Summary
The fundamental process of polarized exocytosis requires the interconnected activity of molecular motors trafficking vesicular cargo within a dynamic cytoskeletal network. In plants, few mechanistic details are known about how molecular motors, such as myosin XI, associate with their secretory cargo to support the ubiquitous processes of polarized growth and cell division.
Live-cell imaging coupled with targeted gene knockouts and a high-throughput RNAi assay enabled the first characterization of the loss of Rab-E function. Yeast two-hybrid and subsequent in silico structural prediction uncovered a specific interaction between Rab-E and myosin XI that is conserved between P. patens and A. thaliana.
Rab-E co-localizes with myosin XI at sites of active exocytosis, and at the growing tip both proteins are spatiotemporally coupled. Rab-E is required for normal plant growth in P. patens and the rab-E and myosin XI phenotypes are rescued by A. thaliana’s Rab-E1c and myosin XI-K/E, respectively. Both PpMyoXI and AtMyoXI-K interact with PpRabE14, and the interaction is specifically mediated by PpMyoXI residue V1422. This interaction is required for polarized growth.
Our results suggest the interaction of Rab-E and myosin XI is a conserved feature of polarized growth in plants.
Keywords: Arabidopsis thaliana, myosin XI, Physcomitrella patens, polarized growth, Rab-E GTPase, vesicle trafficking
Introduction
Polarized exocytosis requires precisely coordinated vesicle trafficking to support the essential function of polarized growth across eukaryotes, in addition to more plant-specific functions such as pathogen defense and cell wall biogenesis (Robatzek, 2007; Bove et al., 2008; Ebine and Ueda, 2015; Bibeau et al., 2018). Plant tip growth exploits turgor pressure and active transport of secretory vesicles to drive rapid anisotropic expansion (Hepler et al., 2001; McKenna et al., 2009). At present, a mechanistic understanding of how secretory vesicles are trafficked for exocytosis in plants is lacking.
Polarized delivery of secretory vesicles is the rate limiting step for tip-growth in plants, which is dependent upon the molecular motor myosin XI and exquisite regulation of endomembrane identity (Woollard and Moore, 2008; Vidali et al., 2010; Madison and Nebenfuhr, 2013; Rounds and Bezanilla, 2013). Rab GTPases can impart membrane specificity through interactions of divergent effectors, and these effectors are discriminated by dynamic structural rearrangements of the Rab in a nucleotide-dependent manner (Khan and Menetrey, 2013; Pfeffer, 2017). In yeast, Myo2’s C-terminal globular tail sequentially interacts with vesicle-localized Rab GTPases Ypt31/32 and Sec4 to faithfully transport vesicles to the growing bud tip and sites of cytokinesis in a stereotypical “transport cascade” (Schott et al., 1999; Pashkova et al., 2006; Lipatova et al., 2008; Jin et al., 2011; Donovan and Bretscher, 2012). Myo2 and Sec4 interact and localize to secretory vesicles, and both interact with the plasma membrane localized, hetero-octameric tethering complex called exocyst (Jin et al., 2011; Donovan and Bretscher, 2012). Sec4 is homologous to the Rab8 subfamily in mammals, and despite evolutionary distance the subfamily’s interaction with class V myosin is conserved from yeast to human (Welz and Kerkhoff, 2017). An analogous Rab cascade in mammalian cells (Knodler et al., 2010; Mizuno-Yamasaki et al., 2012), suggests the presence of a conserved eukaryotic polarized exocytosis mechanism.
The plant Rab-E subfamily is homologous at the amino acid level to Sec4/Rab8 (Rutherford and Moore, 2002), suggesting the presence of an analogous Rab-mediated transport model of secretory vesicles by myosin XI. Both functional and live-cell imaging studies demonstrated Rab-E’s involvement in anterograde transport to the plasma membrane from the Golgi, and a general requirement for plant growth and cell division (Zheng et al., 2005; Speth et al., 2009; Ahn et al., 2013). Recently, Rab-E was identified as functioning in concert with both the SCD and exocyst complexes (Mayers et al., 2017). The SCD complex functions upstream of Rab-E and is a candidate guanine exchange factor (GEF), thereby suggesting an activated Rab-E in proximity to the exocyst. This finding tentatively places Rab-E in a transport pathway similar to Myo2/Sec4/exocyst (Heider and Munson, 2012); however, the presence of a molecular motor to actively transport secretory cargo in plants remained elusive. Moreover, no study has systematically removed all endogenous Rab-E isoforms to evaluate the loss of function phenotype. Here, we investigate the function of Rab-E and explore the possibility of cross-species functional conservation of Rab-E and myosin XI to control polarized growth in plants.
Materials and Methods
Yeast two-hybrid assay
A Y2H screen was performed using the services of Hybrigenics (France), and we partnered with them to construct their Physcomitrella patens cDNA library. We cultured moss protonemata on PpNO3 and PpNH4 medium under standard conditions (Vidali et al., 2007) for 1-week, then harvested the juvenile tissue and isolated total RNA according to the manufacture’s recommendations (Zymo Research Corp. Cat#R1056). The RNA was immediately frozen, then shipped to Hybrigenics where they constructed the cDNA library. The MyoXIa-CCT bait fragment corresponds to residues 1061–1536, was cloned into pB27 (Hybrigenics), and was screened by 132 million pairwise interactions. Positively interacting pairs were identified by growth on SD -Trp -Leu -His.
We validated and performed directed Y2H experiments by cloning into the bait and prey plasmids from Hybrigenics and transforming them into their corresponding Y2H strains, L40 and Y187, respectively. Additional RabEs were inserted into the Y2H prey vector via restriction-based cloning using BamHI and NotI. The A. thaliana XIK-CCT Y2H bait vector was created by amplification of the CCT region using the primers in Table S1, then PCR product and pB27 were digested with EcoRI/NotI and ligated. The MyoXIa-CCT bait was mutagenized with the Q5 Site-Directed Mutagenesis Kit (New England BioLabs) to create the V1418R and V1422R mutants. All constructs were sequence verified and all necessary primers are in supplemental table 1. Bait and prey pairs were mated, then grown in SD -Trp -Leu until ~2 OD. All strains were normalized to 0.3 OD, then serially diluted and spotted on SD -Trp -Leu, and SD -Trp -Leu -His plates.
Construction of fluorescently tagged Rab-E moss lines
Creation of the 3xmEGFP tagged Rab-E12 construct occurred by a two-element LR Gateway® reaction (ThermoFisher) of entry clones pENT-L1–3xmEGFP-L5r and pENT-L5-RabE12-L2, and the destination vector pTHUbi-gate (Vidali et al., 2007). The Rab-E12 entry clone was generated by amplification of cDNA using primers AttB2-PpRabE12Rev and AttB5-PpRabE12For (Table S1) and a BP reaction with the resulting PCR product and pDONR 221 P5P2. cDNA was created from RNA isolated from 1-week old moss protonemal tissue using the SuperScript III Reverse Transcriptase (ThermoFisher) according to manufacturer’s protocol. The 3xmCherry-Rab-E14 construct was generated in a similar manner using Rab-E14 cDNA, 3xmCherry entry clone, and the destination vector pTKUbi-gate (Wu and Bezanilla, 2014). The moss expression construct pTHUbi-gate:3xmEGFP-RabE12 was transformed into the Gransden wild-type laboratory strain (Liu and Vidali, 2011) and selected for stable transformants. pTKUbi-gate:3xmCherry-Rab-E14 was transformed into a previously created line that contains myosin XIa endogenously tagged with 3xmEGFP (Sun et al., 2018), as well as wild-type. This procedure yielded multiple independent transformants that were individually screened for fluorescence.
Live-cell confocal imaging and kymograph analysis
All moss tissue was cultured as previously described (Furt et al., 2013). All moss lines were imaged with a SP5 confocal microscope (Leica) using the 488 nm and 561 laser lines at 5% power, with the emission bandwidth gated to 499–547 nm light for GFP and 574–652 nm for mCherry. For imaging of cell division, all images (512×512) were acquired simultaneously at 400 Hz using a HCX-PL-Apo, 63x, NA1.4 lens (Leica) with a zoom of 6 and a bit depth of 12-bit. All images were single medial slices with the confocal aperture at 1 AU, taken at 10 second intervals, and a line average of 2 was used to increase signal to noise. Images were processed in ImageJ by Gaussian blurring (Sigma radius 1), background subtracted (200-pixel ball radius), and contrast enhanced (normalized using stack histogram and 0.1% saturated pixels).
