ABSTRACT
Background
The literature on associations of circulating concentrations of minerals and vitamins with risk of colorectal cancer is limited and inconsistent. Evidence from randomized controlled trials (RCTs) to support the efficacy of dietary modification or nutrient supplementation for colorectal cancer prevention is also limited.
Objectives
To complement observational and RCT findings, we investigated associations of genetically predicted concentrations of 11 micronutrients (β-carotene, calcium, copper, folate, iron, magnesium, phosphorus, selenium, vitamin B-6, vitamin B-12, and zinc) with colorectal cancer risk using Mendelian randomization (MR).
Methods
Two-sample MR was conducted using 58,221 individuals with colorectal cancer and 67,694 controls from the Genetics and Epidemiology of Colorectal Cancer Consortium, Colorectal Cancer Transdisciplinary Study, and Colon Cancer Family Registry. Inverse variance-weighted MR analyses were performed with sensitivity analyses to assess the impact of potential violations of MR assumptions.
Results
Nominally significant associations were noted for genetically predicted iron concentration and higher risk of colon cancer [ORs per SD (ORSD): 1.08; 95% CI: 1.00, 1.17; P value = 0.05] and similarly for proximal colon cancer, and for vitamin B-12 concentration and higher risk of colorectal cancer (ORSD: 1.12; 95% CI: 1.03, 1.21; P value = 0.01) and similarly for colon cancer. A nominally significant association was also noted for genetically predicted selenium concentration and lower risk of colon cancer (ORSD: 0.98; 95% CI: 0.96, 1.00; P value = 0.05) and similarly for distal colon cancer. These associations were robust to sensitivity analyses. Nominally significant inverse associations were observed for zinc and risk of colorectal and distal colon cancers, but sensitivity analyses could not be performed. None of these findings survived correction for multiple testing. Genetically predicted concentrations of β-carotene, calcium, copper, folate, magnesium, phosphorus, and vitamin B-6 were not associated with disease risk.
Conclusions
These results suggest possible causal associations of circulating iron and vitamin B-12 (positively) and selenium (inversely) with risk of colon cancer.
Keywords: Mendelian randomization, genes, nutrition, supplements, colorectal cancer
Introduction
Colorectal cancer was the third most common cancer worldwide in 2018 (1). Diet and nutrition have an important role in the development of colorectal cancer. A higher consumption of red and processed meat has been linked to a higher risk of colorectal cancer, whereas a higher intake of fiber, milk, and whole grains has been associated with a lower risk, with reasonable consistency in prospective cohort studies (2, 3).
Most of the evidence regarding the nutritional epidemiology of colorectal cancer comes from observational studies that often rely on food frequency questionnaires (FFQs) to measure the consumption of foods and nutrients. This approach is prone to measurement error, because it is based on participants’ self-reports often provided at 1 point in time and the conversion of foods consumed into nutrient intake based on food composition databases that might be inaccurate (4). Furthermore, individuals who follow different diets might also vary in other characteristics, which are not always adequately controlled for in statistical analyses. In addition, evidence from randomized controlled trials (RCTs) to support the efficacy of dietary modification or nutrient supplementation for colorectal cancer prevention is lacking because few adequately powered trials exist, and those that do exist have in general failed to support protective associations (5–8). The molecular epidemiology literature on associations of circulating concentrations of minerals and vitamins with risk of colorectal cancer is generally less extensive and inconsistent (2, 3).
Our aim was to complement findings from observational research and RCTs, and investigate whether circulating concentrations of micronutrients are associated with risk of colorectal cancer using Mendelian randomization (MR) to improve causal inference in observational epidemiology. MR uses genetic variables as instrumental variables to assess the association of the genetically predicted component of the micronutrient biomarkers with colorectal cancer (9). We estimated the relations of single nucleotide polymorphisms (SNPs) associated with circulating concentrations of 11 systematically selected micronutrients (β-carotene, calcium, copper, folate, iron, magnesium, phosphorus, selenium, vitamin B-6, vitamin B-12, and zinc) with the risk of colorectal cancer and its subsites (colon, rectum, and proximal and distal colon). We used summary genetic association data for colorectal cancer and its subsites from 3 consortia: the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO), the Colorectal Cancer Transdisciplinary Study (CORECT), and the Colon Cancer Family Registry (CCFR).
Methods
Data for the genetic epidemiology of circulating micronutrient concentrations
We initially identified 20 micronutrients—β-carotene, calcium, copper, folate, iron, magnesium, phosphorus, potassium, retinol, selenium, sodium, zinc, and vitamins B-1, B-2, B-6, B-12, C, D, E, and K—for which associations with colorectal cancer have been reported in the literature (3). We conducted a search of published genome-wide association studies (GWASs) performed among individuals of European ancestry on circulating concentrations of these minerals and vitamins in the GWAS catalog and PubMed (last search performed in October 2019). Vitamin D was subsequently excluded from our analysis because recently published MR studies have already investigated the role of circulating vitamin D concentrations in relation to risk of colorectal cancer (10, 11). Potassium, sodium, and vitamins B-1, B-2, C, and K were also excluded because either no GWAS has been conducted or no genome-wide significant results have been reported (12, 13). GWASs for circulating vitamin E and retinol concentrations were not used because they adjusted for BMI (14, 15), which may cause collider bias in GWAS and MR estimates (16). After exclusions, published GWASs for 11 micronutrients were retrieved: β-carotene, calcium, copper, folate, iron, magnesium, phosphorus, selenium, vitamins B-6 and B-12, and zinc (13, 17–25). Two separate GWASs were used to instrument calcium concentrations (24, 25), the more recent of which was conducted in UK Biobank samples and published on a preprint server in June 2019 (25). SNPs that were associated with the circulating concentrations of these micronutrients at a genome-wide significance level (P < 5 × 10−8) and were not in linkage disequilibrium (linkage disequilibrium r2 ≤ 0.01) were selected. We used summary estimates for 3 (rs1800562, rs1799945 and rs855791) out of the 5 (rs1800562, rs1799945, rs855791, rs7385804, and rs8177240) available genome-wide significant SNPs for serum iron, because these 3 SNPs showed a concordant effect on serum iron, ferritin, transferrin, and transferrin saturation, and have been associated with an overall increased systemic iron status (17, 26). Three SNPs with minor allele frequency (MAF) <5% (rs12272669, rs2336573, rs6859667) in the GWASs for selenium and vitamin B-12 were excluded because their association estimates with the micronutrients might be imprecise. In total, summary genetic association data for 253 common (MAF ≥ 0.05) SNPs robustly associated with the 11 micronutrient concentrations were obtained. The selected GWASs included data from 12 European countries and the United States and most of the participants were women, with percentages ranging from 55% to 69% of total sample size. Supplemental Table 1 provides detailed information on the selected genetic variants.
Data for the genetic epidemiology of colorectal cancer
A recently published large GWAS of almost 126,000 participants of European ancestry from the GECCO, CORECT, and CCFR consortia provided the genetic effects of the selected instruments on risk of colorectal (58,221 cases and 67,694 controls), colon (31,083 cases), rectal (15,775 cases), proximal colon (13,857 cases), and distal colon (15,306 cases) cancer (27). These endpoints were predeclared and did not change during the analyses. The regression models were adjusted for age, sex, study, and genetic principal components to account for population structure. Out of the 253 SNPs, rs1550532 and rs780094 associated with calcium, rs855791 associated with iron, rs602662 and rs1801222 associated with vitamin B-12, and rs2120019 associated with zinc concentrations were also nominally statistically significantly associated with risk of colorectal cancer (P value range: 5.89 × 10−4 to 0.03). Supplemental Table 2 provides detailed information on the association of the genetic instruments with risk of colorectal cancer and its subsites.
