Graphical abstract
Keywords: rRT-PCR, COVID-19, Diagnosis, Screening, Coronavirus
Abstract
Objectives
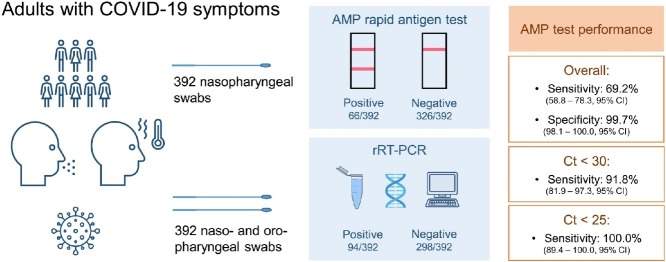
Quick and inexpensive SARS-CoV-2 screening and frontline testing are in growing demand. Our study aimed to evaluate the performance of the immunochromatographic AMP rapid antigen test (AMP RAT) compared to the gold-standard real-time reverse transcription PCR (rRT-PCR) in a hospital cohort.
Methods
A total of 392 patients, who presented consecutively with COVID-19 symptoms in our emergency department, were included in this retrospective study. Two swabs were collected per patient: a nasopharyngeal for the RAT and a combined naso- and oropharyngeal for the rRT-PCR. A positive rRT-PCR (defined as cycle threshold (Ct) < 40) was found in 94 (24%) patients.
Results
In our cohort with a median patient age of 70, overall sensitivity and specificity of the AMP RAT was 69.2% (58.8–78.3, 95% CI) and 99.7% (98.1–100.0, 95% CI), respectively. In patients with a Ct value < 25 and < 30, higher sensitivities of 100.0% (89.4–100.0, 95% CI) and 91.8% (81.9–97.3%, 95% CI) were observed.
Conclusions
The AMP RAT showed a high sensitivity in patients with a Ct value < 25 and < 30 and might be helpful for frontline testing whenever rRT-PCR is not readily available.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the cause of a broad clinical spectrum known as coronavirus disease 2019 (COVID-19) and was first identified in December 2019 in Wuhan, China (Zhu et al., 2020, Zhou et al., 2020). Within a short time, a worldwide spread led to the current pandemic that will presumably remain the leading infectious disease topic in 2021 (WHO, 2020a).
Detection of the virus through nucleic acid amplification tests such as real-time reverse transcription PCR (rRT-PCR) is the gold standard for the diagnosis of COVID-19 (WHO, 2020b, CDC, 2020c). The Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO) recommend nasopharyngeal and/or oropharyngeal swabs as the most sensitive specimen types for rRT-PCR testing (CDC, 2020b, WHO, 2020b). rRT-PCR is a cost- and labor-intensive technology which requires trained personnel (Corman et al., 2020). Furthermore, specimen transport and the inherent logistics are often time-consuming (CDC, 2020b). Although point-of-care testing (POCT) platforms allow a faster and easier preparation for rRT-PCR, they can still be expensive, and their output is limited. POCT is therefore not ideal for screening nor disease outbreaks, especially in low-income countries (Pai et al., 2012).
To dampen and control the spread of this virus, a rapid, cheap, reliable, and easy-to-handle identification test is needed for swift isolation or surveillance of patients as well as broad population screening. Rapid antigen tests (RAT) meet all these criteria and play a central role in the context of acute viral infections (Lai et al., 2020, Clerc and Greub, 2010). Hence, high sensitivity and specificity are crucial (CDC, 2020a). In this study, we set out to evaluate the diagnostic performance of the commercially available AMP SARS-CoV-2 Rapid Antigen Test (AMP Diagnostics, 2020) in comparison with rRT-PCR.
Methods
Clinical specimens
A total of 392 patients with COVID-19 symptoms presented consecutively to the emergency department of the Klinik Landstrasse from November 2nd, 2020, through December 29th, 2020, and were included in this retrospective study. The Klinik Landstrasse is a tertiary hospital, belonging to the Vienna Healthcare Group in Vienna, Austria.