For long-term imaging required for kymograph analysis, all images (1024 × 256 pixels) were acquired simultaneously at 400 Hz using with a zoom of 3. Nine optical slices, 1 μm apart, were collected using at five-second intervals to capture the entire volume of the apical cytosol. Total laser power was maintained at 5% for both laser lines. The confocal aperture was opened to 2 AU. For creation of live-cell movies of growing tips, all images were post-processed in ImageJ with blurring (Sigma radius 1) and the red channel was contrast enhanced (normalized per frame using 0.2% saturated pixels) to adjust for acquisition photobleaching.
For cross-correlation analysis, all data were processed and analyzed in an analogous manner to previous work (Furt et al., 2013). To reduce manual ROI selection and to ensure proper ROI orientation for kymograph analysis, as well as to facilitate data processing and visualization, we implemented a custom MATLAB (MathWorks) routine that is available upon request.
Homology modeling and interface prediction
Homology models of the P.patens myosin XI cargo-binding domain (MyoXI-CBD) and RabE14 were generated using the SWISS-MODEL server (Waterhouse et al., 2018). The MyoXI-CBD model was created using residues 1131–1537 from full-length myosin XIa and supplying yeast Myo2p (PDB:2f6h) as the template. The Rab-E14 model uses the entire protein sequence and is based upon the Sec4 GTP-bound structure (PDB: 1g17).
Subsequent molecular visualization and manipulation was performed using UCSF Chimera (Pettersen et al., 2004). MyoXI-CBD and active Rab-E14 were structurally aligned with the myosin Vb-Rab11a co-crystal structure (PDB: 4lx0) using the MatchMaker tool with default parameters. The template was removed, and putative interface residues were extracted through identification of atoms whose respective van der Waals surfaces were within 1 Å. This process was repeated with Myo2p and active Sec4 to assess the ability of the method to recapitulate a known myosin V:Rab8-like interaction, irrespective of the template containing a Rab11 member.
We also employed the template-based docking algorithm, PRISM (Tuncbag et al., 2011; Baspinar et al., 2014). With this approach, a predicted interface is generated based on mapping template-to-target interfaces, thereby bypassing other structural elements unrelated to the interface. We used our homology models as the two target proteins and specified the co-crystal structure of human myosin Vb and active Rab11a as the template.
CRISPR knockout of Rab-E14 and Rab-E15
The double Δrab-E14/15 mutant was created by gene targeting through CRISPR-Cas9 (Collonnier et al., 2017). Selection of appropriate guide sequences was done with the CRISPOR platform (Haeussler et al., 2016). The final sgRNAs were chemically synthesized (Integrated DNA Technologies, Coralville, IA, USA). Each sgRNA cassette includes the P. patens U6 promoter, the guide sequence target, the tracrRNA scaffold, and are flanked by Gateway® attB sites for BP cloning into pDONR207 (ThermoFisher). All sgRNA constructs were co-transformed with pAct-Cas9 into wild-type P. patens protoplasts and selected for transient transformants on 50 mg L−1 G418. Successful genomic editing events were verified by genotyping (Table S1) and sequencing.
RNAi growth assays
The loss of Rab-E phenotype was elucidated using a recently developed RNAi method that allowed us to use the Δrab-E14/15 line without the need for reporter insertion (Orr et al., 2020b). To maximize silencing efficacy, our target was 401 bp of the Rab-E11 coding sequence (CDS) (RabE11i), which shares 88% and 90% identity with Rab-E12 and Rab-E13, respectively. The Rab-E1c expression construct contains the cDNA of A. thaliana Rab-E1c cloned into the pTHUbi-gate vector. RNAi and expression constructs were transformed into either the wild-type or Δrab-E14/15 protoplasts, regenerated for 4-days, transferred to growth medium supplemented with 1.25 μg mL−1 2-FA for 4-days. Plants were stained with calcofluor and imaged 8-days post-transformation to measure morphometric parameters.
Long-term imaging of RNAi-treated plants initially proceeded as described above, but at 5-days post-transformation a sub-sample of plants from the wild-type and rab-E conditions were transferred to special agar plates. Plants were grown in the central region of the plate, which contained a thin layer of agar to facilitate imaging with an inverted microscope, while the periphery contained a thick agar layer to prevent rapid evaporation of the center. These agar plates contained the same concentration of 2-FA and were supplemented with 5 μM calcofluor. At day 7 post-transformation, these cultures were imaged every 10–15 minutes with a 10X A-Plan (0.25 NA) objective of an epifluorescent microscope (Zeiss Axiovert 200M) coupled to a CCD camera (Zeiss AxioCam MRm) over 9–24 hours. The determination of growth rates was restricted to in-focus apical cells, and was quantified via kymograph analysis using the ‘Multi Kymograph’ tool in FIJI.
The effects on plant growth of myosin XI chimeras and myosin XI point mutants were investigated using a previously developed RNAi methodology (Bezanilla et al., 2005). Myosin XI chimeras were generated via Gateway® cloning. Briefly, an entry clone containing the P. patens Head-Neck-CC domains (pL1-MyoHeadNeckCC-R5) was combined with an entry clone containing either XIK-CCT (pL5-XIK_CCT-L2) or XIE-CCT (pL5-XIK_CCT-L2) via an LR Reaction. Myosin XI-CBD mutant expression constructs were created via site-directed mutagenesis of wild-type myosin XIa-CBD (Table S1). The mutant and wild-type CBDs were concatenated with the myosin XIa head, neck, and coiled-coil domains through an LR reaction (ThermoFisher) into the moss expression construct, pTHUbi-gate. The full-length myosin XI mutant constructs leave a small linker between the two fused pieces, so we created an otherwise wild-type control construct to test for possible deleterious effects of the linker fragment, named ‘P.p. MyoXI.’ The myosin XI-CBD expression constructs were co-transformed (Liu and Vidali, 2011) into the NLS-4 line (Bezanilla et al., 2003) with a previously developed myosin XIa+b 5’UTR silencing construct (Vidali et al., 2010) to evaluate the capacity of the mutant MyoXI-CBDs to rescue polarized growth. Plants were imaged 1-week post-transformation for polarized growth and size defects (Vidali et al., 2007; Galotto et al., 2019).
Real-time quantitative PCR
Total RNA was extracted from RNAi-treated, 8-day old P. patens tissue using the Quick-RNA Microprep kit (Zymo Research) and following the manufacturer’s protocol. cDNA was synthesized with the iScript™ cDNA Synthesis Kit (Bio-Rad) using 200 ng total RNA as the template for all conditions. RT-qPCR experiments were performed with iTaq SYBR Green Supermix (Bio-Rad) and the primers listed in Table S1. The amplicons for all primer pairs were confirmed to have similar PCR efficiencies between 95% and 105%. Relative expression was determined using the 2-ΔΔCT method (Taylor et al., 2019), with Ubiquitin 10 serving as the normalization gene. Three independent experiments were performed.
Statistical analyses
All statistical analyses were performed using GraphPad Prism. All assumptions for each statistical test were validated before performing the statistical test, except for one analysis. Plant area is not normally distributed, so all plant area data was log-transformed to satisfy the normal distribution criterion of the parametric statistical tests we used. All error bars for main text figures represent a 95% confidence interval. For all experiments, statistical significance was determined by either a one-way ANOVA with a post-hoc Tukey test or the Student’s t-test (P < 0.01).
Results
Rab-E Localizes to Sites of Polarized Secretion and Colocalizes with Myosin XI
We hypothesized a homologous polarized trafficking mechanism exists between opisthokonts and plants, whereby plant specific myosin XI associates with Rab-E to support polarized growth. Previous work in plants implicates Rab-E in polarized exocytosis (Zheng et al., 2005; Speth et al., 2009; Ahn et al., 2013; Mayers et al., 2017), and work with homologous Rabs in yeast and mammals directly couples Rabs to the motor protein myosin V to promote polarized vesicle deposition (Jin et al., 2011; Donovan and Bretscher, 2012). To determine the subcellular localization of Rab-E, and test colocalization with myosin XI, we generated a constitutive expression construct of 3mCherry-Rab-E14 (Cherry-Rab-E14) and transformed this into a P. patens line with myosin XI tagged with 3xmEGFP at the endogenous locus (Sun et al., 2018). Transgenic plants that expressed the fluorescent Rab-E protein and showed no growth phenotype were used for subsequent analysis.
