Abstract
Point mutation in alcohol dehydrogenase 2 (ALDH2), ALDH2*2 results in decreased catalytic enzyme activity and has been found to be associated with different human pathologies. Whether ALDH2*2 would induce cardiac remodeling and increase the attack of atrial fibrillation (AF) remains poorly understood. The present study evaluated the effect of ALDH2*2 mutation on AF susceptibility and unravelled the underlying mechanisms using a multi-omics approach including whole-genome gene expression and proteomics analysis. The in-vivo electrophysiological study showed an increase in the incidence and reduction in the threshold of AF for the mutant mice heterozygous for ALDH2*2 as compared to the wild type littermates. The microarray analysis revealed a reduction in the retinoic acid signals which was accompanied by a downstream reduction in the expression of voltage-gated Na+ channels (SCN5A). The treatment of an antagonist for retinoic acid receptor resulted in a decrease in SCN5A transcript levels. The integrated analysis of the transcriptome and proteome data showed a dysregulation of fatty acid β-oxidation, adenosine triphosphate synthesis via electron transport chain, and activated oxidative responses in the mitochondria. Oral administration of Coenzyme Q10, an essential co-factor known to meliorate mitochondrial oxidative stress and preserve bioenergetics, conferred a protection against AF attack in the mutant ALDH2*2 mice. The multi-omics approach showed the unique pathophysiology mechanisms of concurrent dysregulated SCN5A channel and mitochondrial bioenergetics in AF. This inspired the development of a personalized therapeutic agent, Coenzyme Q10, to protect against AF attack in humans characterized by ALDH2*2 genotype.
Keywords: Aldehyde dehydrogenase 2, Transcriptome, Proteomics, ALDH2*2, SCN5A, Coenzyme Q10
1. Introduction
Mitochondrial isoform of acetaldehyde dehydrogenase (ALDH), ALDH2, is the key metabolic enzyme involved in the detoxification of toxic aldehydes, particularly acetaldehyde [1, 2]. A reduced activity of ALDH2 has been known to cause poor metabolism of ethanol, which further contributes to alcohol flushing reaction and organ damage [3, 4]. A coding variant of ALDH2, ALDH2*2, involves a single-nucleotide polymorphism (SNP) that results in Glu-to-Lys substitution at position 504 (E504K). This SNP has been found to affect ~8% of the human population worldwide, with major predominance in East Asia, where it affects ~40% of the population [5]. The ALDH2*2 variant encodes for a protein with decreased catalytic activity and increased turnover rate [6]. In addition to the ethanol-derived acetaldehyde detoxification, ALDH2 also plays an important role in the oxidation of cytotoxic lipid aldehydes that are generated from lipid peroxidation during oxidative stress [2, 3, 7]. This includes 4-hydroxy-2-nonenal (4-HNE), malondialdehyde (MDA) and environmental aldehydes like acrolein. The polymorphism of ALDH2*2 has also been found to be associated with several cardiovascular diseases, including coronary artery disease, myocardial infarction, reperfusion-related ventricular arrhythmia, heart failure, and hypertension [7].
Atrial fibrillation (AF) is the most common cardiac arrhythmia that particularly contributes to higher risk of stroke, heart failure, and cardiovascular mortality [8]. The presence of interaction between AF events and ethanol-metabolism is suggestive of a complex link between ALDH2*2 and AF [9–11]. The atrial expression of ALDH2 in patients with AF was lower as compared to those with sinus rhythm [9], suggesting that ALDH2 dysfunction increased the risk of AF attack. ALDH2 deficiency increased AF incidence in mice exposed to chronic ethanol intoxication through the accumulation of 4-HNE and increased oxidative stress [10]. However, the prevalence of dysfunctional ALDH2*2 carriers was contrarily lower in AF patients, as compared to those without AF [11]. The casual link between ALDH2*2 and AF pathogenesis is needed to established to solve this paradox with reasonable explanation of underlying pathophysiology mechanisms.
The present study aimed to test the hypothesis that the presence of dysfunctional ALDH2*2 could lead to an increased risk of AF induction and clarify the mechanisms by a multi-omics approach. In the knock-in murine model for ALDH2*2 (E504K mutation) [12], an increased susceptibility to AF was observed in ALDH2*2 genotype. The multi-omics approach showed concurrent dysregulation of SCN5A channel and mitochondrial bioenergetics as the unique pathophysiology mechanisms underlying AF attack in ALDH2 deficiency. A personalized therapeutic agent, Coenzyme Q10, was identified to protect against AF attack in humans characterized by ALDH2*2 genotype.
2. Materials and methods
2.1. Mouse model for atrial arrhythmia
In the present study, the knock-in mouse model for ALDH2*2 was constructed by homologous recombination mediated replacement of wild type ALDH2*1 allele with ALDH2*2 allele containing E504K substitution [12]. The study included two groups of 12–15 weeks old mice, the wild type male littermates (referred to as ALDH2*1) and mice heterozygous for ALDH2*2 (referred to as ALDH2*2). For the evaluation of left ventricular ejection fraction and cardiac structure, electrocardiogram (ECG) was performed prior to electrophysiology (EP) study. The EP study involved surface ECG recordings and epicardial stimulation as previously described by Weng et al. [13]. Post EP analysis, the hearts were removed, weighed, and homogenized to isolate protein and RNA for further analysis.
2.2. Mouse EP study
The mice were anesthetized by the inhalation administration of isoflurane. Post anesthetization, the mice were mechanically ventilated at a controlled temperature of 37°C ± 0.5°C. ECG recordings were performed prior to EP study. The surface ECG (channel I, II, and aVF) was recorded by needle electrodes inserted subcutaneously in the limbs. A mini-thoracotomy was performed in the right parasternal area. Following this, the epicardial recording and stimulation electrodes were positioned on the right atrium and the cardiac rhythm was continuously recorded. For each animal, all the frontal axes (P and QRS) and time intervals (PR, QRS, QT, and RR) of ECG were calculated. The pacing thresholds (in milliamperes) were determined for right atrial stimulation using 1 ms pulse widths at twice the diastolic capture threshold. AF was induced by burst atrial pacing (S1S1), from 50 ms down to a minimum coupling interval of 10 ms, repeated for five cycles in each mouse [13]. The longest atrial burst pacing interval that induced AF was defined as the induction threshold. Longer pacing intervals required for the induction of AF correlated with a greater ease of AF induction. The incidence of AF attack (AF incidence), number of AF attack/burst pacing cycles (AF vulnerability), and duration of induced AF episodes were calculated.