Statistical power
Power calculations were performed using an online tool available at http://cnsgenomics.com/shiny/mRnd/ (28). The statistical power to capture an OR for colorectal cancer of 1.10 or 0.90 per SD change in the circulating concentrations of the micronutrients ranged from 0.39 for folate to 0.99 for vitamin B-12, and the statistical power was >0.80 for 6 of the 11 instruments tested, namely calcium (UK Biobank), copper, iron, selenium, vitamin B-12, and zinc. Supplemental Table 3 shows detailed power calculations for all outcomes and Supplemental Table 4 shows the minimum detectable ORs for 80% power.
MR analysis
A 2-sample MR using summary association data from GWASs of circulating micronutrients (first sample) and colorectal cancer risk (second sample) was performed. MR uses genetic variants as instruments to measure the genetically predicted component of the micronutrient concentrations and estimates the association of this component with colorectal cancer risk (29). In the case of β-carotene, where only 1 SNP was available, the effect estimate was calculated as the ratio of the SNP-outcome divided by the SNP–nutrient association (29), whereas the fixed-effects inverse variance-weighted (IVW) method was implemented when the instruments consisted of multiple SNPs. The IVW analysis can be thought of as a meta-analysis of single SNP effects (30). The β estimates and SEs from the regressions for circulating concentrations of β-carotene, copper, selenium, vitamin B-6, and zinc were transformed from the logarithmic scale provided in the published GWAS to the natural scale using a published formula (31). All reported associations correspond to an OR for risk of colorectal cancer and its subsites per SD change in the genetically predicted circulating concentrations of the nutrients.
Methods to assess the robustness of MR findings
To produce valid results, the IVW method requires that all genetic instruments are associated with the micronutrient concentrations (relevance assumption), but not directly with colorectal cancer (only via the micronutrients; exclusion restriction), nor any confounders of the relation between the micronutrient concentrations and colorectal cancer (independence assumption) (9). The strength of each instrument in relation to the circulating concentrations of the micronutrients (relevance assumption) was measured using the F statistic with the formula: F = R2(n − 2)/(1 − R2), where R2 is the proportion of the variance of the micronutrient concentration explained by each genetic instrument and n is the sample size of the GWAS for the SNP–micronutrient association (32). The F statistics ranged from 16 to 2407 for all genetic instruments, implying an absence of weak instruments because all values were >10 (Supplemental Table 1) (32).
Descriptive and statistical analyses were performed to examine the robustness of the MR results to potential violation of the exclusion restriction and independence assumptions. We used diagnostic plots (scatter plots, forest plots, and funnel plots), the Cochran's Q statistical test for heterogeneity, and the I2 statistic to evaluate the extent to which any differences in the individual effect sizes among the selected genetic instruments may be related to pleiotropic effects rather than chance (33). Horizontal pleiotropy is defined as: where 1 genetic variant has independent effects on multiple traits, and is the main reason for potential violation of the exclusion restriction MR assumption. We further evaluated whether the selected genetic instruments were associated with secondary phenotypes in PhenoScanner (http://www.phenoscanner.medschl.cam.ac.uk/) and the GWAS catalog (34, 35). For valid instruments, little heterogeneity across the SNP instruments would be expected because they all estimate similar associations, resulting in uniform plots and small values for the statistical tests and metrics of heterogeneity. The presence and magnitude of any heterogeneity, identified through either visual inspection of the plots or high values for the Q test or the I2 metric, may thus be used to estimate the presence and magnitude of horizontal pleiotropy that may be biasing the MR estimate. When there was evidence of such heterogeneity, we also performed a random-effects IVW MR analysis to account for the additional heterogeneity in the estimation of the SEs (36).
Where the number of genetic instruments was ≥3, robust MR analyses that allow for horizontal pleiotropy were performed, namely the MR-Egger regression, weighted median, and weighted mode methods. The intercept from MR-Egger regression is a statistical test for horizontal pleiotropy, whereas the slope can be interpreted as the circulating nutrient effect on colorectal cancer adjusted for horizontal pleiotropy (37). This method assumes, however, that the pleiotropic effects are independent of the instrument strength (InSIDE assumption). Another limitation is that the MR-Egger method is subject to low power, particularly when using a small number of SNPs (e.g., <10). The weighted median estimator provides a valid causal estimate when at least half of the instruments are valid (38). The estimate from the weighted mode analysis is valid when the largest group of instruments with consistent MR estimates is valid (39). The MR pleiotropy residual sum and outlier test (MR-PRESSO) was also implemented to identify outlying genetic variants and analyses were rerun after excluding these variants (40). P values < 0.05 were considered nominally significant, whereas high-confidence findings were those that survived multiple-testing adjustment with a Bonferroni-corrected threshold of 0.0045. All analyses were prespecified, and implemented in the statistical software R (R Core Team, 2020) version 3.4.3 using the MendelianRandomization package and in Stata (StataCorp. 2013) version 13 using the MRrobust package.
Results
Figure 1 shows all associations using the IVW fixed- and random-effects models. Figures 2–4 depict associations using the MR sensitivity analyses methods for each of the 5 cancer sites studied (colorectum, colon, rectum, and proximal and distal colon). Supplemental Tables 5–7 detail analyses using sensitivity analyses methods; Supplemental Figures 1A–21E show diagnostic plots for all associations.
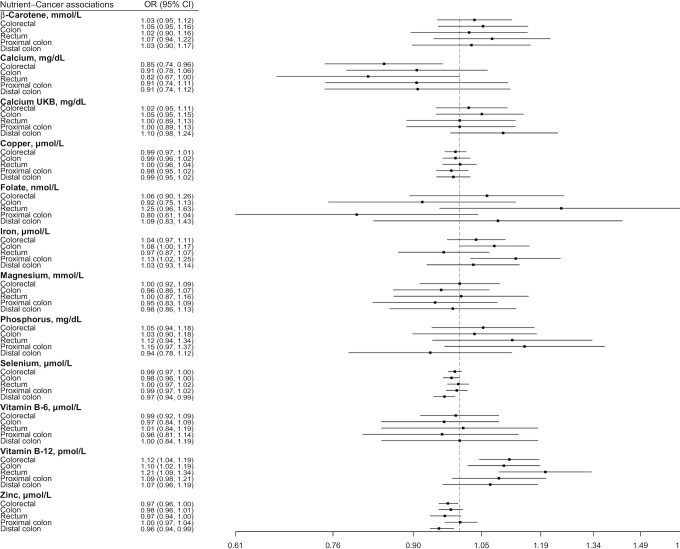
FIGURE 1.
Fixed-effects inverse variance–weighted Mendelian randomization analyses of 11 micronutrient concentrations and risk of colorectal cancer and its subsites. UKB, UK Biobank.
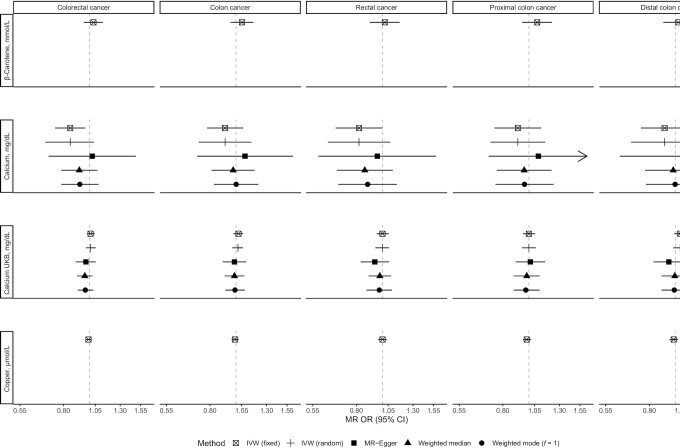
FIGURE 2.
