Abstract
Background
In mid-December 2020, Israel started a nationwide mass vaccination campaign against coronavirus disease 2019 (COVID-19). In the first few weeks, medical personnel, elderly citizens, and patients with chronic diseases were prioritized. As such, patients with primary and secondary immunodeficiencies were encouraged to receive the vaccine. Although the efficacy of RNA-based COVID-19 vaccines has been demonstrated in the general population, little is known about their efficacy and safety in patients with inborn errors of immunity (IEI).
Objective
Our aim was to evaluate the humoral and cellular immune response to COVID-19 vaccine in a cohort of patients with IEI.
Methods
A total of 26 adult patients were enrolled, and plasma and peripheral blood mononuclear cells were collected from them 2 weeks following the second dose of Pfizer-BioNTech COVID-19 vaccine. Humoral response was evaluated by testing anti–SARS-CoV-2 spike (S) receptor-binding domain and antinucleocapsid antibody titers and evaluating neutralizing ability by inhibition of receptor-binding domain–angiotensin-converting enzyme 2 binding. Cellular immune response was evaluated by using ELISpot, estimating IL-2 and IFN-γ secretion in response to pooled SARS-CoV-2 S- or M-peptides.
Results
Our cohort included 18 patients with a predominantly antibody deficiency, 2 with combined immunodeficiency, 3 with immune dysregulation, and 3 with other genetically defined diagnoses. Twenty-two of them were receiving immunoglobulin replacement therapy. Of the 26 patients, 18 developed specific antibody response, and 19 showed S-peptide–specific T-cell response. None of the patients reported significant adverse events.
Conclusion
Vaccinating patients with IEI is safe, and most patients were able to develop vaccine-specific antibody response, S-protein–specific cellular response, or both.
Key words: Inborn errors of immunity, IEI, primary immunodeficiency disorders, PIDD, SARS-CoV-2, COVID-19, vaccine, Pfizer-BioNTech, CVID, XLA, NFKB1, STAT1-GOF, STAT3-LOF, HIES, inhibiting antibodies
Abbreviations used: ACE2, Angiotensin-converting enzyme 2; ALPS, Autoimmune lymphoproliferative syndrome; anti-N, Antinucleocapsid; anti-S, Antispike; AU, Arbitrary unit; AUC, Area under the curve; CID, Combined immunodeficiency; COVID-19, Coronavirus disease 2019; CVID, Common variable immunodeficiency; IEI, Inborn errors of immunity; IVIG, Intravenous immunoglobulin; NF-κB1, Nuclear factor-κB1; NFKB1-HI, Nuclear factor-κB1 haploinsufficiency; RBD, Receptor-binding domain; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; STAT, Signal transducer and activator of translation; STAT1-GOF, STAT1 gain-of-function; XLA, X-linked agammaglobulinemia
In late 2019 the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified and described as causing pneumonia outbreak, known as coronavirus-induced disease-19 (COVID-19).1 Emerging as a local outbreak in Wuhan, China, it had soon spread to cause a pandemic of acute respiratory syndrome that can result in significant morbidity and mortality.2 So far, despite recommendations for potential intervention strategies,3 severely affected patients benefit mostly from supportive treatment. Therefore, mass vaccination using highly effective anti–SARS-CoV-2 vaccines remains the best hope to protect against severe disease,4 , 5 limit viral spread,6 , 7 and hopefully end the pandemic.
While data are accumulating to support the effectiveness and safety of the newly developed anti–SARS-CoV-2 vaccines, initial studies enrolled mostly healthy volunteers, whereas data regarding defined patient populations are still being gathered. As such, patients with inborn errors of immunity (IEI) are of special interest for several reasons. First, recent reports have suggested that patients with IEI might be at increased risk of developing severe COVID-198 , 9 and could therefore benefit from a more “aggressive” immunization effort. On the other hand, their underlying immune abnormality might impair the ability of IEI patients to respond to the vaccine and develop anti–SARS-CoV-2 protective immunity, thus leading to questions regarding the benefit of the vaccination approach. In view of this, characterizing the immune response of IEI patients following SARS-CoV-2 vaccination is crucial, both for understanding their degree of protection and for formulating an optimal immunization regimen. Moreover, data gathered from analyses of the immune response of IEI patients to the anti–COVID-19 vaccine could be relevant to other patient populations, especially those with secondary and acquired immunodeficiency.
Anti–SARS-CoV-2 antibodies developed following infection have been shown to possess neutralizing activity both in vitro and in vivo,10, 11, 12, 13 and a potential benefit of using neutralizing mAbs in patients with mild-to-moderate disease has been suggested.14 , 15 Nevertheless, the longevity of the antibody response and, thus, the duration of humoral protection are as yet unknown, with several studies suggesting a trend toward declining antibody levels over time16 and others showing persistence of receptor-binding domain (RBD)-specific memory B cells.17 , 18 In parallel, several studies have highlighted the role of cellular immunity and the long-term anti–SARS-CoV-2 T-cell response.19 As such, effective T-cell response was shown to be associated with milder COVID-19,20 , 21 and robust and increasing cellular response was shown to develop over time, with specific antiviral T cells detected 6 to 8 months after infection.18 , 22 Interestingly, T cells against the closely related SARS-CoV virus were detected as long as 11 years after recovery,23 whereas no SARS-CoV antigen–specific memory B cells or antibodies were detected 6 years after infection.24 Therefore, although neutralizing antibodies play a role in protection against SARS-CoV-2 and an orchestrated adaptive immunity can limit disease severity,21 when estimating vaccine immunogenicity, one should evaluate both the humoral and cellular antivaccine immune response. Such an evaluation is of even greater importance in patients with IEI, as most of them show some level of impaired antibody production, and evaluation of their postvaccine antibody titers might give a false impression of unresponsiveness, thereby preventing patients from getting vaccinated.
In this article we have aimed to study a group of adult patients with IEI and evaluate their early humoral and cellular immune response to the Pfizer-BioNTech anti–COVID-19 vaccine. On the basis of our analyses, we were able to conclude that most patients with IEI respond safely to the vaccine and should therefore be vaccinated.