The effect of the treatment with Coenzyme Q10 was studied in 12–15 weeks old ALDH2*2 mice. The mutant mice were randomly divided into two groups, treatment group and control group. Mice in the treatment group received oral treatment of 45mg/kg/day dose of Coenzyme Q10 (NDM-02680, Nature Made, West Hills, CA, USA) dissolved in olive oil as vehicle (dose of olive oil 10ml/kg/day) via nasogastric tube every day for 4 weeks. For the control group, only control vehicle i.e. olive oil was administered in the same volume as mentioned for the treatment group. The EP study was performed 4 weeks post the oral administration of Coenzyme Q10. The animal studies were performed according to the NIH guidelines (Guide for the Care and Use of Laboratory Animals) and all the experimental procedures were approved by the Institutional Animal Use and Care Committee at Taipei Veterans General Hospital.
2.3. Western blot analysis
The western blots analysis was performed according to the previously described protocol [13]. Mouse atrial tissues were homogenized in 1X CHAPS lysis buffer (S7705, Millipore, Billerica, MA, USA) supplemented with protease and phosphatase inhibitors (78442, Thermo Fisher Scientific). The atrial tissue homogenates were centrifuged at 13,000 rpm for 20 min at 4°C. The resulting supernatants were collected and protein concentration was determined by BCA protein estimation assay (23225, Thermo Fisher Scientific) according to the manufacturer’s instructions. The denatured proteins (30 μg/lane) were separated on a NuPAGE 4%–12% Bis-Tris protein gels (NP0335BOX, Invitrogen). The resulting protein bands were transferred on to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). For immunodetection, PVDF membranes were blocked with 5% non-fat dry milk and incubated overnight with primary antibodies, including rabbit anti-MDA (1:1000, ab6463, Abcam), mouse anti-4-HNE (1:1000, ab48506, Abcam), mouse anti-ALDH2 (1:1000, sc-48837, Santa Cruz), and mouse anti-GAPDH (1:5000, MA5-15738, Thermo Fisher Scientific). Further, the membranes were incubated with HRP-conjugated secondary anti-mouse (1:10000, AP124P, Millipore) and anti-rabbit (1:10000, 211-032-171, Jackson ImmunoResearch, Pennsylvania, USA) antibodies for 1 hour at room temperature. The unbound secondary antibodies were removed by washing with PBST and PBS as mentioned above. Chemiluminescent detection of protein expression by western blot analysis was performed using ECL western blot detection reagents (WBKLS0500, Millipore, Billerica, MA, USA) and analyzed using AlphaEaseFC 4.0 (Alpha Innotech, San Leandro, CA, USA).
2.4. Purification and proteomics analysis for mitochondrial proteins
The mitochondrial proteins present in the left atrium of mice from both the groups, the wild type group containing ALDH2*1 allele and mutant group with ALDH2*2 allele, were isolated and purified using Qproteome® Mitochondria isolation kit (Qiagen, Hilden, Germany) [14]. Briefly, atrial tissue was freshly excised from 2-3 mice for each proteomics sample, and washed with 1 ml of 0.9% (w/v) sodium chloride solution. Following this, the tissue was cut into small pieces of size 1–2 mm3 and lysed using 500 μl lysis buffer supplemented with protease inhibitors. The resulting samples were homogenized using the Tissue Ruptor rotor-stator homogenizer set at lowest possible speed for 10 s. To this, 1.5 ml of lysis buffer supplemented with protease inhibitor solution was added further. The resulting homogenates were incubated on an end-over-end shaker at 4°C for 10 min. Further, the samples were the centrifuged at 1000 × g for 10 min at 4°C. The supernatant was carefully removed and the resulting cell pellet was resuspended in 1.5 ml of ice-cold disruption buffer. The samples were properly mixed by pipetting up and down using a 1 ml pipette tip. To ensure proper resuspension of the pellet, the samples were further mixed using a blunt-end needle and a syringe. The resulting cell lysate was centrifuged at 1000 × g for 10 min at 4°C. The supernatant was collected in fresh tube and centrifuged again at 6000 × g for 10 min at 4°C. The final pellet containing mitochondrial proteins was collected and stored at −20°C until used for proteomics analysis.
For proteomics analysis, 50 μg of purified mitochondrial proteins from each sample were used. Proteomic analysis was performed in repeats of three for independent samples obtained from both ALDH2*1 and ALDH2*2 mice. The protein lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Post separation, the protein bands present on SDS-PAGE gel were visualized by staining with Coomassie blue (J.T.Baker, USA). These protein bands were divided into 10 parts/fragments (average) according to the distribution of the stain. Following this, these gel pieces were subjected to in-gel digestion as previously described by Chi et al. [15]. Briefly, each gel fragment was cut into small pieces of size 1 mm3 and de-stained with 25 mM ammonium bicarbonate. The de-stained fragments were further reduced by dithiothreitol (DTT, Sigma, USA) and alkylated using iodoacetamide (IAA, Sigma, USA). Following this, the samples were subjected to trypsin (sequencing grade, Promega, USA) digestion. For peptide extraction, the samples were incubated with 60% acetonitrile and sonicated. This was followed by desalting of peptides with 0.1% trifluoroacetic acid (TFA, Sigma, USA). The samples were dried using vacuum centrifuge and analyzed by LC-MS/MS analysis (LTQ-FT LC/MS/MS, Thermo Electron). For peptide analysis, spectrums were searched against Mascot engine (Matrix Science) to match protein ID according to UniProt reference proteome database. The non-labeling protein expression quantification was calculated by Maxquant [16].