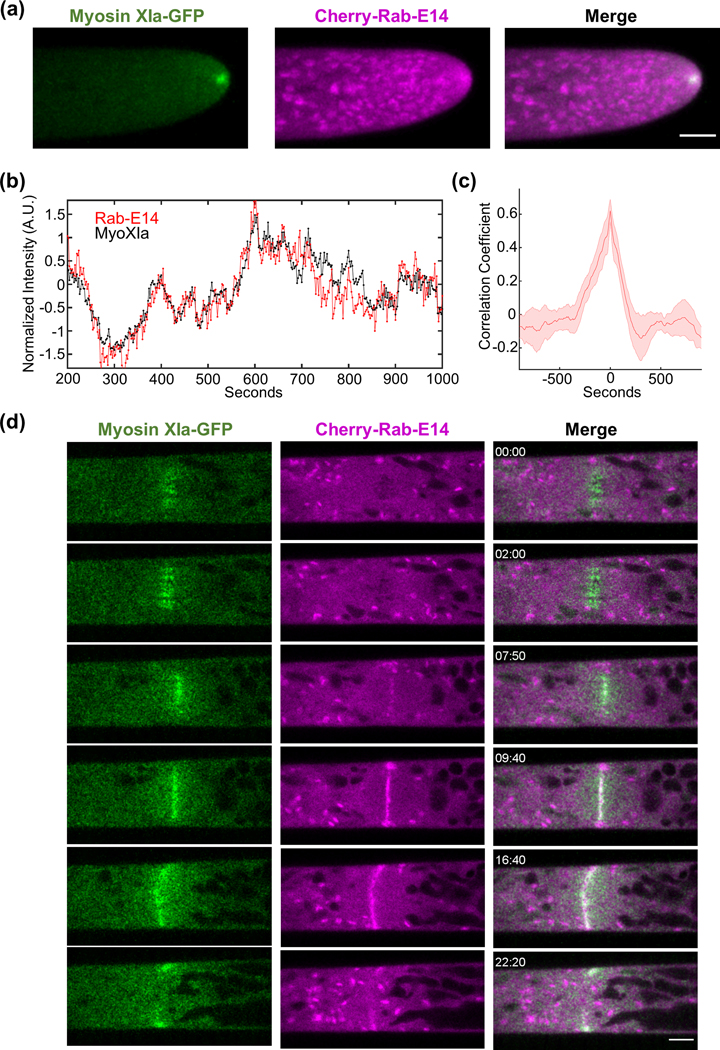
We observed, using confocal laser scanning microscopy, that Cherry-Rab-E14 localizes to discrete, endomembrane compartments, and predominantly accumulates at the extreme apex of growing caulonemal cells (Fig. 1a, Fig. S1a). The immediate subapical Rab-E decorated structures are likely Golgi, as our observations are consistent with data in Arabidopsis showing Rab-E localizing to the Golgi (Speth et al., 2009) and data in P. patens demonstrating a characteristic exclusion of Golgi from the extreme apical region (Furt et al., 2012) where myosin XI, F-actin, and vesicles are localized (Furt et al., 2013). Furthermore, both myosin XI and Rab-E colocalized at the growing tip (Fig. 1a, Video S1). Importantly, myosin XI and Rab-E show synchronized stochastic fluctuations of their fluorescent intensity over time within the apex of the growing moss cell (Fig. 1b). Cross-correlation analysis of the myosin XI and Rab-E time series reveals that these two proteins are in-phase (Fig. 1c). This result is commensurate with our previous data demonstrating a tight spatiotemporal correlation with myosin XI and VAMP-labeled vesicles (Furt et al., 2013), thus supporting the inference that Rab-E is at least partially vesicle-localized. We investigated the subcellular localization of another Rab-E isoform, Rab-E12, and found similar dynamic localization to the growing tip (Fig. S1b, Video S2), suggesting functional redundancy. The strong spatial and temporal correlation between myosin XI and Rab-E is consistent with a direct interaction, as is found in yeast in mammals. However, we cannot conclude with this approach whether myosin and Rab-E interact.
Figure 1. Rab-E co-localizes with myosin XI at sites of polarized exocytosis in Physcomitrella patens.
(A) Subcellular localization of 3xmCherry-Rab-E14 and MyosinXIa-3xmEGFP in an apical caulonemal cell by confocal microscopy. Images are maximum projections of 8 confocal slices at 1 μm spacing to visualize the apical volume. Scale bars are 5 μm.
(B) Intensity fluctuations of myosin XI and Rab-E at the apex of the growing caulonemal cell. Intensity values were obtained through kymograph analysis of time series (Video S1).
(C) Maximum correlation coefficient of myosin XI and Rab-E intensity fluctuations. Solid line indicates the average correlation coefficient of 5 cells. The shaded region represents the standard error of the mean. Scale bar, 5 μm.
(D) An apical caulonemal cell of the double myosinXI-GFP and Cherry-Rab-E14 line was imaged during cell division approximately 1-minute post nuclear envelope breakdown. Scale bar, 5 μm.
Plants use the exocytosis machinery to transport secretory vesicles and other cargoes to build the cell plate in a mechanism similar to tip growth (Fendrych et al., 2010; Wu and Bezanilla, 2014; Mayers et al., 2017). Therefore, we predicted that myosin XI and Rab-E function together, not only at the tip for cell growth, but also at the cell plate for cytokinesis. Consistent with recent work (Abu-Abied et al., 2018; Sun et al., 2018), we observed that myosin XI localized to the cell’s division zone immediately following nuclear envelope breakdown (Fig. 1d, Video S3). Rab-E is noticeably absent from the division zone until approximately 8 minutes following nuclear envelope breakdown, which is consistent with the emergence of the phragmoplast (Sun et al., 2018). Upon Rab-E arrival at the division zone following chromosome separation, Rab-E and myosin XI display coordinated behavior at the nascent and expanding cell plate (Fig. 1, Video S3). Rab-E initially localizes as discrete puncta and then expands outward toward the cell wall (Fig. 1, Video S3). Interestingly, unlike myosin XI, intense foci of Rab-E are present on the opposing plasma membranes that the cell plate-localized Rab-E grows towards. These foci manifest via the coalescence of Rab-E-decorated endomembrane structures (Video S3). Upon fusion of plasma membrane-localized Rab-E and cell plate-localized Rab-E, coincident with myosin XI reaching the plasma membrane, the bright foci dissipate and a near homogeneous cytoplasmic signal of Rab-E remains (Fig. 1). Although we cannot conclude Rab-E and myosin XI function in cytokinesis, these observations establish that Rab-E and myosin XI are spatiotemporally coupled in polarized transport processes.
Rab-E is Required for Polarized Growth in P. patens, and A. thaliana Rab-E1c Rescues the rab-E Mutant.
Our data and previous studies strongly implicate Rab-E in supporting plant growth and development (Zheng et al., 2005; Speth et al., 2009; Ahn et al., 2013; Mayers et al., 2017). However, to date no study has investigated Rab-E in the context of a comprehensive loss-of-function (rab-E) background.
