Abstract
Vigorous vaccination programs against SARS-CoV-2-causing Covid-19 are the major chance to fight this dreadful pandemic. The currently administered vaccines depend on adenovirus DNA vectors or on SARS-CoV-2 mRNA that might become reverse transcribed into DNA, however infrequently. In some societies, people have become sensitized against the potential short- or long-term side effects of foreign DNA being injected into humans. In my laboratory, the fate of foreign DNA in mammalian (human) cells and organisms has been investigated for many years. In this review, a summary of the results obtained will be presented. This synopsis has been put in the evolutionary context of retrotransposon insertions into pre-human genomes millions of years ago. In addition, studies on adenovirus vector-based DNA, on the fate of food-ingested DNA as well as the long-term persistence of SARS-CoV-2 RNA/DNA will be described. Actual integration of viral DNA molecules and of adenovirus vector DNA will likely be chance events whose frequency and epigenetic consequences cannot with certainty be assessed. The review also addresses problems of remaining adenoviral gene expression in adenoviral-based vectors and their role in side effects of vaccines. Eventually, it will come down to weighing the possible risks of genomic insertions of vaccine-associated foreign DNA and unknown levels of vector-carried adenoviral gene expression versus protection against the dangers of Covid-19. A decision in favor of vaccination against life-threatening disease appears prudent. Informing the public about the complexities of biology will be a reliable guide when having to reach personal decisions about vaccinations.
Keywords: Adenovirus DNA integration into mammalian genomes, adenovirus vector DNA integration into human genome, expression of adenoviral genes in vector DNA, adenovirus vector DNA & RNA-based SARS-CoV-2 vaccines, epigenetic consequences of foreign DNA integration, retrotranscription of SARS-CoV-2 RNA into DNA
1. Background
Several of the presently approved vaccines against SARS-Coronavirus-2 (AstraZeneca/Oxford University, Johnson & Johnson's Janssen COVID-19 Vaccine, and Sputnik V) are based on adenovirus DNA vectors as carriers for the genetic information for the SARS-COV-2 spike glycoprotein. The vaccines produced by BioNTech/Pfizer or Moderna contain the messenger RNA (mRNA) for the synthesis of this protein. Upon vaccine injection, the mRNA will trigger the synthesis of the viral spike protein in vaccinees directly. Hence, public interest in the fate of foreign (adenoviral or SARS-CoV-2 reverse transcribed) DNA in mammalian (human) cells and organisms has become acute and widespread. Peoples’ concerns have been phrased in occasional personal encounters like – “does the vaccine enter my genes”? The author submits to the concept that the public is entitled to the full scope of scientific information on these issues. The here presented account of previous experimental work about foreign DNA integration and its consequences will provide a useful update on these issues of public concern. This synopsis describes the fate of foreign DNA in mammalian cells, its possible integration into the host genome and the potential consequences for the transgenomic cells (Doerfler, 2000, Doerfler et al., 2018). In the face of the still expanding worldwide SARS-CoV-2 pandemic, this information has to be counterbalanced vis-à-vis the benefits of an adenovirus DNA vector-based or an mRNA vaccine against life-threatening Covid-19 (Coronavirus Disease 2019). There is no evidence for human adenoviruses to be causally involved in human tumorigenesis, but this possibility cannot be categorically excluded either, in particular for the vector-based vaccines.
Moreover, in Ad12-induced hamster tumors, persistence of the Ad12 genome in tumor cells is not required for the maintenance of the transformed state of Ad12-induced tumors (Kuhlmann et al., 1982). As discussed below, epigenetic factors might play a major role in adenovirus tumorigenesis and for their effect to dominate, long-standing persistence of the transgenome is not essential (Doerfler et al., 2018, Heller et al., 1995).
The possible long-term sequelae of adenoviral vector DNA or reverse transcribed SARS-CoV-2 messenger RNA integration for the functionality and survival of vector transgenic cells cannot be predicted with certainty in individual cases. Nevertheless at this stage in the Covid-19 pandemic, our activities will have to be focused on fighting the pandemic with adequate vaccines and future therapeutic measures.
2. Evolutionary background – transposable elements in the human genome
Almost 50% of the 3 × 109 nucleotide-pair human genome represent transposable elements and 8% constitute endogenous retroviral genomes. The significance of this genetic fact has not been actively reflected in research in molecular genetics. Apparently, a major part of today's human genome has been inserted gradually during evolutionary times by the integration of foreign DNA or of retro-transcribed DNA from RNA retroviral genomes. More recent analyses revealed that repetitive elements might amount to 66%-69% of the human genome (De Koning et al 2011). Foreign DNA sequences have become frequently inactivated through epigenetic mechanisms, i. e. by foreign DNA methylation and altered histone methylation strategies. It is estimated that these ancient integration events date back to >60 to 70 million years ago, possibly longer. Their existence and massive extent support the notion that the insertion of foreign DNA into established genomes was facilitated by an elementary mechanism that emerged in early evolutionary times and very likely has played a major role in driving evolution. Even today, selected parts of these ancient integrates can be transcribed, and there are inter-individual differences as to the extent of their expression levels. Moreover, regulatory elements present in endogenous retroviral elements and recognition sites for DNA-protein interactions serve functions that are poorly understood. Their importance for the human organism can only be speculated about. Some parts of endogenous retroviral sequences can become overexpressed in human tumor and autoimmune diseases as well as viral infections, including those by SARS-CoV-2. There is evidence that endogenous retroviral elements are deregulated during aging. An overview on the biology of these sequences that often have erroneously been underestimated as junk DNA, has been presented recently (Geis and Goff, 2020).
3. Synopsis of previous analyses
Work on and with adenovirus vectors is premised on the unproven claim that these vectors are safe because adenovirus DNA would "not integrate"into the recipient cells’ genomes. There is no solid evidence for this interpretation. Hence, the following synopsis of our work and that of others takes issue with this notion (Doerfler et al., 1984). The problems encountered in gene therapeutic procedures of the past (Samanathan et al, 2020) and presently with adenovirus vector-based SARS-CoV-2 vaccines will require a thorough re-evaluation of the drawbacks that adenovirus vector systems entail. Of course, the fate of intact adenoviral DNA and that of partly defective vector DNA is different in some respects, although both are subject to the same cellular integrative recombination mechanisms. Moreover, the amounts of adenovirus vector DNA administered in vaccination programs (about 2.5 µ) are lower than those in gene therapy proccedures. Adenovirus vector DNA is thought to reach primarily cells of the liver (Stephen et al, 2010) and probably enters also cells of the immune system.
