Dear Editor,
We read with interest the work published by Lui and colleagues in the Journal of Infection1, in which the dynamics of SARS-CoV-2 RNA at different body sites was investigated in a rather small cohort including 5 patients with critical/severe COVID-19. The authors supported the assumption that viral loads in lower respiratory tract (LRT) better reflected clinical progression in severe disease than those in upper respiratory tract (URT) samples. To further address this issue, we conducted an observational study (approved by the Ethics Committee of Hospital Clínico Universitario INCLIVA in May,2020) aimed at characterizing the kinetics of SARS-CoV-2 RNA load in the LRT and plasma (viral RNAemia) and assessing how these relate to the inflammatory state and mortality of critically ill COVID-19 patients. Seventy-three consecutive patients (51 males and 22 females; median age, 65 years; range, 21 to 80 years) were recruited during ICU stay (median,18 days; range, 2–67 days), between October 2020 and February 2021 (Supplementary Table 1). Patients were admitted to ICU at a median of 9 days (range, 2–25) after onset of symptoms. Sixty-four patients underwent mechanical ventilation, from whom 165 tracheal aspirates (TA) were collected (median of 2 specimens/patient; range, 1–11). A total of 340 plasma specimens (median, 4 samples/patient; range, 1–16) were available from the 73 patients. SARS-CoV-2 RNA quantitation in TA and plasma was carried out by the Abbott RealTime SARS-CoV-2 assay Abbott Molecular (Des Plaines, IL, USA) (See Supplementary Material). SARS-CoV-2 viral loads (in copies/ml) were estimated using the AMPLIRUN® TOTAL SARS-CoV-2 RNA Control (Vircell SA, Granada, Spain). The analytical sensitivity of the RT-PCR assay in TA and plasma specimens was 100 copies/ml (95%) for both matrices.
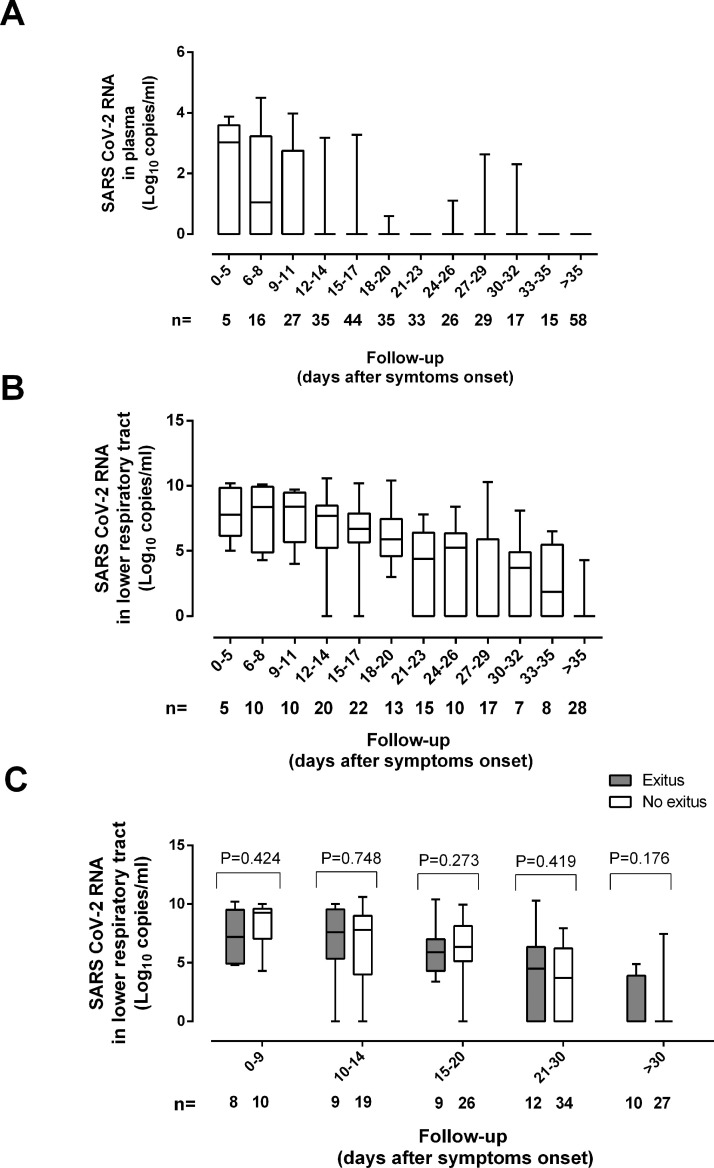
SARS-CoV-2 RNA (median, 6.5 log10 copies/ml; range, 3.03–10.6 log10) was detected in 109 TA from 56 patients (91.8%). Viral load remained relatively stable across the first two weeks from symptoms onset and began to decrease afterwards (Fig. 1 A). No patient tested positive for SARS-CoV-2 RNA in TA beyond day 42. As reported for URT2 , 3, neither remdesivir nor tocilizumab administration appeared to have a major impact on the dynamics of SARS-CoV-2 RNA load in TA (Supplementary Table 2).