2.5. Microarray
RNA was isolated from the left atrium of mouse (ALDH2*1, n=3; ALDH2*2, n=4) using RNeasy Mini kit (Qiagen, USA). For transcriptome microarray analysis, 100 ng of total RNA was used (GeneChip™ Mouse Gene 2.0 ST Assay, Thermo, USA). The reads were normalized and analyzed by GeneSpring software (Agilent Tech, USA). Statistical inference was calculated using independent t-test. For the analysis of signaling pathways, a total of 790 differentially expressed genes (fold change ≥ 1.5 times with P-value < 0.05) between wild type and ALDH2*2 mutant mice were used (Supplementary data 1). The differentially expressed genes were sorted and analyzed using ingenuity pathway analysis (IPA, Qiagen, USA).
2.6. The transcriptomics and network analysis of ALDH2 dysfunction
The canonical pathways for 790 differentially expressed genes were analyzed using IPA for both ALDH2*1 wild type and ALDH2*2 mutant animals. The cardiac toxicity signals correlating to arrhythmia were also collected from IPA database. For the co-expression networks for cardiac toxicity signals, Pearson correlation coefficients (PCC) were calculated for the expressions of retinoic acid receptors (RARs), retinoid X receptors (RXRs), and relevant cardiac toxicity genes associated with ALDH2*1 and ALDH2*2 mice. The protein-protein interaction [14] networks for cardiac toxicity signals were constructed using PPI database STRING [14, 17].
2.7. Retinoic acid (RA) concentration in serum
RA concentration in mice serum samples was determined using the RA ELISA Kit (MBS705877, MyBioSource, USA). The blood sample collection from mouse orbital sinus was performed using cotton, anesthetic agent, blood sample collection tubes, and capillary tube [18]. The samples were centrifugated at 1000 × g for 15 minutes and diluted at the ratio of 1:500 using Sample Diluent before the experiment. All the reagents and working standards were prepared according to the manufacturer’s instructions. A microtiter plate pre-coated with RA specific antibody was provided in the kit. To each well, 50 μl of standards or diluted samples were added. This was immediately followed by the addition of 50 μl Horseradish Peroxidase-conjugated RA (1x), except to the Blank well. For luminescent reaction, 90 μl TMB Substrate was added to each well and incubated at 37°C for 20 minutes in dark. The concentration of RA in each sample was measured in terms of absorbance at 450 nm using a microplate reader. The absorbance was measured within 5 minutes of the addition of stop solution.
2.8. Multi-omics and network analysis for the mitochondrial gene expression in mice atria
According to the DAVID database [19], 86 of the 790 differentially expressed genes obtained from the microarray analysis and 102 of the 170 differentially expressed proteins (fold change ≥ 1.5 times or ≤ 0.5 times) obtained from the proteomics analysis were annotated mainly to localize in the mitochondria (Supplementary data 2 and 3). These were further subjected to gene ontology (GO) functionality enrichment analysis.
For both the cellular component and biological process, the DAVID gene enrichment analysis was carried out on 182 mitochondrial genes/proteins. These 182 genes/proteins included differentially expressed genes and proteins identified in the transcriptomics and proteomics experiments, respectively, as mentioned above. The resulting enriched GO terms for cellular components (CC) were visualized using cytoscape app “ClusGO” [20]. The enriched functions (GO CC) for clusters with similar functions were shown in different colors. A total of nine clusters, characterized by at least two functional terms (i.e. cluster members), were generated. For cluster #1, the term present at the lowest GO level was chosen as the cluster representative, whereas the functional term with the smallest P-value was chosen as the cluster representative for other clusters. The 59 genes/proteins annotated with the cluster-representative GO term were further subjected to PPI network analysis.
2.9. Real-time quantitative polymerase chain reaction (PCR)
The SuperScript® III First-Strand Synthesis System (18080051, Invitrogen, Carlsbad, CA, USA) was used for cDNA synthesis from the extracted RNA samples. Real-time quantitative PCR was performed using Applied Biosystems™ StepOnePlus real-time PCR system with TaqMan probe assays (4331182, Thermo Fisher Scientific, USA). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control. All the genes described in the probe assays are listed in Supplementary Table 1.
2.10. Neonatal rat cardiomyocyte isolation and treatment
Neonatal rat ventricular myocytes (NRVMs) were isolated from 1–2 days old neonatal rat pups as previously described by Kapoor et al. [21]. The isolated NRVMs were cultured in M199 medium supplemented with 10 mM HEPES, 0.1 mM non-essential amino acids, 3.5 mg/mL glucose, 2 mM L-glutamine, 4 μg/ml vitamin B12, 100 U/ml penicillin, and heat-inactivated FBS. The cells were treated with 1μM acetaldehyde (402788, Sigma-Aldrich) and 10μM BMS 493 (B6688, Sigma-Aldrich) for 48 hours using in 2% FBS supplemented medium. The cells treated with ddH2O or DMSO were used as controls. Total RNA isolation for real-time PCR analysis was performed using RNeasy® Mini Kit (74106, Qiagen, Venlo, Netherlands).
2.11. Statistical analysis
Statistical analysis for all the assays was performed using PSAW SPSS 18.0 software. All the data were presented/expressed as mean ± SEM. For the identification of possible differences present between the two groups, Student’s t-test was used. A chi-square test was used to evaluate differences in the categorical variables of the two groups. For all the statistical analysis, a P-value < 0.05 was considered to be statistically significant.