3.1. Adenovirus type 12 genomes in hamster tumor and transformed cells
-
•
Human adenovirus type 12 (Ad12) induces primitive neuroectodermal neoplasias in 70 to 90% of virus-inoculated newborn Syrian hamsters, Mesocricetus auratus (Trentin et al. 1962; Hohlweg et al. 2003). Ad12 DNA in Ad12-induced hamster tumor cell line T637 was integrated by heterologous recombination in 10 to 12 copies, frequently at one randomly selected genomic site (Fig. 1 A, green, white arrow). There is, however, no evidence that any of the human adenoviruses are implicated in human tumorigenesis.
-
•
In 58 independently Ad12-induced hamster tumors, multiple copies of viral DNA were integrated at one randomly chosen cellular integration site, although the genomic locations of these sites differed among individual tumors of clonal origin. In two additional tumors, Ad12 DNA integrates were located each at two different sites in the tumor cell genome. In each tumor, the integrated Ad12 DNA had become de novo CpG-methylated (Hilger-Eversheim and Doerfler, 1997).
-
•
Major emphasis in the analyses of adenovirus-transformed or Ad12-induced hamster tumor cells was placed on the molecular cloning and sequencing of numerous junction sites between integrated adenovirus DNA and the adjacent cellular DNA to ascertain covalent linkage between viral and cellular DNA of the integrates (Deuring et al., 1981a; Stabel, Doerfler, 1982; Gahlmann et al. 1982; (Deuring and Doerfler, 1983)Doerfler et al., 1984, Schulz et al., 1987). The insertion mechanism appeared akin or identical to that of non-homologous recombination. Short, often patchy sequence homologies existed between the termini of adenovirus DNA and the targeted cellular sequence at the abutting sites of insertion. In some instances, the recipient cellular DNA sequence adjacent to the integrated viral DNA was transcriptionally active (Schulz et al. 1987). In a pilot study, the insertional recombination between viral and cellular DNAs was reconstructed in a cell-free system from hamster nuclear extracts by using a previously identified preinsertion cellular DNA sequence and terminal adenovirus DNA segments as recombination partners (Jessberger et al. 1989). The recombination products were analyzed upon re-cloning from the reaction and showed characteristics akin to those from adenovirus-transformed cells (Tatzelt et al. 1992).
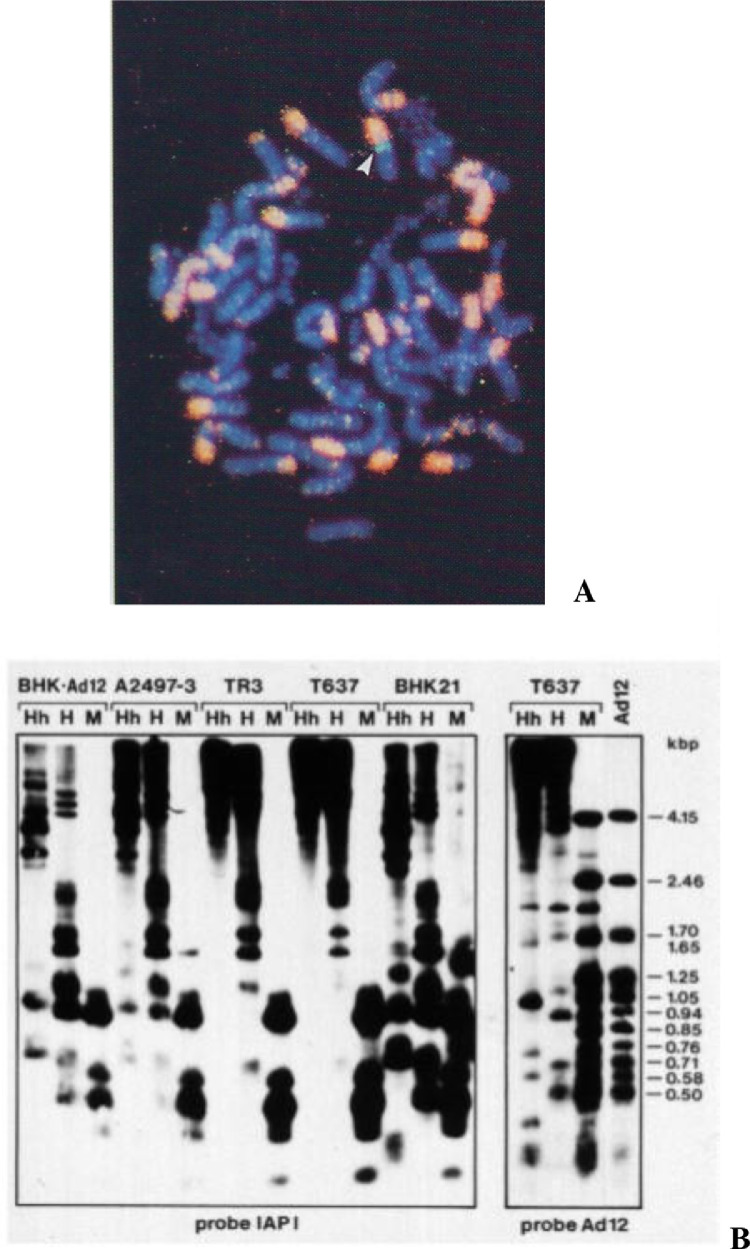
Fig. 1.
Foreign DNA insertion into hamster genome alters profiles of cellular DNA methylation in the ~ 900 copies of IAP retrotransposon sequences. Altered methylation patterns are maintained even after loss of the transgenomes.
[A] In situ hybridization of Ad12-transformed hamster cell nucleus of a T637 cell visualizes about 10 to 12 copies of chromosomally integrated Ad12 DNA (Ad12 DNA probe, green, white arrow); IAP probe (pink) ~ 900 copies of Intracisternal A Particle retrotransposon genomes that are frequently located on short arms of hamster chromosomes; DAPI-staining (blue) of cellular DNA.
