Abstract
The sudden increase in coronavirus disease 2019 (COVID-19) cases puts high pressure on healthcare services worldwide. At this stage, fast, accurate, and early clinical assessment of the disease severity is vital. In general, there are two issues to overcome: (1) Current deep learning-based works suffer from multimodal data adequacy issues; (2) In this scenario, multimodal (e.g., text, image) information should be taken into account together to make accurate inferences. To address these challenges, we propose a multi-modal knowledge graph attention embedding for COVID-19 diagnosis. Our method not only learns the relational embedding from nodes in a constituted knowledge graph but also has access to medical knowledge, aiming at improving the performance of the classifier through the mechanism of medical knowledge attention. The experimental results show that our approach significantly improves classification performance compared to other state-of-the-art techniques and possesses robustness for each modality from multi-modal data. Moreover, we construct a new COVID-19 multi-modal dataset based on text mining, consisting of 1393 doctor–patient dialogues and their 3706 images (347 X-ray 2598 CT 761 ultrasound) about COVID-19 patients and 607 non-COVID-19 patient dialogues and their 10754 images (9658 X-ray 494 CT 761 ultrasound), and the fine-grained labels of all. We hope this work can provide insights to the researchers working in this area to shift the attention from only medical images to the doctor–patient dialogue and its corresponding medical images.
Keywords: COVID-19 diagnose, Knowledge attention mechanism, Knowledge-based representation learning, Knowledge embedding
1. Introduction
The pandemic of the coronavirus disease 2019 (COVID-19) has brought unprecedented disaster to humans’ lives. Facing the ongoing outbreak of COVID-19, viral nucleic acid diagnoses using real-time polymerase chain reaction (RT-PCR) is the accepted standard diagnostic method to find the crowd of infestor [1], [2], [3], [4]. However, due to political and economic reasons, many hyper-endemic regions and countries cannot use the RT-PCR method to detect tens of thousands of suspected patients. On the other hand, due to its high false-negative rate, repeat testing of RT-PCR might be needed to achieve an accurate diagnosis of COVID-19. The chest X-ray, ultrasound, and computed tomography (CT) of imaging tools frequently have been used for diagnosing other diseases. It is fast and easy to operate and has become a widely used diagnostic tool [5], [6]. Researchers are studying how to distinguish COVID-19 from chest X-ray, ultrasound images, or CT scans to solve the lack of reagents [7], [8], [9], [10]. Also, medical COVID-19 data [11], [12], [13], [14] consists of chest X-ray images, ultrasound images, and CT images (i.e., slices) and mostly multi-modal.
The great success of deep learning methods in pneumonia diagnosis tasks has inspired many researchers [15], [16], [17], [18]. The deep-learning-based COVID-19 diagnosis methods are emerging one after another. Nevertheless, extensive medical data is typically required to train these high-quality deep learning-based models. Typically, deep learning models for training this high-performance classification require large amounts of medical COVID-19 data. Besides, medical data on patients with confirmed or suspected COVID-19 might infrequently appear in the public dataset. Thus, it is tough to exploit limited and restricted medical data to train reliable diagnostic models.
On the other hand, in the real world, doctors recommend that physicians communicate with patients (i.e., doctor–patient dialogues) before performing radiological examinations and obtain the patient’s past medical history, current medical history, etc. doctor–patient dialogue is one of the most common forms of consultation [19], [20], [21], [22]. However, this information is not included in the existing medical image dataset. Moreover, in the COVID-19 epidemic, most patients have a past medical history, and chronic medical history [23], [24]. For example, the physician needs to learn about the patients’ history of previous exposure and previous symptoms through a dialogue between the patient and the physician. Also, the physician and the government need to identify relevant close contacts of this patient based on dialogue with the patient and the patient’s recollections to implement effective prevention and control measures.
During the COVID-19 pandemic, even though traditional doctor–patient conversations are at risk of close contact, doctor–patient conversations through Internet video calls and real-time Internet chats are on the rise. The doctor–patient dialogue over the Internet is gradually becoming one of the primary instruments of consultation. Therefore, it is urgent and essential that the deep-learning-based model shifts the attention from only medical images to the doctor–patient dialogue and its corresponding medical images.
In short, there are two main challenges in the task of COVID-19 diagnosis:
(1) Multi-modal information of COVID-19 infestors, including doctor–patient dialogue and different modality medical images, must be jointly considered to make accurate inferences about COVID-19;
(2) Limited multi-modal data makes it challenging to design effective diagnostic models.
Inspired by the success of graph-based attention models (e.g., graph attention network [25]), which capture entities (i.e., nodes) as well as their relationships (i.e., edges) with each other, we focus on the strategy of graph-based attention models to solve these above two issues [26], [27], [28]. While existing work about graph attention mechanisms all considers knowledge graphs, which is a type of heterogeneous graph from multi-modal data and aims to make the full & joint use of multi-modal data, they do not differentiate between the different kinds of links and nodes. This is important as approaches based on heterogeneous graphs have been shown to outperform approaches that assume that graphs only have a single type of link/edge. Therefore, how can attention mechanisms be designed to leverage and jointly exploit the multi-modal data?
Moreover, data and feature representations of a priori knowledge focus on projecting data and relationships between data into a low-dimensional continuous space [29]. Most approaches aim to learn representations with a priori knowledge that represent relationships between data [30]. In this way, we may use the limited multimodal data to train robust deep networks [31], [32]. Why not use knowledge-based representation learning to make representations of attentional mechanisms as joint representations of a priori medical knowledge and deep network?
Motivated by these observations, to tackle all the problems mentioned above, in this paper, we propose a novel multi-modal knowledge embedding-based graph attention model for the COVID-19 diagnosis task. During this process, the model makes use of medical knowledge. Notably, we firstly propose the multimodal attention mechanism that is able to learn the medical knowledge-based representations about the prior multimodal information. Secondly, we get the multimodal medical information embeddings. Thirdly, we create the temporal convolutional self-attention network and obtain the pivotal features of prior multimodal information. Finally, our framework relates feature maps to the embeddings about multimodal medical and explains the classification of COVID-19 diagnosis. Experimental results demonstrate that the proposed approach has higher performance expressively than the state-of-the-art methods in the COVID-19 diagnosis task.
Moreover, we propose a new multi-modal information dataset about COVID-19, which contains doctor–patient dialogue and their corresponding multi-modal medical images ( X-ray images, axial CT images, and ultrasound images) with the text-mined fine-grained disease labels during the ongoing outbreak of COVID-19, mined from the text radiological reports. All in all, the main contributions of this paper are summarized as follows:
We propose a robust and end-to-end multimodal knowledge embedding-based graph attention model to classify COVID-19 multimodal data. To the extent of our knowledge, it is the first attempt to investigate multimodal attentional mechanisms based diagnostic approach about COVID-19.
We construct an effective multi-modal attention mechanism that dramatically improves the performance of the proposed approach. What is more, we design a novel cross-level modality attention to combine single-modality and multiple-modality information.
We present a novel temporal convolutional self-attention network to extract and learn the discriminative features on the datasets. The qualitative discussion demonstrates that this strategy achieves competitive performance over other temporal convolutional network-based methods.
A new dataset about multi-modal information is constructed for the task of COVID-19 diagnosis. This dataset contains doctor–patient dialogues and their images ( X-ray CT ultrasound) about COVID-19 patients and non-COVID-19 patient dialogues and their images ( X-ray CT ultrasound), and the fine-grained labels of all.
2. Previous COVID-19 diagnosis
Radiological diagnosis is a convenient medical technique for patients with COVID-19 who are suspected of urgently needing a risk area diagnosis [33], [34]. X-ray CT scans and ultrasonography are widely used to provide compelling evidence for the analysis of radiologists. To achieve higher accuracy for radiological diagnosis, using either X-ray or CT as the acquisition method, many works have been proposed for COVID-19 diagnosis. Also, benefit from ultrasonography convenience, some works have been proposed for COVID-19 diagnosis via ultrasound images.
Based on chest X-ray images, there are many discussions of the classification between COVID-19 and other non-COVID-19 subjects, including other pneumonia subjects and healthy subjects. Zhang et al. [35] present a ResNet based model to classify COVID-19 and non-COVID-19 X-ray images for COVID-19 diagnose. They use X-ray images from seventy COVID-19 patients and one thousand and eight non-COVID-19 pneumonia patients, and they achieve 96.0% sensitivity and 70.7% specificity along with an AUC of 0.952. A deep CNNs based architecture called COVID-Net [36] is presented for COVID-19 diagnosis from X-ray images. Utilizing their self-built COVIDx dataset, the COVID-Net achieves the testing accuracy of 83.5%. Considering the difficulty of a systematic collection of chest X-ray images for deep neural network training, s patch-based convolutional neural network approach that requires a relatively small number of trainable parameters for COVID-19 diagnosis is proposed by Oh et al. [37]. Also, there have been efforts made for the classification of COVID-19 from non-COVID-19 based on CT scans. Jin et al. [38] build a chest CT dataset consisting of four hundred and ninety-six COVID-19 positive cases and one thousand three hundred and eighty-five negative cases. They propose a two-dimensional CNN-based model for lung segmentation and a COVID-19 diagnosis model. Experimental results show that the proposed model achieves a sensitivity of 94.1%, a specificity of 95.5%, and an AUC of 0.979. To comprehensively explore the description of multiple features of CT images from different perspectives, Shen et al. [39] propose a method for learning a unified potential representation that fully encodes information from different facets of the features and has a promising class structure that enables detachability. Ouyang et al. [40] propose a convolutional network (CNN) with a new online attention module to target the region of lung infection when reaching diagnostic decisions. Based on multi-centre CT data for COVID-19, their algorithm can identify the COVID-19 images with the F1-score of 0.82, the specificity of 0.901, the sensitivity of 0.869, the accuracy of 0.875, and the AUC of 0.944. Furthermore, ultrasonography is another useful technique for diagnosing COVID-19, which is non-invasive, cheap, portable, and available in almost all medical facilities. Roy et al. [41] present a spatial transformer style network with weakly-supervised learning, which localizes the pathological artefacts and predicts the disease severity score. Besides several deep models, they release a novel ultrasound image dataset with fully-annotated labels at a frame-level, video-level, and pixel-level to represent the degree of disease severity.
In summary, lots of studies have been proposed for X-ray-based, CT-based, and ultrasound-based COVID-19 diagnosis. However, most of the recent works only focus on diagnosis tasks in one medical imaging modality and consider less about the doctor–patient conversations before radiological examinations, which also is the crucial part for disease diagnosis through the whole medical healing procedures.
3. Multi-modal COVID-19 pneumonia dataset
In this section, we first describe how to build our proposed Multi-Modal COVID-19 Pneumonia Dataset and introduce the structure of our proposed dataset. Then we make a comparison with existing public available COVID-19 datasets.
3.1. Dataset creation and structure
Medical dialogues are part of the traditional medical procedure when potential patients come to the hospital for professional consultation. Commonly, to avoid a failed diagnosis, doctors always ask patients to make more detailed examinations after necessary doctor–patient dialogues. However, most recent works for COVID-19 diagnosis only pay attention to medical images of COVID-19 without any dialogues, which may lead to biased diagnostic results when information is incomplete. To address this issue, in this paper, we propose a multi-modal dataset for COVID-19 pneumonia, which consists of both images and doctor–patient dialogues.
The image subset mainly contains three medical imaging modalities: X-ray, CT, and ultrasound by collecting them from public radiology medical reports and patient follow-up records. In detail, X-ray images, CT images, and ultrasound images of COVID-19 are collected from radiological reports published by online hospitals and radiology medical centres in China [15]. All X-ray images are posteroanterior (PA) or anteroposterior (AP) views, and salient axial slices of different CT volumes are collected for CT images. Most of the ultrasound images are in a convex view, and the rest are in linear view. Following the work of Chest-X-ray-8 [42], we use the technology of text mining and natural language processing (NLP) to get disease findings and decide the labels of all images. The dialogue subset is assembled from the same websites [11], and the labels of all dialogues are extracted from the disease description part by using natural language processing (NLP) toolkit [43]. Also, we provide text-mined fine-grained disease labels of each image in our dataset, including patient sex, patient age, which can be found in our Supplemental Materials. The image subset has 2D images including X-ray images, axial CT images and ultrasound images. In particular, X-ray images, CT images, and ultrasound images of COVID-19 have been assembled in our proposed image subset. Basic statistics for each class of our proposed image subset are shown in Table 1. Besides, the dialogue subset contains more than thousand sentences between doctors and patients. For both image subset and dialogue subset, two main categories are shared: COVID-19 and Non-COVID-19. The Non-COVID-19 label means other different types of community-acquired pneumonia (CAP) except normal case [40]. We use this strategy to collect a total of 2000 doctor–patient dialogues, of which there are doctor–patient dialogues about COVID-19 patients and non-COVID-19 patient dialogues. We use the NLP toolkit [43] to count the frequency of the keywords of the two kinds of dialogues and formed two kinds of word clouds for the top words, which are shown in Fig. 1. From Fig. 1, we can clearly see that the symptoms of COVID-19 patients and non-patients are significantly different. Different from these existing public datasets or challenges, we focus on the analysis of different patterns between types of pneumonia with diverse causes; that is why there are no normal cases in our dataset.
Table 1.
Basic statistics of the image subset in our proposed multi-modal COVID-19 pneumonia dataset.
| X-ray | CT | Ultrasound | Total | |
|---|---|---|---|---|
| COVID-19 | 340 | 2598 | 761 | 3706 |
| Non-COVID-19 | 9658 | 494 | 599 | 10754 |
| 9998 | 3092 | 1360 | 14450 | |
Fig. 1.
Comparison about COVID-19 and non-COVID-19 dialogues.