Methods
Study design
This study was approved by the institutional review board of the Tel Aviv Sourasky Medical Center and registered under ClinicalTrials.gov identifier NCT04724642. All adult patients with an established IEI diagnosis, who visited our clinic between the end of January and mid-March 2021 were offered the opportunity to join the study. After providing written informed consent, IEI patients and healthy controls were included in the study. Participating donors received 2 doses of the mRNA-based Pfizer-BioNTech COVID19 vaccine 3 weeks apart, and samples were collected 2 weeks after the second vaccine dose. For convalescent donors, samples were collected 2 weeks after they recovered from COVID-19 (defined as 10 days following a positive SARS-CoV-2 RT-PCR test and no symptoms for at least 3 days). All subjects who gave consent were included in this report; no subject was excluded.
PBMC isolation and stimulation
PBMCs were isolated by using Ficoll gradient density. Following isolation, cells were stored in liquid nitrogen for later use, including flow cytometry–based B-cell staining, and intracellular cytokine staining or ELISpot assay for evaluation of peptide-induced cytokine production.
Evaluation of humoral response
Serology
The presence of anti–SARS-CoV-2 IgG antibodies was evaluated by using a commercial automated SARS-CoV-2 IgG assay (Abbott, Sligo, Ireland). The chemiluminescent microparticle immunoassay provided qualitative and quantitative determination of anti–SARS-CoV-2 RBD IgG antibody levels (SARS-CoV-2 IgG II Quant, catalog no. 6S60, Abbott) or qualitative detection of antinucleocapsid antibodies (SARS-CoV-2 IgG, catalog no. 6R86, Abbott). To differentiate between vaccinated and convalescent donors, plasma samples were evaluated for the presence of both antispike (anti-S) IgG antibodies and antinucleocapsid (anti-N) IgG antibodies. The results were provided in arbitrary units (AU) per milliliter, as defined by the manufacturer as ranging between 0 and 40,000 AU/mL for anti-S antibodies (a level of >150 AU/mL was considered positive), and relative light units (RLU) for anti-N antibodies (a level of >1.4 RLU was considered positive).
Serum ELISA
For serum ELISA, high-binding 96-well ELISA plates (Corning, c9018) were coated with 1 μg/mL of RBD antigen in PBS 1× and held overnight at 4°C. The following day, the coating was discarded and washed with “wash buffer” containing PBS 1× and 0.05% Tween20, after which the plates were blocked for 2 hours at room temperature (RT) with 200 μL of “blocking buffer” containing PBS 1×, 3% BSA (MP Biomedicals, Santa Ana, Calif), 20 mM EDTA, and 0.05% Tween20 (Sigma-Aldrich, St Louis, Mo). Plasma samples were diluted 4-fold in blocking buffer, starting from 1:100 with 7 consecutive dilutions (1:100, 1:400, 1:1600, 1:6400, etc), and incubated for 1 hour at RT. The plates were then washed 3 times with washing buffer before the addition of secondary anti-IgG antibodies (Jackson ImmmunoResearch, West Grove, Pa, 109-035-088) conjugated to horseradish peroxidase diluted 1:5000 in blocking buffer and incubated for 45 minutes at RT. Following 4 additional washes with wash buffer and a final wash with PBS 1×, 100 μL of 3,3’,5,5’-tetramethylbenzidine (Abcam, Cambridge, United Kingdom) diluted 1:2 in double deionized water (DDW) was added to each well; absorbance was read after 25 minutes at 650 nm (using a BioTek 800 TS absorbance reader). Positive and negative controls were added to each plate, and the signal was normalized to the controls for each plate.
RBD-ACE2 inhibition assay
For angiotensin-converting enzyme 2 (ACE2)-RBD inhibition ELISA, high-binding 96-well plates were coated with 2 μg/mL of human ACE2 in PBS 1× and held overnight at 4°C. The next day, the plates were washed and blocked with blocking buffer for 2 hours at RT. Plasma samples were diluted 2-fold in blocking buffer starting from 1:10 and incubated with biotinylated RBD for 30 minutes at RT. The RBD-plasma mix was then applied to the ACE2-coated plates and incubated for 20 minutes, followed by 3 washes. Biotinylated RBD was detected with streptavidin conjugated to horseradish peroxidase (Jackson ImmmunoResearch, 016-030-084).
Flow cytometry anti-RBD B-cell receptor staining
PBMCs were rapidly thawed at 37°C and washed in 50 mL of warm RPMI 1640 medium. The cells were resuspended in 1% BSA in PBS 1× and 2 mM EDTA, and stained with anti–CD19 fluorescein isothiocyanate (FITC) (Miltenyi Biotec, Bergisch Gladbach, Germany, 130-113-645), anti–IgG phycoerythrin (PE) (Miltenyi Biotec, 130-119-878), anti–IgA VioBlue (Miltenyi Biotec, 130-113-479), and labeled biotinylated RBD via streptavidin-allophycocyanin (APC) (Miltenyi Biotec, 130-106-792).
Evaluation of cellular response: ELISpot assay
Following initial evaluation of cytokine production by flow cytometry and with the understanding that stimulation with a pooled M-peptide mix induces the strongest IFN-γ and IL-2 production (see Fig E3, A and B in this article's Online Repository at www.jacionline.org), further evaluation of the anti-S (antivaccine) cellular response was evaluated by using ELISpot assay for detection of peptide-induced IFN-γ and IL-2 secretion (Human IFN-γ/IL-2 Dual ELISpot, catalog no. 874.040.005S, Diaclone, Besançon, France) according to the manufacturer's instructions. For this purpose, donor cells were plated at 100,000 cells/100 μL and stimulated with the relevant peptides for 19 hours at 37°C. The cells were stimulated with spike (S) glycoprotein peptides for evaluation of antivaccine response (peptide concentration 0.9 nmol/mL), membrane (M) glycoprotein peptides for evaluation of previous exposure to SARS-CoV-2 and detection of convalescent samples (peptide concentration 0.9 nmol/mL), or phorbol myristate acetate/ionomycin as a control to confirm cell viability and responsiveness (phorbol myristate acetate concentration 5 ng/mL, ionomycin concentration 500 μg/mL) in addition to prestimulation trypan blue staining. Cytokine detection was evaluated by manual spot counting and confirmed by using ImageJ/Fiji software (https://imagej.net/Fiji). The peptides used for stimulation included a pool of lyophilized peptides of the viral S-glycoprotein or M-glycoprotein (Miltenyi’s PepTivator SARS-CoV-2 Prot_S and Prot_M). Per the manufacturer's information, these peptide pools consisted of 15-mer sequences with 11–amino acid overlap, covering the immunodominant sequence domains of the S-glycoprotein (amino acids 304-338, 421-475, 492-519, 683-707, 741-770, and785-802, and the sequence end 885-1273), and the complete sequence of the M-glycoprotein. On the basis of 2 control groups of prevaccinated individuals (n = 8) and convalescent mildly affected individuals (n = 4), we set the threshold for positive cellular response at 4 spots per well.