[B] Southern blot hybridization of DNAs from (right to left) Ad12 (human adenovirus type 12); T637 (Ad12-transformed BHK21 hamster cells); BHK21 (baby hamster kidney cell line); T637; TR3 (a revertant of T637 cell line whose genome had lost all of the originally 10 to 12 integrated viral genome copies (PCR-confirmed); A2497-3 is an Ad12-transformed hamster cell line different from T637 cells; BHK˖Ad12, BHK21 hamster cells abortively infected with Ad12 (Doerfler, 1969, Hochstein et al., 2008). The 32P-labeled DNA hybridization probes were designated at the bottom of the electropherogram, Ad12 or IAP (from right to left).
Levels of CpG DNA methylation in the different DNA samples were assessed by differential cleavage with methylation-sensitive (HpaII, HhaI) or methylation-insensitive restriction endonucleases (MspI): MspI does cleave 5´-CCGG-3´ and 5´-CmCGG-3 sequences (methylation-insensitive), HpaII and HhaI (methylation-sensitive) do not cut 5´-CmCGG-3´ sites. The results document distinct increases of CpG methylated sequences in the Ad12-transgenic cell lines T637 and A-2497-3 as evidenced by the large amounts of HpaII- and HhaI-uncut IAP DNA (top of display). The revertant cell line TR3, derived from T637 cells, also shows increased IAP DNA methylation, although it had lost all copies of the Ad12 transgenomes (PCR confirmed). Apparently, the epigenetic effect (increased IAP DNA methylation) of transgene insertion on the IAP retrotransposon DNA persisted even after the loss of the transgenes that had elicited the trans epigenetic effects. These figures were taken from (Heller et al., 1995).
3.2. A model system to study Ad12 DNA integration in abortively infected hamster cells
-
•
The Ad12-hamster tumor system was recognized at the time as a powerful model to investigate Ad12-host cell interactions. Ad12 infects hamster cells abortively with a complete block of viral DNA replication and a paucity of viral genomes reaching the cells’ nuclei (Doerfler, 1969; Zock and Doerfler, 1990, 1995; Hochstein et al., 2008]. Thus, due to this block in viral DNA replication and late gene expression, Ad12 was incapable of replicating in hamster cells, which fulfilled an essential precondition for viral DNA integration and tumorigenesis. In a series of model experiments [Doerfler, 1968, 1970], hamster cells with 5-bromodeoxuridine (5-BU)-pre-labeled cellular DNA were infected with huge inocula of 3H-thymidine-DNA-labeled Ad12 virus. By alkaline (pH >13) CsCl-equilibrium buoyant density gradient centrifugation, light Ad12 DNA and 5-BU-labeled heavy cellular DNA could be physically separated. Starting at about 12 h post-infection, 3H-thymidine-DNA-labeled Ad12 started to shift from the light density position of Ad12 DNA to the position of heavy cellular DNA, and progressively so with time after infection. The Ad12 authenticity of the density-shifted DNA was ascertained by DNA-DNA hybridization. Moreover, ultrasonic treatment of the DNA from Ad12-infected hamster cells disrupted the alkali-stable (pH 13) bond between the 3H-thymidine-tagged Ad12 and 5-BU heavy cellular hamster DNA. The 3H-labeled Ad12 DNA then relocated to or close to the viral density position indicating that the 3H-labeled Ad12 DNA was released from its bond to 5-Bu-heavy cellular DNA. These results were the first to document recombinatorial integration between Ad12 and mammalian cell DNAs (Doerfler, 1968, 1970).
3.3. What happens in adenovirus productively infected human cells?
-
•
Infections of human cells with adenovirus type 2 (Ad2) or Ad12 were investigated. There is evidence that human adenovirus DNA is capable of recombining with cellular DNA also in productively infected human cells, although true integrates are difficult to document as none of the infected human cells survive productive adenovirus infections. A fast sedimenting, pH >13 alkali-stable form of viral DNA – presumptive recombinants between adenovirus and cellular DNAs – was documented by sequential DNA-DNA hybridization experiments, first to authentic Ad12 DNA followed by hybridization to human cellular DNA (Schick et al. 1976).
-
•
In addition, in Ad12-infected human cells, a naturally arising symmetric recombinant (SYREC) between the left terminal 2,081 nucleotides of Ad12 DNA and a sizable palindrome of cellular DNA was discovered, as documented by nucleotide sequence analyses. The presence of the adenovirus packaging signal in the terminal Ad12 DNA fragment in the hybrid molecule facilitated incapsidation into viral particles. This finding lent unequivocal support for the formation of recombinants between Ad12 and cellular DNA also in productively infected human cells (Deuring et al., 1981b, Deuring and Doerfler 1983). The structure of the SYREC Ad12 DNA – cellular palindrome DNA molecule served as a guide as to how gutless adenovirus vector molecules with high payloads had to be constructed.
-
•
In other laboratories, evidence from in situ hybridization experiments demonstrated that human adenovirus DNA persisted in chronically infected human adenoid cells (Neumann et al. 1987). Additionally, in latently adenovirus-infected human lymphocyte cell lines, adenovirus genomes persisted as well (Zhang et al. 2010). However, in neither system was the molecular status of the persisting viral genomes further investigated.
3.4. Integrated Ad12 DNA becomes de novo CpG methylated – inverse correlations between levels of Ad12 DNA methylation and genetic activities of viral genes
-
•
In Ad12-induced hamster tumor cells and in Ad2- or Ad12-transformed hamster cells, the integrated viral DNA becomes de novo CpG-methylated in specific patterns (Sutter et al. 1978) which tend to spread across the viral integrate in a specific manner (Toth et al. 1989). The demonstration of inverse correlations between levels of DNA methylation and genetic activities of specific regions of the integrated Ad2 or Ad12 genomes were among the first in the literature and marked the beginning of a long-term commitment of our research to elucidate the biological functions of CpG DNA methylation in mammalian cells (Sutter and Doerfler, 1980; Vardimon et al. 1980; Doerfler, 1983). This inverse interrelation between genetic silencing and specific patterns of promoter CpG methylation was subsequently confirmed in many eukaryotic systems to be of general significance.
3.5. Promoter methylation leads to promoter silencing
-
•
In a series of experiments with different eukaryotic promoters and indicator genes, we documented that specific promoter methylation caused inactivation of these promoters (Vardimon et al. 1982; Kruczek and Doerfler, 1983; Langner et al. 1984). Moreover, specific CpG methylation patterns in mammalian genomes can be interpreted as signals for long-term gene inactivation (Doerfler, 1983; 2006). A more detailed description of this research will not be presented here, because it would transgress the scope of this review.