Associations of β-carotene, calcium, and copper with risk of colorectal cancer and its subtypes using main and sensitivity MR analyses. IVW, inverse variance–weighted; MR, Mendelian randomization; UKB, UK Biobank.
FIGURE 4.
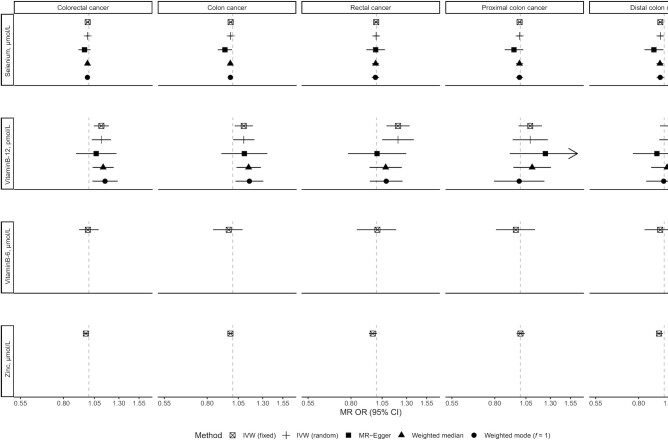
Associations of selenium, vitamin B-6, vitamin B-12, and zinc with risk of colorectal cancer and its subtypes using main and sensitivity MR analyses. IVW, inverse variance–weighted; MR, Mendelian randomization.
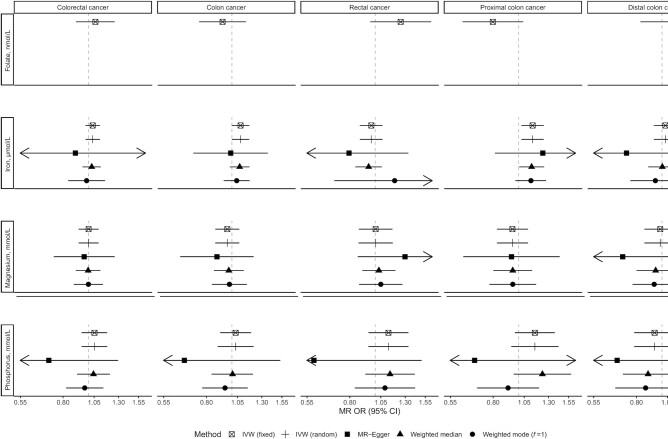
There was evidence that genetically predicted circulating concentrations of iron, selenium, vitamin B-12, and zinc were associated with risk of colorectal cancer or its subsites (Figures 1–4) (presented in detail in what follows). There was little evidence that circulating concentrations of β-carotene, calcium, copper, folate, magnesium, phosphorus, and vitamin B-6 were associated with risk (Figures 1–4).
FIGURE 3.
Associations of folate, iron, magnesium, and phosphorus with risk of colorectal cancer and its subtypes using main and sensitivity MR analyses. IVW, inverse variance–weighted; MR, Mendelian randomization.
Iron and colorectal cancer
A positive nominally significant association was observed for a 1-SD (6.13-µmol/L) increase in genetically predicted iron concentration and risk of colon (OR: 1.08; 95% CI: 1.00, 1.17; P value = 0.05) and proximal colon (OR: 1.13; 95% CI: 1.02, 1.25; P value = 0.01) cancer in the IVW fixed-effects analysis, but there was little evidence of an association for rectal and distal colon cancer (Figure 1). These associations did not survive correction for multiple testing. No heterogeneity was detected in the individual SNPs instrumenting iron and risk of colon (I2: 0%, Cochran's Q test P value = 0.59) and proximal colon (I2: 0%, P value = 0.91) cancer. There was no indication of horizontal pleiotropy based on the MR-Egger intercept test (Supplemental Table 5) (smallest P value = 0.19). Results based on MR-Egger regression were imprecisely estimated (i.e., wide CIs), but the weighted median and weighted mode estimates were consistent with the IVW MR analyses for colon and proximal colon cancer risk (Figures 2–4). The MR-PRESSO analysis did not reveal outlying SNPs (Supplemental Table 6).
Selenium and colorectal cancer
An inverse nominally significant association was observed for a 1-SD (0.53-µmol/L) increase in the genetically predicted selenium concentration and risk of colon (OR: 0.98; 95% CI: 0.96, 1.00; P value = 0.05) and distal colon (OR: 0.97; 95% CI: 0.94, 0.99; P value = 0.005) cancer in the IVW fixed-effects analysis, but there was little evidence of an association for rectal and proximal colon cancer (Figure 1). These associations did not survive correction for multiple testing. No heterogeneity was detected in the association of individual SNPs instrumenting selenium concentrations and risk of colon (I2: 0%, Cochran's Q test P value = 0.38) and distal colon (I2: 0%, P value = 0.54) cancer. There was no indication of horizontal pleiotropy based on the MR-Egger intercept test (Supplemental Table 5) (smallest P value = 0.10), and the associations remained consistent in the MR-Egger regression, the weighted median, and the weighted mode methods compared with the IVW MR results (Figures 2–4). The MR-PRESSO analysis did not reveal outlying SNPs (Supplemental Table 6).
Vitamin B-12 and colorectal cancer
Using the IVW fixed-effects method (Figure 1), a 1-SD (173-pmol/L) increase in the genetically predicted concentration of vitamin B-12 was associated with a 12% (OR: 1.12; 95% CI: 1.04, 1.19; P value = 0.001), 10% (OR: 1.10; 95% CI: 1.02, 1.19; P value = 0.02), and 21% (OR: 1.21; 95% CI: 1.09, 1.34; P value = 0.0003) higher risk of colorectal, colon, and rectal cancer, respectively, but not cancer in other subsites. Moderate heterogeneity was detected in the association of individual SNPs instrumenting vitamin B-12 concentrations with risk of colorectal (I2: 44%, Cochran's Q test P value = 0.11), colon (I2: 37%, P value = 0.19), and rectal (I2: 35%, P value = 0.06) cancer. When the IVW random-effects MR analysis was performed, all associations were still observed (colorectal cancer OR: 1.12; 95% CI: 1.03, 1.21; P value = 0.01; colon cancer OR: 1.10; 95% CI: 1.00, 1.21; P value = 0.04; rectal cancer OR: 1.21; 95% CI: 1.05, 1.39; P value = 0.008), but none survived correction for multiple testing. There was no indication of horizontal pleiotropy based on the MR-Egger intercept test (Supplemental Table 5) (smallest P value = 0.11). The slope of the MR-Egger regression did not yield any associations, but the weighted median and weighted mode estimates were consistent with the IVW MR analyses for colorectal and colon cancer (Figures 2–4). For rectal cancer, all sensitivity analysis MR methods provided little evidence of any association. The MR-PRESSO analysis did not reveal outlying SNPs (Supplemental Table 6).
Zinc and colorectal cancer
Genetically predicted concentrations of zinc were inversely nominally significantly associated with risk of colorectal cancer overall (per SD: 65 µmol/L; OR: 0.97; 95% CI: 0.96, 1.00; P value = 0.02) and distal colon cancer (OR: 0.96; 95% CI: 0.94, 0.99; P value = 0.01), but not in other subsites using the IVW fixed-effects analysis (Figure 1). These associations did not survive correction for multiple testing. Only 2 SNPs were used as instruments for zinc concentrations; thus, sensitivity MR analyses were not performed, but these SNPs have not been associated in GWASs with phenotypes that may indicate horizontal pleiotropy in relation to colorectal cancer (Supplemental Table 7).