Statistical analysis
Continuous variables are presented as means with SDs and were compared by using either the Student t test or ANOVA as described below. A simple linear regression test was used to calculate correlation where indicated. All analyses were calculated using GraphPad software.
Results
Patient characteristics
A total of 26 adult IEI patients who had been followed in a single center at the Tel Aviv Souraski Medical Center were included (for patient characteristics, see Table I ). Patients were recruited after signing an informed consent form, and blood samples for plasma and PBMCs were collected 2 weeks after the second dose of the COVID-19 vaccine. The average patient age was 48.4 years, and the male-to-female ratio was 11:15 (57.6% females). IEI patients were classified into 3 major groups: (a) patients with X-linked agammaglobulinemia (XLA) (n = 4); (b) 17 patients without XLA but with a predominantly antibody deficiency, including 2 with nuclear factor-κB1 (NF-κB1) haploinsufficiency (NFKB1-HI), 3 with hypogammaglobulinemia, 1 with selective IgG2 deficiency, and 1 with combined immunodeficiency (CID); and (c) patients with immune dysregulation and other defined defects, including 2 autoimmune lymphoproliferative syndrome (ALPS)-like patients, 1 signal transducer and activator of translation-1 (STAT1) gain of function (STAT1-GOF) patient, 1 STAT3 loss-of-function (STAT3-LOF) patient, and 1 patient with complete complement C4 deficiency. All the patients with common variable immunodeficiency (CVID) fulfilled the European Society for Immunodeficiencies (ESID) 2019 criteria for diagnosis of probable CVID,25 except for 1 patient who did not have a significant history of infections and was classified as having hypogammaglobulinemia. While the ESID registry criteria require patients with CVID to have either poor antibody response or a low percent of switched memory B cells, we routinely base our diagnosis on an abnormal B-cell immunophenotyping with low switched memory B cells (except for 2 patients in this cohort who were diagnosed many years ago on the basis of very low immunoglobulin levels [IgG level ∼ 150 mg/dL] and recurrent sinopulmonary infections).
Table I.
Patient characteristics
| Patient no. | Age (y) | Sex | Underlying diagnosis | Genetics | Clinical manifestations/complications | IgRT | Immunomodulator/antibiotic |
|---|---|---|---|---|---|---|---|
| 1 | 40 | M | XLA | c.3G>A; p.M1I | Bronchiectasis | Y | None |
| 2 | 51 | M | XLA | c.952T>C; p.S318P | Bronchiectasis and conjunctivitis | Y | None |
| 3 | 49 | M | XLA | Yes∗ | Y | None | |
| 4 | 42 | M | XLA | c.1631+1G>T | Bronchiectasis, Haemophilus influenzae conjunctivitis | Y | None |
| Rit | 56 | F | Hypogammaglobulinemia | No | ↓Ig, B-cell lymphopenia, myasthenia gravis | Y | Rituximab |
| 5 | 37 | M | STAT1-GOF mutation | c.1310C>T; pT437I | CMC, recurrent oral ulcers | N | Ruxolitinib |
| 6 | 21 | F | ALPS-like disease | BCL6B VUS | LAD, ITP, AIN, AIHA | N | Rapamycin |
| 7 | 51 | M | CVID/ALPS-like disease | N | ↓Ig, LAD, pulmonary HTN, s/p splenectomy for ITP | Y | Prophylactic co-trimoxazole |
| 8 | 41 | M | STAT3-LOF mutation (HIES) | c.1144C>T; p.R382W | Pneumatocele, s/p partial lobectomy, after AVR d/t MRSA endocarditis | Y | Co-trimoxazole and azithromycin |
| 9 | 48 | M | CID | Negative | ↓Ig, massive splenomegaly, s/p DLBCL (-3 y) | Y | Prophylactic co-trimoxazole |
| 10 | 32 | F | NFKB1-HI | c.509TinsGGTGCAA; p.L170ins exon 7/24fs | Hypogammaglobulinemia, LAD, AIN | N | None |
| 11 | 72 | M | NFKB1-HI | None | N | None | |
| 12 | 36 | F | Complete C4 deficiency | Yes | Cryoglobulinemia, ↓Ig | Y | Rituximab (-2 y) |
| 13 | 27 | F | Selective IgG2 deficiency | No | ↓Ig, recurrent pneumonia | Y | None |
| 14 | 37 | F | CVID | No | ↓Ig, aHUS, recurrent pneumonia | Y | None |
| 15 | 38 | M | CVID | No | ↓Ig, IBD-like | Y | None |
| 16 | 39 | F | CVID | No | ↓Ig, NRH | Y | None |
| 17 | 45 | F | CVID | No | ↓Ig, T1D, lymphocytic infiltrates on GI biopsy specimens | Y | None |
| 18 | 46 | F | CVID | Negative | ↓Ig, recurrent pneumonia, history of Crohn-like disease | Y | None (azathioprine in the past) |
| 19 | 50 | F | CVID | No | ↓Ig, vitiligo | Y | None |
| 20 | 59 | F | CVID | No | ↓Ig | Y | None |
| 21 | 60 | F | CVID | No | ↓Ig, s/p breast cancer | Y | None |
| 22 | 64 | F | CVID | No | ↓Ig, IBD-like | Y | None |
| 23 | 65 | F | CVID | No | ↓Ig | Y | None |
| 24 | 67 | F | CVID | No | ↓Ig | Y | None |
| 25 | 72 | M | Hypogammaglobulinemia | No | ↓Ig | N | None |
| 26 | 73 | F | CVID | Negative | ↓Ig, lung nodules, sarcoma | Y | None |
| Con | 37 | F | Selective IgG1 and IgG3 deficiency | No | ↓Ig, recurrent pneumonia | Y | None |
aHUS, Atypical hemolytic uremic syndrome; AIHA, autoimmune hemolytic anemia; AIN, autoimmune neutropenia; AVR, aortic valve replacement; CMC, chronic mucocutaneous candidiasis; Con, convalescent; DLBCL, diffuse large B-cell lymphoma; d/t, due to; F, female; GI, gastrointestinal; HIES, hyper IgE syndrome; HTN, hypertension; IBD, inflammatory bowel disease; ↓Ig, hypogammaglobulinemia; IgRT, immunoglobulin replacement therapy; ITP, idiopathic thrombocytopenic purpura; LAD, lymphadenopathy; M, male; N, No; NRH, nodular regenerative hyperplasia; Rit, After rituximab; s/p, status post; STAT3-LOF, STAT3 loss-of-function; T1D, type I diabetes; VUS, Variant of uncertain significance; Y, yes.