3.6. Consequences of foreign DNA integration for cellular genomes
-
•
It has been frequently argued that the integration of foreign DNA into host genomes might lead to the disruption of genes in the host chromosome and thus mutations might arise. Undoubtedly, this is a possibility that has been actually described in some cases. However, considering that the human genome carries 1.1% exons, 24% introns, and 75% intergenic DNA (Venter et al. 2001), integrating foreign DNA has a chance of about 1/100 to hit functional genes. In contrast, in our analyses of the consequences of foreign DNA insertion, we have placed emphasis on the role of epigenetic effects involving sites both close to and remote from the integration locus (see below). This aspect of the sequelae of foreign DNA integration has been the topic of previous work from our laboratory (Heller et al, 1995; Weber et a., 2015, 2016; Doerfler et al, 2018).
-
•
At the sites of viral DNA insertion, the level of CpG DNA methylation in the adjacent cellular DNA became decreased. Hence foreign DNA integration was able to alter cellular DNA epigenetic signals in cis, immediately at the site of insertion (Lichtenberg et al. 1988).
-
•
But alterations can also occur in trans. In Ad12-transformed hamster cells that carry 10 to 12 copies of viral DNA genomically integrated at one chromosomal site [green in Fig. 1 A, highlighted by a white arrow], the levels of CpG methylation in the roughly 900 copies of cellular Intracytoplasmic A Particle (IAP) retrotransposon DNA [pink in Fig. 1 A] were markedly increased in comparison to untransformed hamster cells and to Ad12-infected hamster cells (Fig. 1 B; Heller et al. 1995). Due to the increased levels of CpG DNA methylation in the IAP sequences, their DNA proved resistant to cleavage with methylation-sensitive restriction endonucleases like HpaII and HhaI and failed to migrate in the electrophoresis analysis of cleavage products (Fig. 1 B). The IAP sequences are distributed over many chromosomes and are frequently located on their short arms [pink signals in Fig. 1A]. This finding of a trans-effect of CpG DNA methylation in hamster cells transgenic for Ad12 DNA prompted us to pre-designed investigations into the consequences of foreign DNA insertions in human cells (see below)
-
•
It is important to emphasize that in the experiments described in Fig. 1 B, the transgenomic effect on IAP CpG methylation profiles was not dependent on the continued presence of the Ad12 transgenomes but persisted in cell line TR3, although it had lost all of the previously integrated Ad12 genomes (Fig. 1B; Heller et al. 1995). Hence the argument cannot be made that the lack of finding adenovirus genomes, e.g. in human tumor cells or cells in individuals with chronic ailments, would necessarily imply that adenovirus DNA integration or that of any other foreign DNA had not played a major role in the pathogenesis of these diseases. Persistence of transgenomic DNA is not a necessary precondition for the maintenance of trans effects, e.g. here of the transformed state of the recipient genomes and cells [hit-and-run mechanism].
-
•
For further investigations of epigenetic trans-effects in transgenomic human cells, the following series of pilot experiments was initiated. In clonal lines of human cells transgenomic for a 5.6 kbp bacterial plasmid, transcription and CpG methylation patterns were compared between transgenomic and non-transgenomic cell clones by using gene chip microarray systems. In 4.7% of the 28.869 gene segments analyzed, transcriptional activities were differentially up- or downregulated in the transgenomic clones. Genome-wide profiling demonstrated differential methylation at 3791 of > 480,000 CpG sites that were examined in transgenomic versus non-transgenomic cell clones. These data indicated that the insertion of foreign DNA into an established human genome can markedly alter cellular CpG DNA methylation and transcription profiles. In fact, different cell types seemed to have evolved (Weber et al. 2015; Doerfler et al. 2018). We consider these epigenetic effects of foreign DNA insertion of great significance since they bear on the epigenetic stability and function of the entire (human) genome. Since many experimental approaches in biology and medicine resort to the study of transgenomic cells or organisms, the frequently unsuspected epigenetic effects in the wake of foreign DNA insertion will have to be taken into account in a realistic interpretation of experimental data.
-
•
The afore-described trans effect appeared to be specific, in that it did not affect all parts of the human genome. Methylation profiles in endogenous human retroviral (HERV) sequences, that constitute about 8% of the human genome, remained unaltered in the same cell clones (Weber et al. 2016) that revealed distinct methylation changes in other parts of their genomes (Weber et al. 2015). HERV DNA methylation patterns in transgenic and control cells exposed to the same experimental protocols were almost identical (Weber et al. 2016).
-
•
Extending the studies on the role of foreign DNA in the environment, we explored the fate of isolated foreign DNA that had been fed to laboratory mice. Food-ingested foreign DNA likely constitutes the most abundant and regular encounter of human organs with foreign DNA. Bacteriophage M13 DNA, the plasmid-cloned green fluorescent protein (GFP) gene, or the plant-specific, nuclear DNA-encoded gene for ribulose-1, 5-bisphosphate carboxylase (Rubisco) were administered orally to mice in a large number of different experiments. About 1% of the fed DNA survived the passage through the murine gastro-intestinal tract in fragmented form and could be traced in even lower amounts to different organ systems, notably the spleen and liver of the animals. In one experiment, evidence could be adduced that the test DNA retrieved from spleen cells was linked to an 80 nt DNA segment with 70% homology to the mouse IgE receptor gene (Schubbert et al. 1997). The germ line of mice seemed to be protected from the invasion of food-ingested DNA [ (Hohlweg and Doerfler, 2001)]. Hence it appears likely that the exposure to food-ingested DNA is a continuing challenge to the stability of the genome in stochastically involved cells of the mammalian organism. We have no information about this problem in humans.
3.7. Integration of adenovirus vector DNA
-
•
Stefan Kochanek's laboratory at Ulm University in Germany performed a pilot study to investigate whether adenovirus vector DNA could become chromosomally integrated in the wake of adenovirus vector-mediated gene transfer in mice [Stephen et al. 2010]. Replication-deficient adenovirus vectors carrying different transgenes were injected intravenously into mice. In transgene expressing hepatocytes vector-constructs were found integrated into the mouse genome. The analyses of junction sites between vector and cellular DNA revealed covalent linkage of the vector termini to mouse cellular DNA by heterologous recombination at a frequency of 6.7 × 10−5 transduced hepatocyte. Homologous recombinants were identified at hundredfold lower frequencies (Stephen et al. 2010). In their evaluation of these results the authors suggested that the frequency of heterologous adenoviral vector DNA recombination with the recipient genome was comparable to that of spontaneous mutations in mammalian cells. Of course, it is an open question what factors elicit “spontaneous mutations”. Among other causes, recombination of human DNA with foreign DNA could play an important role in the generation of spontaneous mutations. Lastly, the amount of virion-packaged adenovirus vector DNA that is routinely administered intra-muscularly, e.g. with the AstraZeneca vaccine (50x109 virions per vaccine dose, equivalent to about 2.5µg adenovirus vector DNA per dose), is lower than that in many gene therapy regimens. For details see [https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/963928/UKPAR_COVID_19_Vaccine_AstraZeneca_23.02.2021.pdf].