Calcium and colorectal cancer
Using the IVW fixed-effects method and the older GWAS for calcium concentrations (n = 7 instruments) (Figure 1) (24), a 1-SD (0.48-mg/dL) higher genetically predicted concentration of calcium was nominally significantly associated with a 15% (OR: 0.85; 95% CI: 0.74, 0.96; P value = 0.01) lower risk of colorectal cancer; similar associations were also found for colon and rectal cancer, but these associations did not survive correction for multiple testing. However, when the larger and more recent GWAS using UK Biobank samples was used (n = 207 instruments) (25), little evidence for an association was observed for colorectal cancer (OR per SD: 1.02; 95% CI: 0.95, 1.11; P value = 0.55) or its subsites. There was heterogeneity in the association of individual SNPs instrumenting calcium with risk of colorectal cancer outcomes in analyses using both GWASs (Supplemental Table 5). No indication of horizontal pleiotropy was found in the analyses based on the MR-Egger intercept test (Supplemental Table 5). The slope of the MR-Egger regression, the weighted median, and the weighted mode estimates suggested little evidence of any associations in analyses using both GWASs (Figures 2–4, Supplemental Table 6).
Discussion
Main findings and comparisons with the literature
In this comprehensive MR analysis of 11 circulating micronutrient concentrations and risk of colorectal cancer and its main anatomical subsites, we observed that genetically predicted concentrations of circulating iron and vitamin B-12 were associated with higher risk of colon cancer, whereas selenium concentrations were associated with lower risk of colon cancer. An inverse association was also observed for zinc and colorectal cancer risk, but sensitivity analyses could not be performed. These associations did not survive correction for multiple testing. We observed little evidence that circulating concentrations of any of the other micronutrients (i.e., β-carotene, calcium, copper, folate, magnesium, phosphorus, and vitamin B-6) were associated with risk of colorectal cancer or its subsites.
Iron and colorectal cancer
High iron load has been linked to increased cancer risk in animal models and human experiments (41). The most prominent postulated underlying mechanism is the iron-induced formation of hydroxyl radicals leading to the generation of reactive oxygen species, oxidative tissue damage, and subsequent carcinogenesis (41). However, the epidemiological literature on iron intake and circulating iron biomarkers and colorectal cancer risk is mixed and inconclusive. Whereas heme iron intake, present mostly in red meat, has been positively associated with risk of colorectal cancer in several meta-analyses (42–44), findings for dietary and total iron intake have been mixed, and ferritin (a protein that stores iron) concentrations have been inversely associated with colorectal cancer risk (44). In the current MR study, genetically predicted concentrations of circulating iron were associated with higher risk of colon cancer, and these findings were robust to sensitivity MR methods. This finding was in agreement with another recently published MR study (45), but we used a sample more than double in size, estimated associations with greater precision, and studied associations in colorectal cancer subsites. Three loci were used as genetic instruments: rs1800562 and rs1799945 in the hemochromatosis (HFE) gene and rs855791 in the transmembrane protease serine 6 (TMPRSS6) gene, whose products have recognized roles in iron homeostasis (17). The rs1800562 in HFE has also shown associations with plasma lipids and lipoproteins (Supplemental Table 6) (17), which may indicate horizontal pleiotropy, but lipoprotein particles were not clearly associated with colorectal cancer risk in recent MR studies (46, 47).
Selenium and colorectal cancer
A protective effect of selenium on colorectal cancer has been supported by in vitro and animal studies, and selenium is hypothesized to reduce cancer risk by the antioxidative activity of selenoenzymes (48). However, the evidence from observational studies and RCTs is inconclusive. A meta-analysis of 10 observational studies showed an inverse association between circulating selenium concentrations and risk of colorectal neoplasia, but the association was only present in men (49). The association was no longer observed after excluding studies that measured selenium after cancer diagnosis. Selenium supplementation lowered colorectal cancer incidence by 61% (95% CI: 10%, 83%) in the secondary analysis of an RCT performed among patients with a history of nonmelanoma skin cancer that aimed to study recurrence of nonmelanoma skin cancer (50). In contrast, no such benefit was observed in a prespecified secondary analysis in the large SELECT (Selenium and Vitamin E Cancer Prevention Trial) study that was designed to investigate prostate cancer prevention (HR: 1.05; 95% CI: 0.66, 1.67), where selenium supplementation caused a median 114-µg/L increase in circulating selenium (7). The lower baseline selenium concentrations among participants of the first trial may have contributed to the observed benefit, and this phenomenon has also been shown in observational studies (51). In the current MR study, an increase of 114 µg/L in genetically predicted circulating selenium was associated with lower risk of colon cancer (OR: 0.94; 95% CI: 0.92, 1.00). This finding was in agreement with another recently published MR study (45), and may suggest that early-life effects of selenium play a role in the prevention of colorectal cancer because selenium is known to enhance the DNA damage repair response (52). These potentially early-life effects can be picked up in MR studies but are missed in RCTs.
Vitamin B-12 and colorectal cancer
B vitamins, including vitamin B-12, are essential for DNA methylation, synthesis, stability, and repair (53). Data from both in vitro and animal studies have suggested a protective effect of B vitamins against colorectal carcinogenesis (54), although the associations and mechanisms between the different cofactors of the one-carbon metabolism pathway are complex and have not yet been fully elucidated. No association was observed in the meta-analysis of epidemiological studies for circulating vitamin B-12 concentrations and colorectal cancer risk (per 150 pmol/L RR: 1.02; 95% CI: 0.88, 1.19). Long-term follow-up of participants (n = 2524) in the B-PROOF (B Vitamins for the Prevention of Osteoporotic Fractures) trial, a multicenter, double-blind, placebo-controlled RCT designed to assess the effect of 2–3 y daily supplementation with folic acid (400 mg) and vitamin B-12 (500 mg) compared with placebo on fracture incidence (55), showed that allocation to B vitamins was associated with a higher risk of colorectal cancer (43 compared with 25 cases; HR: 1.77; 95% CI: 1.08, 2.90). The dosage of vitamin B-12 was almost 200 times higher than the recommended intake, and the authors could not rule out that the high dosage of vitamin B-12 supplementation influenced the risk of colorectal cancer in their study. In the current MR study, genetically predicted concentrations of circulating vitamin B-12 were associated with higher risk of colorectal and colon cancer, and these findings were robust to sensitivity MR methods. This finding was in agreement with another recently published MR study (45) and was estimated with much greater precision in our study. Of the 9 loci associated with serum B-12 concentrations, most can be directly linked to the current understanding of B-12 metabolism such as absorption, transport, or enzymatic processes. One of them, fucosyltransferase 2 (FUT2), functions in cell surface glycobiology, and has previously been associated with liver enzymes, cholesterol concentrations, and Crohn disease (Supplemental Table 6), which may indicate horizontal pleiotropy, but when this SNP (i.e., rs602662) was removed from the analyses the results remained very similar.
Other micronutrients and colorectal cancer
In the current MR study, we observed an inverse association between genetically predicted concentrations of zinc and risk of colorectal cancer, but sensitivity analyses could not be performed. There were only 2 genetic instruments available for zinc; thus, we cannot preclude the presence of a potential causal association. Larger GWASs are needed to better understand the genetic regulation of zinc and to better define instrumental variables for MR analysis. Little evidence was observed in the current MR study that genetically predicted concentrations of β-carotene, calcium, copper, folate, magnesium, phosphorus, and vitamin B-6 were associated with risk of colorectal cancer. The observational molecular epidemiology literature for these micronutrient concentrations and risk of colorectal cancer is sparse, but in general the results from the available prospective studies agree with the null results of the current MR study (3, 56–58), with 2 potential exceptions for vitamin B-6 and calcium. A meta-analysis of 4 prospective studies (n = 883 total cases) showed an inverse association (OR: 0.52; 95% CI: 0.38, 0.71) comparing the highest with the lowest category of vitamin B-6 concentrations (59), but this result may be a chance finding given the relatively small sample size. RCTs of calcium supplementation showed a reduction in adenoma recurrence (60–62), but the Women's Health Initiative trial did not find any reduction in colorectal cancer incidence after a mean of 7 y of supplementation with calcium and vitamin D (63). In a reanalysis, a 17% nonsignificant reduction was observed among participants not already taking calcium or vitamin D at randomization (64). In the current MR study, genetically predicted concentrations of circulating calcium were not associated with risk of colorectal cancer or its subsites when the large GWAS on calcium from UK Biobank was used. This analysis used 207 genetic instruments for calcium and yielded good statistical power, but had a higher likelihood of horizontal pleiotropy owing to the large number of instruments. The pleiotropy-robust MR methods also suggested little evidence of association, but had point estimates below unity that fall within the potential effects suggested by the trials. However, the circulating concentrations of calcium are tightly regulated in the human body to maintain homeostasis, which means that large changes in intake will not lead to detectable changes in circulating concentrations; thus, MR estimates should not be interpreted as relevant to dietary intake.