For enrollment in the study, following the infectious disease policy of our hospital, every incoming patient with COVID-19 symptoms had to be tested for SARS-CoV-2 by rRT-PCR and simultaneous RAT to narrow the time until diagnosis. Nasopharyngeal swabs for RAT were collected at the same time as combined naso- and oropharyngeal swabs for rRT-PCR. While RAT was immediately performed and read after 15 min in the emergency department, rRT-PCR was carried out subsequently in our laboratory. Therefore, rRT-PCR results were unknown at the time of RAT administration and reading.
COVID-19 symptoms were grouped according to the WHO classification (WHO 2021) in common (fever, dry cough, fatigue), less common (sore throat, diarrhea, headache or other aches/pains, conjunctivitis, anosmia or ageusia, skin rash, discoloration of fingers or toes) and severe (dyspnea, chest pain, focal neurological deficit).
AMP Rapid Test SARS-CoV-2 Ag
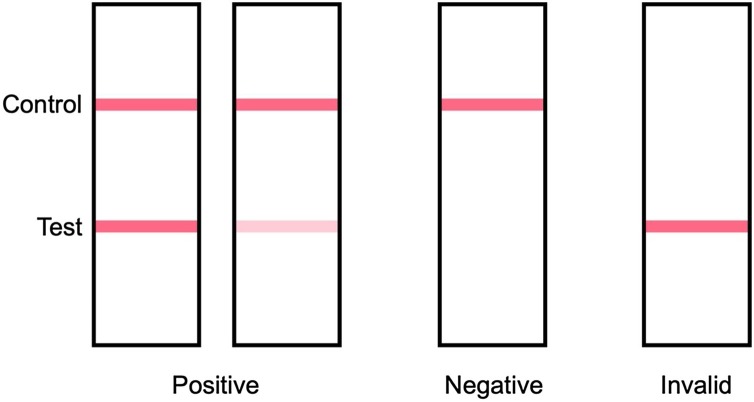
The AMP Rapid Test SARS-CoV-2 Ag (AMP Diagnostics, AMEDA Labordiagnostik GmbH, Graz, Austria), henceforth AMP RAT, is a rapid immunochromatographic test for the qualitative detection of the SARS-CoV-2 nucleocapsid protein antigen in nasopharyngeal swabs (AMP Diagnostics, 2020). The AMP RAT was performed at the emergency unit immediately after collecting nasopharyngeal swabs by specially trained medical staff. Specimen collection and analysis were performed at room temperature. For each RAT, and according to the manufacturer’s instructions, the swab was inserted in the extraction buffer tube and rotated at least six times while gently pressing the flocked head of the swab against the inner wall of the tube. After one minute of incubation, approximately 100 μl (4 drops) of the extraction solution was dropped into the sample well of the cassette, which then migrates by capillary action along the membrane. The SARS-CoV-2 nucleocapsid antigen binds to monoclonal antibodies conjugated with colloid-gold particles. It is then captured by secondary monoclonal antibodies and immobilized in the test region, and a colored line appears. A colored line has to appear in the correspondent internal control (I.C.) region confirming sufficient sample volume and correct test procedure (Figure 1 ). The test results can be read after 15 but no later than 20 min.
Figure 1.
Schematic representation of the AMP test and result possibilities. Positive: two colored stripes appear on the membrane; one stripe appears in the control area and another stripe in the test area; the color intensity of the stripe may vary depending on the concentration of SARS-CoV-2 antigen in the sample; therefore, every shade of color in the test area was regarded as a positive result. Negative: only one colored strip appears in the control area. Invalid: if no colored stripe appears in the control area, the test is invalid.
rRT-PCR
For rRT-PCR, combined naso- and oropharyngeal swabs were collected and inoculated in a sterile 2 ml 0.9% NaCl solution produced by the hospital pharmacy and then sent to our laboratory. All samples were analyzed within less than six hours on one of five different platforms available in our laboratory (Table S1). All rRT-PCR with a cycle threshold (Ct) value < 40 were considered positive. When two Ct values of different target genes were available, a mean Ct value was calculated. In SarbecoV E-gene EAV (TIB Molbiol Syntheselabor GmbH) and cobas® Liat® (Roche Diagnostics), only one Ct value was used. The SarbecoV E-gene EAV reagent targets a single gene (E), and the cobas® Liat® calculates one amplification curve using the same probe for both targeted genes (ORF1a/b and E). Cobas® Liat® only shows the coordinates of the amplification curve and does not give definite Ct values. In this case, Ct values were read and given as integers by two laboratory physicians.