3. Results
3.1. Increased incidence of AF in ALDH2*2 mutant mouse model
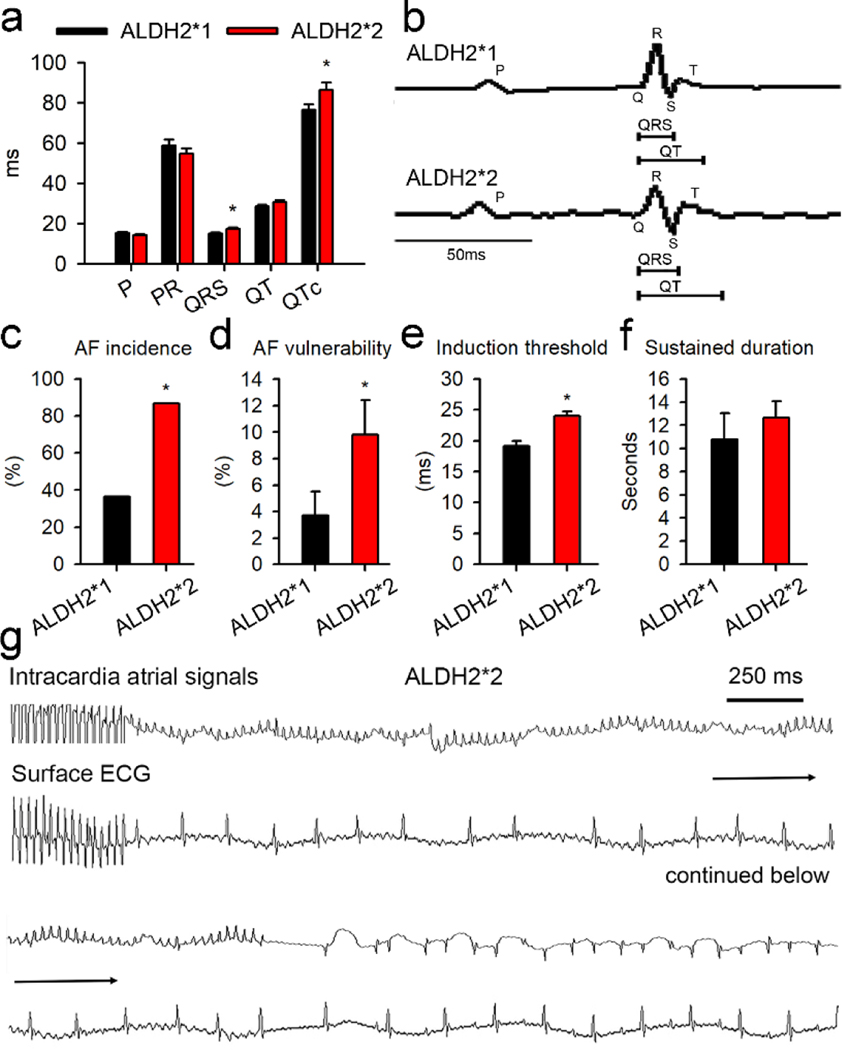
Surface ECG analysis for ALDH2*2 mutant mice showed that the QRS durations and QT intervals were increased as compared to the mice from ALDH2*1 group (Fig. 1a–b). No significant differences were observed in the P-wave duration, P-R interval, body weight, and weight of left atrium for the mice belonging to the two groups (Fig. S1). Echocardiography analysis revealed the absence of any significant differences in the cardiac structure and function for the mice associated with the wild type group and the mutant group. The parameters that were studied included left ventricular anterior wall thickness, left ventricular end-systolic diameter (LVESD), left ventricular end-diastolic diameter (LVEDD), and global cardiac function (Supplementary Fig. 1). As shown in Fig. 1c–d, epicardial atrial electrophysiological stimulation in ALDH2*2 mice resulted in 2.3-fold and 2.6-fold increase in the frequency and vulnerability, respectively, of AF episodes. The threshold for the induction of AF was found to be 20% lower in ALDH2*2 mice as compared to the wild type mice (Fig. 1e). No differences were observed in the sustained duration of AF between the two groups (Fig. 1f). The representative recordings of AF for one of the ALDH2*2 mice are shown in Fig. 1g. In summary, mice from ALDH2*2 group were characterized by higher susceptibility and lower threshold for AF as compared to ALDH2*1 mice.
Fig. 1. ALDH2*2 mediated increase in atrial arrhythmia in transgenic mouse model for ALDH2*2.
(a) The surface ECG measurements, including P-wave, P-R interval, QRS duration, and QT interval, for ALDH2*1 and ALDH2*2 mice. ALDH2*1 wild type group is represented in black (n = 10) and mutant ALDH2*2 group is shown in red (n = 12). (b) Representative ECG morphology for both the groups post atrial burst stimulation. Bar graphs representing (c) AF incidence, the incidence of AF attack, in ALDH2*1 and ALDH2*2 mice; (d) AF vulnerability, the number of AF attack/burst pacing cycles; (e) AF induction threshold. Here, the longest atrial burst pacing interval that induced AF was defined as induction threshold. Longer pacing intervals required for the induction of AF correlated with a greater ease of AF induction; (f) The sustained durations of AF. (g) The representative tracing of AF in an ALDH2*2 mouse. Both intracardiac signals and surface ECG were presented. The burst epicardial pacing was followed by AF attack. The highly fractionated signals occurring in the intracardiac recordings correlated with the fibrillatory atrial waves on the surface ECG and irregular conduction of QRS complex. All these features were compatible with the diagnosis of AF. AF terminated spontaneously in to sinus rhythm, while discrete P wave and QRS complex were observed along with corresponding intracardiac deflection signals. For Fig. 1c–f, ALDH2*1 (n = 11) and ALDH2*2 mice (n = 15). * P-value < 0.05 for was determined by two-tailed t-test for Fig. 1a, d, and e, and for Fig. 1c by chi-square test.