3.8. SARS-CoV-2 RNA reverse transcribed and integrated into the genome?
Motivated by reports about prolonged shedding of SARS-CoV-2 RNA and continued positive RT-PCR tests among survivors of Covid-19, Zhang et al. investigated whether SARS-CoV-2 RNA could be reverse transcribed into DNA that in turn might integrate into the host genome and continue to be transcribed into RNA (Zhang et al., 2021). By nucleotide sequence analyses, the authors observed SARS-CoV-2 DNA copies in the genomes of about 10 to 20% of Covid-19 patients. This SARS-CoV-2 DNA was flanked by target site duplications and consensus LINE-1 endonuclease recognition sequences. The latter two characteristics were not always observed. Thus LINE-1-independent mechanisms for SARS-CoV-2 DNA integration were also conceivable. It was emphasized that only sub-genomic parts of SARS-CoV-2 DNA were found integrated. This type of integrate precludes the possibility of infectious virus being produced by those who carry fragmented viral SARS-CoV-2 DNA. The integrated parts of SARS-CoV-2 DNA were frequently transcribed. The authors concluded that SARS-CoV-2 infection could lead to the synthesis of endogenous LINE-1 retrotransposon-encoded reverse transcriptases. These enzymes generated DNA retro-transcripts. Possibly only fragments of this DNA were inserted into the human genomes and transcribed into SARS-CoV-2 RNA. This finding might have implications in the context of long-term sequelae of SARS-CoV-2 infections for the genomes of Covid-19 patients. Moreover, the RNA-based vaccines reportedly successfully employed around the world will have to be scrutinized in this respect as well. Unexpected findings like the ones described here will have to be reproduced by others. Likely, the here described findings will stimulate investigations on similar mechanisms affecting additional RNA viral pathogens (Zhang et al., 2021).
3.9. Comments on the chimpanzee adenovirus vector used in the AstraZeneca SARS-CoV-2 vaccine ChAdOx1
Details on the structure and viral backbone transcription of the chimpanzee adenovirus vector that has been used in the design of the AstraZeneca SARS-CoV-2 vaccine ChAdOx1, have been recently published (Almuqrin et al. 2021). In this vector, a non-specified part of the E1 region of the adenoviral genome was replaced by the coding sequence of the SARS-CoV-2 spike protein. Hence the vector construct is replication defective. Expression of the SARS-CoV-2 spike glycoprotein in the vector construct was posited under control of the Tet-responsive CMV [cytomegalovirus] promoter (Loew et al. 2010), one of the strong eukaryotic promoters. The vector, however, in addition still carries 28 kbp particularly of late chimpanzee adenovirus genes (Almuqrin et al. 2021). Their transcriptional activity might still be under remote control of segments of the E1 region that possibly remained in the construct. Depending on the human cell line that was control-infected with the vector, adenoviral viral backbone genes were found expressed at different levels. It is presently unknown which genes of the chimpanzee adenovirus are actually expressed in human vaccinees, and how the adenoviral expression profiles vary among different recipients of the vaccine. So far, the causative contribution of adenoviral late gene products to the vaccine's side effects that have been observed worldwide, has not been investigated and cannot be ruled out. Hence, in addition to the possibly long-term problems arising from the use of adenoviral vectors due to their integration and epigenetic sequelae (see above), adenovirus vectors remain problematic since they still express viral gene products (Almuqrin et al. 2021) whose role in eliciting dangerous side effects in human vaccinees has not been critically evaluated.
Information on the extent and nature of ChAdOx1 adenoviral gene expression in human vaccinees has not been presented. In the absence of the trans-activating functions of the ChAdOx1 E1 region, cellular transactivators in vaccinees’ organisms might facilitate the expression of late ChAdOx1 viral genes. Short sequence homologies in these ChAdOx1 genes to human adenovirus genes might suffice to elicit memory immune responses in vaccinees at individually differing strength. The latter might be heightened in young people who are still closer to their childhood experiences with human adenoviral infections. Young women who provide most of the patient and elderly care in society might then be preferentially affected by these immunological overreactions, hence these vaccinees might be more frequently hit by adverse vaccination reactions. Why thrombocytes would be specifically targeted by this type of memory immune responses remains to be investigated. In this context, it will be mandatory to recall that thrombocytopenia was found also in recipients of adenovirus-vector constructs in the course of gene therapeutic applications (Lopez-Gordo et al, 2017).
Researchers who work on adenoviral vectors have been aware of the Jesse Gelsinger incident in 1999 [https://en.wikipedia.org/wiki/Jesse_Gelsinger]. At that time, an 18-year old male who suffered from an X-linked genetic metabolic disease, ornithine transcarbamylase deficiency of the liver, was the first recipient of an adenoviral vector-based gene therapeutic regime and died a few days after application of the recombinant adenovirus. The possible factors involved in the fatal outcome were investigated at the time but could not be clearly identified. The incident terminated the use of adenovirus vectors in gene therapy for a long time. Very recently, a detailed, apparently continuing, investigation of this fatal incident ascribed it to the generation of adenovirus-antibody complexes that contributed to “lethal systemic inflammation” in the recipient's organism (Somanathan et al. 2020). Of course, the human adenovirus type 5 (Ad5) vector that was employed in the 1999 gene therapeutic measure cannot be directly compared to the chimpanzee adenovirus vector used in the SARS-CoV-2 vaccine in 2021. Ad5 derived-vectors are, however, part of other SARS-CoV-2 vaccines. Hence extreme caution should still be observed when injecting adenoviral vectors into humans. Hopefully, catastrophic experiences from the past will not resurface with today's vectors that are, however, not all that different from the older problematic ones. In the long run and upon more careful consideration, conventional vaccines based on recombinant spike protein might have been a safer choice.
