Summary
Fibroblast-growth-factor homologous factor (FHF1) gene variants have recently been associated with developmental and epileptic encephalopathy (DEE). FHF1 encodes a cytosolic protein that modulates neuronal sodium channel gating. We aim to refine the electro-clinical phenotypic spectrum of patients with pathogenic FHF1 variants. We retrospectively collected clinical, genetic, neurophysiologic, neuroimaging data of 17 patients with FHF1-DEE.
Sixteen patients had recurrent heterozygous FHF1 missense variants: fourteen had the recurrent p.Arg114His variant and two had a novel likely pathogenic variant p.Gly112Ser. The p.Arg114His variant is associated with an earlier onset and more severe phenotype. One patient carried a chromosomal microduplication involving FHF1. Twelve patients carried a de novo variant, five (29.5%) inherited from parents with gonadic or somatic mosaicism. Seizure onset was between 1 day and 41 months, in 76.5% it was within 30 days. Tonic seizures were the most frequent seizure type. Twelve patients (70.6%) had drug-resistant epilepsy, 14 (82.3%) intellectual disability, 11 (64.7%) behavioral disturbances. Brain MR showed mild cerebral and/or cerebellar atrophy in 9 patients (52.9%). Overall, our findings expand and refine the clinical, EEG and imaging phenotype of patients with FHF1-DEE which is characterized by early onset epilepsy with tonic seizures, associated with moderate to severe ID and psychiatric features.
Keywords: Developmental and epileptic encephalopathy, FHF1, FGF12, epilepsy, genetic, neonatal onset
Introduction
Developmental and epileptic encephalopathies (DEEs) are clinically and genetically heterogeneous severe neurodevelopmental disorders1. DEEs often start in infancy or early childhood and are severe conditions characterized by multiple seizure types, frequent epileptiform activity on EEG, and developmental slowing or regression2. To date, a genetic etiology can be identified in more than 30% of cases, 60–80% for epilepsies with neonatal onset3,4. Most patients have de novo pathogenic variants in genes encoding neuronal ion channels or proteins involved in synaptic transmission, regulatory, and developmental functions2.
Recently, de novo mutations in fibroblast-growth-factor homologous factor 1 (FHF1) gene, encoding a voltage-gated sodium channel subunit (Nav1.6) binding protein, have been reported in patients with severe epilepsies5–11. Nevertheless, a definite clinical phenotype has not clearly emerged. The recurrent missense variants of FHF1 generally occur mostly as a de novo event in these patients and have been demonstrated to lead to a gain-of-function of the voltage-gated sodium channel (Nav1.6), thus increasing neuronal excitability9,11.
The aim of this study is to report a large series of patients with pathogenic FHF1 variants, including patients already published and new ones, in order to further delineate the phenotypic spectrum of FHF1-DEE, inform management and prognosis, and identify genotype-phenotype correlation.
Methods
This is an international retrospective multicenter study. We ascertained patients with FHF1-related epilepsy, from 14 epilepsy centers (Belgium, Canada, China, France, Italy, Japan, Poland, United Kingdom, and USA). Patients #A-K and #Q have been previously reported and we obtained additional information on all cases5–11. Only the genetic variant has been reported for patient #L12. For each patient, the referring physician completed a detailed medical questionnaire including demographic data, FHF1 variant, family history, age at epilepsy onset, seizure semiology and frequency, EEG features, neurological examination, comorbidities, brain imaging and treatment. We reported all the collected data and we correlated age at epilepsy onset, seizure semiology and SE occurrence, developmental delay, and MR findings with the genotypes: p.Arg114His, c.334G>A and FHF1 microduplication.
The study was approved by the local institutional ethics committees. Written informed consent was obtained from all patients and parents or legal guardians. Epileptic seizures were classified according to the International League Against Epilepsy (ILAE) Classification1,13.
Results
We ascertained 17 patients with FHF1-DEE, including 12 cases previously described. Median age at study was 8.7±8.59 years (range 1 month - 33 years); seven (41.2%) patients were female. All cases were sporadic except for two siblings (#A,B). Clinical, EEG, neuroimaging and genetic details are summarized in Table 1.