3.2. Microarray transcriptome analysis of ALDH2*2 mice atria
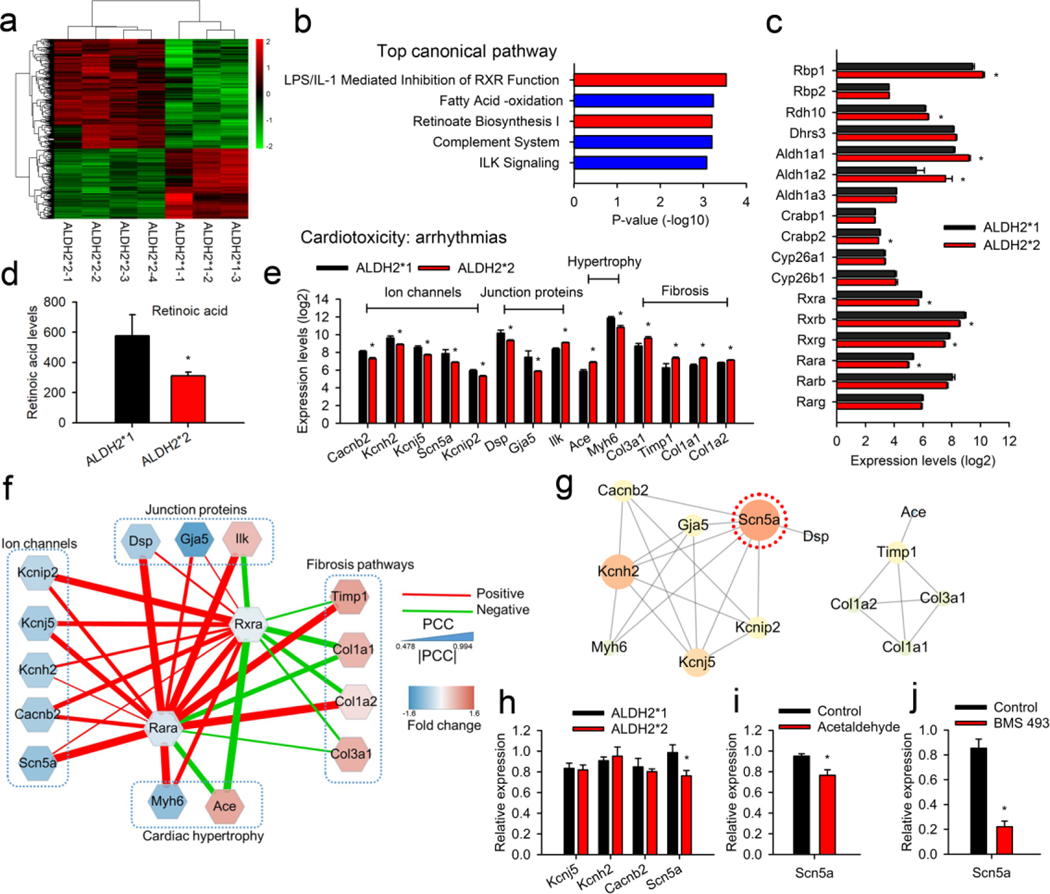
Differential gene expression in the two groups was studied using microarray transcriptome analysis. Microarray transcriptome studies were performed using whole left atrial tissues of ALDH2*1 and ALDH2*2 mice (Fig. 2a, Supplementary data 1). IPA analysis for the canonical pathways revealed the involvement of RA signals in the top two pathways (Fig. 2b). Further, the analysis for the physiological function suggested that the differentially expressed genes were associated with different cardiovascular functions (Supplementary Table 2). Following this, different RA signaling pathways, including biosynthesis, metabolism, and RA receptors, were examined. In general, RA is involved in the regulation of different biological activities in the body via by binding to RARs, RARA, RARB, and RARG (Fig. 2c). Further, the RA/RAR complex heterodimerizes with RXRs, RXRA, RXRB, and RXRG. Post heterodimerization, the resulting complex enters the nucleus and regulates the downstream gene transcripts via retinoic acid response element (RARE) [22]. The expression of RARA, RXRA, RXRB, and RXRG was found to be less in ALDH2*2 mice as compared to ALDH2*1 mice. In comparison to this, the enzymes responsible for the conversion of vitamin A to RA in the biosynthetic pathway, namely RDH10, ALDH1A1, and ALDH1A2 were found to be upregulated in the atrium of ALDH2*2 mice in a compensatory manner. In addition to this, the serum levels of RA in ALDH2*2 mice were found to be less as compared to ALDH2*1 mice (Fig. 2d). These observations were consistent with the downregulation of RA/RAR signaling pathways. In summary, RA levels and its downstream signals were downregulated in the atrium of ALDH2*2 mice.
Fig. 2. The microarray transcriptome analysis for atrial tissue isolated from mutant ALDH2*2 mice.
(a) Heat map showing differential gene expression for the atrial tissue obtained from mice belonging to ALDH2*2 (n=4) and ALDH2*1 (n=3) groups. ALDH2*1 group was used as control, while ALDH2*2 as test group. The details of the gene expressions are provided in Supplementary data 1. The scaled expression values (Z-scores) for different genes were color-coded (represented as a bar on the right side of the map) to generate the heat map. (b) The top 5 canonical pathways identified using IPA. Two pathways (shown in red bar) were related to RA/RAR signaling. (c) Microarray analysis for gene expression involving RA biosynthesis, degradation, and its associated downstream pathways. (d) Bar graph representing serum levels of RA for ALDH2*2 and ALDH2*1 mice. (e) Transcripts of arrhythmia-related genes identified using cardiotoxicity analysis. These genes were selected by IPA and classified into to four subgroups, namely ion channels, junction proteins, cardiac hypertrophy, and fibrosis pathways. (f) The PCC between the expression of RARs, including RARA and RXRA, and arrhythmia-related genes. A positive correlation is represented by red lines, while green lines denote a negative correlation. The width of the line indicates the absolute value of the correlation. The blue nodes indicate downregulated genes in the atrial tissue of ALDH2*2 mice as compared to ALDH2*1. The red nodes represent the upregulated genes. The RA/RAR pathway was characterized by a complex interaction with SCN5A, DSP, MYH6, TIMP1, and COL1A2. (g) PPI network constructed for arrhythmia-related genes using the STRING database. SCN5A, the hub/central protein which is highly connected with other proteins, was circled by red dots. This indicated the central role of SCN5A in the RA/RAR pathway. (h) Quantitative PCR analysis for the expression of transcripts of arrhythmia-related ion channels. The downregulation of SCN5A was observed in the atrial tissues obtained from ALDH2*2 mice (n = 9) as comparted to the wild type ALDH2*1 mice (n = 10). (i) Quantitative PCR analysis for the expression of SCN5A in the mice treated with toxic aldehydes (acetaldehyde, 1μM, n = 7). The treatment of mice with acetaldehyde resulted in a decrease in the expression of SCN5A as compared to the control group receiving only vehicle (n = 7). * P-value < 0.05 was calculated using two-tailed t-test. (j) Quantitative PCR analysis for mice treated with an antagonist of RAR (BMS493, 10μM, n = 7). Control vs. BMS493; 0.9 ± 0.1 vs. 0.2 ± 0.05, P-value < 0.05, n = 7 and 7, respectively. * P-value < 0.05 was calculated by two-tailed t-test for all the tests reported above.