4. Conclusions
Life has developed in a DNA, most likely preceded by an RNA, world. We all live in this DNA world starting in our backyards when we dig the earth with uncounted microorganisms and their DNA buried in age-old debris of decaying animal and plant remnants. Just consider the annually shed foliage. With our daily food supply we take up foreign DNA in huge quantities that is not momentarily digested to mononucleotides but transiently persists in the form of fragments that are capable of entering the human organism and becoming distributed in different human organ systems. DNA is a very stable molecule. In tiny bone splinters from individuals of the Neanderthal population of ~50,000 years ago, Neanderthal DNA fragments have persisted to this day. In excavated bone samples from Neanderthals, that Svante Pääbo and his colleagues investigated, Neanderthal DNA represented only about 0.5% of the DNA from the bone specimens they analyzed (Prüfer et al. 2017). When the occasion arose, foreign DNA and its ability to survive and recombine with DNA from living organisms, might have played a major role in evolution. While we are struggling with the Covid-19 pandemic on planet Earth, rover Perseverance is digging for traces of organic decay on planet Mars.
This brief review was presented here to facilitate an independent and more balanced discussion on the potential risks due to the presence of adenovirus vector DNA (AstraZeneca, Johnson & Johnson, Sputnik V and others) or SARS-CoV-2 RNA (BioNTech/Pfizer, Moderna) in vaccines that are supposed to protect against Covid-19. Of course, injections of vector-based vaccines into human deltoid muscle is a different matter than rare chance events leading to recombination events between foreign and human DNAs in experimental systems as described above. Moreover, neither type nor frequency of consequences of rare vector integration events can be realistically assessed at present. In contrast, the recently published results on the benefits of protection against Covid-19 offered by the BioNTech/Pfizer vaccines are encouraging (Dagan et al. 2021). Granted, the jury is still out to what extent any of the vaccines’ will protect against the more dangerous new SARS-CoV-2 variants from the UK, South Africa, Brazil, India (now termed α, β, γ, δ, respectively) or against unknown variants that might arise in the future given the poorly controlled levels of viral replication around the world (Weber et al., 2021). Lastly, we are ignorant about vaccine protection against the development of prolonged and late-onset symptoms of Covid-19.
The information presented in this review will help future vaccinees to weigh a risk versus benefit assessment, namely the integration events of adenovirus vector or of SARS-CoV-2 RNA reverse transcript DNA at low frequency versus hopefully high vaccine efficacy and protection. Moreover, since SARS-CoV-2 infection by itself can be associated with the integration of reverse transcripts of the viral RNA (Zhang et al., 2021), this series of events might become inescapable in any SARS-CoV-2 infection. Lastly, the extent to which adenoviral gene products might become co-expressed with the SARS-CoV-2 spike glycoprotein upon vector-vaccine injection into human deltoid muscles remains un-investigated. At present we cannot gauge their possible effects on the human organism, if actually expressed. Opportunities and risks, both at the same time, remain beyond our expectations of absolute controls because life and evolution likely have been affected by “chance mechanisms” from the very beginning. Clinical observations on long lasting positive RT-PCR test results that imply SARS-CoV-2 DNA integration into the human genome in the course of some Covid-19 cases, render apprehensions about vaccine-associated integration events unrealistic, when compared to the hoped-for benefits by vaccination against Covid-19. The human population of 2021 faces a biomedical crisis of, in recent times, unprecedented dimensions and will have to accept the presently best available countermeasures against Covid-19 – Vaccination.
4.1. Note added in revision
At the height of an unprecedented pandemic, the rapid development of efficient SARS-CoV-2 vaccines was met with relief and admiration for experimental science (Sahin et al., 2021). Nevertheless, it will prove medically cogent to consider possible sequelae of vaccine injections into billions of humans. One 0.5 ml dose of recombinant adenovirus ChAdOx1-S of AstraZeneca reportedly contains 5 × 1010 particles of chimpanzee adenovirus 26 (Ad26) (https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/963928/UKPAR_COVID_19_Vaccine_AstraZeneca_23.02.2021.pdf) that carry an equivalent of about 2.5µg recombinant Ad26 chimpanzee DNA. Adenovirus particles likely are taken up by cells of the lymphatic system and the liver, and their DNA will be transported to the cells’ nuclei. In this review, the evidence for insertion of adenovirus DNA into recipient genomes and its consequences has been presented.
-
(i)
Adenovirus vector-based vaccines can lead to the integration of adenovirus DNA at unknown frequency and with unpredictable epigenetic consequences. It is conceivable that epigenetic effects might be noticed only years after vaccination.
-
(ii)
The adenoviral genes still present in the adenovirus vector DNA might become trans-activated by cellular factors and lead to individually varying immune memory reactions that are experienced by vaccinees as transient post-vaccination symptoms. Fatal outcomes due to extreme immune reactions have fortunately been very rare events.
-
(iii)
SARS-CoV-2 RNA or segments of it, like the spike gene, can be reverse transcribed by LINE-1-encoded reverse transcriptases or other factors, and the thus synthesized DNA can become integrated at unknown frequencies and locations into the vaccinees’ genomes. Of course, the same is true for all SARS-CoV-2 infections. Hence, the risk of undesired integration events of SARS-CoV-2 RNA reverse transcripts upon SARS-CoV-2 infections appears similar to that upon vaccination with the mRNA-based Covid-19 vaccine.
These mechanisms of basic biology have to be considered in the context of mass vaccination programs of humans around the globe. Whether and how these mechanisms will play a predominant role in medicine in the future will have to be followed by critical meta-analyses. There is at present no viable alternative to the vaccination of billions of humans. The human population presently partakes in the exposure to foreign DNA in a huge experiment. After the completion of worldwide vaccinations, a post-vaccination sentinel program should be set up to monitor the exacerbation of unexpected, possibly novel, human ailments in vaccinated individuals.
Declaration of Competing Interest
The author declares no conflict of interest.