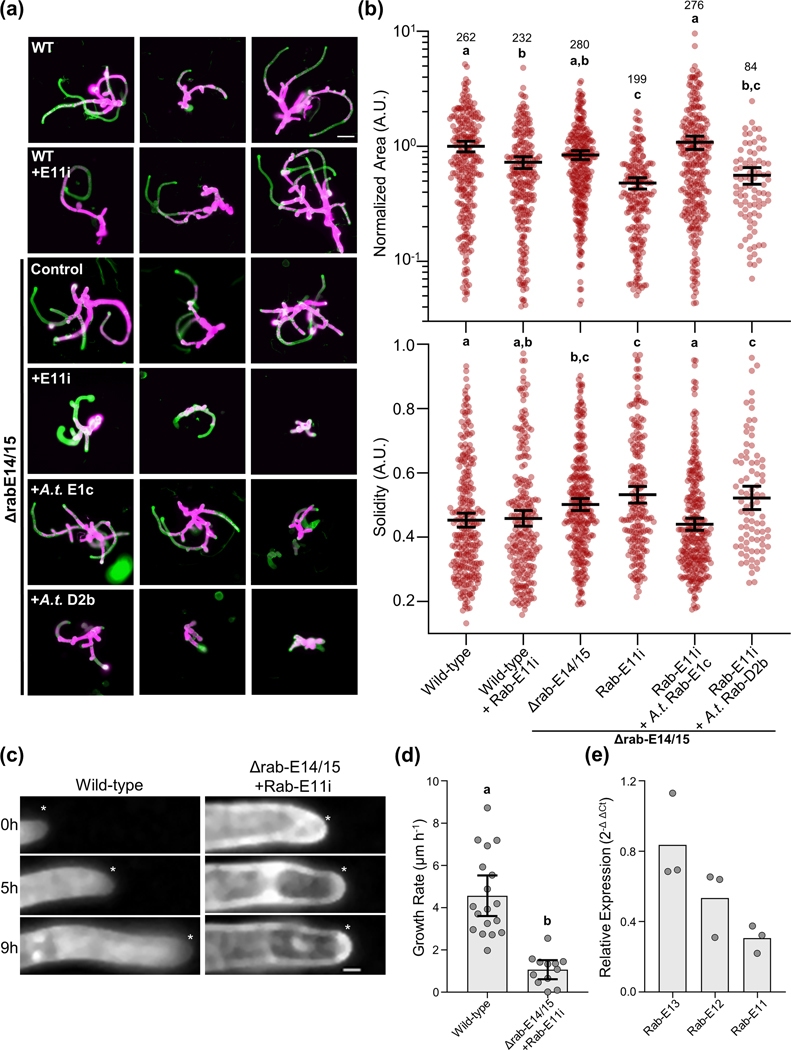
To systematically deplete P. patens of the five-member Rab-E subfamily (Fig. S2, Table S2), we employed an integrative approach. We reasoned complete that removal of Rab-E might result in non-viable plants, so we coupled CRISPR/Cas9 mediated knockouts of two isoforms, Δrab-E14/15 (Fig. S3), with subsequent RNAi to knockdown the remaining three isoforms. To deplete the final three Rab-E isoforms we generated an RNAi construct targeting a conserved ~400 bp region within the Rab-E11 CDS (Rab-E11i), which is 88% and 90% identical to Rab-E12 and Rab-E13, respectively. Additionally, the Rab-E11i targeting sequence was specific to the Rab-E clade, as it shows 54% and 63% identity to representative Rabs of the A and D clades, respectively. To investigate our rab-E plants without the need for a specialized RNAi reporter line we used our recently developed RNAi system that exploits positive selection of an endogenous marker to enable robust, high-throughput phenotyping (Orr et al., 2020b). To characterize the growth phenotype of the mutant moss plants, we quantified two morphological parameters of the plant, area and solidity (Vidali et al., 2010). Solidity is the ratio of the plant area to the convex hull area, which is used as a morphological metric of plant polarization. Upon silencing with the Rab-E11i construct in the Δrab-E14/15 background, we observed morphologically stunted plants at 8-days post-transformation (Fig. 2a,b). Notably, when tested separately, both the Δrab-E14/15 and Rab-E11i conditions resulted in slightly smaller plants, and neither displayed any significant difference in plant solidity. Only when the Rab-E11i construct was introduced into the Δrab-E14/15 background did we observe an additive effect on plant size and reduced solidity, again highlighting the functional redundancy of the Rab-E subfamily (Fig. 2a,b). The rab-E mutant’s diminished size and polarity (Fig. 2b) is consistent with a role in polarized growth, as myosin XI displays a similar, albeit more pronounced, phenotype (Vidali et al., 2010).
Figure 2. The Rab-E GTPases are required for polarized growth and the growth defect is rescued with Rab-E1c from Arabidopsis thaliana.
(A) Representative 8-day old plants regenerated from protoplasts using the APT-based RNAi assay. Plants were stained and imaged with calcofluor, colored in green, and also imaged for chlorophyll autofluorescence shown in magenta. Bar = 100 μm.
(B) Quantification of plants from the APT-based RNAi growth assay. Each data point represents an individual plant. Three independent experiments were performed and pooled. Plant area was normalized to the wild-type condition. Lines and error bars represent the mean and 95% confidence interval for each group, respectively. Shared letters above the bars show those experimental groups that cannot be statistically distinguished. Statistical significance was determined by a one-way ANOVA-Tukey (P<0.01). Numbers above the letters indicate number of plants analyzed per condition.
(C) Time series of representative apical cells used for growth rate determination. The asterisk denotes the cell tip, scale bar = 10 μm
(D) Quantification of growth rate extracted from kymographs of apical cell growth time series, such as in (a). Data points represent in-focus apical cells. Apical cells were pooled across two independent experiments. Bars represent the mean and 95% confidence interval. Significance was tested via a two-tailed T-test, p < 0.001
(E) Relative transcript abundance of Rab-E13, E12, and E11 in Δrab-E14/15 plants treated with an RNAi construct targeting Rab-E11. Transcript abundance was normalized to Ubiquitin10 using the 2-Δ ΔCt method.
We next tested whether our rab-E mutant was Rab-E clade specific, and if rab-E could be rescued by a distantly related Rab-E. This idea was motivated by the fundamental cellular requirement of Rab-E for polarized trafficking, similar Rab-E localization between our observations and other plant species, and protein-level conservation (~90% identity between A. thaliana and P. patens). Furthermore, previous rescue experiments in tobacco using A. thaliana Rabs successfully demonstrated cross-species functionality and clade specificity of Rabs between those two species (Batoko et al., 2000). Strikingly, Rab-E1c from the vascular plant A.thaliana fully rescued our rab-E phenotype in the moss P. patens (Fig. 2a,b). Importantly, the observed phenotype is specific to the plant Rab-E subfamily. Exogenously supplied A. thaliana Rab-D2b failed to rescue the morphological defects (Fig. 2a,b). Also, these results demonstrate that the RNAi-induced phenotype is not likely the consequence of off-target effects, as Rab-E1c from A. thaliana specifically rescues the rabE mutant in P. patens.
Although our plant-level morphological analysis is entirely consistent with Rab-E being required for polarized growth, we sought to directly observe cell growth. We achieved this through long-term (ranging from 9–24 hours) live cell imaging of wild-type and rab-E plants (Fig. 2c). Plants were manipulated identically to those in Figure 2ab, except at day 5 post-transformation the plants of interest were transferred to plates with a thin layer of agar surrounded by a larger agar reservoir, thereby allowing acquisition close to the agar/plate interface. Additionally, the agar contained calcofluor to facilitate automated image acquisition and downstream analysis. This approach revealed a substantial cellular growth phenotype, with the rab-E line growing at an average of 1.1 ± 0.71 μm h−1 compared to the average wild-type growth rate of 4.6 ± 1.9 μm h−1 (Fig. 2d).
Together, these results support our hypothesis that Rab-E is essential for polarized cell growth. However, to rule out the unlikely possibility (given the A. thaliana Rab-E rescue) of an off-target RNAi effect being the cause of the loss of polarized growth phenotype, we performed RT-qPCR on the remaining full-length Rab-E transcripts. This analysis confirmed that Rab-E transcripts were downregulated, specifically Rab-E11, which was reduced by approximately 70% (Fig. 2e). Additionally, Rab-E12 and Rab-E13 had their transcript abundance decreased by 50% and 20%, respectively. This level of transcript reduction is consistent with both the observed reduction in growth rate and our expectation that Rab-E is essential for viability. Based on these results, we conclude that Rab-E is necessary for plant polarized growth. Furthermore, the rescue of the rab-E phenotype with Rab-E1c from A. thaliana supports an evolutionarily conserved function, likely mediating post-Golgi trafficking, and perhaps in concert with myosin XI.