Since AF susceptibility was found to be increased in the case of ALDH2*2 mice, IPA was performed to analyze the signaling pathways associated with cardiac toxicity with respect to arrhythmia (Fig. 2e). For the mutant ALDH2*2 mice, the ion channels (CACNB2, KCNH2, KCNJ5, SCN5A, and KCNIP2), desmosomal protein (DSP), and gap junction proteins (GJA5) expressions were found to be downregulated as compared to the wild type mice. However, the genes related to cardiac hypertrophy (ACE and MYH6) and fibrosis pathways (COL3A1, TIMP1, COL1A1, and COL1A2) were upregulated in ALDH2*2 mice. These changes were possibly associated with cardiac electrical and structural remodeling in AF. The co-expression network analysis showed that RA and its receptors (RAR and RXR) were highly correlated with the cardiac arrhythmia pathways (Fig. 2f). The ECG changes in ALDH2*2 mice were more relevant with respect to the differential changes in the ion channels because SCN5A, an ion channel-related gene, emerged as the tightly connecting node in the PPI network analysis of the cardiac arrhythmia signals (Fig. 2g). In addition to this, RA and its receptors were strongly associated with positive regulation of SCN5A (Fig. 2f). Real-time PCR was further used to validate the expression of genes associated with ion channels. As shown in Fig. 2h, only SCN5A transcripts were found to be downregulated (Fig. 2h). Next, the regulation of RA signals on SCN5A expression was studied in the NRVMs in vitro. The treatment of the cells with a toxic aldehyde, acetaldehyde (1 μM), resulted in a decrease in SCN5A transcripts (Fig. 2i). The cells were also treated with an antagonist of RA/RAR, BMS493, that inhibited the interaction between RA/RAR complex and RARE. BMS493 treatment also resulted in a 5-fold decrease in SCN5A transcripts (Fig. 2j).
3.3. Multi-omics analysis for mitochondrial dysfunction in the atria derived from ALDH2*2 mice
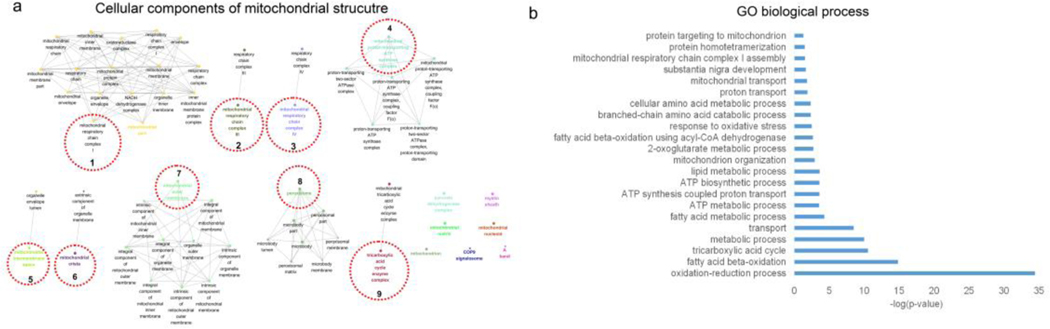
IPA did not identify any mitochondria-related pathways in the whole transcriptome analysis. To further explore the mitochondria-related mechanisms involved in AF, mitochondrial proteins were isolated from the atrial tissues and proteomics analysis was conducted to determine the impact of ALDH2 mutation on the mitochondrial dysfunction. The fractionation and purity of the mitochondrial preparations were confirmed by western blot analysis (Supplementary Fig. 2). The integrative omics approach was utilized to systemically analyze the differentially expressed genes and proteins related to the mitochondria. The GO analysis for cellular component enrichment performed using the integrative multi-omics approach showed the clustering of enriched mitochondrial structures (mainly 9 clusters), including respiratory chain complex I, III, and V, peroxisome, tricarboxylic acid cycle enzyme complexes, and mitochondrial outer membrane proteins (Fig. 3a, Supplementary data 4). Further, GO mapping for biological process reflected the impact of ALDH2*2 mutation on fatty acid β-oxidation and ATP synthesis (Fig. 3b). The identification of top biological processes by GO mapping suggested that these biological processes were associated with energy production, particularly fatty acid β-oxidation, ATP synthesis, and metabolic processes (Fig. 3b, Supplementary data 5). The fatty acid-related processes included oxidation-reduction, fatty acid β-oxidation, fatty acid metabolism, lipid metabolism, and fatty acid β-oxidation using acyl-CoA dehydrogenase. The ATP biosynthesis and metabolism-related processes included ATP metabolism, ATP synthesis coupled proton transport, and ATP biosynthesis. In addition to these, response to oxidative stress was observed as one of the top GO processes. The results of the study revealed only minor involvement of amino acid metabolism.
Fig. 3. The integrated analysis of proteomics and transcriptomics data for differentially expressed mitochondrial genes/proteins in ALDH2*2 mice.