Both medical images and doctor–patient dialogue are tools for physicians to know their patients. For patients with a certain type of disease, their disease characteristics are statistical characteristics [44], [45], [46]. For example, as shown in Fig. 1, the disease characteristics of two diseases are different. Such statistical characteristics are also the basis for physicians to judge the disease [47], [48]. In this way, machine learning models essentially capture these statistical characteristics to be able to classify effectively. The model is trained with the information of a particular individual, not a group of individuals. It is also challenging to capture statistical characteristics with a model that uses this training strategy for classification. Moreover, considering the emergency of diagnosis and different situations for potential patients, the examinations cannot be very comprehensive in a short time when medical resources are extremely saturated [49], [50]. In other words, in a COVID-19 pandemic, medical resources are precious and limited. Most patients do not have medical images of all three modalities (i.e., X-ray, CT, ultrasound). Thus, to simulate this urgent state, the relationship between our proposed image subset and dialogue subset is unpaired. To validate the validity of our proposed dataset, experienced radiologists in our team check all images, including comparing the label of the patient’s medical images with the results of the patient’s RT-PCR, and eliminating images with errors.
3.2. Dataset comparison
For COVID-19, a new type of coronavirus disease that crosses the world, it is essential to collect data for machine learning applications. In recent months, a number of works have been presented on the COVID-19 public dataset [15], [51].
Cohen et al. [52], [53] create an image collection containing 329 images from 183 patients, most of which are chest X-ray images for COVID-19. Based on an early version of the COVID-19 image dataset constructed by the above work, COVID v2.0, and its enriched version [36] adds more bacterial pneumonia chest X-ray images and standard chest X-ray images. Zhao et al. [54] present a publicly available COVID-CT dataset consisted of COVID-19 CT axial images collected from preprinted publications from medRxiv and bioRxiv. They extract figures and captions in conjunction, judging whether a patient is positive for COVID-19 from the associated captions. Besides the X-ray-based image dataset and CT-based image dataset, ultrasound-based image datasets are also reported recently. Jannis et al. [55] propose a lung ultrasound dataset, called POCUS, consisting of one thousand six hundred fifty-four COVID-19 images, two hundred seventy-seven bacterial pneumonia images, and one hundred and seventy-two healthy controls images, which are sampled from sixty-four videos assembled from various online sources. COVIDGR-1.0 dataset is proposed [56] and is organized into two classes: positive and negative. This dataset includes 852 images (426 positive and 426 negative cases). Existing public image datasets only focus on diagnosis tasks using one medical imaging modality but hardly explore the possibility of utilizing different medical imaging modalities together. Furthermore, there are not doctor–patient dialogues in existing public image datasets for precise diagnosis, and the image numbers of existing public image datasets are not enough. To approve the advancement of the image subset in our proposed Multi-Modal COVID-19 Pneumonia Dataset, the X-ray-based part of the image subset is compared to COVID v2.0 dataset [36], the CT-based part is compared to COVID-CT dataset [54] and the ultrasound-based part is compared to POCUS [55]. All of three comparisons are shown in Table 2, Table 3, Table 4.
Table 2.
Comparison of COVID v2.0 dataset [36] and X-ray part of the image subset in our proposed multi-modal COVID-19 pneumonia dataset.
| COVIDx | Normal |
Pneumonia |
COVID-19 |
|---|---|---|---|
| 8066 | 8614 | 190 | |
| Proposed image subset |
Normal |
Non-COVID-19 |
COVID-19 |
| – | 9658 | 340 |
Table 3.
Comparison of COVID-CT dataset [54] and CT part of the image subset in our proposed multi-modal COVID-19 pneumonia dataset.
| COVID-CT | COVID-19 |
Non-COVID-19 |
|---|---|---|
| 349 | 397 | |
| Proposed image subset |
COVID-19 |
Non-COVID-19 |
| 2598 | 494 |
Table 4.
Comparison of POCUS dataset [55] and ultrasound part of the image subset in our proposed multi-modal COVID-19 pneumonia dataset.
| POCUS | Normal |
Bacterial pneumonia |
COVID-19 |
|---|---|---|---|
| 172 | 277 | 654 | |
| Proposed image subset |
Normal |
Non-COVID-19 |
COVID-19 |
| – | 599 | 761 |
As for other formats of the COVID-19 dataset. [57] propose a medical dialogue dataset about COVID-19 and other related pneumonia, which contains more than 1000 consultations. Yet, existing public dialogue datasets only crawl relevant conversations from websites, and there is no precisely fine-grained label for each dialogue.
From the comparisons and the analysis mentioned above, it is evident that our proposed dataset is better than others, the advantages of which can be summarized as follows:
-
•
Our proposed Multi-Modal COVID-19 Pneumonia Data-set considers the exploit of utilizing different medical imaging modalities together.
-
•
With precise labels, our proposed Multi-Modal COVID-19 Pneumonia Dataset considers the fusion of information both from images and doctor–patient dialogues.
-
•
Compared to existing public available COVID-19 data-sets, our proposed Multi-Modal COVID-19 Pneumonia Data-set can be seen as the largest dataset with fine-grained labels.
4. Methodology
In this section, we first describe the notations and the structure of our COVID-19 data. Then, we propose the details of our proposed multi-modal knowledge embedding graph attention model.
4.1. Basic notations
Multimodal Information We suppose there are four kinds of multimodal data: X-ray images, CT images, ultrasound images, and text description of diagnose. The training data is denoted as , where , , , and ; denotes the -th X-ray image, denotes the -th CT image, denotes the -th Ul image, and denotes the -th text data; instead of , , and , they are simplified as ; , , , and mean their corresponding labels are from the set , ; , , , denote the number of corresponding training data and are simply written as ;Testing data is denoted as , , where , , , ; , , , mean the number of corresponding training data and are simply written as .
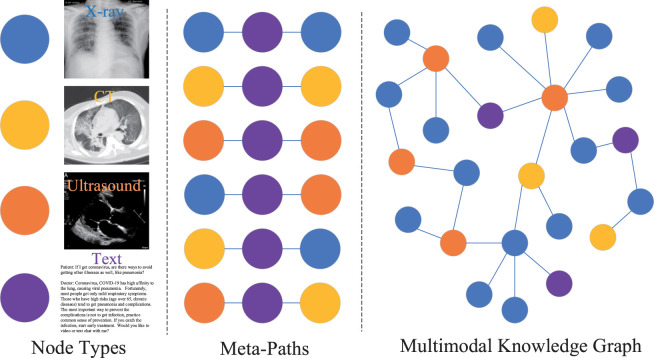
Knowledge Graph We suppose the heterogeneous knowledge graph is built, in which is a link set as well as is an object set, by utilizing the training data. Distinctly, as shown in Fig. 2, the consists of multiple types(i.e., X-ray image, CT image, ultrasound image, and text) and multiple types of links (e.g., X-ray image text X-ray image, X-ray image text CT image). We define a node type mapping function as and the function about link type mapping as , in which and are predefined object types and the sets of link types, respectively. is associated with two these functions, i.e., . We define in as meta-path, where the path can be in the style of and can be abbreviated as . We use the to describe a composite relation between objects and , where means the composition operator on relations. We suppose the nodes set is related with meta-path and node . We define the nodes set, which is the meta-path based neighbours of node including themselves.
Fig. 2.
An illustrative example of a multi-modal knowledge graph. From left to right, these are the four types of nodes (i.e., X-ray, CT, ultrasound, and text description of diagnose), six meta-paths involved in the given knowledge graph, a multi-modal knowledge graph.
Multimodal Graph Attention We denote projected node feature as and initial node feature as . We denote the type-specific transformation matrix as and features of different types of nodes are projected into the same feature space [58]. We design our attention mechanism to deal with all kinds of nodes with type-specific projection operations.
We denote the meta-path based node pair’s weight as to obtain the weight value between node and node , which is associated with (meta-path). We bespeak the single-level modality attention vector about (meta-path) as . We acquire the importance between pairs of nodes based on meta-paths, and then aim to acquire the meta-path-based node pair ’s weight , these node pairs are normalized. We bespeak as single-level modality attention vector about (meta-path). The weight value between pairs of nodes based on meta-paths is learned, and we can obtain the denoted as the meta-path based node pair ’s important. This process is also called the normalization of these nodes. We use the weight of the single-level modality attention to describe the similarity [59] of transformed embedding, denoted as a multiple-level modality attention vector .
Similarly, the weight of each meta-path is defined and denoted as . We can get the each meta-path ’s weight, and obtain the meta-path ’s importance . This process is also called the normalization of these meta-paths.
From these above notations, we input the multimodal knowledge graph , multimodal data , is defined as the number of attention head, the given meta-path set where the number of the given meta-paths are denoted as , and the node feature . As a result, we get the knowledge-based attention feature vector .
4.2. Building multi-modal knowledge graphs
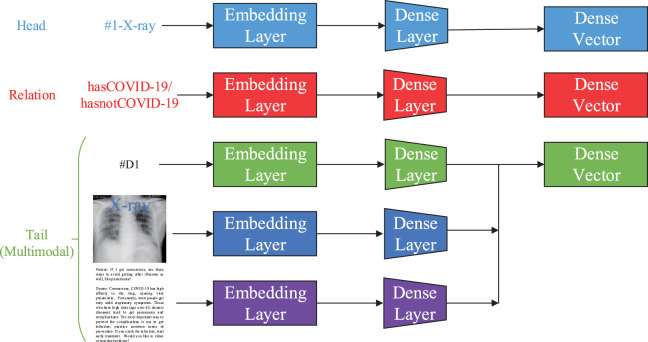
Built upon Pezeshkpour et al.’s work [60], [61], [62], we use a triple of the head, relation, and tail, in order to represent our knowledge graph . Similar to the multi-modal knowledge bases [63], we take advantage of recent advances in deep learning to build embedding layers for these nodes to represent them, in essence offering embeddings for different types of nodes. As shown in Fig. 3, we use different embedding layers to represent each specific data type.
Fig. 3.
Architecture of multi-modal knowledge graph model .
Structured Knowledge Considering a triplet of the head, relation, and tail as independent embedding vectors, we generate dense vectors through embedding layers.
Images A wide variety of models have been devised to represent the semantic information in images compactly and have been successfully applied to tasks such as classification [64], and visual reasoning [65]. To represent the images, we employ the last hidden layer of VGG-16[66] as an embedding layer, which is pre-trained by ImageNet [67].
Texts The doctor–patient dialogues are highly relevant to the text content and can capture the disease status of patients. For these texts, we apply a BERT model [68] to get the weighted word vectors of the sentences as an embedding layer to represent the text features. In this way, it is simple and efficient compared to the conventional LSTM [69].
Finally, as illustrated in Fig. 3, we use dense layers to unify all embedding layers into the same dimension to construct the multi-modal knowledge graph.
4.3. The proposed approach
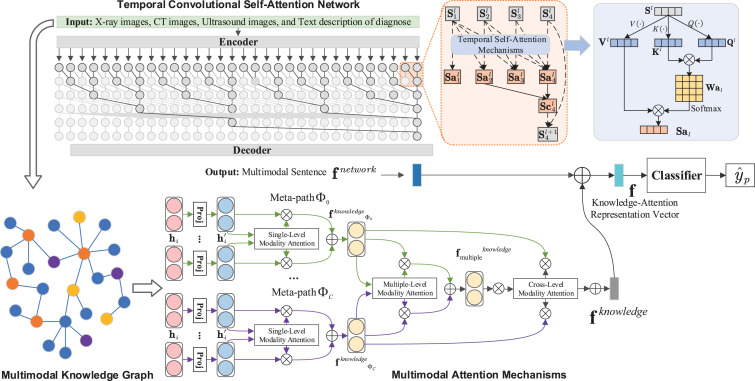
In this paper, the overall framework of our proposed approach is illustrated in Fig. 4. Given knowledge graph , we define the embedding matrix of single-level modality attention as . Similarly, we define the matrix of multiple-level modality attention embeddings as , following the work of graph-based multimodal attention mechanism [58]. Secondly, we create a cross-level modality attention mechanism to fuse the information of single-level modality and multiple-level modality attentions. By this process, we can get the embedding matrix . Thirdly, we propose the Temporal Convolutional Self-Attention Network (TCSAN) to handle the inputted multimodal data and get the multimodal sentence vectors . As a result, we get the knowledge-based attention feature vector , utilizing attention embedding vectors and multimodal sentence vectors, to classify information of the labels, i.e., . Also, our approach only focuses on the interaction between the text of dialogues and the images of three different medical imaging modalities, which means that one dialogue can be combined with two different modality images.
Fig. 4.
The proposed multi-modal knowledge graph attention embedding model. Given multimodal knowledge graph , we propose the multimodal attention mechanisms including three parts: ➀ the single-level modality attention and its results denoted as ; ➁ the multiple-level modality attention and its embedding denoted as ; ➂ cross-level modality attention mechanism that fuse the information of single-level modality and multiple-level modality attentions, and its the embedding matrix denoted as . Meanwhile, we propose the Temporal Convolutional Self-Attention Network (TCSAN) to handle the inputted multimodal data and get the multimodal sentence vectors . Then, we get the knowledge-based attention feature vector . Finally, we use the classifier (in this paper, we use the ResNet-34 [70]) to gain the labels, i.e., .
4.3.1. The multimodal attention mechanism
In this sub-subsection, an exquisite and novel multi-modal attention mechanism with domain knowledge is proposed to deal with multi-modal medical data. Our model consists of three part: Single-Level Modality Attention, Multiple-Level Modality Attention, Cross-Level Modality Attention. Single-level modality attention [58] is designed to obtain the importance of meta-path based neighbours, which can be assembled to obtain the embedding of the single-level modality attention. Similarly, the multiple-level modality attention [58] is utilized to obtain the difference between single-level modality attentions. As a result, we can obtain the embedding of the multiple-level modality attention. For each modality, the multiple complementary separate representations and are obtained and describe the information of the single modality and multiple modality. We design the cross-level modality attention mechanism to fuse the separate representations hierarchically, and get the attention embedding .
Single-Level Modality Attention Due to the heterogeneity of nodes, there are different feature spaces which contain different types of nodes. Before we aggregate the information from the meta-path neighbours of each node (e.g.,node with type ), it has been recognized that different meta-path-based neighbours of each node have varying roles and differing levels of importance in terms of learning embedding. Here, we design single-level modality attention to gain, for each node in , its meta-path-based neighbours’ importance, and fuse these eloquent representations of the neighbours to form the embeddings:
| (1) |
where are performed by the deep neural network, and denotes the concatenate operation.