Strengths and limitations
MR studies can be useful in nutritional epidemiology, because they can avoid biases that are commonly present in traditional observational literature (4). The main challenge of MR studies in this field is to identify genetic variants that are associated with exposures related to diet, specifically for blood concentrations of micronutrients in the current study. Minerals and vitamins are obtained from diet. However, genetic variation in absorption, metabolism, and storage can also be important in determining risk of deficiency and toxicity. MR estimates have a causal interpretation only if the assumptions of the instrumental variable approach hold. Although it is not possible to prove the validity of the assumptions, we performed several sensitivity analyses to detect potential violations. We have taken a conservative approach and only highlighted associations that were robust in sensitivity analyses.
Several limitations should be also considered in interpreting our findings. The summary-level data that we used did not allow for analyses stratified by covariates of interest, such as age, sex, alcohol consumption, dietary intakes, gut flora, or according to whether populations were deficient or not for specific elements. Furthermore, the currently known SNPs associated with folate and vitamin B-6 concentrations account for only a small amount of the variance explained, and the observed nonsignificant associations may be due to low power. In addition, the one-carbon metabolism is a complex web of biochemically interdependent reactions and it may be misleading to examine a single one-carbon nutrient (e.g., folate and vitamins B-6 and B-12) in isolation without considering the others. SNPs for micronutrients predict blood concentrations and genetic factors affecting concentrations in more clinically relevant tissues may differ. Future large pooling consortiums, larger single- and multitrait GWASs of micronutrient concentrations, and MR studies with individual-level data could address some of the latter issues.
Conclusion
In summary, using a comprehensive MR study, we found evidence for possible causal associations of higher circulating concentrations of iron and vitamin B-12 with higher risk of colon cancer, and higher selenium concentrations with lower risk of the disease. These results in combination with previous literature could open up new possibilities for chemoprevention of colorectal cancer using diet, supplements, or other means to modify circulating iron, vitamin B-12, and selenium concentrations.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—KKT and MJG: conceived and designed the study; NP, ND, and KKT: analyzed the data; KKT: wrote the paper taking into account the comments and suggestions of all the coauthors and had primary responsibility for the final content; NP, ND, DG, SJL, RMM, NM, GM, VZ, AJC, KB, DSL, TJK, RCT, AP-C, DJH, FJBvD, DA, VA, SIB, SB, DTB, JB, HB, AB-H, PTC, GC, SC-B, ATC, JC-C, AdlC, JCF, SJG, GGG, PJG, AG, JH, HH, MH, MAJ, TOK, S-SK, SCL, LLM, CIL, LL, AL, V Martín, RLM, V Moreno, HN, RN, PAN, KO, PDPP, EAP, JDP, LQ, GR, LCS, CS, MLS, LS, JS, SNT, CMU, BVG, SH, KV, LV, HW, EW, AW, MOW, AHW, WZ, BB-d-M, M-CB-R, DJH, PJ, TK, DP, ER, ELG, BLB, SBG, UP, and MJG: commented on the analysis and interpretation of the findings; and all authors: read and approved the final manuscript. The authors report no conflicts of interest.
Notes
Supported by World Cancer Research Fund (WCRF) International Regular Grant Programme grant WCRF 2014/1180 (to KKT). Funding statements for the Genetics and Epidemiology of Colorectal Cancer Consortium, Colorectal Cancer Transdisciplinary Study, and Colon Cancer Family Registry are shown in the Supplementary data. DG is supported by the Wellcome 4i Programme at Imperial College London. RMM was supported by Cancer Research UK programme grant C18281/A19169 (the Integrative Cancer Epidemiology Programme) and is part of the Medical Research Council Integrative Epidemiology Unit at the University of Bristol supported by Medical Research Council grants MC_UU_12013/1, MC_UU_12013/2, and MC_UU_12013/3 and the University of Bristol. RMM is also supported by the National Institute for Health Research (NIHR) Bristol Biomedical Research Centre which is funded by the NIHR and is a partnership between University Hospitals Bristol National Health Service Foundation Trust and the University of Bristol. LV was supported by Czech Science Foundation grants 18-09709S and 17-16857S.
The study sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health and Social Care. The authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/WHO.
Supplemental Tables 1–7 and Supplemental Figures 1–21 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: CCFR, Colon Cancer Family Registry; CORECT, Colorectal Cancer Transdisciplinary Study; GECCO, Genetics and Epidemiology of Colorectal Cancer Consortium; GWAS, genome-wide association study; HFE, hemochromatosis; IVW, inverse variance–weighted; MAF, minor allele frequency; MR, Mendelian randomization; MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier test; RCT, randomized controlled trial; SNP, single nucleotide polymorphism.
Contributor Information
Konstantinos K Tsilidis, Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, Ioannina, Greece; Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, United Kingdom.
Nikos Papadimitriou, Section of Nutrition and Metabolism, International Agency for Research on Cancer, Lyon, France.
Niki Dimou, Section of Nutrition and Metabolism, International Agency for Research on Cancer, Lyon, France.
Dipender Gill, Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, United Kingdom.
Sarah J Lewis, Department of Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, United Kingdom; Medical Research Council Integrative Epidemiology Unit, Department of Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, United Kingdom.
Richard M Martin, Department of Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, United Kingdom; Medical Research Council Integrative Epidemiology Unit, Department of Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, United Kingdom; University Hospitals Bristol National Health Service Foundation Trust National Institute for Health Research Bristol Biomedical Research Centre, University of Bristol, Bristol, United Kingdom.
Neil Murphy, Section of Nutrition and Metabolism, International Agency for Research on Cancer, Lyon, France.
Georgios Markozannes, Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, Ioannina, Greece.
Verena Zuber, Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, United Kingdom; Medical Research Council Biostatistics Unit, School of Clinical Medicine, University of Cambridge, Cambridge, United Kingdom.
Amanda J Cross, Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, United Kingdom.
Kimberley Burrows, Department of Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, United Kingdom; Medical Research Council Integrative Epidemiology Unit, Department of Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, United Kingdom.
David S Lopez, Department of Preventive Medicine and Community Health, The University of Texas Medical Branch, Galveston, TX, USA.
Timothy J Key, Nuffield Department of Population Health, University of Oxford, Oxford, United Kingdom.
Ruth C Travis, Nuffield Department of Population Health, University of Oxford, Oxford, United Kingdom.
Aurora Perez-Cornago, Nuffield Department of Population Health, University of Oxford, Oxford, United Kingdom.
David J Hunter, Nuffield Department of Population Health, University of Oxford, Oxford, United Kingdom.
Fränzel J B van Duijnhoven, Division of Human Nutrition and Health, Wageningen University & Research, Wageningen, Netherlands.
Demetrius Albanes, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA.
Volker Arndt, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Sonja I Berndt, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA.
Stéphane Bézieau, Medical Genetics Service, University Hospital Center (CHU) Nantes, Nantes, France.