As rRT-PCR is the diagnostic gold standard for SARS-CoV-2 detection, positive and negative samples were considered true positive and true negative.
Statistics
IBM SPSS Statistics version 22.0.0.1 (IBM Corp., Armonk, NY) and MedCalc version 17.7.2 (MedCalc Software, Ostend, Belgium) were used for statistical analysis, and a significance value of 0.05 was considered in all statistic tests. For test performance statistics, rRT- PCR was presumed the gold standard for true positive and negative values. Of the positive rRT-PCR Ct values, we calculated the mean when there were two genes detected. The Kolmogorow–Smirnow test was used to test the normality of distribution. Not normally distributed variables were reanalyzed after logarithmic transformation. Continuous variables were reported using median, 25th, and 75th percentiles and analyzed using Student’s t-test or the Mann Whitney U test, as appropriate. Categorical variables were expressed in frequencies (with percentages in parenthesis) and tested for significance using the χ² test or the Fisher’s exact test. A univariate and multivariate binary logistic regression model was used to predict RAT positivity (dependent or outcome variable). Mean Ct value and time since symptom onset were used as independent variables, as these have been shown to be associated with positive RAT in other studies (Dinnes et al., 2021).
Patients were also grouped according to two different Ct cut-offs, 25 and 30. Thirty is the most relevant cut-off in Austria as it is used for clinical and epidemiological decision-making. The Austrian Federal Ministry of Social Affairs, Health, Care, and Consumer Protection published a recommendation for discharging patients previously diagnosed with COVID-19 out of isolation according to clinical criteria and if follow-up Ct values are > 30 (Bundesministerium Soziales Gesundheit Pflege und Konsumentenschutz, 2021, Robert Koch Institut, 2021). According to Kim et al., viral culture was positive only in samples with a Ct value of 28.4 or less (Kim et al., 2021).
Results
In this study, samples from 392 symptomatic patients were collected. The median patient age was 70, and no sex differences were noted. Of all patients, 27% presented with severe symptoms, and 33% needed oxygen support (Table 1 ).
Table 1.
Demographic and clinical data of patients.
| All patients n = 392 |
rRT-PCR positive = 94 | rRT-PCR negative = 298 | P value | |
|---|---|---|---|---|
| Age, years | 70 (55–80) | 70 (57–80) | 71 (54–80) | 0.81 |
| Sex, female/male (%) | 192/200 (49/51) | 45/49 (48/52) | 147/151 (49/51) | 0.81 |
| Symptomsa, n (%) | 0.55 | |||
| Most common | 134 (34) | 34 (36) | 100 (34) | |
| Less common | 232 (59) | 50 (53) | 182 (61) | |
| Severe | 106 (27) | 21 (22) | 85 (29) | |
| Oxygen therapy, n (%) | 129 (33) | 32 (34) | 97 (33) | 0.78 |
| Time since symptom onset, days | 1 (0–3) | 2 (0–5) | 1 (0–3) | 0.10 |
Data are given as n (%) or median (25th–75th percentiles).
Symptoms were grouped according to the WHO classification (WHO 2021) in common (fever, dry cough, fatigue), less common (sore throat, diarrhea, headache or other aches/pains, conjunctivitis, anosmia or ageusia, skin rash, discoloration of fingers or toes) and severe (dyspnea, chest pain, focal neurological deficit). These were counted separately, as a combination of symptoms from different groups is possible.
The rRT-PCR results revealed 94 positive samples and 298 negative samples. The median Ct value was 27.6 (range: 14.1–39.9). Ct values did not significantly differ between the different rRT-PCR devices (Figure S1). When rRT-PCR positive and negative patients were compared, no significant differences were observed in age, sex, symptoms, and need for oxygen therapy. Time of presentation since symptom onset ranged from 0 to 60 days in COVID-19 patients and from 0 to 28 days in rRT-PCR negative patients, but this difference did not reach a significant difference (Table 1).