Table 1.
Summary of molecular, clinical, EEG, and neuroimaging features of all published and unpublished patients with FHF1 developmental and epileptic encephalopathies.
| PATIENT ID | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Siekierska et al. 2016 | Siekierska et al. 2016 | Al-mehmadi et al. 2016 | Al-mehmadi et al. 2016 | Al-mehmadi et al. 2016 | Guella et al. 2016 | Guella I. et al. 2016 | Villeneuve et al. 2017 | Shi RM. et al. 2017 | Takeguchi R. et al. 2018 | Takeguchi R. et al. 2018 | Epilepsy Genetics Initiative, 2019* phenotype unpublished | unpublished | unpublished | unpublished | unpublished | Paprocka et al. 2019 |
|
Gender/
Age at last observation/death (cause) |
F/died age 7y (SE) | M/died age 3y6m (unknow cause) | M/3y | F/16y | F/8y | F/3y3m | F/15y | M/9y | M/15y1m | M/33y3m | M/2y6m | M/5y8m | F/1 m | F/13y | M/2y10m | M/4y2m | M/4y6m |
| FHF1 variant, inheritance | c.341G>A* presumed gonadal mosaicism |
c.341G>A* presumed gonadal mosaicism |
c.341G>A de novo | c.341G>A de novo | c.341G>A de novo | c.341G>A de novo | c.341G>A de novo | c.341G>A de novo | arr[hg19] 3q28q29 ×1, 0.58-Mb gain, including FHF1 gene, de novo | c.341G>A^ inherited, see legend |
c.341G>A de novo | c.341G>A de novo | c.341G>A de novo | c.341G>A de novo | c.341G>A$
inherited, see legend |
c.334G>A#
inherited, see legend |
c.334G>A de novo |
| EPILEPSY | |||||||||||||||||
| Epilepsy onset | 14d | 28d | 2d | 42d | 2d | 2d | 2d | 1d | 3y5m | 7d | 1d | 31d | 2d | 3d | 8d | 4m | 4m |
| Seizure type | TS | TS | FTS, FBTCS | FTS, MS, FBTCS | FTS, FS | FTS | FS, FBTCS | AS, FS | GTCS, FS, FTS | TS, ES | FTS, FS, FBTCS | TS | FS FTS |
A, FTS, FBTCS | FS, TS, FBTCS | TS, FBTCS | TS, MS, ES, FTS,FBTCS |
| Epilepsy Type | Combined generalized and focal epilepsy | Combined generalized and focal epilepsy | Focal epilepsy | Combined generalized and focal epilepsy | Focal epilepsy | Focal epilepsy | Focal epilepsy | Combined generalized and focal epilepsy | Combined generalized and focal epilepsy | Focal epilepsy | Focal epilepsy | Combined generalized and focal epilepsy | Focal epilepsy | Combined generalized and focal epilepsy | Unknown | Unknown | Combined generalized and focal epilepsy |
| SE (frequency) | frequent | infrequent | frequent | frequent | frequent | no | n.a. | n.a. | no | monthly | no | yes (twice) | no | frequent | yes (twice) | no | no |
| EEG | |||||||||||||||||
| Interictal EEG | Slow BG, multifocal SW (onset) | Slow BG, multifocal SW (onset) | Slow BG, multifocal SW | Slow BG, multifocal SW | Slow BG, multifocal SW | Discontinuous, multifocal SW (onset); increase of diffuse/multifocal SW; from 10m normal EEG (FU) | Slow BG left temporal SW (onset); slow BG and multifocal SW (FU) | Normal BG, multifocal spikes (onset); focal spikes (FU) | Slow BG, Frontal SW (onset); Slow BG, multifocal SW (FU) | Suppression burst (onset); slow BG with focal spikes (FU) | Slow BG, multifocal and diffuse SW (onset); slower BG multifocal SW (FU) | Normal BG, multifocal SW (onset); slow BG and multifocal SW (FU) | Discontinuous and multifocal SW (onset); slow BG with increase of diffuse/multifocal SW | Slow BG, multifocal SW (onset); Slow BG and multifocal and diffuse SW (FU) | Normal BG multifocal SW (onset) Slow BG and posterior SW (FU) | Normal | Normal (onset); generalized and focal paroxysmal in temporal regions (FU) |