We can get the learned embeddings with repeated the above process for times. These embeddings concatenated and the results is our single-level modality attention embedding:
| (2) |
We input the meta-path set , and obtain the single-level modality attention embedding .
Multiple-Level Modality Attention Each node in a heterogeneous graph includes multiple kinds of semantic information, thus, single-level modality embedding is only able to consider the information of nodes in one way. To this end, we have to integrate multiple semantics with representation of meta-paths. In order to obtain a more widespread embedding, we use the multiple-level modality attention to automatically understand the different meta-path’s importance and fuse them together, as follows:
| (3) |
in which represents a weight matrix as well as represents the bias vector.
Cross-Level Modality Attention For multimodal information in the heterogeneous graph , we realize that it is fundamental to consider the balance between the each node’s importance and the meta-paths ’importance. It is apparent that each modality plays a different role, and therefore, a model of the relation between each modality is needed. To address this problem, we propose cross-level modality attention. We firstly, for different node kinds, fashion single-level modality and multiple-level modality embeddings to learn the well-rounded representation, by a nonlinear transformation (for example, one-layer MLP (Multi-Layer Perception)). Moreover, the weight of all nodes of single-level modality and multiple-level modality embeddings is averaged and is explained as the weight of each modality. We identify the weight of each node as , is calculated as follows:
| (4) |
where two weight matrices and . It is worth mentioning that this kind method follows Ref. [71] However our methodology is based on node types rather than entity types.
We express the each node ’s weight as . To this end, we normalize the above weights for all nodes using the softmax function.
| (5) |
where the above equation could be interpreted as a node’s contribution to a particular task. Each node could have varying weights for different tasks. In addition, the higher , the more important node is.
By fusing the following embeddings, we can obtain the weight of learning as a factor to obtain the final embedding :
| (6) |
4.3.2. The Temporal Convolutional Self-Attention Network
Inspired by the success of temporal convolutional network (TCN) [72], [73], [74] and the self-attention mechanism [75], we design a novel convolutional network, named Temporal Convolutional Self-Attention Network (TCSAN). Similar to most competitive neural sequence transduction models [27], [28], [75], we use the encoder and decoder structure in our network. Encoder maps the input data to its representations . Then we use the causal convolutions as a hidden layer across layers, and the intermediate variable at time step and level ()is divided to four steps, illustrated in Fig. 4:
-
Step 1:
We use encoder to encode the ;
-
Step 2:
The is passed through Temporal Self-Attention Mechanism (TSAM): where means an intermediate variable that contains information, illustrated in Fig. 4;
-
Step 3:
We apply causal convolution on the : where indicates the output of causal convolution. For keeping the same length of each layer, we add zero padding on the left, white blocks in Fig. 4. In this way, the left relevant information of input will gradually accumulate to the right;
-
Step 4:
We can get , when the is passed through the activation function.
A full TCSAN is built by stacking layers of TCSAN block across depth and time, and we use the decoder to decode the , and get the output sequence : .
Temporal Self-Attention Mechanism Temporal Self-Attention Mechanism (TSAM) is illustrated as Fig. 4. Inspired by self-attention structure [75], we use three linear transformations , , and to three different vectors: key , query , value , and the dimension . For computing the weight matrix , we compute the vectors and , and divided each by :
| (7) |
where means the transpose of the matrix .
Given the weights , we can get the weighted output by:
| (8) |
Causal Convolutions TCNs are a peculiar kind of 1D convolutional neural network (CNN), which is a natural way to encode information from a sequence [72]. A 1D convolutional layer can be written as
| (9) |
where we define the size convolution filter as , and the input sequence data as . However, when applied to model sequences [76], one-dimensional CNNs are limited by their reduced output size and limited receptive field, and the TCNs in this paper have the technique to solve these problems, i.e., causal convolution.
The causal convolutional layer is filled at the beginning of the input sequence [76] with a zero connection length. Besides, It ensures that there is no disclosure of information that never came to the past, which is essential for predicting future communication, as follows:
| (10) |
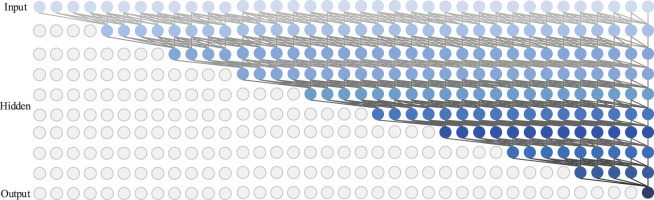
Furthermore, simple standard causal convolution can only add a receptive field with linear size to the network depth. This makes it challenging to handle sequential data. Therefore, we construct the proposed model with an exponentially large receptive field via the use of a dilated causal convolution [72]. Fig. 5 illustrates the dilated causal convolution (we set the filter size to 5). As we can see, a dilated causal convolution is a convolution where the filter is applied over an area larger than its length by skipping input values with a certain step. It is equivalent to a convolution with a larger filter derived from the original filter by dilating it with zeros, but is significantly more efficient since it utilizes fewer parameters. As a result, a dilated causal convolution can better process sequential data without more layers.
Fig. 5.
The illustration of TCN with dilated causal convolutions.
4.3.3. Knowledge-based representation learning
We present a gate mechanism for embedding knowledge representations to strengthen representation learning, with consideration of the suppression of the non-informational features and permission of the informational features to transit under the tutorial of a multi-peaked knowledge graph, similar to Ref. [77], [78], [79], denoted as
| (11) |
where we define the element-wise multiplication operation as , the logistic sigmoid as ; is a neural network that combines features embedded in the final knowledge with features extracted using TCSAN.
4.3.4. Objective function
Depending on the final embedding of a particular task, we can design different loss functions. In this paper, we can choose to minimize the cross-entropy of all labelled nodes between ground-truth and prediction:
| (12) |
Notice that in our implementation, we assume the average of the single cross-entropy errors of all nodes. The Algorithm 1 depicts the overall training process of our approach.
5. Experimental results
In this section, we experiment with the multimodal COVID-19 dataset to assess the performance of the proposed method. In comparison to the state-of-the-art models, our approach would be better in different evaluation strategies.
5.1. Experimental setup
In this subsection, we begin with an overview of the training data setup and the measures used for performance evaluation. Then we describe our experiment’s implementation details.
5.1.1. Evaluation protocol
Training Data Setup We randomly choose the diagnoses and corresponding X-ray images, CT images, ultrasound images from our dataset to construct the training set; we randomly choose the diagnoses and corresponding images from our dataset to construct the validation set; we randomly choose the diagnoses and corresponding images from our dataset to construct the test set; note that we randomly choose ten times as per the above strategy and take the average evaluation criteria for comparison.
We take advantage of the training data in different dataset and constitute the knowledge graph by [61], [62]. There are four modality data: X-ray images, CT images, ultrasound images, and text description of diagnose. Apparently, most of diagnoses includes some images. If a certain diagnose contains no image, we use a zero vector to represent the image. Besides, if a certain diagnose contains images, we extract image features from a -layer pre-trained VGGNet [66] to represent the images, similar to [80]. By this way, we get the feature of X-ray images (XR), CT images (CT) and ultrasound images (UL). We use the sentences in a certain diagnose to represent the text (T). Here we define the meta-path set as {XRTXR, XRTCT, CTTCT, CTTUL, ULTUL, ULTXR} to perform experiments. We define the images as the image-type node features , , . We use the BERT method [68] to deal with the text samples, and then conduct the text-type node features . After these, we obtain the node features .
Evaluation Measures We use the accuracy, precision, F1-score, sensitivity, specificity, and area under the receiver operator curve (AUC) to assess the performance of all models. More precisely, we use sensitivity and specificity to denote the number of positive and negative samples correctly identified, respectively. Besides, we use AUC to measure the overall classification performance, which is sensitive to the imbalance among multiple classes.
5.1.2. Implementation details
All experiments are performed this way with a 4-core PC with four 12 GB NVIDIA TITAN XP GPUs, 16 GB RAM, and Ubuntu 16. In all models, we set the number of epochs to . We input the knowledge-based attention feature vector into the COVID-19 classifier and then acquire the final classification results. We perform COVID-19 classification with ResNet-34 classifier [70]. The individual state of the knowledge attention mechanism and TCSAN implementation is as follows:
Temporal Self-Attention Network We utilize layers of temporal convolutional networks [72] to serve as our network architecture. For each layer, the hidden node has a value of , and the kernel size has a value of . By extending the dropout to all nonlinear layers, we have a probability of 0.5. Adam optimizer is used to optimize our model. In addition, our is set to 0.9, and the setting of is set to 0.999, in which the weights of decay to . For classification, the initial learning rate has a value of . The batch size is set to . Besides, for multimodal sentences, the final representation vector ’s length is set to .
The Multimodal Attention Mechanism We randomly initialize the parameters. For optimization of the model we use Adam [81]. The attention ’s dropout has a value of 0.6, and we define the number of attentional heads as , the regularization parameter as 0.001, and the learning rate as 0.005. We set the dimension of (the multiple-level modality attention vector) to . In addition, the final embedding dimension is set to . We put in , , and for training or testing of attentional mechanisms for experiments on our dataset.
5.2. Comparison with state-of-the-art methods
We present a comparison of state-of-the-art methods with our dataset. In this subsection, “Ours w/o Knowledge” denotes a variant of Ours, involving the use of only network representations of learning without the use of multimodal attention mechanisms.“Ours w/o TSAM” indicates a variant of Ours with TCN only and no TCSAN. “XR” means X-ray modality, “CT” means CT modality, “UL” means ultrasound modality and “T” represents text modality; “XRCTUL” stands for the combination of X-ray modality, CT modality and ultrasound modality, and “XRCTULT” means combining X-ray modality, CT modality, ultrasound modality and text modality.
Baselines on Our Dataset We compare against various state-of-the-art baselines on our dataset, including CAAD [35], COVID-CAPS [82], COVID-DA [83], COVID-ResNet [84], DCSL [85], COVNet [86], AnoDet [87], DLS [88], DeCovNet [89], DLQCTM [90], DenseNet-161[91], ResNet-34[70], VGG-19[66], ResNet-18[70], VGG-16[66], DGLM [92], LM3FT [93], MMCL [94], MEC [95], and GMMF [96].
Effect of Proposed Multimodal Attention Mechanisms. To estimate the performance of our methodology, we present a comparison of the results reported in the “Ours w/o Knowledge” rows and the “Ours” in rows in Table 5. Our approach utilizes the fair comparison of the same loss functions and features in the “Ours w/o Knowledge” row. Drawing from Table 5, we observe that our methodology continuously increases performance in all cases. In particular, from “Ours w/o Knowledge” to Ours, the number of parameters of models changes from 71.2M to 75.6M with three modal data (i.e., XRCTUL). Similarly, for the case of four modal data (i.e., XRCTULT), the number of parameters of models changes from M to 76.4M. Moreover, the batch time of the trained models all decrease in the testing stage, after “Ours w/o Knowledge” is added the multimodal attention mechanisms. In terms of all evaluation measures (i.e., accuracy, precision, sensitivity, specificity, F1-score, AUC), the performance of Ours all increased compared to “Ours w/o Knowledge”. It is clear that the design of multimodal attention mechanisms can enhance the effectiveness of our model.
Table 5.
Classification results of each model on our dataset. “M” means . Batch time means the runtime of each batch in the model testing.