(a) The clustered expressions of mitochondrial genes/proteins for corresponding structural components. GO analysis for cellular component enrichment identified 182 mitochondrial genes/proteins clustered using “ClusGO” (9 clusters for corresponding structural components, adjusted P-value < 0.05). GO terms sharing at least one gene were connected. The GO term with smallest P-value in each cluster was highlighted with a different color and the cluster representative was indicated by red dotted circles. The edges connecting the nodes represented PPI. (b) Enriched GO biological process terms (P-value < 0.05) identified for 182 differentially expressed genes/proteins. These biological terms are ranked in terms of −log (P-value).
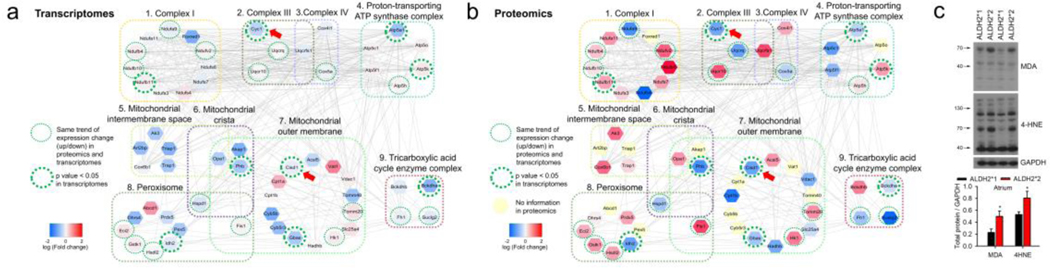
The comparison of the degree of translational and transcriptional changes within the clustered expressions of mitochondrial structure revealed a simultaneous downregulation of Cytochrome c1 (CYC1), mitochondrial complex V (ATP5A1), and CDGSH iron-sulfur domain 1 (CISD1) in the mutant ALDH2*2 mice (Fig. 4a and b, Supplementary data 6). The downregulation of CYC1 is generally associated with a decrease in electron transport and ATP production, and increase in reactive oxygen species (ROS) generation. Similarly, downregulation of CISD1 has been reported to be associated with an increase in ROS and lipid peroxidation [23]. The analysis of lipid peroxidation revealed an increase in the levels of lipid peroxidation end-products, including protein adducts of 4-HNE and MDA in the atrium of ALDH2*2 mice (Fig. 4c). These results correlated well with the findings of the GO analysis and changes associated with transcription and translation processes.
Fig. 4. The link of oxidative stress to differential expressions of mitochondrial transcripts and proteins.
(a) and (b) The color-coded fold changes obtained from the differential expressions of transcriptomics (a) and proteomics analysis (b), respectively. Representative functions (in GO CC terms) for the 9 highly-connected broad clusters were identified for both proteomics and transcriptomics analysis (59 genes, Supplementary data 6). The simultaneous downregulation of CYC1 and CISD1(arrow head) in ALDH2*2 mice is linked to increased oxidative stress. The red colored nodes indicate that the expression of the corresponding gene was higher in ALDH2*2 mice as compared to ALDH2*1 mice, while the blue colored nodes indicate a lower expression of the associated gene. The yellow nodes correspond to the proteins for which no information was available from the proteomics experiments. The dotted green circles indicated that the trend for the change (i.e. up or down) in the expression for ALDH2*2 and ALDH2*1 mice was same for the results obtained from transcriptomics and proteomics analysis. Further, the thick green circles indicated a significant difference in the transcriptomics data for ALDH2*2 and ALDH2*1 mice (P-value < 0.05). The edges connecting the nodes represented PPI. (c) Western blot analysis for aldehyde-modified proteins (4-HNE and MDA) isolated from left atrial tissue of ALDH2*2 and ALDH2*1 mice. Increased expressions of 4-HNE and MDA suggest of increased oxidative stress and lipid peroxidation. Upper panel shows representative blots for 4-HNE and MDA. Lower panel includes the bar graph showing the expression of 4-HNE and MDA-modified proteins. Mutant ALDH2*2 mice (4-HNE n = 4; MDA n = 4); Wild type ALDH2*1 mice (4-HNE, n = 5; MDA n = 4), * P-value < 0.05 determined by two-tailed t-test.
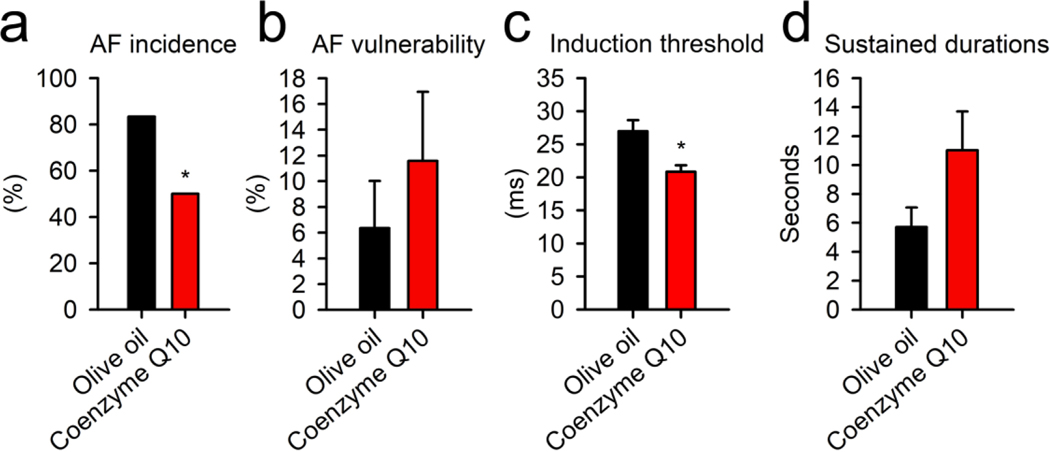
The combined analysis of all the important links for all the signaling pathways (fatty acid β-oxidation, TCA cycle, ATP production, ROS, and lipid peroxidation) revealed a common intersection point through Coenzyme Q/complex III. For ATP synthesis, the electron transport from either TCA cycle or fatty acid β-oxidation would come across mitochondrial respiratory complex III with electron transfer flavoprotein-ubiquinone oxidoreductase and Coenzyme Q interaction. In addition to this, Coenzyme Q and complex III also determine the production of ROS in the mitochondria and subsequent lipid peroxidation. Thus, the effect of supplementation with Coenzyme Q on the susceptibility to AF was studied in ALDH2*2 mice. The treatment of ALDH2*2 mice with Coenzyme Q10 resulted in a 30% reduction in the incidence of AF attack (AF incidence). This was accompanied by a significant decrease in the AF threshold (Fig. 5, n = 6 treatment vs. control). However, the treatment of the mutant mice with Coenzyme Q10 showed no significant effects on AF duration and vulnerability. This suggested the existence of a critical link between mitochondrial dysfunction and AF in the mutant ALDH2*2 mice. In addition to this, these results highlighted the suitability of Coenzyme Q10 to be used as a potential pharmacological agent to reduce the risk of AF.