| Ictal EEG | Generalized onset (tonic seizure) | Generalized onset (tonic seizure) | Focal to bilateral t-c | n.a. | n.a. | Generalized onset | Generalized onset (tonic seizure) | Generalized onset (tonic seizure) | Generalized onset (AS, TS, GTCS) Focal seizures: R or L hemisphere | n.a. | Generalized onset. Seizure activity migrated from one region to another | Diffuse onset | n.a. | L hemisphere; R frontal/frontotemporal | R central | n.a. | Generalized polyspikes, spike-SW complexes |
| TREATMENT | |||||||||||||||||
| ASM | PB, VPA, PHT, GVG, TPM, CZP; PN | PB, VPA, PHT, GVG, TPM | LEV, PB, KD | PHT, PER, VNS | PHT, PRG, PER, VNS | PB, LEV, TPM, PHT, CBZ | PB, TPM, LTG, RUF | PB, GVG, CBZ, CLB, ESM, KD | PB, CLB, VPA, KBr, PHT, LEV, NZP | VPA, PB, PHT, CZP, AZA, PHT, GBP | PB, PN, CBZ, CLB, VPA, ZNS, LEV, KBr, PHT | PB, PHT, CBZ, LEV, RTG, VPA | PB, LEV, PN/PLP, TPM, CBZ, PHT | PB, LEV, PHT, OXC, LCM, CLZ, PER, TPM, VNS, KD | VPA, CZP, LEV, GVG, CBZ, PN, PLP, PB, Folic Acid, Biotine | LEV, VPA, LTG | PB, CBZ, VPA, LEV, GVG, steroids, PHT |
| ASM efficacy | Resistant to ASMs; best response to PHT | Resistant to ASMs; best response to PHT | Resistant to ASMs | Resistant to ASMs; partially responsive to PHT and VNS | Resistant to ASMs | Responsive to PHT and CBZ | Responsive: to RUF and LTG, | Partially responsive to CBZ | Best response to high dose of PHT | Partially responsive to PHT, CZP and VPA | Best response to PHT and high-dose of PB | Partially responsive to VPA | Resistant to ASMs | Resistant to ASMs | ASMs-responsive | ASMs-responsive | Best response to PB and PHT |
| DEVELOPMENT | |||||||||||||||||
| ID | severe | severe | severe | severe | moderate | no | moderate | mild | severe | severe | severe | moderate | moderate | moderate | moderate | no | moderate |
| ASD and other disturbances | stereotypies, absent eye contact, acquired microcephaly | stereotypies, absent eye contact, acquired microcephaly | n.a. | n.a. | yes | no | yes | very tight | yes, stereotypies, absent eye contact | yes, stereotypies, absent eye contact | No, poor eye contact, congenital microcephaly | yes, stereotypies, absent eye contact | No, rapid mood swings, congenital microcephaly | yes, severe obsessive behaviour | yes, stereotypies, absent eye contact | no | yes |
| BRAIN MR | |||||||||||||||||
| First brain MR/age | Normal/6m | Normal/4m | Normal/5d | Normal/1y | Normal/21d | Normal/3d | Mild Chiari I /14d | Tight T2 weighted hyper intensity of the parietal region, cerebellum and brain stem/15d | Mild cerebral and cerebellar atrophy/3y | Mild enlargement of lateral ventricle/7y | Mild cerebral atrophy/6m | Mild cerebral atrophy/4m | Normal/5d | Normal/1y | Normal/21d | Normal/4m | Normal/4m |
| Second brain MR/age | Cerebellar atrophy/6y | Cerebellar atrophy/3y | Cerebral atrophy/2y | Cerebellar atrophy/8y | Bilateral mesial temporal sclerosis (R>L), mild prominence of cerebellar folia/12y | No | Mild Chiari I /2y | n.a. | Mild cerebral and cerebellar atrophy/8y | Mild enlargement of lateral ventricle/13y | Diffuse cerebral atrophy/1y7m | Mild cerebral atrophy/2y9m | Normal/10d | Normal/4y | Normal/2y | Normal/3y4m | No |
Table Legend:
= presumed gonadal mosaicism in unaffected parent;