| Method | Modality | Accuracy | Precision | Sensitivity | Specificity | F1-score | AUC | Parameters (a.k.a., model size) | Batch time |
|---|---|---|---|---|---|---|---|---|---|
| CAAD [35] | XR | 0.7394 | 0.7430 | 0.7659 | 0.7791 | 0.7543 | 0.7488 | 138.3M | 2.06 s |
| COVID-CAPS [82] | XR | 0.7413 | 0.7450 | 0.7689 | 0.7844 | 0.7568 | 0.7540 | 23M | 2.09 s |
| COVID-DA [83] | XR | 0.7446 | 0.7471 | 0.7692 | 0.7844 | 0.7580 | 0.7593 | 33M | 2.32 s |
| COVID-ResNet [84] | XR | 0.7466 | 0.7497 | 0.7747 | 0.7856 | 0.7620 | 0.7702 | 60.2M | 2.31 s |
| DCSL [85] | XR | 0.7486 | 0.7510 | 0.7777 | 0.7870 | 0.7641 | 0.7709 | 125M | 2.54 s |
| COVNet [86] | CT | 0.7518 | 0.7512 | 0.7805 | 0.7880 | 0.7656 | 0.7812 | 130.8M | 2.18 s |
| AnoDet [87] | CT | 0.7552 | 0.7518 | 0.7853 | 0.7907 | 0.7682 | 0.7843 | 125.8M | 2.19 s |
| DLS [88] | CT | 0.7607 | 0.7535 | 0.7878 | 0.7958 | 0.7703 | 0.7878 | 179.1M | 2.18 s |
| DeCovNet [89] | CT | 0.7613 | 0.7573 | 0.7947 | 0.8022 | 0.7755 | 0.7885 | 93M | 2.12 s |
| DLQCTM [90] | CT | 0.7663 | 0.7648 | 0.7989 | 0.8081 | 0.7815 | 0.7896 | 103M | 2.26 s |
| DenseNet-161[91] | UL | 0.7029 | 0.7229 | 0.7394 | 0.7517 | 0.7311 | 0.7285 | 18.8M | 2.12 s |
| ResNet-34[70] | UL | 0.7154 | 0.7288 | 0.7514 | 0.7553 | 0.7400 | 0.7287 | 63.6M | 0.80 s |
| VGG-19[66] | UL | 0.7214 | 0.7367 | 0.7525 | 0.7605 | 0.7445 | 0.7307 | 144M | 2.82 s |
| ResNet-18[70] | UL | 0.7278 | 0.7395 | 0.7575 | 0.7708 | 0.7484 | 0.7434 | 33.3M | 0.56 s |
| VGG-16[66] | UL | 0.7365 | 0.7420 | 0.7633 | 0.7777 | 0.7525 | 0.7474 | 140M | 2.48 s |
| DGLM [92] | XRCTUL | 0.8070 | 0.7842 | 0.8322 | 0.8591 | 0.8075 | 0.8067 | 130.2M | 2.08 s |
| LM3FT [93] | XRCTUL | 0.8110 | 0.7984 | 0.8323 | 0.8676 | 0.8150 | 0.8120 | 126M | 2.16 s |
| MMCL [94] | XRCTUL | 0.8120 | 0.8010 | 0.8351 | 0.8682 | 0.8177 | 0.8154 | 129.3M | 2.34 s |
| MEC [95] | XRCTUL | 0.8122 | 0.8079 | 0.8370 | 0.8716 | 0.8222 | 0.8186 | 131.7M | 2.22 s |
| GMMF [96] | XRCTUL | 0.8127 | 0.8082 | 0.8378 | 0.8741 | 0.8227 | 0.8202 | 130.3M | 2.53 s |
| Ours w/o knowledge | XRCTUL | 0.8160 | 0.8108 | 0.8378 | 0.8741 | 0.8413 | 0.8234 | 71.2M | 1.74 s |
| Ours w/o TSAM | XRCTUL | 0.8308 | 0.8121 | 0.8494 | 0.8898 | 0.8491 | 0.8293 | 70M | 1.12 s |
| Ours w/o knowledge | XRCTULT | 0.8389 | 0.8293 | 0.8297 | 0.8292 | 0.8292 | 0.8319 | 73M | 1.77 s |
| Ours w/o TSAM | XRCTULT | 0.8679 | 0.8688 | 0.8641 | 0.8909 | 0.8664 | 0.8675 | 72M | 1.14 s |
| Ours | XRCTUL | 0.9371 | 0.9209 | 0.8884 | 0.9805 | 0.9498 | 0.9171 | 75.6M | 1.12 s |
| Ours | XRCTULT | 0.9810 | 0.9889 | 0.9861 | 0.9859 | 0.9875 | 0.9908 | 76.4M | 1.14 s |
Effect of Proposed Temporal Self-Attention Mechanism. When our model and its variants use the XRCTULT, “Ours” is 0.1131, 0.1202, 0.1220, 0.0951, 0.1211, 0.1232 higher than “Ours w/o TSAM”, in term of accuracy, precision, sensitivity, specificity, F1-Score, AUC. Similarly, with XRCTUL, “Ours” is 0.1063, 0.1089, 0.0390, 0.0908, 0.1006, 0.0878 higher than “Ours w/o TSAM”, in term of accuracy, precision, sensitivity, specificity, F1-Score, AUC. These improvements demonstrate that learning by the temporal self-attention mechanism for the better performance of COVID-19 case diagnoses. With almost unchanged batch time in the test phase, such a large performance improvement can be obtained at the cost of less than 6M increase in the model size. It once again shows that our model is effective.
Effect of Doctor–Patient Dialogues. From Table 5, the performance of all models improved after adding the text from the doctor–patient dialogues to the training of our model and its variants. This emphasizes the significance of the doctor–patient dialogues for COVID-19 diagnosis.
Effect of Our Approach. Looking at Table 5, it’ s patently apparent that our method is superior to others. Particularly, ours is 0.2416, 0.2397, 0.2364, 0.2344, 0.2324, 0.2292, 0.2258, 0.2203, 0.2197, 0.2147, 0.2781, 0.2656, 0.2596, 0.2532, 0.2445, 0.1741, 0.1700, 0.1690, 0.1688, and 0.1683 higher than CAAD [35], COVID-CAPS [82], COVID-DA [83], COVID-ResNet [84], DCSL [85], COVNet [86], AnoDet [87], DLS [88], DeCovNet [89], DLQCTM [90], DenseNet-161[91], ResNet-34 [70], VGG-19 [66], ResNet-18 [70], VGG-16 [66], DGLM [92], LM3FT [93], MMCL [94], MEC [95], and GMMF [96], in the light of accuracy, respectively. In the light of precision, sensitivity, specificity, F1-Score, AUC, there are similar scenarios as the above. From above, our approach has more robust performances than the state-of-the-art approaches on our dataset. With the best performance, in terms of model size, none of our models exceeds M, and the batch time is around 1.14 seconds(s). In other words, our model processes a medical image of a lung in about 70 ms on average. This demonstrates that our model has good application perspectives, although it may seem a bit redundant and heavy. For above, these mean our approach can detect COVID-19 diagnose effectively.
5.3. Ablation study
In order to verify the reasonableness and effectiveness of each component of our attention machine, we develop the ablation experiment.
In Table 6, “Ours without Single” means a variant of Ours, which assigns the same weight to each neighbour and removes single-level modality attention; “Ours without Multiple” means a variant of Ours, which removes multiple-level modality attention and assigns the same weight to each meta-path; “Ours without Cross” means a variant of Ours, which assigns the same importance to each node and removes cross-level modality attention. We analyse the following two aspects:
Table 6.
The results of ablation study.
| Method | Accuracy | Precision | Sensitivity | Specificity | F1-score | AUC |
|---|---|---|---|---|---|---|
| Ours w/o cross | 0.9444 | 0.9462 | 0.9338 | 0.9442 | 0.9399 | 0.9577 |
| Ours w/o multiple | 0.9608 | 0.9523 | 0.9691 | 0.9659 | 0.9606 | 0.9818 |
| Ours w/o single | 0.9520 | 0.9513 | 0.9634 | 0.9622 | 0.9573 | 0.9768 |
| Ours | 0.9810 | 0.9889 | 0.9861 | 0.9859 | 0.9875 | 0.9908 |
Compared with “Ours”. From Table 5, it is quite apparent that our approach has better performances than others. In particular, ours is 0.0366, 0.0202, and 0.0290 better than “Ours w/o Cross”, “Ours w/o Multiple”, and “Ours w/o Single”, in term of accuracy, respectively. In terms of precision, sensitivity, specificity, F1-Score, AUC, there are similar scenarios as the above. As we can see, “Ours” is better than others. These suggest making full use of multimodal information helps us to improve COVID-19 diagnosis.
Compared with “Ours without Cross”. “Ours without Cross” is 0.0164 and 0.0076 lower than “Ours w/o Multiple”, and “Ours w/o Single”, in term of accuracy, respectively. In terms of precision, sensitivity, specificity, F1-Score, and AUC, there are similar scenarios. As we can see, “Ours without Cross” is worse than others. These suggest the importance of making joint use of the single-level modality and multiple-level modality information.
From the above, we get the conclusion in the following two aspects:
(1) It is apparent that the design of our attention mechanisms improves COVID-19 diagnosis.
(2) It is evident that the design of our cross-level modality mechanisms is better than our other attention mechanisms. This suggests that the design of cross-level modality mechanisms is robust and effective.
5.4. Discussion about knowledge attention mechanisms
In this subsection, we compare with some state-of-the-art graph attention mechanisms, including network embedding approach and graph neural network-based methodology, to validate the effectiveness of the presented attention mechanisms. Firstly, we introduce state-of-the-art graph attention mechanisms. Then, we analyse discussion experiment results.
5.4.1. State-of-the-art graph attention mechanisms
We review the theory and implementation of state-of-the-art graph attention mechanisms, like the following:
DeepWalk [97] is a random walk-based network embedding method. Here, we perform DeepWalk on the entire graph and ignore the nodes. Obviously, this method is able to be designed for homogeneous graphs.
ESim [98] captures multi-modal information from multiple meta-paths and is a graph embedding method. We assign the weights from our attention mechanisms to ESim in our discussion because it is hard to search for weights for the meta-paths set.
metapath2vec [99] performs a random walk based on a meta-path and uses skip-gram to embed knowledge graphs. This is a graph embedding method. Here, we report the best performance in the discussion and test all meta-paths.
HERec [100] devises a constraint policy for filtering node sequences. Besides, this is a graph embedding method and uses a skip-gram embedding knowledge graph. Here, we report the best performance in the discussion and test all the meta paths for this method.
GCN [101] designs for the graphs and is a semi-supervised graph convolutional network. Here we report the best performance and test all the meta-paths for GCN.
GAT [25] considers the attention mechanism on the graphs and is a semi-supervised neural network. Here we report the best performance in our discussion and test all the meta-paths for this method.
MAGNN [102] maps the heterogeneous node attribute information to the vector space of the same hidden layer, uses the attention mechanism to consider the semantic information inside the meta path, and applies the attention mechanism to aggregate information from multiple meta paths. Here we report the best performance in our discussion and test all the meta-paths for this method.
RGCN [103] applies the GCN framework to relational data modelling and employs sharing parameters techniques and a sparse matrix multiplications implementation in multiple graphs with a large number of relations. Obviously, this method is able to be designed for multiple homogeneous graphs. However, our graph attention mechanism is to consider building a relational model at a given heterogeneous graph. Here we report the best performance and test all the meta-paths for RGCN.
GATNE [104] classifies all node embeddings of heterogeneous graphs into three categories: base embeddings, edge embeddings, and attribute embeddings. Base embeddings and attribute embeddings are shared among different types of edges, while edge embeddings are computed through the aggregation of neighbourhood information and self-attention mechanisms. Different from the proposed graph attention mechanism that focuses on the relationship between data modalities, GATNE focuses on the relationship between different node embeddings and different attributes between nodes. Here we report the best performance and test all the meta-paths for GATNE.
HGAN [105] is a heterogeneous GNN. It learns metapath-specific node embeddings from different metapath-based homogeneous graphs and leverages the attention mechanism to combine them into one vector representation for each node. Different from the proposed graph attention mechanism that focuses on the relationship of cross-modalities, HGAN focuses only on the relationship of multiple modalities as a whole. Here we report the best performance and test all the meta-paths for HGAN.
HetGNN [106] first samples a fixed number of neighbours in the vicinity of an object via random walk with a restart. Then it performs within-type aggregation of these neighbours and designs a type-level attention mechanism for type-level aggregation. If we regard a type as a modality, different from the proposed graph attention mechanism that focuses on the relationship of cross-modalities, HetGNN focuses only on intra-modalities. Here we report the best performance and test all the meta-paths for HetGNN.
HGT [107] is a heterogeneous method that considers all possible by computing all possible meta-path based graphs and then performs graph convolution on the resulting graphs. Unlike the proposed graph attention mechanism driven by some meta-paths, HGT focuses on all relationships from all meta-paths. If meta-paths contain noise or bias, HGT may not work well. Here we report the best performance and test all the meta-paths for HGT.
MMGCN [108] is a graph-based algorithm. It devises a model-specific bipartite graph based on user–item interactions for each modality to learn representations of user preferences on different modalities. After that, it aggregates all model-specific representations to obtain the representations of users or items for prediction. Different from the proposed graph attention mechanism that focuses on the relationship of cross-modalities, MMGCN focuses on the relationship of model-specific modalities. Here we report the best performance and test all the meta-paths for MMGCN.
5.4.2. Analysis of attention mechanisms
In this sub-sub section, Ours(DeepWalk) means a variant of Ours, which only using DeepWalk and not using our graph attention mechanisms; Ours(ESim) means a variant of Ours, which only using ESim and not using our graph attention mechanisms; Ours(metapath2vec) means a variant of Ours, which only using metapath2vec and not using our graph attention mechanisms; Ours(HERec) means a variant of Ours, which only using HERec and not using our graph attention mechanisms; Ours(GCN) means a variant of Ours, which only using GCN and not using our graph attention mechanisms; Ours(GAT) means a variant of Ours, which only using GAT and not using our graph attention mechanisms; Ours(MAGNN) means a variant of Ours, which only using MAGNN and not using our graph attention mechanisms; Ours(RGCN) means a variant of Ours, which only using RGCN and not using our graph attention mechanisms; Ours(GATNE) means a variant of Ours, which only using GATNE and not using our graph attention mechanisms; Ours(HGAN) means a variant of Ours, which only using HGAN and not using our graph attention mechanisms; Ours(Het-GNN) means a variant of Ours, which only using HetGNN and not using our graph attention mechanisms; Ours(HGT) means a variant of Ours, which only using HGT and not using our graph attention mechanisms; Ours(MMGCN) means a variant of Ours, which only using MMGCN and not using our graph attention mechanisms.
Based on Table 7, we can see that our attention mechanisms achieve the best performance. Specifically, Our is 0.1347, 0.1322, 0.1311, 0.1287, 0.1144, 0.1140, 0.1120, 0.1094, 0.1074, 0.1045, 0.0474, 0.0206, and 0.0177 higher than Ours(DeepWalk), Ours(Esim), Ours(metapath2vec), Ours(HE-Rec), Ours(GCN), Ours(GAT), Ours(MAGNN), Ours (RGCN), Ours (GATNE), Ours (HGAN), Ours (HetGNN), Ours (HGT), and Ours (MMGCN), in term of accuracy, respectively. In terms of precision, sensitivity, specificity, F1-Score, AUC, there are similar scenarios as the above. On the other hand, from Table 7, we compare the model size of our knowledge attention mechanism with the model size of others: in the case of best performance, the model size of our knowledge attention mechanism does not exceed M. Compared to the heaviest model (i.e., Ours (HGAN)), our model parameters are about M smaller, and the performance of our model increases all by almost 0.1. Besides, the batch time of our model (i.e., per image) is not the optimal one, but it can satisfy the requirements of real-time applications (less than 100 ms to process each image [112]).
Table 7.