Fig. 1.
Kinetics of SARS-CoV-RNA load in tracheal aspirates (A) and plasma (B) of critically ill patients undergoing invasive ventilation. Panel C shows the kinetics of SARS-CoV-2 RNA load in the lower tracheal aspirates in patients who either died or
SARS-CoV-2 RNAemia (median, 3.03 log10 copies/ml; range, 1.69 to 5.27 log10) was detected in 37 plasma specimens from 26 patients (35.6%). Median time to first detection of viral RNA in plasma was 10 days after symptoms onset (range, 3–32 days) Viral RNA cleared faster in plasma than in TA (Fig. 1B). Previous studies using a droplet-based digital PCR, which seemingly outperforms conventional RT-PCR assays in terms of sensitivity, reported higher rates of viral RNAemia detection in ICU patients (77% in4 and 88% in5) than found in the current study. This discrepancy could also be related to different timing of sample collection across studies.
A moderate yet significant correlation was found between SARS-CoV-2 RNA levels in TA and in paired plasma specimens (rho, 0.41; p < 0.001). SARS-CoV-2 RNA load in TA was significantly higher (p < 0.001) in presence than absence of concomitant viral RNAemia (median, 9.5 log10 copies/ml; range, 4.3 to 10.4 log10 copies/ml vs. median, 6.2 log10 copies/ml; range, 3.0–10.6 log10 copies/ml), this suggesting that LRT may be a substantial source of SARS-CoV-2 RNA.
Plasma levels of ferritin, lactose dehydrogenase (LDH), but not interleukin-6 (IL-6), C-reactive protein (CRP), or D-Dimer (D-D), were significantly higher when SARS-CoV-2 RNA was detected in paired TA or plasma specimens (Table 1 ), yet SARS-CoV-2 RNA loads in these specimens correlated modestly (Rho < 0.31) with plasma levels of ferritin and LDH (Supplementary Fig 1). Lymphocyte counts were significantly lower in the presence of SARS-CoV-2 RNA in TA and plasma (Table 1). Nevertheless, the level of correlation (inverse) between SARS-CoV-2 RNA load in TA and plasma and lymphocyte counts was modest ((rho, -0.43; p < 0.01 and rho, -0.25, p < 0.01, respectively). A significant association between SARS-CoV-2 RNAemia detection and blood levels of IL-6, interleukin-10, CRP, ferritin, D-D and LDH was previously reported4 , 6. In these studies, a single time point specimen per patient collected at ICU admission was considered for the analyses, as opposed to the serial specimens used herein.
Table 1.
Qualitative detection of SARS-CoV-2 RNA in the lower respiratory tract or plasma or SARS-CoV-2 N protein in plasma and blood levels of biomarkers of COVID-19 severity.
| Qualitative result of a given virological parameter |
no. of paired specimens | Parameter (Median range) |
p value | ||
|---|---|---|---|---|---|
| SARS-CoV-2 RNA load in tracheal aspirates | Pos | 23 | IL-6 in pg/ml. | 111.4 (4–3,548) | 0.84 |
| Neg | 2 | 142 (22–262) | |||
| Pos | 82 | Ferritin in ng/ml. | 805.5 (69–6,440) | 0.01 | |
| Neg | 34 | 421.5 (46–2,659) | |||
| Pos | 101 | D-D in ng/ml. | 1,730 (270–29,940) | 0.87 | |
| Neg | 49 | 1,790 (270–16,160) | |||
| Pos | 105 | LDH in UI/l. | 687 (93–2,132) | 0.001 | |
| Neg | 51 | 520 (214–1,395) | |||
| Pos | 107 | CRP in mg/l. | 35 (1–746) | 0.62 | |
| Neg | 54 | 32.85 (1–606.7) | |||
| Pos | 74 | Lymphocytes in cell/µl | 0.96 (0.02–3.73) | <0.001 | |
| Neg | 42 | 1.40 (0.44–2.43) | |||
| SARS-CoV-2 RNAemia | Pos | 9 | IL-6 in pg/ml. | 111.4 (10.8–1363.) | 0.92 |
| Neg | 40 | 141.8 (4–3,548) | |||
| Pos | 31 | Ferritin in ng/ml. | 1,176 (147–6,440) | <0.001 | |
| Neg | 211 | 590 (42–5,847) | |||
| Pos | 36 | D-D in ng/ml. | 1,535 (320–9,170) | 0.17 | |
| Neg | 274 | 1740 (270–60,000) | |||
| Pos | 36 | LDH in UI/l. | 765.5 (329–1,720) | 0.002 | |
| Neg | 281 | 637 (58–2,132) | |||
| Pos | 36 | CRP in mg/l. | 47.95 (1.2–459) | 0.38 | |
| Neg | 299 | 30.7 (1–746) | |||
| Pos | 26 | Lymphocytes in cell/µl | 0.72 (0.02–3.13) | <0.001 | |
| Neg | 209 | 1.13 (0.17–3.73) | |||
CRP, C-reactive protein; D-D, Dimer-D; IL-6, interleukin-6; LDH, lactose dehydrogenase.