Myosin XI is a conserved effector of Rab-E, and A. thaliana myosin isoforms XI-K and XI-E rescue loss endogenous myosin XI in P. patens
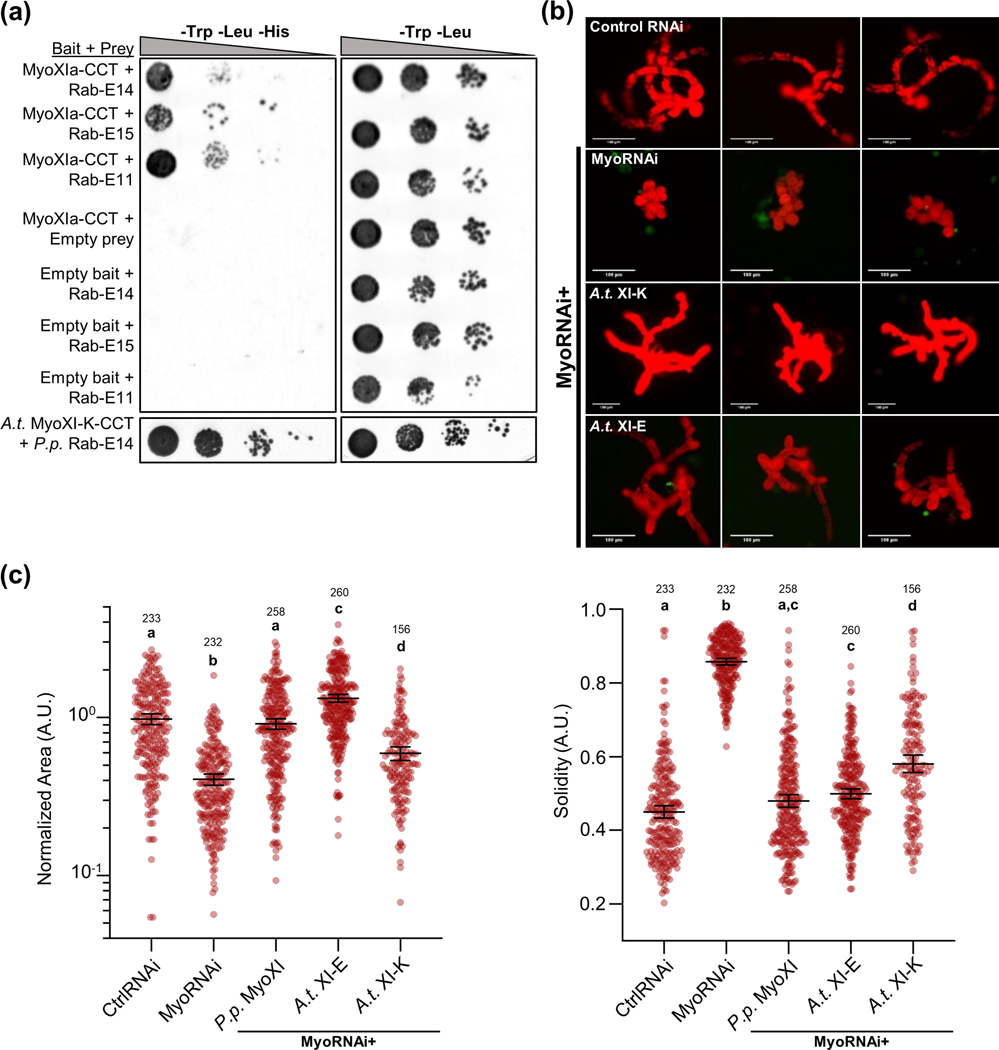
Our data demonstrated a strong relationship between Rab-E and myosin XI in polarized growth, therefore we investigated the existence of a direct interaction. We hypothesized that myosin XI directly interacts with Rab-E, and that this interaction is required for polarized growth. In parallel with our directed investigation, we performed a yeast two-hybrid screen in conjunction with Hybrigenics (France) as an unbiased approach to identify putative P. patens myosin XI interactors. The myosin XI bait construct contained the canonical C-terminal myosin XI cargo-binding domain plus a small N-terminal extension into the coiled-coil domain, hereafter called MyoXI-CCT. Consistent with our previous data and hypothesis, our Y2H screen identified a full-length Rab-E GTPase protein, Rab-E14 (Pp3c6_11710). As yeast two-hybrid screens can be prone to false-positives, we independently verified this putative interaction with a directed Y2H (Fig. 3a). Almost all sequence diversity between the Rab-E isoforms is located within the hypervariable C-terminal domain, while the canonical effector-determining regions localized within and near the switch regions maintain high sequence identity (Fig. S2a). Therefore, we predicted that myosin XI binding is a shared function within the Rab-E subfamily. The most similar isoform, Rab-E15, and divergent isoform, Rab-E11, both show a positive interaction with myosin XI (Fig. 3a). We interpret this as the Rab-E subfamily being functionally redundant with respect to myosin XI binding, and is consistent with other identified Rab-E effectors (Camacho et al., 2009; Mayers et al., 2017).
Figure 3. P. patens Rab-E subfamily interacts with both moss myosin XI and Arabidopsis myosin XI-K isoform, and A.t. XI-K/E rescue loss of endogenous P.p. myosin XI.
(A) Directed Y2H using the P.p. myoXIa-CCT and A.t. XI-K-CCT bait constructs and full-length Rab-E14, Rab-E11, and Rab-E15 results in growth on synthetic defined (SD) medium lacking histidine. Empty bait and prey plasmids were tested with their respective partners to test for autoactivation of HIS3.
(B) Representative 1-week old RNAi plants—all plants except ‘CtrlRNAi’ are silencing expression of both endogenous myosin XIs. All plants except ‘MyoRNAi’ were co-transformed with the myosin XI silencing construct and another construct expressing either WT myosin XI or mutant myosin XI. Bar = 100 μm.
(C) Quantification of plants from the myosin XI-chimera growth assay. Each data point represents an individual plant. Three independent experiments were performed and pooled. Plant area was normalized to the wild-type condition. Lines and error bars represent the mean and 95% confidence interval for each group, respectively. Shared letters above the bars show those experimental groups that cannot be statistically distinguished. Statistical significance was determined by a one-way ANOVA-Tukey (P<0.01). Numbers above the letters indicate number of plants analyzed per condition.
Our results demonstrating cross-species functionality of Rab-E and the pervasiveness of homologous myosin:Rab interactions across eukaryotes motivated us to explore the possibility of a conserved interaction in plants. To that end, we tested and confirmed that the cargo-binding domain of A. thaliana myosin XI-K interacts with P. patens Rab-E (Fig. 3a). Taken together with the literature showing similar subcellular co-localization of XI-K with vesicles at the growing root hair tip and XI-K’s general requirement for polarized growth, we hypothesized the CBD of XI-K from A. thaliana would rescue P. patens plants depleted of endogenous myosin XI. To test this, we generated a full-length chimeric myosin XI protein containing the CCT of A.t. XI-K, while maintaining the head and neck domains of P. patens myosin XIa. In this way, we are directly investigating the same domain of XI-K for our in planta assay as tested in the Y2H. We found that A.t. XI-K and another Arabidopsis isoform, XI-E, are sufficient to promote polarized growth in P. patens plants lacking native myosin XI (Fig. 3b,c). These results support the hypothesis of a conserved mechanism of polarized growth, likely mediated by an interaction between myosin XI and Rab-E.
Myosin XI:Rab-E Interface Prediction
To gain molecular insights of the MyoXI:Rab-E interaction, we hypothesized that the binding interface is structurally homologous to that of the well characterized class V myosin binding to Rab8/Sec4 (Goldenring et al., 2012; Welz and Kerkhoff, 2017). A multiple sequence alignment of moss Rab-E14, Sec4 from Saccharomyces cerevisiae, and Rab8a from Homo sapiens strikingly illustrates the extent of conservation within the canonical Rab effector-binding regions (Fig. S4a). Rabs undergo a dynamic GTP-dependent rearrangement of two structural regions, named “switch-I” and “switch-II,” which defines their “active” GTP-bound state and effector recognition that typically involves an invariant hydrophobic triad (Stroupe and Brunger, 2000; Lee et al., 2009; Khan and Menetrey, 2013; Pylypenko et al., 2018). Given the near perfect identity within the switch regions, we speculated that structural models of myosin XI-tail and Rab-E14, based on the well-characterized Sec4 and Myo2 interaction, could assist in mapping the functional interface.