The results of discussion about knowledge attention mechanisms. “M” means . Batch time means the runtime of each batch in the model testing.
| Method | Accuracy | Precision | Sensitivity | Specificity | F1-score | AUC | The model size of knowledge attention mechanisms | Batch time |
|---|---|---|---|---|---|---|---|---|
| Ours (DeepWalk [97]) | 0.8463 | 0.8549 | 0.8506 | 0.8867 | 0.8528 | 0.8507 | 23M | 0.40 s |
| Ours (Esim [98]) | 0.8488 | 0.8567 | 0.8545 | 0.8869 | 0.8556 | 0.8552 | 24.3M | 0.35 s |
| Ours (metapath2vec [99]) | 0.8499 | 0.8628 | 0.8554 | 0.8912 | 0.8591 | 0.8568 | 24M | 0.34 s |
| Ours (HERec [100]) | 0.8523 | 0.8637 | 0.8638 | 0.8938 | 0.8638 | 0.8658 | 23.75M | 0.32 s |
| Ours (GCN [101]) | 0.8666 | 0.8701 | 0.8703 | 0.8991 | 0.8702 | 0.8678 | 25.4M | 0.46 s |
| Ours (GAT [25]) | 0.8670 | 0.8715 | 0.8724 | 0.8992 | 0.8719 | 0.8766 | 25.4M | 0.97 s |
| Ours (MAGNN [102]) | 0.8690 | 0.8764 | 0.8736 | 0.8992 | 0.8750 | 0.8772 | 37.2M | 1.48 s |
| Ours (RGCN [103]) | 0.8716 | 0.8716 | 0.8714 | 0.8913 | 0.8814 | 0.8744 | 44M | 1.24 s |
| Ours (GATNE [104]) | 0.8736 | 0.8726 | 0.8734 | 0.8955 | 0.8839 | 0.8764 | 44.6M | 1.54 s |
| Ours (HGAN [105]) | 0.8765 | 0.8746 | 0.8745 | 0.8960 | 0.8852 | 0.8772 | 47.3M | 2.27 s |
| Ours (HetGNN [106]) | 0.9336 | 0.8764 | 0.9090 | 0.9344 | 0.9045 | 0.9459 | 42.1M | 1.35 s |
| Ours (HGT [107]) | 0.9604 | 0.8764 | 0.9444 | 0.9331 | 0.9039 | 0.9536 | 41M | 1.50 s |
| Ours (MMGCN [108]) | 0.9633 | 0.8764 | 0.9466 | 0.9371 | 0.9057 | 0.9539 | 46.7M | 2.13 s |
| Ours | 0.9810 | 0.9889 | 0.9861 | 0.9859 | 0.9875 | 0.9908 | 39.4M | 1.14 s |
From the above observation, for traditional graph embedding methods, ESim performs better than metapath2vec. Besides, it is known that ESim is able to take multiple meta-paths. In general, graph neural network-based methods combine feature and structure information, such as GCN, GAT, and MAGNN. This method generally performs better. To delve into these methods, MAGNN can accurately weigh information and improve learning embedding performance compared to just the average node neighbour. Compared to MAGNN, our attention mechanism can capture the more valuable multi-modal information successfully and shows its superiority. Compared to some advanced models, including RGCN, GATNE, HGAN, HetGNN, HGT, and MMGCN, our attention mechanism can capture the cross-modalities information successfully and shows its superiority. Besides, according to Table 6, without single-level modality attention (“Ours without Single”), multiple-level modality attention (“Ours without Multiple”), cross-level modality attention (“Ours without Cross”), the performance of these becomes worse than ours, which indicates the importance of modelling the attention mechanism on both of the single-level modality and multiple-level modality information, and joint of multi-modal information.
Through the above analysis, we can find that the proposed knowledge attention mechanisms achieve the best performance among the state-of-the-art graph attention mechanisms. The experimental results also demonstrate that it is essential to capture and joint the importance of single-level modality information and multiple-level modality information in a multi-modal knowledge graph.
5.5. Compared to different temporal convolution networks
In this subsection, we compare with some state-of-the-art temporal convolution networks to verify the effectiveness of the proposed temporal convolution networks. Firstly, we introduce state-of-the-art temporal convolution networks. Then, we analyse the comparison experiment results.
State-of-The-Art Temporal Convolution Networks We review the theory and implementation of state-of-the-art temporal convolution networks, like the following:
TCN [72] use dilated convolutions to solve the global information of the entire input sequence and sets the Residual block for further feature extraction.
TrellisNet [109] is similar to TCN, but it is different in the weight sharing mechanism and hidden layer state calculation process. Each layer of TrellisNet can be regarded as performing a one-dimensional convolution operation on the hidden state sequence and then convolution. The output is passed to the activation function.
SA-TCN [110] is a TCN-based model embedded with a temporal self-attention block. Each block extracts a global temporal attention mask from the hidden representation laying between the encoder and decoder. Instead, our model utilizes information from other blocks to boost the representation of one block. Obviously, there is a clear difference between these two models.
TCAN [111] is also combines temporal convolutional network and attention mechanism. Its temporal attention can integer internal correlative features under the condition of satisfying sequential characteristics. We have a similar idea to TCAN, but the implementation is slightly different. TCAN uses a conventional convolution operation to extract features and then employs a residual network to augment the features. Instead, we adopt causal convolutions to extract and boost the features from one block.
Analysis of Temporal Convolution Networks In this subsection, Ours(TCN) means a variant of Ours, which only using TCN and not using our temporal convolution networks; Ours(TrellisNet) means a variant of Ours, which only using TrellisNet and not using our temporal convolution networks; Ours(SA-TCN) means a variant of Ours, which only using SA-TCN and not using our temporal convolution networks; Ours(TCAN) means a variant of Ours, which only using TCAN and not using our temporal convolution networks.
From Table 8, it is apparent that our approach has better performances than others. Specifically, ours is 0.1131, 0.1022, 0.0956, and 0.0940 higher than Ours (TCN), Ours (TrellisNet), Ours (SA-TCN), and Ours (TCAN), in term of accuracy, respectively. In terms of precision, sensitivity, specificity, F1-Score, AUC, there are similar scenarios as the above. On the other hand, we compare the model size of our temporal convolution network with the model size of others: in the case of best performance, the model size of our temporal convolution network is required as M, which is the second smallest. Besides, the batch time of our model is the smallest. Thus, our method has more convincing performance than other state-of-the-art temporal convolution networks for COVID-19 diagnosis.
Table 8.
The results of discussion about temporal convolution networks. “M” means . Batch time means the runtime of each batch in the model testing.
| Method | Accuracy | Precision | Sensitivity | Specificity | F1-score | AUC | The model size of temporal convolution networks | Batch time |
|---|---|---|---|---|---|---|---|---|
| Ours (TCN [72]) | 0.8679 | 0.8688 | 0.8641 | 0.8909 | 0.8664 | 0.8675 | 28.6M | 1.77 s |
| Ours (TrellisNet [109]) | 0.8788 | 0.8824 | 0.8761 | 0.9036 | 0.8792 | 0.8842 | 87M | 1.69 s |
| Ours (SA-TCN [110]) | 0.8854 | 0.8995 | 0.8837 | 0.9087 | 0.9040 | 0.8869 | 54M | 1.31 s |
| Ours (TCAN [111]) | 0.8870 | 0.9302 | 0.8981 | 0.9248 | 0.9275 | 0.8895 | 33M | 1.26 s |
| Ours | 0.9810 | 0.9889 | 0.9861 | 0.9859 | 0.9875 | 0.9908 | 32M | 1.14 s |
5.6. Compared to different spatial–temporal networks
In this subsection, we compare with some state-of-the-art spatial–temporal networks to verify the effectiveness of the proposed model. Firstly, we introduce state-of-the-art spatial–temporal networks. Then, we analyse the comparison experiment results.
State-of-The-Art spatial–temporal Networks We review the theory and implementation of state-of-the-art spatial–temporal networks, like the following:
DyHAN [113] is a dynamic heterogeneous graph embedding method with hierarchical attention that learns node embeddings leveraging both structural heterogeneity and temporal evolution.
CE-LSTM [114] is an event-flow serializing method to learn the representation from heterogeneous spatial–temporal graph through encoding the evolution of dynamic heterogeneous graph into a special language pattern such as word sequence in a corpus.
Analysis of spatial–temporal Networks From Table 9, it is apparent that our approach has better performances than others. Specifically, ours is 0.0802 and 0.0649 higher than DyHAN and CE-LSTM, in terms of accuracy, respectively. In terms of precision, sensitivity, specificity, F1-Score, AUC, there are similar scenarios as the above. On the other hand, we compare our model’s model size with the model size of others: in the case of best performance, the model size and batch time of our model are the smallest. Therefore, our method has more effective performance than other state-of-the-art spatial–temporal networks for COVID-19 diagnosis.
Table 9.
The results of discussion about spatial–temporal networks. “M” means . Batch time means the runtime of each batch in the model testing.
5.7. Parameters experiments
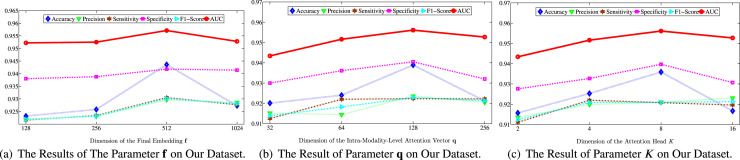
In this subsection, we investigate the sensitivity of parameters and report the performance results on our dataset with various settings in Fig. 6.
Fig. 6.
The result of parameters experiments.
Different Dimension of The Final Embedding For testing the final embedding of , the results are shown in Fig. 6(a). We can see that as the embedding dimension grows, the performance first goes up and then starts to decrease slowly. This is because our method requires a suitable dimension to encode multimodal information, and larger dimensions may introduce additional redundancy.
Different Dimension of Multiple-Level Modality Attention Vector As multimodal attention’s capability is influenced by the dimensions of the multimodal attention vector , we expose experimental results for different dimensions. The results are shown in Fig. 6(b). We can see that when the dimension of is set to , the performance of our method reaches its best performance as the number of dimensions of the multilevel modal attention vector grows. Afterward, as the performance of our method begins to degrade, overfitting may occur.
Number of attention head To examine the effects of multi-head attention, we investigate the performance of various attention head approaches. The results are shown in Fig. 6(c). From the results, it can be seen that the number of attention heads usually improves the performance of our method. is the best performance. After that, the performance gradually decreases.
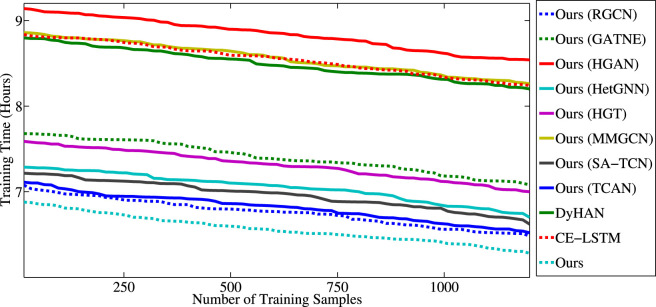
5.8. Scalability analysis
In this subsection, we investigate our model’s scalability and compared methods deployed on different numbers of training samples for optimization. Fig. 7 shows the speedup, w.r.t., the number of training samples on the proposed dataset.
Fig. 7.
Scalability. The training time decreases as the number of training samples increases. Ours takes less training time to converge compared with others.
From Fig. 7, our model is entirely scalable as the training time decreases significantly when we add up the number of training samples, and finally, the proposed model takes less than six hours to converge with 1200 training samples. We also find that our method’s training speed increases almost linearly as the number of training samples increases, while other methods converge slower. Besides the state-of-the-art performance, ours is also scalable enough to be adopted in practice.
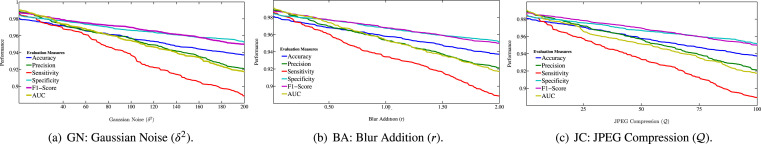
5.9. Robustness analysis
In this subsection, in order to investigate the robustness of our model under study, we consider the alterations for all medical chest images in our proposed dataset. In other words, we think the most common alterations that can occur when working on digital images in the medical sector:
Gaussian Noise (GN) simulates the possible effect of a wrong manipulation of the microscopic slide (e.g., too much dye has been used for contrast) [115]. We considered different values for the variance of the noise.
Blur Addition (BA) may occur due to a small move of the tool causing a focus loss. We vary the radius of blurring.
JPEG Compression (JC) may occur when images are transferred in a lossy manner. We vary compression value .
From Fig. 8, the performance profiles of our model are similar under different noisy data conditions. Moreover, the performance degradation does not exceed 0.12. It suggests that our model has a good ability to resist noise. Further, it demonstrates the robustness of our model.
Fig. 8.
The result of robustness analysis. Performance modification using the altered data input over our model .
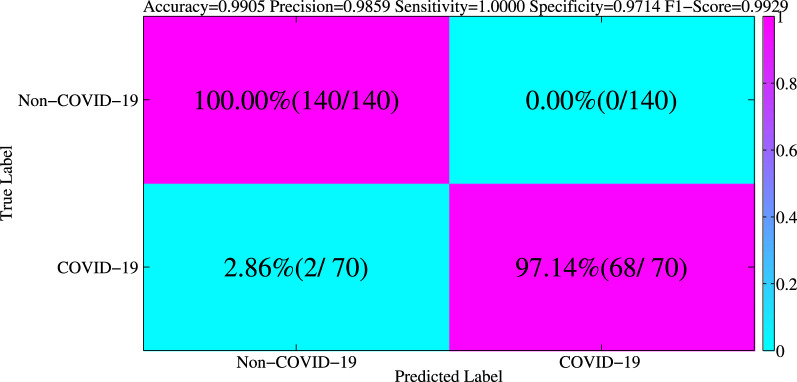
5.10. Generalization about asymptomatic infection cases
A rising number of asymptomatic patients with a confirmed diagnosis of COVID-19 has been reported. Asymptomatic infections are those patients who do not have clinical symptoms associated with COVID-19 (e.g., fever, cough, sore throat, etc.) but who test positive for antibodies on RTPCR or in specimens such as the respiratory tract [116]. Asymptomatic patients can be the source of infection and carry some risk of transmission. Therefore, it is urgent to recognize asymptomatic infected patients from non-COVID-19 patients. Asymptomatic patients with COVID-19 pneumonia have unilateral ground-glass opacities on medical imaging of the lungs [117]. Therefore, the chest images of asymptomatic COVID-19 patients have several imaging features, and these images are of significant diagnostic value in close contact with an infected person.