Acknowledgments
I thank Rudolf Jaenisch, Whitehead Institute for Biomedical Research, Cambridge, MA, USA, for making their manuscript Zhang et al., Proc. Natl. Acad. Sci., USA, 2021, online, available to me prior to publication. At different times, research in W.D.’s laboratory has been supported by the Deutsche Forschungsgemeinschaft in Bonn-Bad Godesberg (SFB 74 and SFB 274), by the Center for Molecular Medicine Cologne (CMMC, TP13), by the Thyssen Foundation in Cologne (Az. 10.07.2.138 plus a stipend to A. Naumann Az. 40.12.0.029.), by the Staedtler Stiftung in Nürnberg (WW/eh 01/15), by the Dr. Robert Pfleger Stiftung in Bamberg and by the continued support of W.D.’s Epigenetics Group at the Institute for Clinical and Molecular Virology, Friedrich-Alexander University, Erlangen-Nürnberg. Our very recent SARS-CoV-2 mutant analyses (Weber et al. 2020, 2021), have in part been supported by the Dr. Robert Pfleger Stiftung.
References
- Almuqrin A., et al. SARS-CoV-2 vaccine ChAdOx1 nCoV-19 infection of human cell lines reveals low levels of viral backbone gene transcription alongside very high levels of SARS-CoV-2 S glycoprotein gene transcription. Genome Med. 2021 doi: 10.1186/s13073-021-00859-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan N., et al. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12) doi: 10.1371/journal.pgen.1002384. Epub 2011 Dec 1. PMID: 22144907; PMCID: PMC3228813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R., Doerfler W. Proof of recombination between viral and cellular genomes in human KB cells productively infected by adenovirus type 12: structure of the junction site in a symmetric recombinant (SYREC) Gene. 1983;26:283–289. doi: 10.1016/0378-1119(83)90198-1. [DOI] [PubMed] [Google Scholar]
- Deuring R., Klotz G., Doerfler W. An unusual symmetric recombinant between adenovirus type 12 DNA and human cell DNA. Proc. Natl. Acad. Sci. U.S.A. 1981;78:3142–3146. doi: 10.1073/pnas.78.5.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R., Winterhoff U., Tamanoi F., Stabel S., Doerfler W. Site of linkage between adenovirus type 12 and cell DNAs in hamster tumour line CLAC3. Nature. 1981;293:81–84. doi: 10.1038/293081a0. [DOI] [PubMed] [Google Scholar]
- Doerfler W. The fate of the DNA of adenovirus type 12 in baby hamster kidney cells. Proc. Natl. Acad. Sci. U.S.A. 1968;60:636–643. doi: 10.1073/pnas.60.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. Nonproductive infection of baby hamster kidney cells (BHK21) with adenovirus type 12. Virology. 1969;38:587–606. doi: 10.1016/0042-6822(69)90179-2. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Integration of the deoxyribonucleic acid of adenovirus type 12 into the deoxyribonucleic acid of baby hamster kidney cells. J. Virol. 1970;6:652–666. doi: 10.1128/JVI.6.5.652-666.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Ann. Rev. Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Gahlmann R., Stabel S., et al. On the mechanism of recombination between adenoviral and cellular DNAs: the structure of junction sites. Curr. Top. Microbiol. Immunol. 1984;109:193–228. doi: 10.1007/978-3-642-69460-8_9. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Wiley-VCH; Weinheim, New York, Chichester, Brisbane, Singapore, Toronto: 2000. Foreign DNA in Mammalian Systems. [Google Scholar]
- Doerfler W. DNA methylation: De novo methylation, long-term promoter silencing, DNA methylation patterns and their changes. Curr. Top. Microbiol. Immunol. 2006;301:125–175. doi: 10.1007/3-540-31390-7_5. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Weber S., Naumann A. Inheritable epigenetic response towards foreign DNA entry by mammalian host cells: a guardian of genomic stability. Invited Review, Epigenetics. 2018;13:1141–1153. doi: 10.1080/15592294.2018.1549463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann R., Leisten R., Vardimon L., Doerfler W. Patch homologies and the integration of adenovirus DNA in mammalian cells. EMBO J. 1982;1:1101–1104. doi: 10.1002/j.1460-2075.1982.tb01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis F.K., Goff S.P. Silencing and transcriptional regulation of endogenous retroviruses: An Overview. Viruses. 2020;12(8):884. doi: 10.3390/v12080884. PMID: 32823517; PMCID: PMC7472088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller H., Kämmer C., Wilgenbus P., Doerfler W. Chromosomal insertion of foreign (adenovirus type 12, plasmid, or bacteriophage lambda) DNA is associated with enhanced methylation of cellular DNA segments. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5515–5519. doi: 10.1073/pnas.92.12.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger-Eversheim K., Doerfler W. Clonal origin of adenovirus type 12-induced tumors: non-specific chromosomal integration sites of viral DNA. Cancer Res. 1997;57:3001–3009. [PubMed] [Google Scholar]
- Hochstein N., Webb D., Hösel M., Seidel W., Auerochs S., Doerfler W. Human CAR gene expression in non-permissive hamster cells boosts entry of type 12 adenovirions and nuclear import of viral DNA. J. Virol. 2008;82:4159–4163. doi: 10.1128/JVI.02657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohlweg U., et al. Intraperitoneal dissemination of Ad12-inducced un-differentiated neuroectodermal hamster tumors: de novo methylation and transcription patterns of integrated viral and cellular genes. Virus Res. 2003;98:45–56. doi: 10.1016/j.virusres.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Hohlweg U., Doerfler W. On the fate of plant or other foreign genes upon the uptake in food or after intramuscular injection into mice. Mol. Genet. Genomics. 2001:225–233. doi: 10.1007/s004380100450. [DOI] [PubMed] [Google Scholar]
- Jessberger R., Heuss D., Doerfler W. Recombination in hamster cell nuclear extracts between adenovirus type 12 DNA and two hamster preinsertion sequences. EMBO J. 1989;8:869–878. doi: 10.1002/j.1460-2075.1989.tb03448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruczek I., Doerfler W. Expression of the chloramphenicol acetyltransferase gene in mammalian cells under the control of adenovirus type 12 promoters: effect of promoter methylation on gene expression. Proc. Natl. Acad. Sci. U.S.A. 1983;80:7586–7590. doi: 10.1073/pnas.80.24.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann I., Achten S., Rudolph R., Doerfler W. Tumor induction by human adenovirus type 12 in hamsters: loss of the viral genome from adenovirus type 12-induced tumor cells is compatible with tumor formation. EMBO J. 1982;1:79–86. doi: 10.1002/j.1460-2075.1982.tb01128.x. PMID: 7188179; PMCID: PMC552999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner K.D., Vardimon L., Renz D., Doerfler W. DNA methylation of three 5′ C-C-G-G 3′ sites in the promoter and 5′ region inactivates the E2a gene of adenovirus type 2. Proc. Natl. Acad. Sci. U.S.A. 1984;81:2950–2954. doi: 10.1073/pnas.81.10.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenberg U., Zock C., Doerfler W. Integration of foreign DNA into mammalian genome can be associated with hypo-methylation at site of insertion. Virus Res. 1988;11:335–342. doi: 10.1016/0168-1702(88)90006-8. [DOI] [PubMed] [Google Scholar]