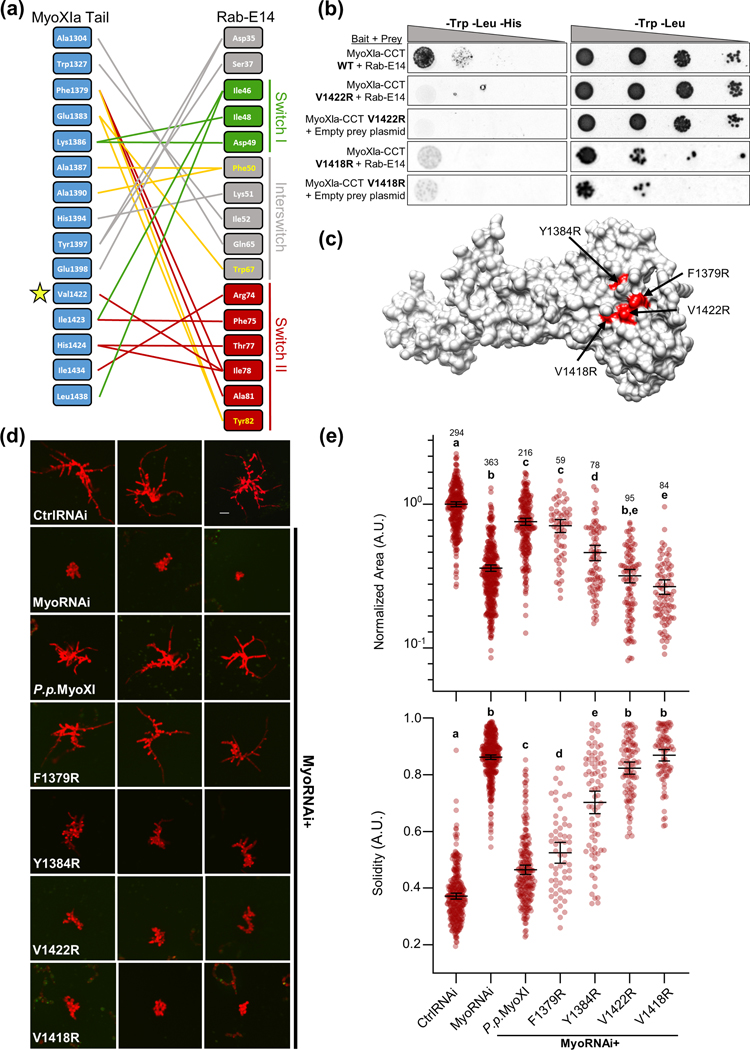
To identify the putative interface between Rab-E14 and myosin XI, we employed a homology-based in silico approach. We first generated homology models of P. patens myosin XI CBD and Rab-E14 using yeast Myo2-CBD and Sec4-GTP as templates (Fig. S5). We leveraged these homology models to computationally predict the binding interface between myosin XI and Rab-E14 using both structural super-positioning (Fig. S4b) and the template-based algorithm, PRISM (Tuncbag et al., 2011; Baspinar et al., 2014) (Fig. 4a). At the amino acid level, almost all predicted associations occur within the switch-I, interswitch, switch-II region (Fig. 4a) Both interface prediction methods and myosin templates identified the myosin XI residue V1422 as a putative interface residue (Fig. S6a). V1422 of myosin XI structurally aligns with Q1447 of Myo2p (Fig. S6b); Q1447 was previously determined to reside in the yeast secretory vesicle binding site (Pashkova et al., 2006) and later shown to directly associate with the Rabs Ypt31/32 (Lipatova et al., 2008) and Sec4 (Jin et al., 2011). Altogether, these results support a remarkable degree of structural conservation, and suggest that Rab-E functions as a myosin XI receptor on secretory vesicles in plants.
Figure 4. Structure-guided mutagenesis of predicted Myosin XI-CBD:Rab-E14 binding interface reveals polarized growth mutants in Physcomitrella patens.
(A) Contact map of modeled myosin XI:Rab-E14 (−17.8 kcal mol-1) determined by template-based docking algorithm PRISM (Tuncbag et al., 2011). Hydrophobic triad residues and their putative contacts colored in yellow. Colors of lines representing contacts reflect the domain within Rab-E14 that the contact belongs. Green=Switch I, Red= Switch II, Grey=Interswitch, Yellow=Hydrophobic triad.
(B) Directed Y2H with the P.p. myoXI-CCT WT bait fragment from (Fig. 3) and bait fragments containing mutations in the myosin XI cargo-binding domain that are required for polarized growth and predicted to interact with Rab-E14. All yeast strains were grown on the same SD plates, but rearranged for clarity.
(C) Homology model of P.p. myosin XI-tail is shown, with the candidate interface residues predicted to mediate Rab-E interactions in red.
(D) Representative 1-week old RNAi plants—all plants except ‘CtrlRNAi’ are silencing expression of both endogenous myosin XIs. All plants except ‘MyoRNAi’ were co-transformed with the myosin XI silencing construct and another construct expressing either WT myosin XI or mutant myosin XI. Bar = 100 μm.
(E) Quantification of the morphological parameters of solidity and area extracted from images in (d). Error bars indicate standard error of the mean. Shared letters above the bars show those experimental groups that cannot be statistically distinguished. Statistical significance was determined by a one-way ANOVA-Tukey (P<0.01). Numbers above the letters indicate number of plants analyzed per condition.
Structure-based Mutants Disrupt the MyoXI:Rab-E Interaction, Resulting in a Loss of Polarized Growth
Prediction of the putative MyoXI:Rab-E interaction identified several candidate residues on myosin XI-CBD that could mediate an interaction with Rab-E, with residue V1422 emerging as the primary candidate. To test if V1422 is required for myosin XI-CBD to interact with Rab-E14, we performed a directed Y2H assay in which V1422 was mutated to arginine. We generated an additional myosin XI-CBD mutant, V1418R, to probe the region near V1422 that is bounded by our predicted interface. Consistent with our interface prediction, both mutants eliminated the interaction with Rab-E14 (Fig. 4b). Furthermore, western blot analysis of the positive Y2H interactions confirmed the presence of the myosin XI mutants at levels equivalent to wild-type (Fig. S7). However, we suspected that the V1418R mutant resulted in a misfolded protein, as evidenced by a slight cytotoxic effect (Fig. 4b), its buried side chain relative to V1422 (7.7 Å2 and 36.2 Å2 solvent accessible surface area, respectively), and low solubility when purified recombinantly (Fig. S8c). Taken together, these data support our in silico predicted interface, such that the V1422R mutation within myosin XI’s cargo-binding domain specifically abolishes Rab-E14 binding.
Based on our previous results demonstrating cross-species complementarity and interaction between myosin XI and Rab-E, we predict the MyoXI:Rab-E interaction to be a fundamental component of the polarized transport process. Therefore, specific disruption of this interaction should manifest as loss of polarized growth. To perturb the predicted MyoXI:Rab-E interaction, we generated a panel of single-residue myosin XI-CBD mutants within the full-length CDS, which are highlighted in the model (Fig. 4c). As expected, silencing of endogenous myosin XI results in small and round plants, as determined by significantly reduced area and solidity, respectively (Fig. 4d,e). All the mutant plants show a statistically significant increase in solidity compared to control RNAi, with the Y1384R, V1422R, and V1418R mutants also showing a statistically significant decrease in normalized plant area (Fig. 4e). The V1422R and V1418R mutants are morphologically equivalent to silencing myosin XI, Y1384 shows an intermediate phenotype, and F1379 displays a near wild-type morphology (Fig. 4d,e). The dramatic loss of a polarized growth phenotype observed in myosin XI-CBD mutants demonstrates that these individual residues are required for proper functioning of myosin XI in polarized growth.
To exclude the possibility that our observed mutant phenotypes were a consequence of absent or reduced myosin XI expression or stability, we created GFP and His-tagged constructs for the myosin XI mutants (Methods S1). Our GFP reporters confirmed myosin XI mutant expression in protoplasts at wild-type levels (Fig. S8a,b), and purification of the V1422R mutant suggests normal solubility and stability (Fig. S8c). Together, these results indicate that V1422 is required to mediate the polarized growth process, and the V1422R mutation specifically abrogates myosin XI’s biological function through defective binding to Rab-E.
Discussion
Direct evidence supporting a myosin:Rab-driven transport model in plants, as in budding yeast, was lacking despite multiple independent lines of research demonstrating the individual requirements for myosin XI and Rab-E in polarized trafficking, and the sequence homology to myosin V and Sec4 (Zheng et al., 2005; Peremyslov et al., 2008; Speth et al., 2009; Vidali et al., 2010; Peremyslov et al., 2012; Park and Nebenführ, 2013; Peremyslov et al., 2013; Ryan and Nebenfuhr, 2018). By exploiting the reduced gene family of myosin XI and absence of confounding cytoplasmic streaming in the moss Physcomitrella patens, in combination with Y2H, live-cell imaging, cross-species complementation, and computationally directed mutant analysis, we identified functional homology between myosin XI/myosin V and Rab-E/Sec4.