In this subsection, to better verify the robust performance of our algorithm, we extend our model to a specific classification task, where we use the trained model to directly classify the asymptomatic infection (COVID-19) cases and non-COVID-19 cases. To this end, we collect cases from asymptomatic infection (COVID-19) patients with the similar methods mentioned in Section 3, and we randomly choose non-COVID-19 cases from our test dataset. In this way, we get the new test set in this generalization experiment. We analyse the generalization results shown in Fig. 9, in terms of accuracy, precision, F1-score, sensitivity, specificity. From Fig. 9, our model considers only asymptomatic infection cases as non-COVID-19 patients, with a 97.14% specificity. It demonstrates the good performance of our model on this particular task. Furthermore, it suggests that our model has good generalization performance.
Fig. 9.
The confusion matrix of classifying the asymptomatic infection (COVID-19) cases and non-COVID-19 cases.
5.11. Error analysis
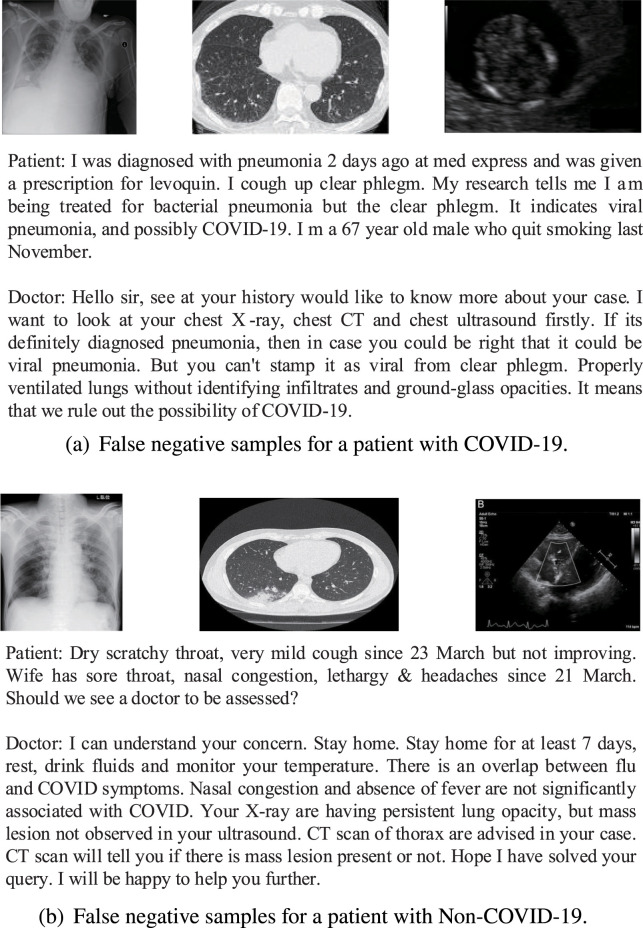
We conducted error analysis on our model results on the test data and identified two types of dominating errors as shown in Fig. 10.
Fig. 10.
Error analysis.
In Fig. 10(a), our model predicts that the patient has COVID-19 because there are words in the doctor–patient dialogues: ground-glass opacities, infiltrates, and viral pneumonia. These words are closely related to COVID-19, although some of them are denial terms, which leads to confusion and errors in our model.
As shown in Fig. 10(b), our model falsely identifies this as a non-COVID-19 patient. In this kind of case, we find that there are words in doctor–patient dialogues: not observed, not significantly. In this case, we also observe that the doctor gives words like lung opacity, which may cause our model not to identify and classify it correctly. In summary, these errors provide suggestions for future work on our model.
6. Conclusion
In this paper, we propose a novel COVID-19 diagnosis approach, which can fully take advantage of multi-modal medical information to build up the performance. Our approach gains the precise embedding of multi-modal medical information and exports medical knowledge directly from a deep learning-based network through learning from a given knowledge graph. With the learned deep learning-based network and medical embedding, our approach can yield the knowledge-based attention feature vector that can mainly contribute to the improved performance of diagnostic models. Experimental results demonstrate the effectiveness and robustness of our approach for the task of COVID-19 diagnosis. We believe and hope this work can provide insights to the researchers working in this area to shift the attention from only medical images to the doctor–patient dialogue and its corresponding medical images.
CRediT authorship contribution statement
Wenbo Zheng: Conceptualization, Methodology, Software, Validation, Investigation, Writing - original draft, Writing- review & editing. Lan Yan: Methodology, Investigation, Writing - original draft. Chao Gou: Conceptualization, Resources, Writing- review & editing, Project administration, Funding acquisition. Zhi-Cheng Zhang: Data curation,Validation, Software, Resources. Jun Jason Zhang: Supervision, Software, Validation, Investigation. Ming Hu: Data curation, Validation, Software, Resource. Fei-Yue Wang: Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank the General Hospital of the People’s Liberation Army and Wuhan Pulmonary Hospital for medical data and helpful advice on this research. This work is supported in part by the National Key R&D Program of China (2020YFB1600400), in part by the National Natural Science Foundation of China (61806198, 61533019, U1811463), in part by the Key Technologies Research and Development Program of Guangzhou, China (202007050002), and in part by the National Key Research and Development Program of China (No. 2018AAA0101502).
Biographies

Wenbo Zheng received his bachelor degree in software engineering from Wuhan University of Technology, Wuhan, China, in 2017. He is currently a PhD candidate in the School of Software Engineering, Xi’an Jiaotong University as well as the State Key Laboratory for Management and Control of Complex Systems, Institute of Automation, Chinese Academy of Sciences. His research interests include computer vision and machine learning.

Lan Yan received her bachelor degree from the University of Electronic Science and Technology of China in 2017. She is currently a PhD candidate in the School of Artificial Intelligence, University of Chinese Academy of Sciences as well as the State Key Laboratory for Management and Control of Complex Systems, Institute of Automation, Chinese Academy of Sciences. Her research interests include computer vision and pattern recognition.

Chao Gou received the B.S. degree from the University of Electronic Science and Technology of China, Chengdu, China, in 2012 and the Ph.D. degree from the University of Chinese Academy of Sciences (UCAS), Beijing, China 2017. From September 2015 to January 2017, he was supported by UCAS as a joint-supervision Ph.D. student in Rensselaer Polytechnic Institute, Troy, NY, USA. He is currently an Assistant Professor with the School of Intelligent Systems Engineering, Sun Yat-Sen University. His research interests include computer vision and machine learning.

Zhi-Cheng Zhang is currently a Senior Associate Doctor with the Seventh Medical Center, General Hospital of People’s Liberation Army, Beijing 100700, China.

Jun Jason Zhang received the B.E. and M.E. degrees in electrical engineering from Huazhong University of Science and Technology, Wuhan, China, in 2003 and 2005, respectively, and the Ph.D. degree in electrical engineering from Arizona State University, USA, in 2008. He is currently a Professor of electrical and computer engineering at Wuhan University. He authored/ co-authored over 70 peers reviewed publications. His research interests include signal processing and its applications.
Ming Hu is currently a Chief Physician and the Director of the Intensive Care Unit, Wuhan Pulmonary Hospital, Wuhan 430030, China.

Fei-Yue Wang received his Ph.D. degree in computer and systems engineering from the Rensselaer Polytechnic Institute, Troy, NY, USA, in 1990. He joined The University of Arizona in 1990 and became a Professor and the Director of the Robotics and Automation Laboratory and the Program in Advanced Research for Complex Systems. In 1999, he founded the Intelligent Control and Systems Engineering Center at the Institute of Automation, Chinese Academy of Sciences (CAS), Beijing, China, under the support of the Outstanding Chinese Talents Program from the State Planning Council, and in 2002, was appointed as the Director of the Key Laboratory of Complex Systems and Intelligence Science, CAS. In 2011, he became the State Specially Appointed Expert and the Director of the State Key Laboratory for Management and Control of Complex Systems. His current research focuses on methods and applications for parallel intelligence, social computing, and knowledge automation. He is a fellow of IEEE, INCOSE, IFAC, ASME, and AAAS. In 2007, he received the National Prize in Natural Sciences of China and became an Outstanding Scientist of ACM for his work in intelligent control and social computing. He received the IEEE ITS Outstanding Application and Research Awards in 2009 and 2011, respectively. In 2014, he received the IEEE SMC Society Norbert Wiener Award. Since 1997, he has been serving as the General or Program Chair of over 30 IEEE, INFORMS, IFAC, ACM, and ASME conferences. He was the President of the IEEE ITS Society from 2005 to 2007, the Chinese Association for Science and Technology, USA, in 2005, the American Zhu Kezhen Education Foundation from 2007 to 2008, the Vice President of the ACM China Council from 2010 to 2011, the Vice President and the Secretary-General of the Chinese Association of Automation from 2008-2018. He was the Founding Editor-in-Chief (EiC) of the International Journal of Intelligent Control and Systems from 1995 to 2000, the IEEE ITS Magazine from 2006 to 2007, the IEEE/CAA JOURNAL OF AUTOMATICA SINICA from 2014-2017, and the China’s Journal of Command and Control from 2015-2020. He was the EiC of the IEEE Intelligent Systems from 2009 to 2012, the IEEE TRANSACTIONS ON Intelligent Transportation Systems from 2009 to 2016, and is the EiC of the IEEE TRANSACTIONS ON COMPUTATIONAL SOCIAL SYSTEMS since 2017, and the Founding EiC of China’s Journal of Intelligent Science and Technology since 2019. Currently, he is the President of CAA’s Supervision Council, IEEE Council on RFID, and Vice President of IEEE Systems, Man, and Cybernetics Society.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.inffus.2021.05.015.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
Metadata of Our Dataset.
References
- 1.van de Haar J., Hoes L.R., Coles C.E., Seamon K., Fröhling S., Jäger D., Valenza F., de Braud F., De Petris L., Bergh J., Ernberg I., Besse B., Barlesi F., Garralda E., Piris-Giménez A., Baumann M., Apolone G., Soria J.C., Tabernero J., Caldas C., Voest E.E. Caring for patients with cancer in the COVID-19 era. Nature Med. 2020;26:665–671. doi: 10.1038/s41591-020-0874-8. [DOI] [PubMed] [Google Scholar]
- 2.Schett G., Manger B., Simon D., Caporali R. COVID-19 revisiting inflammatory pathways of arthritis. Nat. Rev. Rheumatol. 2020 doi: 10.1038/s41584-020-0451-z. doi: 10.1038/s41584-020-0451-z. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palayew A., Norgaard O., Safreed-Harmon K., Andersen T.H., Rasmussen L.N., Lazarus J.V. Pandemic publishing poses a new COVID-19 challenge. Nat. Hum. Behav. 2020 doi: 10.1038/s41562-020-0911-0. doi: 10.1038/s41562-020-0911-0. URL: [DOI] [PubMed] [Google Scholar]
- 4.Robilotti E.V., Babady N.E., Mead P.A., Rolling T., Perez-Johnston R., Bernardes M., Bogler Y., Caldararo M., Figueroa C.J., Glickman M.S., Joanow A., Kaltsas A., Lee Y.J., Lucca A., Mariano A., Morjaria S., Nawar T., Papanicolaou G.A., Predmore J., Redelman-Sidi G., Schmidt E., Seo S.K., Sepkowitz K., Shah M.K., Wolchok J.D., Hohl T.M., Taur Y., Kamboj M. Determinants of COVID-19 disease severity in patients with cancer. Nature Med. 2020 doi: 10.1038/s41591-020-0979-0. doi: 10.1038/s41591-020-0979-0. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z., Zhao W., Shi F., Qi L., Xie X., Wei Y., Ding Z., Gao Y., Wu S., Shi Y., Shen D., Liu J. A novel multiple instance learning framework for COVID-19 severity assessment via data augmentation and self-supervised learning. Med. Image Anal. 2021;69 doi: 10.1016/j.media.2021.101978. URL: https://www.sciencedirect.com/science/article/pii/S1361841521000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng N., Du S., Wang J., Zhang H., Cui W., Kang Z., Yang T., Lou B., Chi Y., Long H., Ma M., Yuan Q., Zhang S., Zhang D., Ye F., Xin J. Predicting COVID-19 in China using hybrid AI model. IEEE Trans. Cybern. 2020;50:2891–2904. doi: 10.1109/TCYB.2020.2990162. [DOI] [PubMed] [Google Scholar]
- 7.Ikizler T.A., Kliger A.S. Minimizing the risk of COVID-19 among patients on dialysis. Nat. Rev. Nephrol. 2020 doi: 10.1038/s41581-020-0280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39:2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 9.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nature Med. 2020 doi: 10.1038/s41591-020-0944-y. doi: 10.1038/s41591-020-0944-y. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Müller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.-E., Eils R. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nature Biotechnol. 2020 doi: 10.1038/s41587-020-0602-4. doi: 10.1038/s41587-020-0602-4. URL: [DOI] [PubMed] [Google Scholar]
- 11.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jing L., Zhang M., Huang X., Xiao Y., Cao H., Chen Y., Ren T., Wang F., Xiao Y., Huang S., Tan X., Huang N., Jiao B., Cheng C., Zhang Y., Luo A., Mombaerts L., Jin J., Cao Z., Li S., Xu H., Yuan Y. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020 doi: 10.1038/s42256-020-0180-7. doi: 10.1038/s42256-020-0180-7. URL: [DOI] [Google Scholar]
- 12.Abdel-Basset M., Ding W., Abdel-Fatah L. The fusion of internet of intelligent things (ioit) in remote diagnosis of obstructive sleep apnea: A survey and a new model. Inf. Fusion. 2020;61:84–100. doi: 10.1016/j.inffus.2020.03.010. URL: http://www.sciencedirect.com/science/article/pii/S1566253519307043. [DOI] [Google Scholar]
- 13.Zhou L., Li Z., Zhou J., Li H., Chen Y., Huang Y., Xie D., Zhao L., Fan M., Hashmi S., Abdelkareem F., Eiada R., Xiao X., Li L., Qiu Z., Gao X. A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 diagnosis. IEEE Trans. Med. Imaging. 2020;39(8):2638–2652. doi: 10.1109/TMI.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Z., Wei B., Hong Y., Li T., Cong J., Zhu X., Wei H., Zhang W. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans. Med. Imaging. 2020;39:2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 15.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2021;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 16.Bi W.L., Hosny A., Schabath M.B., Giger M.L., Birkbak N.J., Mehrtash A., Allison T., Arnaout O., Abbosh C., Dunn I.F., Mak R.H., Tamimi R.M., Tempany C.M., Swanton C., Hoffmann U., Schwartz L.H., Gillies R.J., Huang R.Y., Aerts H.J.W.L. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA: Cancer J. Clin. 2019;69:127–157. doi: 10.3322/caac.21552. URL: https://acsjournals.onlinelibrary.wiley.com/doi/abs/10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mei X., Lee H.-C., Diao K.-y., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., Bernheim A., Mani V., Calcagno C., Li K., Li S., Shan H., Lv J., Zhao T., Xia J., Long Q., Steinberger S., Jacobi A., Deyer T., Luksza M., Liu F., Little B.P., Fayad Z.A., Yang Y. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nature Med. 2020 doi: 10.1038/s41591-020-0931-3. doi: 10.1038/s41591-020-0931-3. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang W., Yao J., Chen A., Lv Q., Zanin M., Liu J., Wong S., Li Y., Lu J., Liang H., Chen G., Guo H., Guo J., Zhou R., Ou L., Zhou N., Chen H., Yang F., Han X., Huan W., Tang W., Guan W., Chen Z., Zhao Y., Sang L., Xu Y., Wang W., Li S., Lu L., Zhang N., Zhong N., Huang J., He J. Early triage of critically ill COVID-19 patients using deep learning. Nature Commun. 2020;11:3543. doi: 10.1038/s41467-020-17280-8. doi: 10.1038/s41467-020-17280-8. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enea H., Colby K.M. Proceedings of the 3rd International Joint Conference on Artificial Intelligence. Morgan Kaufmann Publishers Inc.; San Francisco, CA, USA: 1973. Idiolectic language-analysis for understanding doctor-patient dialogues; pp. 278–284. [Google Scholar]
- 20.Quiroz J.C., Laranjo L., Kocaballi A.B., Berkovsky S., Rezazadegan D., Coiera E. Challenges of developing a digital scribe to reduce clinical documentation burden. Npj Digit. Med. 2019;2:114. doi: 10.1038/s41746-019-0190-1. doi: 10.1038/s41746-019-0190-1. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furfaro F., Vuitton L., Fiorino G., Koch S., Allocca M., Gilardi D., Zilli A., D’Amico F., Radice S., Chevaux J.-B., Schaefer M., Chaussade S., Danese S., Peyrin-Biroulet L. SFED Recommendations for IBD endoscopy during COVID-19 pandemic: Italian and french experience. Nature Reviews Gastroenterology & Hepatology. 2020 doi: 10.1038/s41575-020-0319-3. doi: 10.1038/s41575-020-0319-3. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mei Q., Li J., Du R., Yuan X., Li M., Li J. Assessment of patients who tested positive for COVID-19 after recovery. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30433-3. URL: http://www.sciencedirect.com/science/article/pii/S1473309920304333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butow P.N., Dunn S.M., Tattersall M.H.N., Jones Q.J. Computer-based interaction analysis of the cancer consultation. Br. J. Cancer. 1995;71:1115–1121. doi: 10.1038/bjc.1995.216. doi: 10.1038/bjc.1995.216. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drach L.L., Hansen D.A., King T.M., Sibinga E.M.S. Communication between neonatologists and parents when prognosis is uncertain. Journal of Perinatology. 2020 doi: 10.1038/s41372-020-0673-6. doi: 10.1038/s41372-020-0673-6. URL: [DOI] [PubMed] [Google Scholar]
- 25.P. Veličković, G. Cucurull, A. Casanova, A. Romero, P. Liò, Y. Bengio, Graph attention networks, in: International Conference on Learning Representations, 2018, URL: https://openreview.net/forum?id=rJXMpikCZ.