- Loew R., Heinz N., Hampf M., Bujard H., Gossen M. Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 2010;10:81. doi: 10.1186/1472-6750-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gordo E., Doszpoly A., Duffy M.R., et al. Defining a novel role for the coxsackievirus and adenovirus receptor in human adenovirus serotype 5 transduction in vitro in the presence of mouse serum. J Virol. 2017;91(12) doi: 10.1128/JVI.02487-16. e02487-16PMID: 28381574; PMCID: PMC5446653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann R., Genersch E., Eggers H.J. Detection of adenovirus nucleic acid sequences in human tonsils in the absence of infectious virus. Virus Res. 1987;7:93–97. doi: 10.1016/0168-1702(87)90060-8. [DOI] [PubMed] [Google Scholar]
- Prüfer K., de Filippo C., Grote S., et al. A high-coverage Neandertal genome from Vindija cave in Croatia. Science. 2017;358:655–658. doi: 10.1126/science.aao1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin, U., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;(May 27) doi: 10.1038/s41586-021-03653-6. Epub ahead of print. PMID: 34044428, In press. [DOI] [PubMed] [Google Scholar]
- Schick J., et al. Intracellular forms of adenovirus DNA: Integrated form of adenovirus DNA appears early in productive infection. Proc. Natl. Acad. Sci. U.S.A. 1976;73:1043–1047. doi: 10.1073/pnas.73.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert R., Renz D., Schmitz B., Doerfler W. Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA. Proc. Natl. Acad. Sci. U.S.A. 1997;94:961–966. doi: 10.1073/pnas.94.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M., Freisem-Rabin U., Jessberger R., Doerfler W. Transcriptional activities of mammalian genomes at sites of recombination with foreign DNA. J. Virol. 1987;61:344–353. doi: 10.1128/JVI.61.2.344-353.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanathan S., Calcedo R., Wilson J.M. Adenovirus-antibody complexes contributed to lethal systemic inflammation in a gene therapy trial. Mol. Ther. 2020;28:784–793. doi: 10.1016/j.ymthe.2020.01.006. Epub 2020 Feb 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabel S., Doerfler W. Nucleotide sequence at the site of junction between adenovirus type 12 DNA and repetitive hamster cell DNA in transformed cell line CLAC1. Nucleic Acids Res. 1982;10:8007–8023. doi: 10.1093/nar/10.24.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S.L., et al. Chromosomal integration of adenoviral vector DNA in vivo. J. Virol. 2010;84:9987–9994. doi: 10.1128/JVI.00751-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter D., Westphal M., Doerfler W. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell. 1978;14:569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc. Natl. Acad. Sci. U. S. A. 1980;77:253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatzelt J., Scholz B., Fechteler K., Jessberger R., Doerfler W. Recombination between adenovirus type 12 DNA and a hamster preinsertion sequence in a cell-free system. Patch homologies and fractionation of nuclear extracts. J. Mol. Biol. 1992;226:117–126. doi: 10.1016/0022-2836(92)90128-7. [DOI] [PubMed] [Google Scholar]
- Toth M., Lichtenberg U., Doerfler W. Genomic sequencing reveals a 5-methylcytosine free domain in active promoters and the spreading of pre-imposed methylation patterns. Proc. Natl. Acad. Sci. U.S.A. 1989;86:3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentin J.J., Yabe Y., Taylor G. The quest for human cancer viruses. Science. 1962;137:835–841. doi: 10.1126/science.137.3533.835. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Neumann R., Kuhlmann I., Sutter D., Doerfler W. DNA methylation and viral gene expression in adenovirus-transformed and -infected cells. Nucl. Acids Res. 1980;8:2461–2473. doi: 10.1093/nar/8.11.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardimon L., Kressmann A., Cedar H., Maechler M., Doerfler W. Expression of a cloned adenovirus gene is inhibited by in vitro methylation. Proc. Natl. Acad. Sci. U.S.A. 1982;79:1073–1077. doi: 10.1073/pnas.79.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C., et al. The sequence of the human genome. Science. 2001;291:1304. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Weber S., Hofmann A., Herms S., Hoffmann P., Doerfler W. Destabilization of the human epigenome: consequences of foreign DNA insertions. Epigenomics. 2015;7:745–755. doi: 10.2217/epi.15.40. [DOI] [PubMed] [Google Scholar]
- Weber S., Jung S., Doerfler W. DNA methylation and transcription in HERV (K, W, E) and LINE sequences remain unchanged upon foreign DNA insertions. Epigenomics. 2016;8:157–165. doi: 10.2217/epi.15.109. [DOI] [PubMed] [Google Scholar]
- Weber S., Ramirez C., Doerfler W. Signal hotspot mutations in SARS-CoV-2 genomes evolve as the virus spreads and actively replicates in different parts of the world. Virus Research. 2020;289 doi: 10.1016/j.virusres.2020.198170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S., Ramirez C.M., Weiser B., Burger H., Doerfler W. SARS-CoV-2 worldwide replication drives rapid rise and selection of mutations across the viral genome: A time-course study - Potential challenge for vaccines and therapies. EMBO Mol. Med. 2021:e14062. doi: 10.15252/emmm.202114062. PMID: 33931941, In this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang W., Ornelles D.A., Gooding L.R.J. Modeling adenovirus latency in human lymphocyte cell lines. J. Virol. 2010;84:8799–8810. doi: 10.1128/JVI.00562-10. Epub 2010 Jun 23.PMID: 20573817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. U S A. 2021;118(21) doi: 10.1073/pnas.2105968118. PMID: 33958444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zock C., Doerfler W. A mitigator sequence in the downstream region of the major late promoter of adenovirus type 12 DNA. EMBO J. 1990;9:1615–1623. doi: 10.1002/j.1460-2075.1990.tb08281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zock C., Doerfler W. Investigations on virus-host interactions: an abortive system. Methods in Molecular Genetics. 1995;7:167–192. [Google Scholar]