= inherited from healthy mother (blood leukocyte with a variant allele fraction of 11.7% – 11/94 clones);
= inherited from affected father with onset of drug-resistant epilepsy at age 8 months (blood leukocyte mutant allele fraction of 7% – 10/1479 reads);
= inherited from affected mother who had epilepsy during infancy (blood leukocyte mutant allele fraction of 52% – 178/338 reads); A= Absences; AS= atonic seizure; ASD= autism spectrum disorder; ASM= anti-seizure medication; AZA= acetazolamide; BG= background activity; CBZ= carbamazepine; CZP= clonazepam; d= days; ES= epileptic spasm; ESM= etosuximide; F= female; FS= focal seizure; FTS=focal tonic seizure; FBTCS=focal to bilateral tonic clonic seizures; FU= follow-up; GBP= gabapentin; GTCS= generalized tonic-clonic seizure; GVG= vigabatrin; ID= intellectual disability; KBr= potassium bromide; KD= ketogenic diet; L= left; LEV= levetiracetam; LTG= lamotrigine; M= male; m= months; MS= myoclonic seizure; n.a.= not available; NZP= nitrazepam; OXC= oxcarbazepine; PB= phenobarbital; PER= perampanel; PHT= phenytoin; PLP= pyridoxal-5-phosphate; PN= pyridoxine; PRG= pregabalin; R= right; RTG= retigabine; RUF= rufinamide; SE=status epilepticus; SW= spike and wave; TPM= topiramate; TS= tonic seizure; VNS= vagal nerve stimulation; VPA= valproate; y=years; ZNS= zonisamide.
FHF1 pathogenic variants (NM_021032.4)
Two heterozygous missense mutations were identified: three recurrent c.341G>A, p.(Arg114His) was present in 14 patients, whereas two unrelated patients had a novel missense variant: c.334G>A, p.(Gly112Ser) (#P,Q). They were localized in the same protein domain. In silico predictions for the p.(Gly112Ser) variant were very similar to the recurrent p.(Arg114His): CADD=28.3, Polyphen-2=Pathogenic, Mutation Taster = Pathogenic, and SIFT=Tolerated. According to the standards and guidelines for the interpretation of sequence variants of the ACMG, this variant could be classified as likely pathogenic (PS2, PP3, PM2, PP5). It was absent from the gnomAD database of control individuals but had be reported once in ClinVAr as likely pathogenic in a patient with developmental and epileptic encephalopathy (RCV000626031.1).
In addition, one patient (#I) carried a chromosomal microduplication including the whole FHF1 gene (0.58-Mb gain, arr[hg19] 3q28q29 (191876978_192454675)x1). FHF1 pathogenic variants were de novo in 12 patients. Five patients (29.5%) inherited the variant from a parent, unaffected or affected with a milder epilepsy phenotype, who had the variant present but with mosaicism with variant allele fractions 0 (germline mosaicism) to 52%. Patient #J inherited the FHF1 variant from his unaffected mother who had somatic mosaicism (blood leukocytes showed a variant allele fraction of 11.7% - 11/94 clones). Two patients inherited the variant from their affected parents: patient #O from his 23 year-old father who had a drug-resistant epilepsy since the age of 8 months, carrying a somatic mosaicism (blood leukocyte variant allele fraction of 7% - 10/1479 reads); and patient #P from his mother who had epilepsy during infancy (blood leukocyte variant allele fraction of 52% - 178/338 reads). In one family (#A,B), paternal germline mosaicism was presumed based on the occurrence of one epileptic seizure when the father was 5 years old (Table 1).