Genetically confirmed but patient preferred to not publish the pathogenic variant.
In addition to the 26 patients with IEI, we also tested 1 non-IEI vaccinated patient who had been treated with rituximab, and 1 convalescent unvaccinated patient with selective antibody (IgG1 and IgG3) deficiency.
Of the 26 patients, 22 were receiving immunoglobulin replacement therapy (5 with subcutaneous immunoglobulin and 17 with intravenous immunoglobulin [IVIG]), including 1 patient with STAT3-LOF pathogenic variant and 1 ALPS-like patient. Preferably, patients on IVIG received their COVID-19 vaccine at least 1 week apart from their IVIG infusion (either before or after).
Safety
The vaccines were generally well tolerated, with limited injection site pain being the most common reported adverse event in 9 of 26 patients following the first dose of the vaccine. Following the second dose of the vaccine, 3 patients reported fever (including a patient with ALPS-like disease who experienced a body temperature of up to 39°C for 3 days). One patient with CVID reported unilateral axillary lymphadenopathy that lasted 5 days. The adverse events were similar to those previously described,4 and none of the patients reported long-lasting adverse effects.
Humoral response
Humoral response was evaluated by using commercially available serology assays, ELISA assays for detecting anti-RBD antibodies and neutralizing activity, and flow cytometry using fluorophore-conjugated recombinant RBD for detecting specific anti-RBD B cells.
Serology
To differentiate between vaccinated and convalescent donors, serum samples were tested for the presence of both anti-S and anti-N antibodies, under the assumption that convalescent individuals would test positive for both antibodies whereas vaccinated individuals would test positive for anti-S antibodies only. Four recently recovered mildly affected convalescent patients, 11 prevaccinated patients, and 11 healthy vaccinated donors were included.
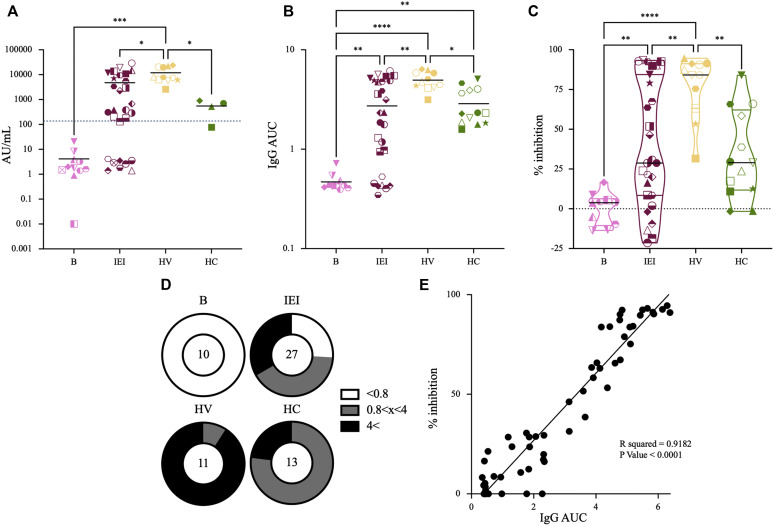
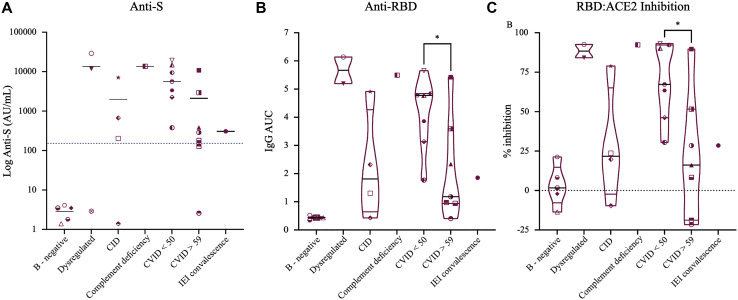
As can be seen in Fig 1 , A, vaccinated individuals had overall higher titers of anti-S antibodies compared with convalescent controls (11906.2 ± 7909.2 AU/mL vs 549.9 ± 350.9 AU/mL [P = .0149]). IEI patients showed a wide range of anti-S antibody titers, ranging from undetectable levels to normal-to-high titers (Figs 1, A and 2 , A and see Table E1 in this article's Online Repository at www.jacionline.org). Of the 26 patients with IEI, 18 tested positive for anti-S antibodies. As expected, “B-cell–negative” patients with XLA (and a non-IEI rituximab-treated patient ) failed to produce anti-S antibodies (2.86 ± 1.18 AU/mL). After exclusion of the 4 patients with XLA, 18 of 22 patients (81.8%) developed anti-S antibodies. Of those 18 patients, 13 were receiving immunoglobulin replacement therapy for diagnosis of CVID. The 4 non-XLA IEI patients who were tested negative for anti-S antibodies included 1 patient with ALPS-like disease, 1 patient with CID, and 2 patients with CVID. Analyzing the different groups of patients with IEI (Fig 2, A) showed that patients with CVID had lower anti-S antibody levels than did healthy vaccinated controls (4993.5 ± 6239.8 AU/mL vs 11906.15 ± 7909.2 AU/mL [P = .0225]). Further analysis showed that within the group of patients with CVID/hypogammaglobulinemia, there was a trend toward lower anti-S antibody titers in older patients (aged >59 years [n = 7]) compared with younger (aged <50 years [n = 7]) individuals (2105.7 ± 3978.3 AU/mL vs 7888.4 ± 7005.3 AU/mL [P = .082]). This trend became statistically significant when healthy vaccinated individuals were compared with older CVID patients (11906.15 ± 7909.2 AU/mL vs 2105.7 ± 3978.3 AU/mL, respectively [P = .0081]), nearly significant when older healthy vaccinated donors (n = 4) were compared with older CVID patients (6547.7 ± 872.0 AU/mL vs 2105.7 ± 3978.3 AU/mL [P = .0594]), but nonsignificant when younger healthy vaccinated donors (n = 7) were compared with younger CVID patients (14968.4 ± 8590.6 AU/mL vs 7881.4 ± 7005.3 AU/mL [P = .116]). However, comparing younger and older healthy vaccinated donors showed a similar trend, with older individuals having a tendency toward lower anti-S titers than younger healthy individuals (14968.4 ± 8590.6 AU/mL vs 6547.7 ± 872.0 AU/mL [P = .088]). Although not statistically significant, this trend could suggest that older individuals respond with lower antibody production than do younger individuals, independent of their immunologic background. When anti-N antibody titers were tested, all of the individuals other than convalescent donors were negative, with a single convalescent IEI patient (selective IgG1 and IgG3 deficiency) testing negative to anti-N antibodies (see Table E1 and Fig E1 in this article's Online Repository at www.jacionline.org).