D Timothy Bishop, , Leeds Institute of Cancer and Pathology, University of Leeds, Leeds, United Kingdom.
Juergen Boehm, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA; Department of Population Health Sciences, University of Utah, Salt Lake City, UT, USA.
Hermann Brenner, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany; Division of Preventive Oncology, German Cancer Research Center (DKFZ) and National Center for Tumor Diseases (NCT), Heidelberg, Germany; German Cancer Consortium (DKTK), German Cancer Research Center (DKFZ), Heidelberg, Germany.
Andrea Burnett-Hartman, Institute for Health Research, Kaiser Permanente Colorado, Denver, CO, USA.
Peter T Campbell, Behavioral and Epidemiology Research Group, American Cancer Society, Atlanta, GA, USA.
Graham Casey, Center for Public Health Genomics, University of Virginia, Charlottesville, VA, USA.
Sergi Castellví-Bel, Gastroenterology Department, Hospital Clinic, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Biomedical Research Network Center for Liver and Digestive Diseases (CIBEREHD), University of Barcelona, Barcelona, Spain.
Andrew T Chan, Division of Gastroenterology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA; Channing Division of Network Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA; Broad Institute of Harvard and MIT, Cambridge, MA, USA; Department of Epidemiology, Harvard TH Chan School of Public Health, Harvard University, Boston, MA, USA; Department of Immunology and Infectious Diseases, Harvard TH Chan School of Public Health, Harvard University, Boston, MA, USA.
Jenny Chang-Claude, Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany; University Medical Centre Hamburg-Eppendorf, University Cancer Centre Hamburg, Hamburg, Germany.
Albert de la Chapelle, Department of Cancer Biology and Genetics, The Ohio State University, Columbus, OH, USA; Comprehensive Cancer Center, The Ohio State University, Columbus, OH, USA.
Jane C Figueiredo, Department of Medicine, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Steven J Gallinger, Lunenfeld Tanenbaum Research Institute, Mount Sinai Hospital, University of Toronto, Toronto, Ontario, Canada.
Graham G Giles, Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia; Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Melbourne, Victoria, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, Victoria, Australia.
Phyllis J Goodman, SWOG Statistical Center, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Andrea Gsur, Institute of Cancer Research, Department of Medicine I, Medical University Vienna, Vienna, Austria.
Jochen Hampe, Department of Medicine I, University Hospital Dresden, Dresden University of Technology (TU Dresden), Dresden, Germany.
Heather Hampel, Division of Human Genetics, Department of Internal Medicine, The Ohio State University, Columbus, OH, USA.
Michael Hoffmeister, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Mark A Jenkins, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Melbourne, Victoria, Australia.
Temitope O Keku, Center for Gastrointestinal Biology and Disease, University of North Carolina, Chapel Hill, NC, USA.
Sun-Seog Kweon, Department of Preventive Medicine, Chonnam National University Medical School, Gwangju, Republic of Korea; Jeonnam Regional Cancer Center, Chonnam National University Hwasun Hospital, Hwasun, Republic of Korea.
Susanna C Larsson, Institute of Environmental Medicine, Karolinska Institute, Stockholm, Sweden.
Loic Le Marchand, University of Hawaii Cancer Center, Honolulu, HI, USA.
Christopher I Li, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Li Li, Department of Family Medicine, University of Virginia, Charlottesville, VA, USA.
Annika Lindblom, Department of Clinical Genetics, Karolinska University Hospital, Stockholm, Sweden; Department of Molecular Medicine and Surgery, Karolinska Institute, Stockholm, Sweden.
Vicente Martín, CIBER of Epidemiology and Public Health (CIBERESP), Madrid, Spain; Biomedicine Institute (IBIOMED), University of León, León, Spain.
Roger L Milne, Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia; Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Melbourne, Victoria, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, Victoria, Australia.
Victor Moreno, CIBER of Epidemiology and Public Health (CIBERESP), Madrid, Spain; Oncology Data Analytics Program, Catalan Institute of Oncology–Bellvitge Biomedical Research Institute (IDIBELL), L'Hospitalet de Llobregat, Barcelona, Spain; Department of Clinical Sciences, Faculty of Medicine, University of Barcelona, Barcelona, Spain; ONCOBEL Program, Bellvitge Biomedical Research Institute (IDIBELL), L'Hospitalet de Llobregat, Barcelona, Spain.
Hongmei Nan, Department of Epidemiology, Richard M Fairbanks School of Public Health, Indiana University, Indianapolis, IN, USA; IU Melvin and Bren Simon Cancer Center, Indiana University, Indianapolis, IN, USA.
Rami Nassir, Department of Pathology, School of Medicine, Umm Al-Qura'a University, Mecca, Saudi Arabia.
Polly A Newcomb, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; School of Public Health, University of Washington, Seattle, WA, USA.
Kenneth Offit, Clinical Genetics Service, Department of Medicine, Memorial Sloan-Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Paul D P Pharoah, Department of Public Health and Primary Care, University of Cambridge, Cambridge, United Kingdom.
Elizabeth A Platz, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
John D Potter, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Centre for Public Health Research, Massey University, Wellington, New Zealand.
Lihong Qi, Department of Public Health Sciences, School of Medicine, University of California Davis, Davis, CA, USA.
Gad Rennert, Department of Community Medicine and Epidemiology, Lady Davis Carmel Medical Center, Haifa, Israel; Ruth and Bruce Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Haifa, Israel; Clalit National Cancer Control Center, Haifa, Israel.
Lori C Sakoda, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Division of Research, Kaiser Permanente Northern California, Oakland, CA, USA.
Clemens Schafmayer, Department of General Surgery, University Hospital Rostock, Rostock, Germany.
Martha L Slattery, Department of Internal Medicine, University of Utah, Salt Lake City, UT, USA.
Linda Snetselaar, Department of Epidemiology, University of Iowa College of Public Health, Iowa City, IA, USA.
Jeanette Schenk, SWOG Statistical Center, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Stephen N Thibodeau, Division of Laboratory Genetics, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN, USA.
Cornelia M Ulrich, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA; Department of Population Health Sciences, University of Utah, Salt Lake City, UT, USA.
Bethany Van Guelpen, Department of Radiation Sciences, Oncology Unit, Umeå University, Umeå, Sweden.
Sophia Harlid, Department of Radiation Sciences, Oncology Unit, Umeå University, Umeå, Sweden.
Kala Visvanathan, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
Ludmila Vodickova, Department of Molecular Biology of Cancer, Institute of Experimental Medicine of the Czech Academy of Sciences, Prague, Czech Republic; Institute of Biology and Medical Genetics, First Faculty of Medicine, Charles University, Prague, Czech Republic; Faculty of Medicine and Biomedical Center in Pilsen, Charles University, Pilsen, Czech Republic.
Hansong Wang, University of Hawaii Cancer Center, Honolulu, HI, USA.
Emily White, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Department of Epidemiology, University of Washington, Seattle, WA, USA.
Alicja Wolk, Institute of Environmental Medicine, Karolinska Institute, Stockholm, Sweden.
Michael O Woods, Discipline of Genetics, Memorial University of Newfoundland, St. John's, Newfoundland, Canada.
Anna H Wu, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Wei Zheng, Division of Epidemiology, Department of Medicine, Vanderbilt-Ingram Cancer Center, Vanderbilt Epidemiology Center, Vanderbilt University School of Medicine, Nashville, TN, USA.
Bas Bueno-de-Mesquita, Formerly, Department for Determinants of Chronic Diseases (DCD), National Institute for Public Health and the Environment (RIVM), Bilthoven, Netherlands.
Marie-Christine Boutron-Ruault, Faculty of Medicine, CESP, University of Paris-Sud, Faculty of Medicine UVSQ, INSERM, University of Paris-Saclay, Villejuif, France; Centre for Research in Epidemiology and Population Health (CESP), Gustave Roussy, Villejuif, France.