Epilepsy
Seizure onset ranged from 1 day to 3 years 5 months. In thirteen patients (76.5%), seizures onset was within 30 days. Tonic seizures were the most common seizure type (15/17, 88.2%), and were associated with autonomic signs such as apnea (5/15, 33.3%) and bradycardia (2/15, 13.3%). Fourteen patients developed additional seizures, including focal to bilateral tonic-clonic (n=11), myoclonic (n=2), atonic (n=2), epileptic spasms (n=2), absence (n=1) and generalized tonic-clonic (n=1) after the age of 3 years and 5 months. Status epilepticus (SE) occurred in 8 (47%) patients. Seizure frequency was highly variable, from multiple per day to long seizure-free periods lasting up to several years (12 years for patient #J). Following ILAE classification1, eight patients had combined generalized and focal epilepsy, seven presented focal epilepsy, and two unknown epilepsy.
Twelve patients (70.6%) had drug resistant epilepsy, while five (#F,G,O,P,Q) achieved good seizure control with different drug combinations. Drugs most commonly used were phenobarbital and phenytoin. Twelve patients were treated with phenytoin with a transient effect in six (#A,B,D,F,I,K). Data on response to other drugs (phenobarbital, rufinamide, lamotrigine, carbamazepine and vigabatrin) were not conclusive. Ketogenic diet and vagal nerve stimulation were beneficial in patients #N and #D respectively.
Two siblings (#A,B) died at age 7 and 3.5 years from SE and unknown cause, respectively.
EEG studies
At seizure onset, EEG was available in 13 patients: background activity was slow in six (46.1%), discontinuous in two (15.4%), normal in four (30.8%), one patient at onset had a suppression-burst pattern (7.7%). Nine patients (69.2%) had multifocal spikes, and two (15.4%) had focal spikes. Discontinuous and suppression burst EEG patterns were seen between 2 and 7 days of life. During follow-up, 12 patients (70.6%) showed increased background slowing with marked suppression in one case (#J), 12 patients showed increased interictal focal or multifocal spikes. Four patients also had diffuse spike and wave discharges.
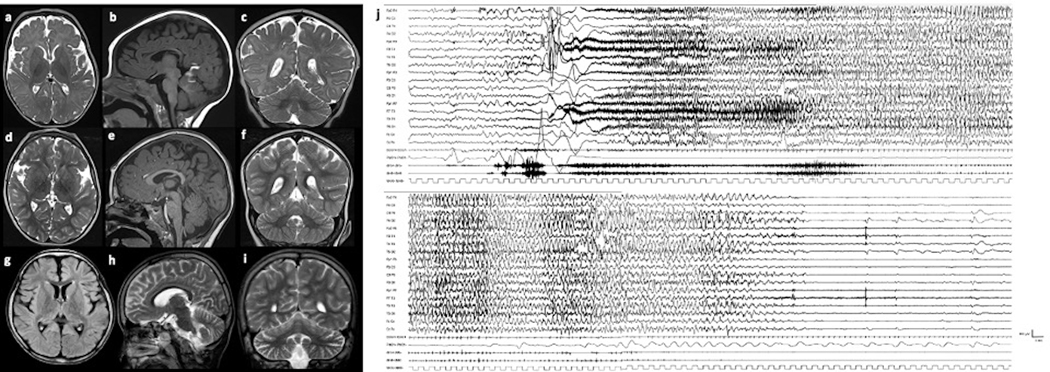
Ictal EEGs were reported as focal (with rapid spread to bilateral regions) or generalized with low voltage fast activity, followed by diffuse rhythmic spikes and postictal suppression (Fig.1).
Figure 1. Brain MR of patients #L and #I (a-i), and Ictal EEG of patient #L (j).