- 26.Lee J.B., Rossi R.A., Kim S., Ahmed N.K., Koh E. Attention models in graphs: A survey. ACM Trans. Knowl. Discov. Data. 2019;13 doi: 10.1145/3363574. doi: 10.1145/3363574. [DOI] [Google Scholar]
- 27.Zhang Z., Cui P., Zhu W. Deep learning on graphs: A survey. IEEE Trans. Knowl. Data Eng. 2020 doi: 10.1109/TKDE.2020.2981333. [DOI] [Google Scholar]
- 28.Wu Z., Pan S., Chen F., Long G., Zhang C., Yu P.S. A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2020:1–21. doi: 10.1109/TNNLS.2020.2978386. [DOI] [PubMed] [Google Scholar]
- 29.Xie R., Liu Z., Jia J., Luan H., Sun M. Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence. AAAI Press; 2016. Representation learning of knowledge graphs with entity descriptions; pp. 2659–2665. URL: http://dl.acm.org/citation.cfm?id=3016100.3016273. [Google Scholar]
- 30.Cai H., Zheng V.W., Chang K.C. A comprehensive survey of graph embedding: Problems, techniques, and applications. IEEE Trans. Knowl. Data Eng. 2018;30:1616–1637. doi: 10.1109/TKDE.2018.2807452. [DOI] [Google Scholar]
- 31.Peng Y.-x., Zhu W.-w., Zhao Y., Xu C.-s., Huang Q.-m., Lu H.-q., Zheng Q.-h., Huang T.-j., Gao W. Cross-media analysis and reasoning: advances and directions. Front. Inf. Technol. Electron. Eng. 2017;18:44–57. doi: 10.1631/FITEE.1601787. doi: 10.1631/FITEE.1601787. URL: [DOI] [Google Scholar]
- 32.Nguyen H.L., Vu D.T., Jung J.J. Knowledge graph fusion for smart systems: A survey. Inf. Fusion. 2020;61:56–70. doi: 10.1016/j.inffus.2020.03.014. URL: http://www.sciencedirect.com/science/article/pii/S1566253519307729. [DOI] [Google Scholar]
- 33.Ding W., Abdel-Basset M., Hawash H., Pedrycz W. Multimodal infant brain segmentation by fuzzy-informed deep learning. IEEE Trans. Fuzzy Syst. 2021:1. doi: 10.1109/TFUZZ.2021.3052461. [DOI] [Google Scholar]
- 34.W. Zheng, L. Yan, C. Gou, Z.-C. Zhang, J.J. Zhang, M. Hu, F.-Y. Wang, Learning to learn by yourself: Unsupervised meta-learning with self-knowledge distillation for COVID-19 diagnosis from pneumonia cases, International journal of Intelligent Systems, n/a, URL: https://onlinelibrary.wiley.com/doi/abs/10.1002/int.22449, 10.1002/int.22449, arXiv:https://onlinelibrary.wiley.com/doi/pdf/10.1002/int.22449. [DOI] [PMC free article] [PubMed]
- 35.Zhang J., Xie Y., Pang G., Liao Z., Verjans J., Li W., Sun Z., He J., Li Y., Shen C., Xia Y. Viral pneumonia screening on chest X-rays using confidence-aware anomaly detection. IEEE Trans. Med. Imaging. 2020:1. doi: 10.1109/TMI.2020.3040950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L., Wong A. 2020. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39:2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 38.Jin C., Chen W., Cao Y., Xu Z., Zhang X., Deng L., Zheng C., Zhou J., Shi H., Feng J. 2020. Development and evaluation of an AI system for COVID-19 diagnosis. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang H., Xia L., Yan F., Wan Z., Shi F., Yuan H., Jiang H., Wu D., Sui H., Zhang C., Shen D. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imaging. 2020;39:2606–2614. doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 40.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B., Shi F., Yuan H., Wei Y., Cao X., Gao Y., Wu D., Wang Q., Shen D. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39:2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 41.Roy S., Menapace W., Oei S., Luijten B., Fini E., Saltori C., Huijben I., Chennakeshava N., Mento F., Sentelli A., Peschiera E., Trevisan R., Maschietto G., Torri E., Inchingolo R., Smargiassi A., Soldati G., Rota P., Passerini A., Van Sloun R.J.G., Ricci E., Demi L. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/TMI.2020.2994459. (in press) [DOI] [PubMed] [Google Scholar]
- 42.X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri, R.M. Summers, Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 2097–2106.
- 43.Manning C.D., Surdeanu M., Bauer J., Finkel J., Bethard S.J., McClosky D. Association for Computational Linguistics (ACL) System Demonstrations; 2014. The Stanford CoreNLP Natural Language Processing Toolkit; pp. 55–60. URL: http://www.aclweb.org/anthology/P/P14/P14-5010. [Google Scholar]
- 44.Dhamodharavadhani S., Rathipriya R., Chatterjee J.M. COVID-19 mortality rate prediction for India using statistical neural network models. Frontiers in Public Health. 2020;8:441. doi: 10.3389/fpubh.2020.00441. URL: https://www.frontiersin.org/article/10.3389/fpubh.2020.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okereafor K., Ekong I., Okon Markson I., Enwere K. Fingerprint biometric system hygiene and the risk of COVID-19 transmission. JMIR Biomed. Eng. 2020;5 doi: 10.2196/19623. URL: http://biomedeng.jmir.org/2020/1/e19623/ [DOI] [Google Scholar]
- 46.Dotolo S., Marabotti A., Facchiano A., Tagliaferri R. A review on drug repurposing applicable to COVID-19. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa288. doi: 10.1093/bib/bbaa288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z., He G., Huang N., Chen H., Zhang S., Zhao Z., Zhao Y., Yang G., Yang S., Xiong H., Karuppiah T., Kumar S.S., He J., Xiong C. Impact of the inflow population from outbreak areas on the COVID-19 epidemic in Yunnan province and the recommended control measures: A preliminary study. Front. Public Health. 2020;8:860. doi: 10.3389/fpubh.2020.609974. URL: https://www.frontiersin.org/article/10.3389/fpubh.2020.609974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Q., Liu W., Sha D., Kumar S., Chang E., Arora V., Lan H., Li Y., Wang Z., Zhang Y., Zhang Z., Harris J.T., Chinala S., Yang C. An environmental data collection for COVID-19 pandemic research. Data. 2020;5 doi: 10.3390/data5030068. URL: https://www.mdpi.com/2306-5729/5/3/68. [DOI] [Google Scholar]
- 49.Kraemer M.U.G., Yang C.-H., Gutierrez B., Wu C.-H., Klein B., Pigott D.M., du Plessis L., Faria N.R., Li R., Hanage W.P., Brownstein J.S., Layan M., Vespignani A., Tian H., Dye C., Pybus O.G., Scarpino S.V. The effect of human mobility and control measures on the COVID-19 epidemic in China. Science. 2020;368:493–497. doi: 10.1126/science.abb4218. URL: https://science.sciencemag.org/content/368/6490/493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heymann D.L. Data sharing and outbreaks: best practice exemplified. Lancet. 2020;395:469–470. doi: 10.1016/S0140-6736(20)30184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W., Zhou T., Lu Q., Wang X., Zhu C., Sun H., Wang Z., Lo S.K., Wang F.-Y. Dynamic fusion-based federated learning for COVID-19 detection. IEEE Internet Things J. 2021:1. doi: 10.1109/JIOT.2021.3056185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. arXiv preprint arXiv:2003.11597. [Google Scholar]
- 53.Maguolo G., Nanni L. A critic evaluation of methods for COVID-19 automatic detection from X-ray images. Inf. Fusion. 2021;76:1–7. doi: 10.1016/j.inffus.2021.04.008. URL: https://www.sciencedirect.com/science/article/pii/S1566253521000816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J., Zhang Y., He X., Xie P. 2020. COVID-CT-Dataset: A CT scan dataset about COVID-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- 55.Born J., Brändle G., Cossio M., Disdier M., Goulet J., Roulin J., Wiedemann N. 2020. POCOVID-Net: Automatic detection of COVID-19 from a new lung ultrasound imaging dataset (POCUS) pp. 2676–2687. [DOI] [Google Scholar]
- 56.Tabik S., Gómez-Ríos A., Martín-Rodríguez J.L., Sevillano-García I., Rey-Area M., Charte D., Guirado E., Suárez J.L., Luengo J., Valero-González M.A., García-Villanova P., Olmedo-Sánchez E., Herrera F. COVIDGR Dataset and COVID-Sdnet methodology for predicting COVID-19 based on chest X-ray images. IEEE J. Biomed. Health Inf. 2020;24:3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang W., Zeng G., Tan B., Ju Z., Chakravorty S., He X., Chen S., Yang X., Wu Q., Yu Z., et al. 2020. On the generation of medical dialogues for COVID-19. arXiv preprint arXiv:2005.05442. [Google Scholar]
- 58.Wang X., Ji H., Shi C., Wang B., Ye Y., Cui P., Yu P.S. 2019. Heterogeneous Graph Attention Network; pp. 2022–2032. URL: [DOI] [Google Scholar]
- 59.Huang X., Ye Z., Peng Y. In: Image and Graphics. Zhao Y., Kong X., Taubman D., editors. Springer International Publishing; Cham: 2017. Attention-sharing correlation learning for cross-media retrieval; pp. 477–488. [Google Scholar]
- 60.P. Pezeshkpour, L. Chen, S. Singh, Embedding multimodal relational data for knowledge base completion, in: Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2018, pp. 3208–3218.