The I.C. was positive in all 392 performed RAT and so no test had to be repeated. The AMP RAT showed an overall sensitivity of 69.2% and specificity of 99.7%, with a single false positive (Table 2 ). As expected, binary logistic regression could identify the mean Ct value as an independent predictor for a positive RAT in the COVID-19 (rRT-PCR positive) group. In contrast, time since symptom onset did not predict RAT positivity (Table 3 ).
Table 2.
Performance of the rapid antigen test in comparison with rRT-PCR in all samples and according to mean Ct values.
| All samples n = 392 |
Ct < 25 n = 33 | Ct < 30 n = 61 | |
|---|---|---|---|
| True positive, n | 65 | 33 | 56 |
| True negative, n | 297 | – | – |
| False-positive, n | 1 | – | – |
| False negative, n | 29 | 0 | 5 |
| Sensitivity, % (95% CI) | 69.15 (58.78–78.27) | 100.00 (89.42–100.00) | 91.80 (81.90–97.28) |
| Specificity, % (95% CI) | 99.66 (98.14–99.99) | – | – |
Table 3.
Binary logistic regression to estimate RAT positivity in rRT-PCR positive patients.
| rRT-PCR positive n = 94 |
||||
|---|---|---|---|---|
| Univariate OR (95% CI) | P | Multivariate OR (95% CI) | P | |
| Mean Ct value | 0.680 (0.581–0.796) | <0.001 | 0.678 (0.578–0.795) | <0.001 |
| Time since symptom onset | 0.982 (0.932–1.035) | 0.50 | 1.014 (0.940–1.094) | 0.73 |
For further analysis, COVID-19 patients were grouped according to a mean Ct value < 30 and < 25, in which case RAT sensitivity increased to 91.8% and 100.0%, respectively (Table 2).
Discussion
Different SARS-CoV-2 RAT with distinct handlings and test performances are available (Dinnes et al., 2021). Here, we set out to compare the AMP RAT with rRT-PCR in a hospital cohort. This is the first AMP RAT study to deliver real-life data. Many studies evaluating RAT have been published using transport media (e.g., universal transport medium, viral transport medium, and phosphate-buffered saline) as a proxy for direct testing after sample collection (Kruttgen et al., 2021, Mak et al., 2020, Porte et al., 2020). However, most of the available tests, including the AMP RAT, are only approved to be performed directly after swabbing. A strength of our study relies on the fact that the RAT were performed strictly according to the manufacturer’s instructions in a point-of-care setting.
In direct comparison with the rRT-PCR, the AMP RAT showed a sensitivity of 100.0% for Ct values < 25 and 91.8% for Ct values < 30. As expected, and in accordance with previous studies, sensitivity dropped progressively with higher Ct values (Kruttgen et al., 2021, Mak et al., 2020, Porte et al., 2020, Scohy et al., 2020).
One possible study limitation is that Ct values had to be manually read from the cobas® Liat® rRT-PCR system as in this case only indexed amplification curves, but no given Ct values were produced. To minimize observational bias, Ct values were read by two physicians as integers. We consider that the data presented here would not change significantly with automatically given Ct values. Another inherent limitation to our study is the retrospective character of data collection.
The AMP RAT product information discloses an overall sensitivity of 97.3%, which differs considerably from the sensitivity of 69.2% reported here. This could be due to different patient selection as, in our cohort, a wide range of Ct values, including 33 samples with a Ct value of ≥ 30, were found. Another relevant issue is that the viral load of the nasopharyngeal swab does not necessarily reflect the viral load of the upper respiratory tract due to an inherent preanalytical variance concerning the quality of the swabbing method (e.g., nasal polyps, anatomical differences, patient discomfort). Further, the SARS-CoV-2 RAT landscape has been recently analyzed in an extensive Cochrane systematic review and revealed widely divergent overall sensitivities (range: 0–96%) (Dinnes et al., 2021).