Patient #L at the age of 2 years 3 months (a-c) and 2 years 7 months (d-f): cerebral atrophy, with enlarged subarachnoid spaces around the frontal and insular lobes, without significant progression of the atrophy. Patient #I at the age of 8 years (g-i): mild cerebral and cerebellar atrophy with enlarged subarachnoid spaces around the frontal and insular lobes and cerebellar folia. Ictal EEG of patient #L at the age of 42 days (j). Ictal discharge starts with diffuse bilateral, symmetrical, low-voltage fast activity, increasing in amplitude and decreasing in frequency. The patient has a massive tonic contraction with perioral cyanosis and sialorrhea. Polygraphic recording shows ictal bradycardia at seizure onset for about 5 seconds concomitant with the beginning of the tonic phase (see bilateral contraction of upper limb in deltoids). Afterwards, the patient appears floppy and pale associated with a brief compensatory tachycardia. The seizure spontaneously ends after 86 seconds.
Other neurologic findings
Intellectual disability (ID) was severe in seven patients, moderate in seven and mild in one; nine were non-verbal. Out of 14 patients older than 18 months, nine walked independently, while six of them were ataxic, and five never walked. Three patients had cortical visual impairment (#A,B,C), one nystagmus (#G). In 14 out of 17 patients with neonatal onset epilepsy developmental delay was not noted before seizure onset, in the remaining three patients with onset between 4 months and 3 years and 5 months the development was reported as normal before seizure onset.
Brain MR findings
Brain MR was normal in 11/15 patients who had imaging within the first year of life. The remaining four patients had: a mild Chiari I malformation (#G), T2 weighted hyperintensity of the parietal areas, cerebellum and brainstem (#H), and mild cerebral atrophy (#K,L) (Fig.2). During follow-up, in four patients MR remained normal. Five patients developed cerebellar atrophy and one of them also bilateral mesial temporal sclerosis. Two patients (#I,J) had their first brain MR later in life, between the age of 3 and 7 years, revealing mild cerebral and cerebellar atrophy (Fig.2)
Overall nine out of 17 patients (52.9%) had cerebral (n=5) and/or cerebellar (n=5) atrophy on brain MR, which was progressive in six patients.
Genotype-phenotype correlation
Seizures onset was before the age of 42 days in all 14 patients with the p.Arg114His variant (13 within 30 days, and 10 out of them within eight days of life). Seizure onset was at 4 months in both children with the p.Gly112Ser variant (#P-Q). Patient (#I) with FHF1 duplication developed seizures at age 3 years 5 months.
Twelve out of 14 patients (85.7%) with the p.Arg114His variant had moderate to severe ID, while the two patients with the p.Arg114His mutation had normal development (#P) and one moderate ID (#O). Brain MR abnormalities were recurrent in patients with p.Arg114His variant, while both patients with p.Gly112Ser variant had normal brain MR.
Discussion
We present a large cohort of individuals with FHF1-DEE, incorporating twelve previously published case reports, and refine the phenotypic spectrum.
FHFs are small cytosolic proteins that interact with the cytoplasmic tails of voltage-gated sodium channels (Nav1.6), encoded by SCN8A, and promote excitability by elevating the voltage dependence of neuronal sodium channel fast inactivation9.
Siekierska et al. were the first ever to link the FHF1 gene to an early-onset epileptic encephalopathy, also demonstrating a gain-of-function effect of the FHF1 mutant protein (p.Arg114His)9. The prevalence of FHF1-DEE has not been estimated, however it seems to be rare as only 12 patients have been reported since 20165–11 and we collected only 5 additional new patients. Interestingly, complex chromosomal rearrangements involving 9p deletion and the 3q28q29 microduplication involving FHF1 have been recently described in 16 patients. However, only 3 out of 16 had epilepsy14, far less than patients with FHF1 missense variants.
As previously reported, the FHF1 p.Arg114His missense mutation is a hotspot locus and acts in a gain-of-function fashion on voltage-gated sodium channels9. A gain-of-function mechanism has also been invoked for the p.Gly112Ser variant and FHF1 duplication10,14.