Fig 1.
Humoral anti-S response: general. Anti–SARS-CoV2 humoral response was evaluated 2 weeks following a second vaccine dose. A, Titers of prevaccinated donors (B) (pink [n = 11]), donors with IEI (IEI) (bordeaux [n = 26]), healthy vaccinated donors (HV) (yellow [n = 11]), and recently convalescent mildly affected healthy donors (HC) (green [n = 4]) are shown. Dotted line marks a titer threshold of 150 AU/mL, with higher titers considered positive. Of note, the anti-S antibody titers of vaccinated individuals were higher than those of convalescent donors, whereas the samples from patients with IEI showed significant variability. B, Values of the AUC for anti-RBD IgG are shown. Here too, vaccinated individuals had higher anti-RBD titers than did convalescent donors (n = 13 HC). C, Inhibition percentage of RBD-ACE2 binding by donors’ sera. Zero inhibition was set on the basis of the average value of the patients before vaccination. The 1-way ANOVA test was performed for statistical analysis. D, Pie charts representing the division of each group according to different anti-RBD IgG titers. E, Correlation between IgG AUC and sera inhibition for all donors is shown. Correlation was calculated by a simple linear regression test. All analyses were calculated using GraphPad software.
Fig 2.
Groups of patients with IEI and a humoral anti-S response. A, Subdividing IEI according to different disease categories showed that as expected, all B-negative donors (4 donors with XLA and 1 post–rituximab treatment donor) tested negative for anti-S antibodies. In addition, younger patients with CVID (n = 7) showed a trend toward higher titers than older patients with CVID (n = 7), and older patients with CVID had lower titers of anti-S antibodies than healthy vaccinated donors. B and C, Values of the AUC for anti-RBD IgG and inhibition percentage of RBD-ACE2 binding by donors’ sera are shown. Zero inhibition was set on the basis of the average value of the patients before vaccination. For simplicity, data for only postvaccination IEI donors are shown. Here too, younger patients with CVID had higher anti-RBD titers and showed higher inhibition compared with older patients with CVID. For statistical analysis, an unpaired Student t test between younger and older groups of CVID patients was performed.
ELISA
The SARS-CoV-2 Spike protein RBD is the part of the virus responsible for viral entry through attachment to ACE2 receptor.26 In view of this fact, RBD was previously identified as one of the key determinants for neutralizing antibodies, with high correlation between anti-RBD antibody titers and serum neutralization.27 , 28 Therefore, we next sought to determine the levels of anti-RBD IgG in the plasma of donors with IEI. For this purpose, serially diluted serum samples were incubated with plate-bound RBD, and the area under curve (AUC) was calculated for each sample. In accordance with the commercial anti-S test results, healthy vaccinated individuals (n = 11) had higher titers than did convalescent donors (n = 12); this was true for patients recovering from severe, moderate, and mild COVID-19 (Fig 1, B and D) (an AUC for healthy vaccinated individuals of 4.954 vs an AUC for convalescent individuals of 2.872 [P = .0101]). Similarly, healthy vaccinated individuals also had higher average titers compared with IEI donors (an AUC for healthy vaccinated individuals of 4.954 vs an AUC for individuals with IEI of 2.723 [P = .0011]), although the samples from IEI patients showed a high distribution range. Here too, analysis of CVID samples according to patient age showed higher titers in younger CVID patients compared with older patients with CVID (average titers of 4.118 vs 2.122 [P = .036]) (Fig 2, B). Next, we tested the ability of plasma samples to inhibit SARS-CoV-2 attachment to host cells by assessing the level of serum antibodies that interfere with RBD-ACE2 interaction.13 As we previously reported, anti-RBD IgG level correlated well with the ability of serum samples to inhibit RBD-ACE2 binding in a competitive ELISA assay (Figs 1, C and E and 2, C). When patients with XLA were excluded, 18 of 21 IEI patients (85.7%) showed some level of RBD-ACE2 inhibition, suggesting that the majority of vaccinated IEI patients produced specific anti-RBD antibodies that were able to block SARS-CoV-2 infection. Here too, younger CVID patients showed higher average RBD-ACE2 inhibition than did older patients with CVID (average 68.98% vs 21.98%, [P = .019]) (Fig 2, C).
Specific anti-RBD B cells
To estimate the level of B-cell response following vaccination, we stained PBMC samples of IEI patients and healthy controls for the general B-cell markers CD19, IgG, IgA, as well as for fluorophore-conjugated RBD to detect RBD-specific memory B cells. Representative images of the gating strategy are shown in Fig E2 (in this article's Online Repository at www.jacionline.org). As can be seen, RBD-binding B cells (IgG+ and IgA+) were detected in convalescent individuals, healthy vaccinated donors, and patients with CVID. Because of the limited number of PBMCs, we were not able to complete this assay on all samples from IEI patients.