- 61.Oñoro-Rubio D., Niepert M., García-Durán A., Gonzalez R., López-Sastre R.J. 2017. Representation learning for visual-relational knowledge graphs.arXiv:1709.02314 Arxiv abs/1709.02314, http://arxiv.org/abs/1709.02314. [Google Scholar]
- 62.Garcia-Duran A., Dumančić S., Niepert M. Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics; Brussels, Belgium: 2018. Learning sequence encoders for temporal knowledge graph completion; pp. 4816–4821. URL: https://www.aclweb.org/anthology/D18-1516. [Google Scholar]
- 63.Zheng W., Yan L., Gou C., Wang F.-Y. KM4: Visual reasoning via knowledge embedding memory model with mutual modulation. Inf. Fusion. 2021;67:14–28. doi: 10.1016/j.inffus.2020.10.007. URL: https://www.sciencedirect.com/science/article/pii/S1566253520303766. [DOI] [Google Scholar]
- 64.W. Zheng, L. Yan, C. Gou, F.Y. Wang, Graph attention model embedded with multi-modal knowledge for depression detection, in: 2020 IEEE International Conference on Multimedia and Expo (ICME), 2020, pp. 1–6, 10.1109/ICME46284.2020.9102872. [DOI]
- 65.W. Zheng, L. Yan, C. Gou, F.Y. Wang, Webly supervised knowledge embedding model for visual reasoning, in: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2020, pp. 12442–12451, 10.1109/CVPR42600.2020.01246. [DOI]
- 66.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition.arXiv:1409.1556 arXiv e-prints, arXiv:1409.1556, http://arxiv.org/abs/1409.1556. [Google Scholar]
- 67.J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li, L. Fei-Fei, ImageNet: A Large-Scale Hierarchical Image Database, in: CVPR09, 2009.
- 68.Devlin J., Chang M.-W., Lee K., Toutanova K. 2019. BERT: Pre-training of deep bidirectional transformers for language understanding; pp. 4171–4186. URL: https://www.aclweb.org/anthology/N19-1423. [DOI] [Google Scholar]
- 69.Guo J., Lu S., Cai H., Zhang W., Yu Y., Wang J. Proceedings of the AAAI Conference on Artificial Intelligence, vol. 32. 2018. Long text generation via adversarial training with leaked information. URL: https://ojs.aaai.org/index.php/AAAI/article/view/11957. [Google Scholar]
- 70.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, in: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016.
- 71.Trivedi R., Sisman B., Dong X.L., Faloutsos C., Ma J., Zha H. Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (vol. 1: Long Papers) Association for Computational Linguistics; Melbourne, Australia: 2018. LinkNBed: Multi-graph representation learning with entity linkage; pp. 252–262. URL: https://www.aclweb.org/anthology/P18-1024. [Google Scholar]
- 72.A. van den Oord, S. Dieleman, H. Zen, K. Simonyan, O. Vinyals, A. Graves, N. Kalchbrenner, A. Senior, K. Kavukcuoglu, WaveNet: A generative model for raw audio, in: 9th ISCA Speech Synthesis Workshop, 2016, pp. 125.
- 73.Zhang T., Wang X., Xu X., Chen C.L.P. GCB-Net: Graph convolutional broad network and its application in emotion recognition. IEEE Trans. Affect. Comput. 2019:1. doi: 10.1109/TAFFC.2019.2937768. [DOI] [Google Scholar]
- 74.Liu Z., Chen C.L.P., Feng S., Feng Q., Zhang T. Stacked broad learning system: From incremental flatted structure to deep model. IEEE Trans. Syst. Man Cybern. A. 2021;51:209–222. doi: 10.1109/TSMC.2020.3043147. [DOI] [Google Scholar]
- 75.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser L.u., Polosukhin I. 2017. Attention is All You Need; pp. 5998–6008. URL: http://papers.nips.cc/paper/7181-attention-is-all-you-need.pdf. [Google Scholar]
- 76.Bai S., Kolter J.Z., Koltun V. 2018. An empirical evaluation of generic convolutional and recurrent networks for sequence modeling.arXiv:1803.01271 Arxiv abs/1803.01271 http://arxiv.org/abs/1803.01271. [Google Scholar]
- 77.T. Chen, W. Yu, R. Chen, L. Lin, Knowledge-embedded routing network for scene graph generation, in: Conference on Computer Vision and Pattern Recognition, 2019.
- 78.Chen T., Wu W., Gao Y., Dong L., Luo X., Lin L. Proceedings of the 26th ACM International Conference on Multimedia. ACM; New York, NY, USA: 2018. Fine-grained representation learning and recognition by exploiting hierarchical semantic embedding; pp. 2023–2031. URL: http://doi.acm.org/10.1145/3240508.3240523. [DOI] [Google Scholar]
- 79.X. Lin, C. Ding, J. Zeng, D. Tao, GPS-Net: Graph property sensing network for scene graph generation, in: Conference on Computer Vision and Pattern Recognition, 2020.
- 80.T. Gui, L. Zhu, Q. Zhang, M. Peng, X. Zhou, K. Ding, Z. Chen, Cooperative multimodal approach to depression detection in Twitter, in: Proceedings of the Thirty-Second AAAI Conference on Artificial Intelligence,(AAAI-19), 2019, pp. 110–117.
- 81.Kingma D.P., Ba J. 2014. Adam: A method for stochastic optimization.arXiv:1412.6980 arXiv e-prints, arXiv:1412.6980, http://arxiv.org/abs/1412.6980. [Google Scholar]
- 82.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit. Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. URL: https://www.sciencedirect.com/science/article/pii/S0167865520303512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., Niu S., Qiu Z., Wei Y., Zhao P., Yao J., Huang J., Wu Q., Tan M. 2020. COVID-DA: Deep domain adaptation from typical pneumonia to COVID-19. arXiv preprint arXiv:2005.01577. [Google Scholar]
- 84.Farooq M., Hafeez A. 2020. Covid-resnet: A deep learning framework for screening of covid19 from radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
- 85.Li T., Han Z., Wei B., Zheng Y., Hong Y., Cong J. 2020. Robust screening of COVID-19 from chest X-ray via discriminative cost-sensitive learning. arXiv preprint arXiv:2004.12592. [Google Scholar]
- 86.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. doi: 10.1148/radiol.2020200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pang G., Shen C., van den Hengel A. 2019. Deep anomaly detection with deviation networks; pp. 353–362. URL: [DOI] [Google Scholar]
- 88.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J., Lang G., Li Y., Zhao H., Liu J., Xu K., Ruan L., Sheng J., Qiu Y., Wu W., Liang T., Li L. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6:1122–1129. doi: 10.1016/j.eng.2020.04.010. URL: https://www.sciencedirect.com/science/article/pii/S2095809920301636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39:2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 90.Shi W., Peng X., Liu T., Cheng Z., Lu H., Yang S., Zhang J., Li F., Wang M., Zhang X., et al. Deep learning-based quantitative computed tomography model in predicting the severity of COVID-19: A retrospective study in 196 patients. Lancet. 2020 doi: 10.2139/ssrn.3546089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.G. Huang, Z. Liu, L. Van Der Maaten, K.Q. Weinberger, Densely connected convolutional networks, in: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017, pp. 2261–2269.
- 92.Lin G., Liao K., Sun B., Chen Y., Zhao F. Dynamic graph fusion label propagation for semi-supervised multi-modality classification. Pattern Recognit. 2017;68:14–23. doi: 10.1016/j.patcog.2017.03.014. URL: http://www.sciencedirect.com/science/article/pii/S0031320317301139. [DOI] [Google Scholar]
- 93.Luo Y., Wen Y., Tao D., Gui J., Xu C. Large margin multi-modal multi-task feature extraction for image classification. IEEE Trans. Image Process. 2016;25:414–427. doi: 10.1109/TIP.2015.2495116. [DOI] [PubMed] [Google Scholar]
- 94.Gong C., Tao D., Maybank S.J., Liu W., Kang G., Yang J. Multi-modal curriculum learning for semi-supervised image classification. IEEE Trans. Image Process. 2016;25(7):3249–3260. doi: 10.1109/TIP.2016.2563981. [DOI] [PubMed] [Google Scholar]
- 95.Illendula A., Sheth A. Companion Proceedings of the 2019 World Wide Web Conference. Association for Computing Machinery; New York, NY, USA: 2019. Multimodal emotion classification; pp. 439–449. URL: [DOI] [Google Scholar]
- 96.Angelou M., Solachidis V., Vretos N., Daras P. Graph-based multimodal fusion with metric learning for multimodal classification. Pattern Recognit. 2019;95:296–307. doi: 10.1016/j.patcog.2019.06.013. URL: http://www.sciencedirect.com/science/article/pii/S0031320319302444. [DOI] [Google Scholar]
- 97.Perozzi B., Al-Rfou R., Skiena S. Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM; New York, NY, USA: 2014. Deepwalk: Online learning of social representations; pp. 701–710. URL: http://doi.acm.org/10.1145/2623330.2623732. [DOI] [Google Scholar]
- 98.Shang J., Qu M., Liu J., Kaplan L.M., Han J., Peng J. 2016. Meta-path guided embedding for similarity search in large-scale heterogeneous information networks.arXiv:1610.09769 Arxiv abs/1610.09769, http://arxiv.org/abs/1610.09769. [Google Scholar]
- 99.Dong Y., Chawla N.V., Swami A. KDD ’17. ACM; 2017. Metapath2vec: Scalable representation learning for heterogeneous networks; pp. 135–144. [Google Scholar]
- 100.Shi C., Hu B., Zhao W.X., Yu P.S. Heterogeneous information network embedding for recommendation. IEEE Trans. Knowl. Data Eng. 2019;31:357–370. doi: 10.1109/TKDE.2018.2833443. [DOI] [Google Scholar]
- 101.T.N. Kipf, M. Welling, Semi-supervised classification with graph convolutional networks, in: International Conference on Learning Representations (ICLR), 2017.
- 102.Fu X., Zhang J., Meng Z., King I. Proceedings of the Web Conference 2020. Association for Computing Machinery; New York, NY, USA: 2020. MAGNN: Metapath aggregated graph neural network for heterogeneous graph embedding; pp. 2331–2341. URL: [DOI] [Google Scholar]
- 103.Schlichtkrull M., Kipf T.N., Bloem P., van den Berg R., Titov I., Welling M. In: The Semantic Web. Gangemi A., Navigli R., Vidal M.-E., Hitzler P., Troncy R., Hollink L., Tordai A., Alam M., editors. Springer International Publishing; Cham: 2018. Modeling relational data with graph convolutional networks; pp. 593–607. [Google Scholar]
- 104.Cen Y., Zou X., Zhang J., Yang H., Zhou J., Tang J. Association for Computing Machinery; New York, NY, USA: 2019. Representation Learning for Attributed Multiplex Heterogeneous Network; pp. 1358–1368. URL: [DOI] [Google Scholar]
- 105.Wang X., Ji H., Shi C., Wang B., Ye Y., Cui P., Yu P.S. The World Wide Web Conference. Association for Computing Machinery; New York, NY, USA: 2019. Heterogeneous graph attention network; pp. 2022–2032. URL: [DOI] [Google Scholar]
- 106.Zhang C., Song D., Huang C., Swami A., Chawla N.V. Association for Computing Machinery; New York, NY, USA: 2019. Heterogeneous Graph Neural Network; pp. 793–803. URL: [DOI] [Google Scholar]
- 107.Hu Z., Dong Y., Wang K., Sun Y. Association for Computing Machinery; New York, NY, USA: 2020. Heterogeneous Graph Transformer; pp. 2704–2710. URL: [DOI] [Google Scholar]
- 108.Wei Y., Wang X., Nie L., He X., Hong R., Chua T.-S. Proceedings of the 27th ACM International Conference on Multimedia. Association for Computing Machinery; New York, NY, USA: 2019. MMGCN: Multi-modal graph convolution network for personalized recommendation of micro-video; pp. 1437–1445. URL: [DOI] [Google Scholar]
- 109.S. Bai, J.Z. Kolter, V. Koltun, Trellis networks for sequence modeling, in: International Conference on Learning Representations (ICLR), 2019.
- 110.R. Dai, L. Minciullo, L. Garattoni, G. Francesca, F. Bremond, Self-Attention Temporal Convolutional Network for Long-Term Daily Living Activity Detection, in: 2019 16th IEEE International Conference on Advanced Video and Signal Based Surveillance (AVSS), 2019, pp. 1–7, 10.1109/AVSS.2019.8909841. [DOI]
- 111.Hao H., Wang Y., Xia Y., Zhao J., Shen F. 2020. Temporal convolutional attention-based network for sequence modeling. arXiv preprint arXiv:2002.12530. [Google Scholar]
- 112.Armbrust M., Das T., Torres J., Yavuz B., Zhu S., Xin R., Ghodsi A., Stoica I., Zaharia M. Proceedings of the 2018 International Conference on Management of Data. Association for Computing Machinery; New York, NY, USA: 2018. Structured streaming: A declarative api for real-time applications in apache spark; pp. 601–613. URL: [DOI] [Google Scholar]
- 113.Yang L., Xiao Z., Jiang W., Wei Y., Hu Y., Wang H. In: Advances in Information Retrieval. Jose J.M., Yilmaz E., Magalhães J., Castells P., Ferro N., Silva M.J., Martins F., editors. Springer International Publishing; Cham: 2020. Dynamic heterogeneous graph embedding using hierarchical attentions; pp. 425–432. [Google Scholar]
- 114.Li Y., Zhu Z., Kong D., Xu M., Zhao Y. Learning heterogeneous spatial-temporal representation for bike-sharing demand prediction. Proc. AAAI Conf. Artif. Intell. 2019;33:1004–1011. doi: 10.1609/aaai.v33i01.33011004. URL: https://ojs.aaai.org/index.php/AAAI/article/view/3890. [DOI] [Google Scholar]
- 115.Chatterjee S. Artefacts in histopathology. J. Oral Maxillofac Pathol. 2014;18(4):111–116. doi: 10.4103/0973-029X.141346. URL: https://www.jomfp.in/article.asp?issn=0973-029X;year=2014;volume=18;issue=4;spage=111;epage=116;aulast=Chatterjee;t=6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.u. Kim G., Kim M.-J., Ra S., Lee J., Bae S., Jung J., Kim S.-H. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin. Microbiol. Infect. 2020;26:948.e1–948.e3. doi: 10.1016/j.cmi.2020.04.040. URL: https://www.sciencedirect.com/science/article/pii/S1198743X20302688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang R., Gui X., Xiong Y. Comparison of clinical characteristics of patients with asymptomatic vs symptomatic coronavirus disease 2019 in Wuhan, China. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.10182. doi: 10.1001/jamanetworkopen.2020.10182. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metadata of Our Dataset.