In terms of genotype-phenotype correlation, we found that patients with the recurrent p.Arg114His missense mutation had an earlier epilepsy onset, mainly in the neonatal period, with severe developmental impairment and psychiatric features. Patients with the p.Gly112Ser variant and the FHF1 duplication, had later epilepsy onset, after the age of four months, and a milder clinical phenotype in terms of development. Conversely, there were no significant differences between the two missense variants, in terms of seizure semiology. Evolution to SE was frequent and, in about one third of patients, SE was recurrent during life.
The clinical outcome of patients with FHF1 pathogenic variants was poor in most of patients. ID was moderate or severe in 76,47% of patients, and a developmental regression was observed in 47.1%. Patients with the FHF1 p.Arg114His mutation had even poorer outcome with a moderate-severe ID in 78.6% of cases, while patients with the p.Gly112Ser mutation had normal development or moderate ID. Because of neonatal seizures onset, in most of patients it was difficult to ascertain the role of epilepsy in developmental delay.
The retrospective nature of this study does not allow to draw any firm conclusions about the efficacy of specific anti-seizure medications (ASM). However, it is worth noting that phenytoin, was effective in six patients (35.3%), confirming what was observed in single cases10,11. The efficacy of sodium channel blocking drugs, such as phenytoin, could be explained by the interaction between FHF1 and the NaV1.6 sodium channel subunit, encoded by SCN8A. In SCN8A-DEE, epilepsy is responsive to sodium channel blockers15–16.
Brain MR showed no specific abnormalities at seizure onset, while 52.9% of patients had cerebral and/or cerebellar atrophy on brain MR during follow-up, which was progressive in six patients, mostly in patients with p.Arg114His mutation.
The overall mortality (11.8%) is high compared with other epilepsies, but comparable with SCN8A-DEE and SCN2A-DEE17. Two patients of our cohort died during infancy: one died during SE, while the other of unknown cause, consistent with possible SUDEP. Both of them carried the recurrent p.Arg114His mutation, which is associated with the poorer phenotype.
With regard to inheritance, as observed in other gene related DEEs18, the majority of mutations arise de novo; although it is remarkable that in 29.5% of cases, somatic or gonadal mosaicism was reported in a parent. Because of the possibility of undetected gonadal or somatic mosaicism, it is critical to offer reproductive counseling to couples who have a child with FHF1-DEE regardless of whether the disease-causing mutation has been detected in a parent or not18.
Comparing FHF1-DEE with the other neonatal onset genetic DEEs (such as SCN2A, KCNQ2, SCN8A), they share a high occurrence of tonic seizures. Cerebellar atrophy has previously been reported in DEEs associated with mutations in several other genes including those encoding voltage-gated sodium channel subunits such as SCN8A15. Among other early onset DEEs, SCN8A-DEE seems to be the condition which FHF1-DEE share more features, such as seizure type, response to drugs and cerebellar atrophy and these common features might be due to the interaction between FHF1 and the NaV1.6 sodium channel subunit, encoded by SCN8A.
Overall, we described the phenotype of 17 patients with FHF1-DEE collected through an international multicenter collaboration. Most of the patients presented with drug-resistant epilepsy and, even if most of the drugs were not effective, a slight improvement was reached with the use of sodium channel blockers. EEG showed mainly multifocal abnormalities, not specific for this condition.
Our findings expand and refine the clinical, EEG and imaging phenotype of patients with FHF1-DEE which is characterized by an early onset epilepsy with tonic seizures, associated with moderate to severe ID and psychiatric features. Further experimental studies are needed to shed light on the underlying pathophysiology of FHF1-DEE in order to inform management and treatment, define the natural history, and prognostic factors for outcome.
Acknowledgments
We would like to thank the patients and their families for participation in this study.