Taken together, these results suggest that majority of vaccinated IEI patients are able to produce a significant antibody response, including non-XLA patients with predominantly antibody deficiency.
Cellular response
We next studied the antivaccine cellular response by stimulating PBMCs with different SARS-CoV-2 pooled peptide mixes of either the S- or the M-glycoproteins, and evaluating IL-2 and IFN-γ production by using ELISpot. Although anti-N antibodies were used for serologic evaluation of prevaccine viral exposure, we previously observed that stimulation of convalescent PBMCs with pooled peptide mix of the nucleocapsid protein (N) resulted in lower cytokine production than did stimulation with M- or S-peptides (evaluated by intracellular staining for IL-2 and IFN-γ [see the Methods section and Fig E3, A and B in this article's Online Repository at www.jacionline.org]). We therefore chose to use an S-peptide mix for evaluation of antivaccine cellular response, and an M-peptide mix for evaluation of previous exposure and convalescence. On the basis of control groups of prevaccinated individuals (n = 7) and mildly affected convalescent individuals (n = 4), we set the threshold for positive cellular response at 4 spots per well. According to this set threshold, 19 of 26 of patients with IEI (73.1%) had a positive cellular response to S-peptides, including all 4 patients with XLA (Fig 3 , A and B and see Table E1). The 7 nonresponders included 1 patient with ALPS-like disease, 2 patients with NFKB1-HI, 2 young patients with CVID, and 2 older patients with CVID. Only 1 of these nonresponders (the patient with ALPS-like disease) also failed to produce anti-S antibodies. The remaining 6 patients tested positive for anti-S antibodies, although 3 of them had a low-positive anti-S antibody titer of less than 400 AU/mL (see Table E1).
Fig 3.
Cellular response. A, Number of ELISpot dots per well for prevaccinated donors (B) (n = 7), recently convalescent mildly affected donors (n = 4), healthy vaccinated donors (n = 11) (HV), donors with IEI (n = 26), and 1 convalescent patient with IEI are shown. Similar to what was observed with anti-S antibody titers, healthy vaccinated donors showed a trend toward a higher number of IL-2/IFN-γ spots than convalescent individuals. Dotted line marks a threshold of 4 spots per well, which we considered positive on the basis of samples from prevaccinated and convalescent individuals. B, Dividing IEI according to different disease categories showed a stronger cellular response in B-negative patients (4 donors with XLA and 1 post–rituximab treatment donor) compared with healthy vaccinated donors (average 58.6 vs 12.55 [P = .0073]) or with B-cell–positive IEI donors (average 58.6 vs 15.35 [P = .0056]). A 1-way ANOVA test was performed for the statistical analysis. For the purpose of logarithmic presentation, 0 points are presented with a value of 0.01. HC, Recently convalescent mildly affected healthy donor.
Similar to what we observed in the antibody assays, here too, the vaccine induced a strong cellular response with a trend toward statistical significance when convalescent individuals were compared with healthy vaccinated donors (average number of spots 6.25 ± 3.5 vs 13.2 ± 6.79 [P = .079]) (Fig 3, A).
In contrast to what was observed when analyzing the humoral response data, there were no statistically significant differences between healthy vaccinated donors and patients with IEI at any age group, with an average number of spots of 13.2 ± 6.79 vs 23.14 ± 35.36 (P = .356) for all patients with IEI (Fig 3, A), 13.2 ± 6.79 vs 19.42 ± 34.15 (P = .446) for older patients with CVID, and 13.2 ± 6.79 vs 13.57 ± 13.77 (P = .942) for young patients with CVID.
Although the response to M-peptides was mostly limited to convalescent individuals (see Fig E4, A and B in this article's Online Repository at www.jacionline.org), we were able to identify 2 healthy vaccinated individuals who responded to M-peptide stimulation (with 31 and 8 spots, respectively [see Fig 5, A and B in this article's Online Repository at www.jacionline.org]) but tested negative for anti-N antibodies. This response could have been due to cross-reactivity, nonspecific activation, or possible convalescence from asymptomatic COVID-19. In addition, 1 patient with CID had a very strong response to both S- and M- peptides (67 and 161 spots, respectively [see Fig E6 in this article's Online Repository at www.jacionline.org]) but tested negative for both anti-S and anti-N antibodies. This patient has longstanding lymphopenia with an absolute lymphocyte count of approximately 300/μL; therefore, nonspecific activation was suspected, although we did not have an unstimulated control to confirm our suspicion. All 3 subjects denied a history of symptomatic COVID-19 and never had a positive RT-PCR test result for SARS-CoV-2 RNA.
Fig 5.
T-cell immunophenotyping of 5 T-cell nonresponders. T-cell immunophenotyping of prevaccine samples that were available for 5 of the 7 nonresponders IEI patients showed that 4 of 5 donors had an abnormal CD4/CD8 ratio of less than 0.8. All 5 donors had a normal absolute lymphocyte count without CD4 lymphopenia.
Specific patients
Patients with XLA
As expected, all 4 patients with XLA tested negative for anti-S and anti-N antibodies and failed to show IgG or IgM binding to RBD antigen by ELISA. Similarly, 1 patient with hypogammaglobulinemia following anti-CD20 treatment (and an absolute CD19 count of 0/μL) did not show any evidence of humoral response. However, all 5 B-cell–negative patients showed significant cytokine secretion in response to S-peptide stimulation and were able to generate a stronger cellular response even when compared with healthy vaccinated donors (number of spots 58.6 ± 65.15 vs 12.5 ± 6.79 [P = .0073]), or B-positive IEI patients (58.6 ± 65.15 vs 15.35 ± 19.63 [P = .0056]), suggesting that their anti-S cellular response was preserved (Figs 3, B and Fig 4 , A). In addition, all 5 B-negative patients (4 donors with XLA and 1 post–rituximab treatment donor) did not respond to M-peptide stimulation, emphasizing the specificity of their enhanced anti-S T-cell response (Fig 4, A-C and see Fig E4, B).
Fig 4.