Dynamics of SARS-CoV-2 RNA load (initial, peak and trajectory) in TA following ICU admission were comparable across patients who either died or survived (Fig. 1C). Moreover, neither initial nor peak viral load in TA was associated with increased mortality (OR, 0.81; 95% CI, 0.68–2.24; p = 0.68, and OR, 0.39; 95% CI, 0.22–1.82; p = 0.39, respectively) (Supplementary Table 3). Other studies, in contrast, pointed to an association between protracted SARS-CoV-2 RNA clearance in LRT and/or simple presence of SARS-CoV-2 RNA in LRT and increased risk of mortality7, 8, 9. In these studies, a wide variety of LRT specimens were used, and no data proving a dose-dependent relationship between SARS-CoV-2 RNA load in LRT and mortality were provided.
We found a trend towards an association between qualitative detection of SARS-CoV-2 RNA in plasma and increased mortality in adjusted multivariate logistic regression models (OR, 2.82, 95% CI, 0.94–8.47), and failed to demonstrate such a trend for initial or peak viral loads (supplementary Table 3). SARS-CoV-2 RNAemia has been previously associated with poor clinical outcome in series including only ICU patients, in which patients who died displayed higher viral RNA loads in plasma collected at ICU admission than those who survived4.
The main limitation of the current study is its relatively small sample size. Analysis of sequential specimens from patients could be considered a strength of the research.
The current study provides a further insight into the pathogenesis of SARS-CoV-2 infection in ICU patients. In our view, our data fit better with a pathogenetic model, in which SARS-CoV-2 replication in the LRT or its presence in blood at a certain point over the course of ICU stay might not be a major driver of systemic inflammation, lymphopenia, lung dysfunction, multisystemic organ failure and death. This does not invalidate the importance of virus replication rate in the URT in the early stage after infection in determining the clinical course of COVID-1910. Further studies are needed to resolve this issue.
Financial Support
This work received no public or private funds.
Author Contributions
BA, EA, IT, RG-R, RC, JC and JR: Methodology and validation of data. NC, JF and MLB: Medical care of ICU patients. DN: Conceptualization, supervision, writing the original draft. All authors reviewed the original draft.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
No public or private funds were used for the current study. We are grateful to all personnel who work at Clinic University Hospital, in particular to those at Microbiology laboratory and the Intensive Care Unit for their commitment in the fight against COVID-19. Eliseo Albert holds a Juan Rodés Contract (JR20/00011) from the Health Institute Carlos III. Ignacio Torres holds a Río Hortega Contract (CM20/00090) the Health Institute Carlos III.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.05.036.
Appendix. Supplementary materials
References
- 1.Lui G, Ling L, Lai CK, Tso EY, Fung KS, Chan V, et al. Viral dynamics of SARS-CoV-2 across a spectrum of disease severity in COVID-19. J Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg E, Ben Zvi H, Sheena L, Sheena L, Sofer S, Krause I, et al. A real-life setting evaluation of the effect of remdesivir on viral load in COVID-19 patients admitted to a large tertiary center in Israel. Clin Microbiol Infect. 2021 doi: 10.1016/j.cmi.2021.02.029. S1198-743X(21)00113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masiá M, Fernández-González M, Padilla S, Ortega P, García JA, Agulló V, et al. Impact of interleukin-6 blockade with tocilizumab on SARS-CoV-2 viral kinetics and antibody responses in patients with COVID-19: A prospective cohort study. E Bio Med. 2020;60:10299. doi: 10.1016/j.ebiom.2020.102999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bermejo-Martin JF, González-Rivera M, Almansa R, Micheloud D, Tedim AP, Domínguez-Gil M, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24:691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veyer D, Kernéis S, Poulet G, Wack M, Robillard N, Taly V, et al. Highly sensitive quantification of plasma severe acute respiratory syndrome coronavirus 2 RNA sheds light on its potential clinical value. Clin Infect Dis. Clin Infect Dis. 2020:ciaa1196. doi: 10.1093/cid/ciaa1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin Infect Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buetti N, Wicky PH, Le Hingrat Q, Ruckly S, Mazzuchelli T, Loiodice A, et al. SARS-CoV-2 detection in the lower respiratory tract of invasively ventilated ARDS patients. Crit Care. 2020;24:610. doi: 10.1186/s13054-020-03323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z, et al. SARS-CoV-2 viral load in clinical samples from critically Ill patients. Am J Respir Crit Care Med. 2020;201:1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitker L, Dhelft F, Chauvelot L, Frobert E, Folliet L, Mezidi M, et al. Protracted viral shedding and viral load are associated with ICU mortality in Covid-19 patients with acute respiratory failure. Ann Intensive Care. 2020;10:167. doi: 10.1186/s13613-020-00783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alteri C, Cento V, Vecchi M, Colagrossi L, Fanti D, Vismara C, et al. Nasopharyngeal SARS-CoV-2 load at hospital admission as predictor of mortality. Clin Infect Dis. 2020;16:ciaa956. doi: 10.1093/cid/ciaa956. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.