David J Hughes, Cancer Biology and Therapeutics Group, UCD Conway Institute of Biomolecular and Biomedical Research, University College Dublin, Dublin, Ireland.
Paula Jakszyn, Unit of Nutrition and Cancer, Cancer Epidemiology Research Program, Catalan Institute of Oncology– Bellvitge Biomedical Research Institute (IDIBELL), L'Hospitalet de Llobregat, Barcelona, Spain; Blanquerna Faculty of Health Sciences, Ramon Llull University, Barcelona, Spain.
Tilman Kühn, Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Domenico Palli, Cancer Risk Factors and Life-Style Epidemiology Unit, Institute for Cancer Research, Prevention and Clinical Network—ISPRO, Florence, Italy.
Elio Riboli, Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, United Kingdom.
Edward L Giovannucci, Channing Division of Network Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA; Department of Epidemiology, Harvard TH Chan School of Public Health, Harvard University, Boston, MA, USA; Department of Nutrition, Harvard TH Chan School of Public Health, Harvard University, Boston, MA, USA.
Barbara L Banbury, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Stephen B Gruber, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA; USC Norris Comprehensive Cancer Center, University of Southern California, Los Angeles, CA, USA.
Ulrike Peters, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Department of Epidemiology, University of Washington, Seattle, WA, USA.
Marc J Gunter, Section of Nutrition and Metabolism, International Agency for Research on Cancer, Lyon, France.
Data Availability
All data described in the article are provided within the article.
References
- 1. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–91. [DOI] [PubMed] [Google Scholar]
- 2. Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology. 2015;148(6):1244–60.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Cancer Research Fund (WCRF) / American Institute for Cancer Research. Diet, nutrition, physical activity and cancer: a global perspective. Continuous Update Project Expert Report 2018. London: WCRF; 2018. [Google Scholar]
- 4. Schatzkin A, Abnet CC, Cross AJ, Gunter M, Pfeiffer R, Gail M, Lim U, Davey-Smith G. Mendelian randomization: how it can—and cannot—help confirm causal relations between nutrition and cancer. Cancer Prev Res. 2009;2(2):104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The Alpha-Tocopherol Beta Carotene Cancer Prevention Study Group. The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N Engl J Med. 1994;330(15):1029–35. [DOI] [PubMed] [Google Scholar]
- 6. Lappe J, Watson P, Travers-Gustafson D, Recker R, Garland C, Gorham E, Baggerly K, McDonnell SL. Effect of vitamin D and calcium supplementation on cancer incidence in older women: a randomized clinical trial. JAMA. 2017;317(12):1234–43. [DOI] [PubMed] [Google Scholar]
- 7. Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JAet al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA. 2009;301(1):39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beresford SA, Johnson KC, Ritenbaugh C, Lasser NL, Snetselaar LG, Black HR, Anderson GL, Assaf AR, Bassford T, Bowen Det al. Low-fat dietary pattern and risk of colorectal cancer: the Women's Health Initiative Randomized Controlled Dietary Modification Trial. JAMA. 2006;295(6):643–54. [DOI] [PubMed] [Google Scholar]
- 9. Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Davey Smith G. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr. 2016;103(4):965–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dimitrakopoulou VI, Tsilidis KK, Haycock PC, Dimou NL, Al-Dabhani K, Martin RM, Lewis SJ, Gunter MJ, Mondul A, Shui IMet al. Circulating vitamin D concentration and risk of seven cancers: Mendelian randomisation study. BMJ. 2017;359:j4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ong J-S, Gharahkhani P, An J, Law MH, Whiteman DC, Neale RE, MacGregor S. Vitamin D and overall cancer risk and cancer mortality: a Mendelian randomization study. Hum Mol Genet. 2018;27(24):4315–22. [DOI] [PubMed] [Google Scholar]
- 12. Dashti HS, Shea MK, Smith CE, Tanaka T, Hruby A, Richardson K, Wang TJ, Nalls MA, Guo X, Liu Yet al. Meta-analysis of genome-wide association studies for circulating phylloquinone concentrations. Am J Clin Nutr. 2014;100(6):1462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meyer TE, Verwoert GC, Hwang S-J, Glazer NL, Smith AV, van Rooij FJ, Ehret GB, Boerwinkle E, Felix JF, Leak TSet al. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six loci influencing serum magnesium levels. PLos Genet. 2010;6(8):e1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Major JM, Yu K, Wheeler W, Zhang H, Cornelis MC, Wright ME, Yeager M, Snyder K, Weinstein SJ, Mondul Aet al. Genome-wide association study identifies common variants associated with circulating vitamin E levels. Hum Mol Genet. 2011;20(19):3876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mondul AM, Yu K, Wheeler W, Zhang H, Weinstein SJ, Major JM, Cornelis MC, Männistö S, Hazra A, Hsing AWet al. Genome-wide association study of circulating retinol levels. Hum Mol Genet. 2011;20(23):4724–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aschard H, Vilhjálmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet. 2015;96(2):329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, Gogele M, Anderson D, Broer L, Podmore Cet al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2014;5:4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cornelis MC, Fornage M, Foy M, Xun P, Gladyshev VN, Morris S, Chasman DI, Hu FB, Rimm EB, Kraft Pet al. Genome-wide association study of selenium concentrations. Hum Mol Genet. 2015;24(5):1469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evans DM, Zhu G, Dy V, Heath AC, Madden PA, Kemp JP, McMahon G, St Pourcain B, Timpson NJ, Golding Jet al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum Mol Genet. 2013;22(19):3998–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferrucci L, Perry JR, Matteini A, Perola M, Tanaka T, Silander K, Rice N, Melzer D, Murray A, Cluett Cet al. Common variation in the β-carotene 15,15'-monooxygenase 1 gene affects circulating levels of carotenoids: a genome-wide association study. Am J Hum Genet. 2009;84(2):123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grarup N, Sulem P, Sandholt CH, Thorleifsson G, Ahluwalia TS, Steinthorsdottir V, Bjarnason H, Gudbjartsson DF, Magnusson OT, Sparsø Tet al. Genetic architecture of vitamin B12 and folate levels uncovered applying deeply sequenced large datasets. PLos Genet. 2013;9(6):e1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hazra A, Kraft P, Lazarus R, Chen C, Chanock SJ, Jacques P, Selhub J, Hunter DJ. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet. 2009;18(23):4677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kestenbaum B, Glazer NL, Köttgen A, Felix JF, Hwang S-J, Liu Y, Lohman K, Kritchevsky SB, Hausman DB, Petersen A-Ket al. Common genetic variants associate with serum phosphorus concentration. J Am Soc Nephrol. 2010;21(7):1223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Seaghdha CM, Wu H, Yang Q, Kapur K, Guessous I, Zuber AM, Köttgen A, Stoudmann C, Teumer A, Kutalik Zet al. Meta-analysis of genome-wide association studies identifies six new loci for serum calcium concentrations. PLos Genet. 2013;9(9):e1003796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars NJ, Aguirre M, Venkataraman GR, Wainberg M, Ollila HM, Pirruccello JP, Qian Jet al. Genetics of 38 blood and urine biomarkers in the UK Biobank. BioRxiv. 2019. Available from: doi:10.1101/660506. [Google Scholar]