B-negative ELISpot results. Images of S-pool–stimulated (upper images) and M-pool–stimulated (lower images) PBMCs are shown. A, Strong cellular response with a high number of IL-2/IFN-γ spots can be clearly seen in PBMCs samples of XLA1, XLA2, XLA4, and rituximab-treated donors. B and C, Images of 7 younger (aged <50 years) healthy vaccinated (HV) individuals (B) and 5 convalescent (Con) donors (C) are shown as controls. One of the convalescent samples is from a selective IgG1 and IgG3-deficient patient (IEI-Con).
NFΚB1-HI
Our cohort included 2 patients with NFKB1-HI, a 32-year-old female and her 72-year-old father. Although the daughter had a history of lymphadenopathy, autoimmune neutropenia, and mild hypogammaglobulinemia (IgG level ∼ 500 mg/dL with no significant history of infection and currently off immunoglobulin replacement therapy), her father was asymptomatic and was incidentally diagnosed by trio whole exome sequencing while his daughter’s symptoms were being investigated. Both NFΚB1-HI donors failed to show cellular response (both exhibited only 2 S-induced spots [see Fig E6]). Despite that, the father showed a good antibody response (titer of 7102.7 AU/mL), whereas his daughter had a weak positive response (titer of 202.5 AU/mL [positive >150 AU/mL]). While patients with NFΚB1-HI can present with a wide range of clinical symptoms,29 the absent T-cell response in these 2 family members suggests that the vaccine response in patients with NF-κB pathway defects should be evaluated further.
STAT1-GOF
Our cohort included 1 patient with STAT1-GOF pathogenic variant who had a history of chronic mucocutaneous candidiasis and recurrent oral ulcers, and was currently on low-dose ruxolitinib treatment (5 mg twice daily). Evidence suggests that pathogenic STAT1-GOF variants can result in an enhanced type I interferon response30 , 31; therefore, there is a theoretical concern that STAT1-GOF patients would experience more significant adverse events following an mRNA-based vaccine, with exacerbation of underlying autoimmunity or development of new inflammatory symptoms. Despite that, our patient did not report any significant adverse events other than transient mild weakness after the second dose of vaccine. Interestingly, he had the highest anti-S antibody titer in our cohort (>29,000 AU/mL).
T-cell nonresponders
As already mentioned, 7 patients with IEI failed to show an anti-S cellular response on our ELISpot assay (see Figs E6 and E7 in this article's Online Repository at www.jacionline.org). These included 2 patients with NFΚB1-HI, 1 patient with ALPS-like disease, and 4 patients with CVID. There was no common T-cell immunophenotyping feature shared by all the nonresponding individuals other than an inverted CD4/CD8 ratio in 4 of 5 available prevaccine T-cell immunophenotyping samples (all in the presence of normal absolute lymphocyte counts [Fig 5 ]).
Discussion
Immunization is the most efficient intervention for preventing infectious diseases, and mass vaccination against SARS-CoV-2 has proved extremely successful, with the potential to control the COVID-19 pandemic, limit viral spread, and prevent severe illness.32 However, data regarding the effectiveness of anti–SARS-CoV-2 vaccine in immunocompromised populations, including in patients with IEI, are still being collected. In this study we evaluated the humoral and cellular immune response to the mRNA-based Pfizer-BioNTech anti–COVID-19 vaccine in a small cohort of 26 adult patients with IEI. Our data show that the majority of patients with IEI were able to respond to the vaccine with significant levels of neutralizing antibodies, cellular response, or both. In fact, when patients with XLA were excluded, 18 of 22 IEI patients (81.8%) tested positive for anti-S antibodies, and serum samples of 18 of 21 (85.7%) IEI patients showed ability to inhibit RBD-ACE2 interaction, suggesting that these patients were able to produce specific anti-RBD neutralizing antibodies. As for cellular response, 19 of 26 patients (73.1%) showed IL-2/IFN-γ secretion in response to stimulation with pooled S-peptide mix, with patients with XLA (n = 4) showing stronger cytokine secretion.
Our study has several major limitations. First, the small number of patients with diverse underlying diagnoses makes it difficult to draw firm conclusions. In addition, our samples were collected 2 weeks after the second vaccine dose. Therefore, the data are limited to the early postvaccine period, without any ability to predict how long the induced response would last. Finally, the short-term follow-up did not allow us to learn whether the presence of neutralizing antibodies or positive cellular response can confer protection against infection. In fact, 1 of our patients with IEI who did not participate in this study, and for whom we did not have serum or PBMC samples (a 51-year-old female with CID, lymphopenia, and active bowel disease who was receiving IVIG and systemic steroids), got infected with SARS-CoV-2 3 weeks after her second vaccine dose. However, she experienced only mild COVID-19 and was able to clear the virus after 18 days.
Despite those limitations, several points can be made. First, we did not see unusual or severe adverse events in our cohort. Three of the patients were categorized as having “dysregulation,” including 1 patient with STAT1-GOF pathogenic variant and 2 patients with presentation of ALPS-like disease. Our STAT1-GOF patient (patient 5) reported transient mild weakness after the second vaccine dose; 1 of the patients with ALPS-like disease (patient 7) reported no adverse symptoms, whereas the second patient (patient 6) reported fever (ie, body temperatures as high as 39°C for 3 days) and a large local reaction. Nevertheless, her counts remained stable. All other adverse events reported were considered mild and were similar to those described in the general population.
Second, age-dependent vaccine response has been described (reviewed in Gustafson et al33). Similarly, we observed that older patients with CVID had an overall tendency toward lower anti-S antibody titers and lower RBD-ACE2 inhibition. This tendency was not observed when younger and older healthy vaccinated donors were compared. On the basis of previous reports of patients with XLA who were able to clear SARS-CoV-2 and did not experience severe COVID-19,34 , 35 it might be reasonable to assume that humoral response is not central for anti–SARS-CoV-2 activity. Despite this, the lower antibody titers observed in older patients with CVID could imply that a third vaccine dose should be considered in selected patients.
Third, the cellular response observed in patients with XLA is reassuring. Our cohort included only 4 patients with XLA, but all 4 of them developed a robust cellular response with a higher level of cytokine production than that exhibited by other IEI patients , healthy vaccinated individuals, or convalescent patients. A fifth patient, who had rituximab-treated myasthenia gravis (with an absolute B-cell count of 0/μL), also showed strong T-cell response. A similar response of patients with XLA to influenza vaccine has been described before.36 These results suggest that B-cell–negative patients, either because of primary defects or because of B-cell–depleting therapies, can still benefit from vaccination. In addition, the fact that only 1 of our T-cell nonresponders failed to produce anti-S antibodies suggests that absent antibody response does not mean lack of protection.
