int. J. Mass Spectrom, 2015, 377, 0, 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma X; Zhou M; Wysocki VH Surface induced dissociation yields quaternary substructure of refractory phosphorylase B and glutamate dehydrogenase complexes. J. Am. Soc. Mass Spectrom, 2014, 25, 368–379. [DOI] [PubMed] [Google Scholar]
- 42.Zhou M; Wysocki VH Surface induced dissociation: dissecting noncovalent protein complexes in the gas phase. Acc. Chem. Res, 2014, 47, 4, 1010–1018. [DOI] [PubMed] [Google Scholar]
- 43.Quintyn RS; Zhou M; Yan J; Wysocki VH. Surface-Induced Dissociation Mass Spectra as a Tool for Distinguishing Different Structural Forms of Gas-Phase Multimeric Protein Complexes. Anal. Chem, 2015, 87, 11879–11886. [DOI] [PubMed] [Google Scholar]
- 44.Snyder DT; Panczyk EM; Somogyi A; Kaplan D; Wysocki VH Simple and Minimally Invasive SID Devices for Native Mass Spectrometry. Anal. Chem, 2020, 92, 16, 11195–11203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan J; Zhou M; Gilbert JD; Wolff JJ; Somogyi A; Peder RE; Quintyn RS; Morrison LJ; Easterling ML; Paša-Tolić L; Wysocki VH Surface-induced dissociation (SID) of protein complexes in a hybrid Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer. Anal. Chem, 2017, 89, 1, 895–901. [DOI] [PubMed] [Google Scholar]
- 46.Snyder DT; Panczyk E; Stiving AQ; Gilbert JD; Somogyi A; Kaplan D; Wysocki VH Design and Performance of a Second-Generation Surface-Induced Dissociation Cell for Fourier Transform Ion Cyclotron Resonance Mass Spectrometry of Native Protein Complexes. Anal. Chem, 2019, 91, 21, 14049–14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou M; Huang C; Wysocki VH Surface-Induced Dissociation of Ion Mobility-Separated Noncovalent Complexes in a Quadrupole/Time-of-Flight Mass Spectrometer. Anal. Chem, 2012, 84, 14, 6016–6023. [DOI] [PubMed] [Google Scholar]
- 48.Hall Z and Robinson CV Do Charge State Signatures Guarantee Protein Conformations? J. Am. Soc. Mass Spectom, 2012, 23, 1161–1168. [DOI] [PubMed] [Google Scholar]
- 49.Stiving AQ; Gilbert JD; Jones BJ; Wysocki VH A Tilted Surface and Ion Carpet Array for SID. J. Am. Soc. Mass Spectrom, 2020, 31, 2, 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelly RT; Tolmachev AV; Page JS; Tang K; Smith RD The Ion Funnel: Theory, Implementations, and Applications. Mass Spectrom. Rev 2010, 29, 2, 294–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quintyn RS; Yan J; Wysocki VH Surface-Induced Dissociation of Homotetramers with D2 Symmetry Yields their Assembly Pathways and Characterized the Effect of Ligand Binding. Chemistry & Biology, 2015, 22, 5, 583–592. [DOI] [PubMed] [Google Scholar]
- 52.Morsa D; Hanozin E; Eppe G; Quinton L; Gabelica V; De Pauw E Effective Temperature and Structural Rearrangement in Trapped Ion Mobility Spectrometry. Anal. Chem, 2020, 92, 6, 4573–4582. [DOI] [PubMed] [Google Scholar]
- 53.Bleiholder C; Liu FC; Chai M Comment on Effective Temperature and Structural Rearrangement in Trapped Ion Mobility Spectrometry: TIMS Enables Native Mass Spectrometry Applications. Anal. Chem. 2020, Just Accepted, 10.1021/acs.analchem.0c02052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Origin (Pro), Version 9. OriginLab Corporation, Northampton, MA, USA. [Google Scholar]
- 55.Busch F; VanAernum ZL; Ju Y; Yan J; Gilbert JD; Quintyn RS; Bern M; Wysocki VH, Localization of protein complex bound ligands by surface-induced dissociation high-resolution mass spectrometry, Anal Chem 2018, 90, 21, 12796–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harvey SR; Yan J, Brown JM; Hoyes E; Wysocki VH, Extended gas-phase trapping followed by surface-induced dissociation of non-covalent protein complexes, Anal. Chem 2016, 88: 1218–1221. [DOI] [PubMed] [Google Scholar]
- 57.Stiving A; Harvey S; Jones BJ; Bellina B; Brown J; Barran PE; Wysocki V Coupling 193 nm Ultraviolet Photodissociation and Ion Mobility for Sequence Characterization of Conformationally-Selected Peptides. J. Am. Soc. Mass Spectrom 2020, 31, 11, 2313–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.