- 26. Gill D, Benyamin B, Moore LSP, Monori G, Zhou A, Koskeridis F, Evangelou E, Laffan M, Walker AP, Tsilidis KKet al. Associations of genetically determined iron status across the phenome: a mendelian randomization study. PLoS Med. 2019;16(6):e1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, Conti DV, Qu C, Jeon J, Edlund CKet al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51(1):76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Statist Med. 2008;27(8):1133–63. [DOI] [PubMed] [Google Scholar]
- 30. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodríguez-Barranco M, Tobías A, Redondo D, Molina-Portillo E, Sánchez MJ. Standardizing effect size from linear regression models with log-transformed variables for meta-analysis. BMC Med Res Methodol. 2017;17(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burgess S, Thompson SG, CRP CHD Genetics Collaboration . Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–64. [DOI] [PubMed] [Google Scholar]
- 33. Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Statist Med. 2015;34(21):2926–40. [DOI] [PubMed] [Google Scholar]
- 34. MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales Jet al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45(D1):D896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh Jet al. PhenoScanner: a database of human genotype–phenotype associations. Bioinformatics. 2016;32(20):3207–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Statist Med. 2017;36(11):1783–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toyokuni S. Role of iron in carcinogenesis: cancer as a ferrotoxic disease. Cancer Sci. 2009;100(1):9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bastide NM, Pierre FH, Corpet DE. Heme iron from meat and risk of colorectal cancer: a meta-analysis and a review of the mechanisms involved. Cancer Prev Res. 2011;4(2):177–84. [DOI] [PubMed] [Google Scholar]
- 43. Qiao L, Feng Y. Intakes of heme iron and zinc and colorectal cancer incidence: a meta-analysis of prospective studies. Cancer Causes Control. 2013;24(6):1175–83. [DOI] [PubMed] [Google Scholar]
- 44. Fonseca-Nunes A, Jakszyn P, Agudo A. Iron and cancer risk—a systematic review and meta-analysis of the epidemiological evidence. Cancer Epidemiol Biomarkers Prev. 2014;23(1):12–31. [DOI] [PubMed] [Google Scholar]
- 45. Cornish AJ, Law PJ, Timofeeva M, Palin K, Farrington SM, Palles C, Jenkins MA, Casey G, Brenner H, Chang-Claude Jet al. Modifiable pathways for colorectal cancer: a mendelian randomisation analysis. Lancet Gastroenterol Hepatol. 2020;5(1):55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ibáñez-Sanz G, Díez-Villanueva A, Riera-Ponsati M, Fernández-Villa T, Fernández Navarro P, Bustamante M, Llorca J, Amiano P, Ascunce N, Fernández-Tardón Get al. Mendelian randomization analysis rules out disylipidaemia as colorectal cancer cause. Sci Rep. 2019;9(1):13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez-Broadbent H, Law PJ, Sud A, Palin K, Tuupanen S, Gylfe A, Hänninen UA, Cajuso T, Tanskanen T, Kondelin Jet al. Mendelian randomisation implicates hyperlipidaemia as a risk factor for colorectal cancer. Int J Cancer. 2017;140(12):2701–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moghadaszadeh B, Beggs AH. Selenoproteins and their impact on human health through diverse physiological pathways. Physiology (Bethesda). 2006;21:307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takata Y, Kristal AR, King IB, Song X, Diamond AM, Foster CB, Hutter CM, Hsu L, Duggan DJ, Langer RDet al. Serum selenium, genetic variation in selenoenzymes, and risk of colorectal cancer: primary analysis from the Women's Health Initiative Observational Study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2011;20(9):1822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Duffield-Lillico AJ, Reid ME, Turnbull BW, Combs GF Jr, Slate EH, Fischbach LA, Marshall JR, Clark LC. Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiol Biomarkers Prev. 2002;11(7):630–9. [PubMed] [Google Scholar]
- 51. Hughes DJ, Fedirko V, Jenab M, Schomburg L, Méplan C, Freisling H, Bueno-de-Mesquita HB, Hybsier S, Becker N-P, Czuban Met al. Selenium status is associated with colorectal cancer risk in the European prospective investigation of cancer and nutrition cohort. Int J Cancer. 2015;136(5):1149–61. [DOI] [PubMed] [Google Scholar]
- 52. Bera S, De Rosa V, Rachidi W, Diamond AM. Does a role for selenium in DNA damage repair explain apparent controversies in its use in chemoprevention?. Mutagenesis. 2013;28(2):127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Friso S, Udali S, De Santis D, Choi S-W. One-carbon metabolism and epigenetics. Mol Aspects Med. 2017;54:28–36. [DOI] [PubMed] [Google Scholar]
- 54. Giovannucci E. Epidemiologic studies of folate and colorectal neoplasia: a review. J Nutr. 2002;132(8 Suppl):2350S–5S. [DOI] [PubMed] [Google Scholar]
- 55. Oliai Araghi S, Kiefte-de Jong JC, van Dijk SC, Swart KMA, van Laarhoven HW, van Schoor NM, de Groot L, Lemmens V, Stricker BH, Uitterlinden AGet al. Folic acid and vitamin B12 supplementation and the risk of cancer: long-term follow-up of the B Vitamins for the Prevention of Osteoporotic Fractures (B-PROOF) Trial. Cancer Epidemiol Biomarkers Prev. 2019;28(2):275–82. [DOI] [PubMed] [Google Scholar]
- 56. Kabat GC, Kim MY, Sarto GE, Shikany JM, Rohan TE. Repeated measurements of serum carotenoid, retinol and tocopherol levels in relation to colorectal cancer risk in the Women's Health Initiative. Eur J Clin Nutr. 2012;66(5):549–54. [DOI] [PubMed] [Google Scholar]
- 57. Malila N, Virtamo J, Virtanen M, Pietinen P, Albanes D, Teppo L. Dietary and serum α-tocopherol, β-carotene and retinol, and risk for colorectal cancer in male smokers. Eur J Clin Nutr. 2002;56(7):615–21. [DOI] [PubMed] [Google Scholar]
- 58. Stepien M, Jenab M, Freisling H, Becker N-P, Czuban M, Tjønneland A, Olsen A, Overvad K, Boutron-Ruault M-C, Mancini FRet al. Pre-diagnostic copper and zinc biomarkers and colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition cohort. Carcinogenesis. 2017;38(7):699–707. [DOI] [PubMed] [Google Scholar]
- 59. Larsson SC, Orsini N, Wolk A. Vitamin B6 and risk of colorectal cancer: a meta-analysis of prospective studies. JAMA. 2010;303(11):1077–83. [DOI] [PubMed] [Google Scholar]
- 60. Baron JA, Beach M, Mandel JS, van Stolk RU, Haile RW, Sandler RS, Rothstein R, Summers RW, Snover DC, Beck GJet al. Calcium supplements for the prevention of colorectal adenomas. N Engl J Med. 1999;340(2):101–7. [DOI] [PubMed] [Google Scholar]
- 61. Bonithon-Kopp C, Kronborg O, Giacosa A, Räth U, Faivre J; European Cancer Prevention Organisation Study Group . Calcium and fibre supplementation in prevention of colorectal adenoma recurrence: a randomised intervention trial. Lancet. 2000;356(9238):1300–6. [DOI] [PubMed] [Google Scholar]
- 62. Wallace K, Baron JA, Cole BF, Sandler RS, Karagas MR, Beach MA, Haile RW, Burke CA, Pearson LH, Mandel JSet al. Effect of calcium supplementation on the risk of large bowel polyps. J Natl Cancer Inst. 2004;96(12):921–5. [DOI] [PubMed] [Google Scholar]
- 63. Wactawski-Wende J, Kotchen JM, Anderson GL, Assaf AR, Brunner RL, O'Sullivan MJ, Margolis KL, Ockene JK, Phillips L, Pottern Let al. Calcium plus vitamin D supplementation and the risk of colorectal cancer. N Engl J Med. 2006;354(7):684–96. [DOI] [PubMed] [Google Scholar]
- 64. Bolland MJ, Grey A, Gamble GD, Reid IR. Calcium and vitamin D supplements and health outcomes: a reanalysis of the Women's Health Initiative (WHI) limited-access data set. Am J Clin Nutr. 2011;94(4):1144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the article are provided within the article.