Finally, 1 of the criteria for the diagnosis of CVID is poor antibody response to vaccines. In addition, testing antibody response to neoantigen challenges (such as rabies vaccine, typhoid vaccine or ψX174) is being used for evaluating antibody production in patients who are already receiving immunoglobulin replacement therapy. Therefore, evaluating the humoral response to SARS-CoV-2 or the COVID-19 vaccine in convalescent or vaccinated patients with IEI, could have been considered another neoantigen challenge. However, the high percentage of responding IEI patients, including our patients with a well-established diagnosis of CVID, suggests that vaccine response should be interpreted carefully and that a positive antibody response does not rule out clinically significant antibody deficiency. It should be noted that we routinely use B-cell immunophenotyping for establishing a diagnosis of CVID and not evaluation of vaccine response, and therefore, we cannot compare our patients’ response to the COVID-19 vaccine with their response to other vaccine challenges.
Altogether, our results support the safety and efficacy of the anti–SARS-CoV-2 vaccine and argue in favor of vaccinating patients with IEI.
Clinical implications.
The majority of patients with IEI are likely to safely mount a humoral or cellular immune response to the anti–SARS-CoV-2 vaccine. Patients with IEI should be encouraged to get vaccinated.
Acknowledgments
We would like to thank our patients and our dedicated medical teams.
Footnotes
Supported by the Israel Science Foundation (grants 41222/18 and 3711/20 [to N.T.F.]), the Dahlia Greidinger Cancer Fund (to N.T.F.), a Marguerite Stolz Fellowship (N.T.F.), The Campbell Foundation for AIDS Research (to N.T.F.), and the Alrov Foundation (to D.H.).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Supplementary data
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.COVID-19 Treatment Guidelines Panel Coronavirus disease 2019 (COVID-19) treatment guidelines. National Institutes of Health. https://www.covid19treatmentguidelines.nih.gov/ Available at: [PubMed]
- 4.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keehner J., Horton L.E., Pfeffer M.A., Longhurst C.A., Schooley R.T., Currier J.S. SARS-CoV-2 infection after vaccination in health care workers in California. N Engl J Med. 2021;384:1774–1775. doi: 10.1056/NEJMc2101927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine-Tiefenbrun M., Yelin I., Katz R., Herzel E., Golan Z., Schreiber L. Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat Med. 2021;27:790–792. doi: 10.1038/s41591-021-01316-7. [DOI] [PubMed] [Google Scholar]
- 8.Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147:520–531. doi: 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields A.M., Burns S.O., Savic S., Richter A.G,, UK PIN COVID-19 Consortium COVID-19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. J Allergy Clin Immunol. 2021;147:870–875.e1. doi: 10.1016/j.jaci.2020.12.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mor M, Werbner M, Alter J, Safra M, Chomsky E, Lee JC. Multi-clonal SARS-CoV-2 neutralization by antibodies isolated from severe COVID-19 convalescent donors. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb R.L., N ruxolitinab irula A., Chen P., Boscia J., Heller B., Morris J. Effect of Bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Post N., Eddy D., Huntley C., van Schalkwyk, M C I, Shrotri M., Leeman D. Antibody response to SARS-CoV-2 infection in humans: a systematic review. PLoS One. 2020;15 doi: 10.1371/journal.pone.0244126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaebler C., Wang Z., Lorenzi J.C., Muecksch F., Finkin S., Tokuyama M. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 21.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bilich T., Nelde A., Heitmann J.S., Maringer Y., Roerden M., Bauer J. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals [abstract] Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abf7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 25.Seidel M.G., Kindle G., Gathmann B., Quinti I., Buckland M., van Montfrans J. The European Society for Immunodeficiencies (ESID) Registry working definitions for the clinical diagnosis of inborn errors of immunity. J Allergy Clin Immunol Pract. 2019;7:1763–1770. doi: 10.1016/j.jaip.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar E., Kuchipudi S.V., Christensen P.A., Eagar T., Yi X., Zhao P. Convalescent plasma anti-SARS-CoV-2 spike protein ectodomain and receptor-binding domain IgG correlate with virus neutralization. J Clin Invest. 2020;130:6728–6738. doi: 10.1172/JCI141206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, et al. COVID-19 neutralizing antibodies predict disease severity and survival. Cell 2021;184:476-88.e11. [DOI] [PMC free article] [PubMed]
- 29.Kaustio M., Haapaniemi E., Goos H., Hautala T., Park G., Syrjanen J. Damaging heterozygous mutations in NFKB1 lead to diverse immunologic phenotypes. J Allergy Clin Immunol. 2017;140:782–796. doi: 10.1016/j.jaci.2016.10.054. [DOI] [PubMed] [Google Scholar]
- 30.Kaleviste E., Saare M., Leahy T.R., Bondet V., Duffy D., Mogensen T.H. Interferon signature in patients with STAT1 gain-of-function mutation is epigenetically determined. Eur J Immunol. 2019;49:790–800. doi: 10.1002/eji.201847955. [DOI] [PubMed] [Google Scholar]
- 31.Okada S., Asano T., Moriya K., Boisson-Dupuis S., Kobayashi M., Casanova J. Human STAT1 gain-of-function heterozygous mutations: chronic mucocutaneous candidiasis and type I interferonopathy. J Clin Immunol. 2020;40:1065–1081. doi: 10.1007/s10875-020-00847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N Engl J Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gustafson C.E., Kim C., Weyand C.M., Goronzy J.J. Influence of immune aging on vaccine responses. J Allergy Clin Immunol. 2020;145:1309–1321. doi: 10.1016/j.jaci.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Foca E. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinti I., Lougaris V., Milito C., Cinetto F., Pecoraro A., Mezzaroma I. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol. 2020;146:211–213.e4. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Wu Y., Lam K.T., Lee P.P., Tu W., Lau Y.L. Dendritic and T cell response to influenza is normal in the patients with X-linked agammaglobulinemia. J Clin Immunol. 2012;32:421–429. doi: 10.1007/s10875-011-9639-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.