Footnotes
Disclosure
The authors have no conflicts of interest to report. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- 1.Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017;58:512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trivisano M, Specchio N. What are the epileptic encephalopathies? Curr Opin Neurol. 2020. Feb 7: 10.1097/WCO.0000000000000793. doi:. [DOI] [PubMed] [Google Scholar]
- 3.Rochtus A, Olson HE, Smith L, Keith LG, El Achkar C, Taylor A, et al. Genetic diagnoses in epilepsy: The impact of dynamic exome analysis in a pediatric cohort. Epilepsia 2020;61:249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symonds JD, McTague A. Epilepsy and developmental disorders: Next generation sequencing in the clinic. Eur J Paediatr Neurol. 2020;24:15–23. [DOI] [PubMed] [Google Scholar]
- 5.Takeguchi R, Haginoya K, Uchiyama Y, Fujita A, Nagura M, Takeshita E, et al. Two Japanese cases of epileptic encephalopathy associated with an FGF12 mutation. Brain Dev 2018;40:728–32. [DOI] [PubMed] [Google Scholar]
- 6.Villeneuve N, Abidi A, Cacciagli P, Mignon-Ravix C, Chabrol B, Villard L, et al. Heterogeneity of FHF1 related phenotype: Novel case with early onset severe attacks of apnea, partial mitochondrial respiratory chain complex II deficiency, neonatal onset seizures without neurodegeneration. Eur J Paediatr Neurol 2017;21:783–6. [DOI] [PubMed] [Google Scholar]
- 7.Guella I, Huh L, McKenzie MB, Toyota EB, Bebin EM, Thompson ML, et al. De novo FGF12 mutation in 2 patients with neonatal-onset epilepsy. Neurol Genet 2016;2:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Mehmadi S, Splitt M, Ramesh V, DeBrosse S, Dessoffy K, Xia F, et al. FHF1 (FGF12) epileptic encephalopathy. Neurol Genet 2016;2:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siekierska A, Isrie M, Liu Y, Scheldeman C, Vanthillo N, Lagae L, et al. Gain-of-function FHF1 mutation causes early-onset epileptic encephalopathy with cerebellar atrophy. Neurology 2016;86:2162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi RM, Kobayashi T, Kikuchi A, Sato R, Uematsu M, An K, et al. Phenytoin-responsive epileptic encephalopathy with a tandem duplication involving FGF12. Neurol Genet 2017;3:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paprocka J, Jezela-Stanek A, Koppolu A, Rydzanicz M, Kosińska J, Stawiński P, et al. FGF12p.Gly112Ser variant as a cause of phenytoin/phenobarbital responsive epilepsy. Clin Genet 2019;96:274–5. [DOI] [PubMed] [Google Scholar]
- 12.Epilepsy Genetics Initiative. The Epilepsy Genetics Initiative: Systematic reanalysis of diagnostic exomes increases yield. Epilepsia 2019;60:797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher RS, Cross JH, French JA, Higurashi N, Hirsch E, Jansen FE, et al. Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017;58:522–30. [DOI] [PubMed] [Google Scholar]
- 14.Oda Y, Uchiyama Y, Motomura A, Fujita A, Azuma Y, Harita Y, et al. Entire FGF12 duplication by complex chromosomal rearrangements associated with West syndrome. J Hum Genet 2019;64:1005–14. [DOI] [PubMed] [Google Scholar]
- 15.Gardella E, Marini C, Trivisano M, Fitzgerald MP, Alber M, Howell KB, et al. The phenotype of SCN8A developmental and epileptic encephalopathy. Neurology 2018;91(12):e1112–24. [DOI] [PubMed] [Google Scholar]
- 16.Oyrer J, Maljevic S, Scheffer IE, Berkovic SF, Petrou S, Reid CA . Ion Channels in Genetic Epilepsy: From Genes and Mechanisms to Disease-Targeted Therapies. Pharmacol Rev 2018;70:142–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg T, Nickels K, Wirrel C, Geerts AT, Callenbach PM, Arts WF, et al. Mortality risk in new-onset childhood epilepsy. Pediatrics 2013; 132(1):124–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers CT, Hollingsworth G, Muir AM, Schneider AL, Thuesmunn Z, Knupp A, et al. Parental Mosaicism in “De Novo” Epileptic Encephalopathies. The New England Journal of Medicine 2018;378,1646–8. [DOI] [PMC free article] [PubMed] [Google Scholar]