Our live-cell observations of myosin XI and Rab-E established striking cross-correlation at the site of polarized growth. The distinct co-localization of Rab-E and myosin XI to the growing apical zone is reminiscent of commonly observed localization patterns of F-actin, secretory vesicles, myosin XI/V, and other secretory components across plants and fungi (Bi and Park, 2012; Furt et al., 2013; Hepler and Winship, 2015; Bibeau et al., 2018; Riquelme et al., 2018). Additionally, we observed co-localization and co-migration of Rab-E and myosin XI during expansion of the nascent cell plate. Importantly, this result is not only in agreement with previous reports separately investigating myosin XI and Rab-E (Zheng et al., 2005; Chow et al., 2008; Yokota et al., 2009; Abu-Abied et al., 2018; Sun et al., 2018), but is internally consistent with our apical co-localization data and the perspective that plant cell division internally adopts much of the canonical polarized exocytosis machinery (Fendrych et al., 2010; Mayers et al., 2017). Our observations taken together with emerging evidence of interactions between Rab-E and vesicle tethering complexes TRAPPII and the exocyst (Rybak et al., 2014; Mayers et al., 2017; Kalde et al., 2019) supports the hypothesis of a myosin XI:Rab-E centric model of polarized transport and growth. Future experiments are needed to test the dependence of myosin XI on Rab-E subcellular localization, such as through directed re-localization of Rab-E and observing if myosin XI dynamically rearranges its localization.
Our systematic depletion of Rab-E through genetic knockouts and RNAi establishes Rab-E as an essential component of plant growth, with our long-term imaging experiments demonstrating Rab-E is required for polarized growth. Furthermore, cross-species complementation and Y2H experiments with Arabidopsis Rabs and myosin XI isoforms demonstrated functional conservation despite ~450 million years of evolutionary divergence between vascular and non-vascular plants. Our modeling and experimental validation of the putative MyoXI:Rab-E interface identified that residue V1422 of the myosin XI-CBD is required for Rab-E binding and polarized growth. Importantly, V1422 structurally aligns to Q1447 of myosin V in yeast and is required for polarized growth and Rab binding (Schott et al., 1999; Pashkova et al., 2006; Jin et al., 2011). Although we only demonstrated a direct interaction between myosin XI and Rab-E via Y2H, we argue that our specific perturbation of the interaction and the shared loss of polarized growth phenotype between rab-E and the myoXI-V1422R mutant is highly suggestive of an interaction in vivo. Future work is needed to determine if Rab-E functions as the direct vesicle receptor of myosin XI, perhaps through electron microscopy or vesicle purification experiments. If so, the interaction between Rab-E and myosin XI is likely rapid and transient (Bibeau et al., 2020), which may be challenging to verify biochemically. Also, the possible transient nature of the myosin XI:Rab-E interaction would explain why previous Y2H screens in plants failed to identify this interaction. This is consistent with our own Y2H screen that identified only one of the five Rab-E isoforms, with our subsequent directed-Y2H identifying additional isoforms.
Taken together, our results offer an exciting link between the endomembrane system and cytoskeleton in plants, which is a poorly understood but growing area of plant cell biology research (Steiner et al., 2016; Renna et al., 2018; Orr et al., 2020a). Additionally, there existed a substantial gap in our understanding of post-Golgi trafficking to the plasma membrane, with Rab-E speculated to be directly involved in this process (Elliott et al., 2020). These findings offer a mechanistic link between the Rab-E class and polarized growth through an interaction with the molecular motor myosin XI.
Here, we demonstrate functional conservation between the bryophyte P. patens and flowering plant A. thaliana proteins Rab-E and myosin XI. These results coupled with recent discoveries that established functional homology to the well-characterized yeast system, such as the putative Rab-E GEF SCD1/SCD2 and exocyst components, supports the enticing hypothesis of a myosin XI/V-driven, pan-eukaryotic mechanism of polarized exocytosis. We anticipate future work will employ this fundamental framework of polarized exocytosis through leveraging experiments in both flowering and basal plants to build additional complexity.
Supplementary Material
Acknowledgements
The authors are grateful to Fabien Nogué and his laboratory for their assistance in using CRISPR to generate knockout lines, Imogen Sparkes for the A. thaliana myosin XI clones, Jeffrey P. Bibeau for developing the XCorr script, and Miye Jacques for creating the myosin XIa-CCT E. coli expression construct. We also thank Sebastian Bednarek for critical feedback on this work. This work was supported by NSF grant MCB-1253444 to L.V., NIH grant #1R01 GM068803 to M.M., and NIH grant #1R15GM134493 to L.V. and M.M.
References
- Abu-Abied M, Belausov E, Hagay S, Peremyslov V, Dolja V, and Sadot E. (2018). Myosin XI-K is involved in root organogenesis, polar auxin transport, and cell division. Journal of Experimental Botany 69, 2669–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn CS, Han JA, and Pai HS (2013). Characterization of in vivo functions of Nicotiana benthamiana RabE1. Planta 237, 161–172. [DOI] [PubMed] [Google Scholar]
- Baspinar A, Cukuroglu E, Nussinov R, Keskin O, and Gursoy A. (2014). PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res 42, W285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, and Moore I. (2000). A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Pan A, and Quatrano RS (2003). RNA interference in the moss Physcomitrella patens. Plant Physiology 133, 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Perroud P-F, Pan A, Klueh P, and Quatrano RS (2005). An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biology 7, 251–257. [DOI] [PubMed] [Google Scholar]
- Bi EF, and Park HO (2012). Cell Polarization and Cytokinesis in Budding Yeast. Genetics 191, 347–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibeau JP, Kingsley JL, Furt F, Tuzel E, and Vidali L. (2018). F-Actin Mediated Focusing of Vesicles at the Cell Tip Is Essential for Polarized Growth. Plant Physiol 176, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibeau JP, Furt F, Mousavi SI, Kingsley JL, Levine MF, Tüzel E, and Vidali L. (2020). In vivo interactions between myosin XI, vesicles and filamentous actin are fast and transient in Physcomitrella patens. J. Cell Sci 133, jcs234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove J, Vaillancourt B, Kroeger J, Hepler PK, Wiseman PW, and Geitmann A. (2008). Magnitude and direction of vesicle dynamics in growing pollen tubes using spatiotemporal image correlation spectroscopy and fluorescence recovery after photobleaching. Plant physiology 147, 1646–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L, Smertenko AP, Perez-Gomez J, Hussey PJ, and Moore I. (2009). Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. Journal of Cell Science 122, 4383–4392. [DOI] [PubMed] [Google Scholar]
- Chow CM, Neto H, Foucart C, and Moore I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20, 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collonnier C, Epert A, Mara K, Maclot F, Guyon-Debast A, Charlot F, White C, Schaefer DG, and Nogue F. (2017). CRISPR-Cas9-mediated efficient directed mutagenesis and RAD51-dependent and RAD51-independent gene targeting in the moss Physcomitrella patens. Plant Biotechnol J 15, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan KW, and Bretscher A. (2012). Myosin-V Is Activated by Binding Secretory Cargo and Released in Coordination with Rab/Exocyst Function. Developmental Cell 23, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, and Ueda T. (2015). Roles of membrane trafficking in plant cell wall dynamics. Front Plant Sci 6, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott L, Moore I, and Kirchhelle C. (2020). Spatio-temporal control of post-Golgi exocytic trafficking in plants. J. Cell Sci 133, jcs237065. [DOI] [PubMed] [Google Scholar]
- Fendrych M, Synek L, Pečenková T, Toupalová H, Cole R, Drdová E, Nebesářová J, Šedinová M, Hála M, and Fowler JE (2010). The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. The Plant Cell 22, 3053–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furt F, Lemoi K, Tuzel E, and Vidali L. (2012). Quantitative analysis of organelle distribution and dynamics in Physcomitrella patens protonemal cells. BMC Plant Biol. 12, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furt F, Liu YC, Bibeau JP, Tüzel E, and Vidali L. (2013). Apical myosin XI anticipates F-actin during polarized growth of Physcomitrella patens cells. The Plant Journal 73, 417–428. [DOI] [PubMed] [Google Scholar]
- Galotto G, Bibeau JP, and Vidali L. (2019). Automated image acquisition and morphological analysis of cell growth mutants in Physcomitrella patens. Methods Mol Biol 1992, 307–322. [DOI] [PubMed] [Google Scholar]
- Goldenring JR, Roland JT, and Lapierre LA (2012). Rablla, Rab8a and Myosin V: Regulators of Recycling and Beyond. In Rab GTPases and membrane trafficking, Li G. and Segev N, eds (Bentham Science; ), pp. 123–191. [Google Scholar]
- Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, and Concordet JP (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider MR, and Munson M. (2012). Exorcising the exocyst complex. Traffic 13, 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, and Winship LJ (2015). The pollen tube clear zone: Clues to the mechanism of polarized growth. Journal of Integrative Plant Biology 57, 79–92. [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, and Cheung AY (2001). Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17, 159–187. [DOI] [PubMed] [Google Scholar]
- Jin Y, Sultana A, Gandhi P, Franklin E, Hamamoto S, Khan AR, Munson M, Schekman R, and Weisman LS (2011). Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev Cell 21, 1156–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde M, Elliott L, Ravikumar R, Rybak K, Altmann M, Klaeger S, Wiese C, Abele M, Al B, Kalbfuss N, Qi XY, Steiner A, Meng C, Zheng HQ, Kuster B, Falter-Braun P, Ludwig C, Moore I, and Assaad FF (2019). Interactions between Transport Protein Particle (TRAPP) complexes and Rab GTPases in Arabidopsis. Plant J 100, 279–297. [DOI] [PubMed] [Google Scholar]
- Khan AR, and Menetrey J. (2013). Structural biology of Arf and Rab GTPases’ effector recruitment and specificity. Structure 21, 1284–1297. [DOI] [PubMed] [Google Scholar]
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, and Guo W. (2010). Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci U S A 107, 6346–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Mishra A, and Lambright DG (2009). Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic 10, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Tokarev AA, Jin Y, Mulholland J, Weisman LS, and Segev N. (2008). Direct interaction between a myosin V motor and the Rab GTPases Ypt31/32 is required for polarized secretion. Molecular biology of the cell 19, 4177–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, and Vidali L. (2011). Efficient polyethylene glycol (PEG) mediated transformation of the moss Physcomitrella patens. J Vis Exp 50, e2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison SL, and Nebenfuhr A. (2013). Understanding myosin functions in plants: are we there yet? Curr Opin Plant Biol 16, 710–717. [DOI] [PubMed] [Google Scholar]
- Mayers JR, Hu T, Wang C, Cardenas JJ, Tan Y, Pan J, and Bednarek SY (2017). SCD1 and SCD2 form a complex that functions with the Exocyst and RabE1 in exocytosis and cytokinesis. Plant Cell 29, 2610–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna ST, Kunkel JG, Bosch M, Rounds CM, Vidali L, Winship LJ, and Hepler PK (2009). Exocytosis precedes and predicts the increase in growth in oscillating pollen tubes. The Plant Cell 21, 3026–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Rivera-Molina F, and Novick P. (2012). GTPase networks in membrane traffic. Annu Rev Biochem 81, 637–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr RG, Cheng X, Vidali L, and Bezanilla M. (2020a). Orchestrating cell morphology from the inside out - using polarized cell expansion in plants as a model. Curr Opin Cell Biol 62, 46–53. [DOI] [PubMed] [Google Scholar]
- Orr RG, Foley SJ, Sherman C, Abreu I, Galotto G, Liu B, Gonzalez-Guerrero M, and Vidali L. (2020b). Robust survival-based RNA interference of gene families using in tandem silencing of adenine phosphoribosyltransferase. Plant Physiol. 184, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, and Nebenführ A. (2013). Myosin XIK of Arabidopsis thaliana accumulates at the root hair tip and is required for fast root hair growth. PloS one 8, e76745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Jin Y, Ramaswamy S, and Weisman LS (2006). Structural basis for myosin V discrimination between distinct cargoes. The EMBO Journal 25, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Avisar D, and Dolja VV (2008). Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiology 146, 1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Klocko AL, Fowler JE, and Dolja VV (2012). Arabidopsis myosin XI-K localizes to the motile endomembrane vesicles associated with F-actin. Frontiers in plant science 3, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Morgun EA, Kurth EG, Makarova KS, Koonin EV, and Dolja VV (2013). Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. The Plant Cell 25, 3022–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR (2017). Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol Biol Cell 28, 712–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylypenko O, Hammich H, Yu IM, and Houdusse A. (2018). Rab GTPases and their interacting protein partners: Structural insights into Rab functional diversity. Small GTPases 9, 22–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna L, Stefano G, Slabaugh E, Wormsbaecher C, Sulpizio A, Zienkiewicz K, and Brandizzi F. (2018). TGNap1 is required for microtubule-dependent homeostasis of a subpopulation of the plant trans-Golgi network. Nat Commun 9, 5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M, Aguirre J, Bartnicki-Garcia S, Braus GH, Feldbrugge M, Fleig U, Hansberg W, Herrera-Estrella A, Kamper J, Kuck U, Mourino-Perez RR, Takeshita N, and Fischer R. (2018). Fungal Morphogenesis, from the Polarized Growth of Hyphae to Complex Reproduction and Infection Structures. Microbiol Mol Biol Rev 82, e00068–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S. (2007). Vesicle trafficking in plant immune responses. Cell Microbiol 9, 1–8. [DOI] [PubMed] [Google Scholar]
- Rounds CM, and Bezanilla M. (2013). Growth mechanisms in tip-growing plant cells. Annu Rev Plant Biol 64, 243–265. [DOI] [PubMed] [Google Scholar]
- Rutherford S, and Moore I. (2002). The Arabidopsis Rab GTPase family: another enigma variation. Current opinion in plant biology 5, 518–528. [DOI] [PubMed] [Google Scholar]
- Ryan JM, and Nebenfuhr A. (2018). Update on Myosin Motors: Molecular Mechanisms and Physiological Functions. Plant Physiology 176, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak K, Steiner A, Synek L, Klaeger S, Kulich I, Facher E, Wanner G, Kuster B, Zarsky V, Persson S, and Assaad FF (2014). Plant Cytokinesis Is Orchestrated by the Sequential Action of the TRAPPII and Exocyst Tethering Complexes. Developmental Cell 29, 607–620. [DOI] [PubMed] [Google Scholar]
- Schott D, Ho J, Pruyne D, and Bretscher A. (1999). The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J Cell Biol 147, 791–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth EB, Imboden L, Hauck P, and He SY (2009). Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiol 149, 1824–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner A, Rybak K, Altmann M, McFarlane HE, Klaeger S, Nguyen N, Facher E, Ivakov A, Wanner G, Kuster B, Persson S, Braun P, Hauser MT, and Assaad FF (2016). Cell cycle-regulated PLEIADE/AtMAP65–3 links membrane and microtubule dynamics during plant cytokinesis. Plant J 88, 531–541. [DOI] [PubMed] [Google Scholar]
- Stroupe C, and Brunger AT (2000). Crystal structures of a Rab protein in its inactive and active conformations. J Mol Biol 304, 585–598. [DOI] [PubMed] [Google Scholar]
- Sun H, Furt F, and Vidali L. (2018). Myosin XI localizes at the mitotic spindle and along the cell plate during plant cell division in Physcomitrella patens. Biochem Biophys Res Commun 506, 409–421. [DOI] [PubMed] [Google Scholar]
- Taylor SC, Nadeau K, Abbasi M, Lachance C, Nguyen M, and Fenrich J. (2019). The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol 37, 761–774. [DOI] [PubMed] [Google Scholar]
- Tuncbag N, Gursoy A, Nussinov R, and Keskin O. (2011). Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc 6, 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Augustine RC, Kleinman KP, and Bezanilla M. (2007). Profilin is essential for tip growth in the moss Physcomitrella patens. The Plant Cell 19, 3705–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Burkart GM, Augustine RC, Kerdavid E, Tüzel E, and Bezanilla M. (2010). Myosin XI is essential for tip growth in Physcomitrella patens. The Plant Cell 22, 1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, and Schwede T. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz T, and Kerkhoff E. (2017). Exploring the iceberg: Prospects of coordinated myosin V and actin assembly functions in transport processes. Small GTPases 10, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard AA, and Moore I. (2008). The functions of Rab GTPases in plant membrane traffic. Current opinion in plant biology 11, 610–619. [DOI] [PubMed] [Google Scholar]
- Wu SZ, and Bezanilla M. (2014). Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. Elife 3, e03498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E, Ueda S, Tamura K, Orii H, Uchi S, Sonobe S, Hara-Nishimura I, and Shimmen T. (2009). An isoform of myosin XI is responsible for the translocation of endoplasmic reticulum in tobacco cultured BY-2 cells. Journal of Experimental Botany 60, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Camacho L, Wee E, Batoko H, Legen J, Leaver CJ, Malho R, Hussey PJ, and Moore I. (2005). A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17, 2020–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.