In the binary logistic regression analysis, RAT positivity was not associated with the time between symptom onset and SARS-CoV-2 testing, most probably because median time after symptom onset in our study was very low (two days in COVID-19 patients and one day in rRT-PCR negative patients).
From a user-friendly perspective, the AMP RAT is comparable to other commercially available RAT that we tested at our laboratory. An additional incubation step of one minute in the extraction buffer tube is needed and makes testing more demanding. Nonetheless, in our experience, the AMP RAT still has good deployability in an emergency department setting.
In conclusion, the AMP RAT showed good test performance in patients with low Ct values and might be a valuable tool for frontline testing whenever quick rRT-PCR testing is not feasible.
Author contributions
Conceptualization, G.L., A.V-G., A.V.; Methodology, G.L., A.V-G., A.V.; Validation, G.L., A.V-G., R.Z., A.V.; Formal Analysis, G.L., A.V-G., A.V.; Data Curation, all authors; Writing – Original Draft Preparation, G.L., A.V.; Writing – Review & Editing, all authors.; Visualization, all authors; Supervision, A.V-G., A.V.; Project Administration, G.L., A.V-G., A.V.
Funding
This research received no external funding.
Institutional review board statement
We received ethical approval from the ethics committee of the City of Vienna to conduct this study (internal registration number E.K. 21-061-VK).
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.05.063.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- AMP Diagnostics . 2020. AMP Rapid Test SARS-CoV-2 Ag.https://www.amp-med.com/news-rapid-test-sars-cov-2-ag [Google Scholar]
- Bundesministerium Soziales Gesundheit Pflege und Konsumentenschutz . 2021. Empfehlung für die Gesundheitsbehörden zur Entlassung von COVID-19-Fällen aus der Absonderung.https://www.aekktn.at/documents/3b4d12c9-620b-11eb-8af6-52540052f55b [Google Scholar]
- CDC . 2020. Interim guidance for antigen testing for SARS-CoV-2.https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html [Google Scholar]
- CDC . 2020. Interim guidelines for collecting and handling of clinical specimens for COVID-19 testing.https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html [Google Scholar]
- CDC . 2020. Overview of testing for SARS-CoV-2 (COVID-19)https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html [Google Scholar]
- Clerc O., Greub G. Routine use of point-of-care tests: usefulness and application in clinical microbiology. Clin Microbiol Infect. 2010;16:1054–1061. doi: 10.1111/j.1469-0691.2010.03281.x. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnes J., Deeks J.J., Berhane S., Taylor M., Adriano A., Davenport C. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2021;3 doi: 10.1002/14651858.CD013705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.C., Cui C., Shin K.R., Bae J.Y., Kweon O.J., Lee M.K. Duration of culturable SARS-CoV-2 in hospitalized patients with Covid-19. N Engl J Med. 2021;384:671–673. doi: 10.1056/NEJMc2027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruttgen A., Cornelissen C.G., Dreher M., Hornef M.W., Imohl M., Kleines M. Comparison of the SARS-CoV-2 Rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J Virol Methods. 2021;288 doi: 10.1016/j.jviromet.2020.113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Wang C.Y., Ko W.C., Hsueh P.R. In vitro diagnostics of coronavirus disease 2019: technologies and application. J Microbiol Immunol Infect. 2021;54(April (2)):164–174. doi: 10.1016/j.jmii.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai N.P., Vadnais C., Denkinger C., Engel N., Pai M. Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries. PLoS Med. 2012;9 doi: 10.1371/journal.pmed.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert Koch Institut . 2021. COVID-19: Entlassungskriterien aus der Isolierung.https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Entlassmanagement [Google Scholar]
- Scohy A., Anantharajah A., Bodeus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. A coordinated global research roadmap: 2019 novel coronavirus.https://www.who.int/publications/m/item/a-coordinated-global-research-roadmap . [Accessed 24 February 2021] [Google Scholar]
- WHO . 2020. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases.https://www.who.int/publications/i/item/10665-331501 . [Accessed 24 February 2021] [Google Scholar]
- WHO . 2021. Coronavirus, symptoms.https://www.who.int/health-topics/coronavirus#tab=tab_3 [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.