Fig. 5. The treatment of ALDH2*2 mutant mice with Coenzyme Q10.
(a) Bar graph representing AF incidence in Coenzyme Q10 treated and control mice. The incidence of AF attack decreased after treatment with Coenzyme Q10. Bar graphs for (b) AF vulnerability, (c) AF induction threshold, and (d) sustained duration of AF in mice receiving olive oil alone or Coenzyme Q10 dissolved in olive oil. No differences were observed in the AF vulnerability and sustained duration between the mice treatment with the vehicle and Coenzyme Q10. However, the treatment of mice with Coenzyme Q10 increased the induction threshold. These results suggested that Coenzyme Q10 provided protection against AF in ALDH2*2 mutant mice. Vehicle alone (n = 6), and mice receiving Coenzyme Q10 (n = 6). * P-value < 0.05 was calculated using (a) chi-square test and (b–d) two-tailed t-test.
3.4. Independent signaling between transcriptomics and proteomics data
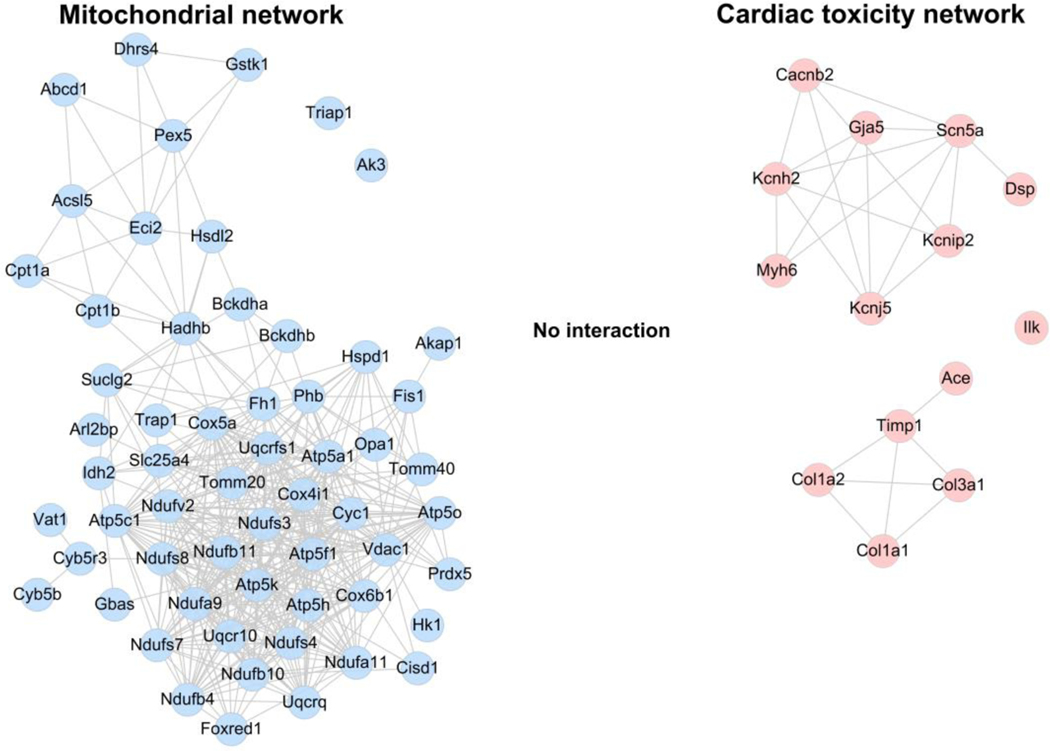
The information of all the possible interactions was extracted from the PPI database for 59 differentially expressed genes/proteins in mitochondria in the cluster analysis (Fig. 3) and 14 genes of cardiac toxicity signals obtained from IPA (Fig. 2). As observed from mitochondria-related and cardiac toxicity-related PPI networks, the PPI networks of mitochondria-related and cardiac toxicity-related genes were devoid of any overlap or crosstalk (Fig. 6), suggesting no interaction between these networks. These results suggested an independent regulation of the mitochondrial dysfunction and RA/RAR-SCN5A pathway.
Fig. 6. The PPI networks for mitochondria-related genes and arrhythmia-related genes.
Left panel represents the mitochondria-PPI network constructed for the representative 59 differentially expressed genes/proteins in the mitochondria (blue node). The right panel includes the PPI network for 14 cardiac toxicity genes (red node). The edges connecting the nodes represented PPIs. There is no edge connecting the blue and red nodes, indicating no interaction between these mitochondria-related and cardiac toxicity-related PPI networks.
4. Discussion
The present study demonstrated that ALDH2*2 mutation in murine model resulted in an increase in the AF susceptibility and reduction in the threshold for AF induction. The comprehensive multi-omics approach revealed ALDH2 deficiency simultaneously induced electrical remodeling (Fig. 7, reduced SCN5A channels, mitochondria-independent) and decreased bioenergetics of cardiomyocytes (mitochondria-dependent). These results suggested of unique pathophysiology mechanisms to initiate AF in the patients characterized by ALDH2*2 genotype. According to ALDH2*2 genotype, a personalized target therapy of Coenzyme Q10 could be used as a future therapeutic agent.
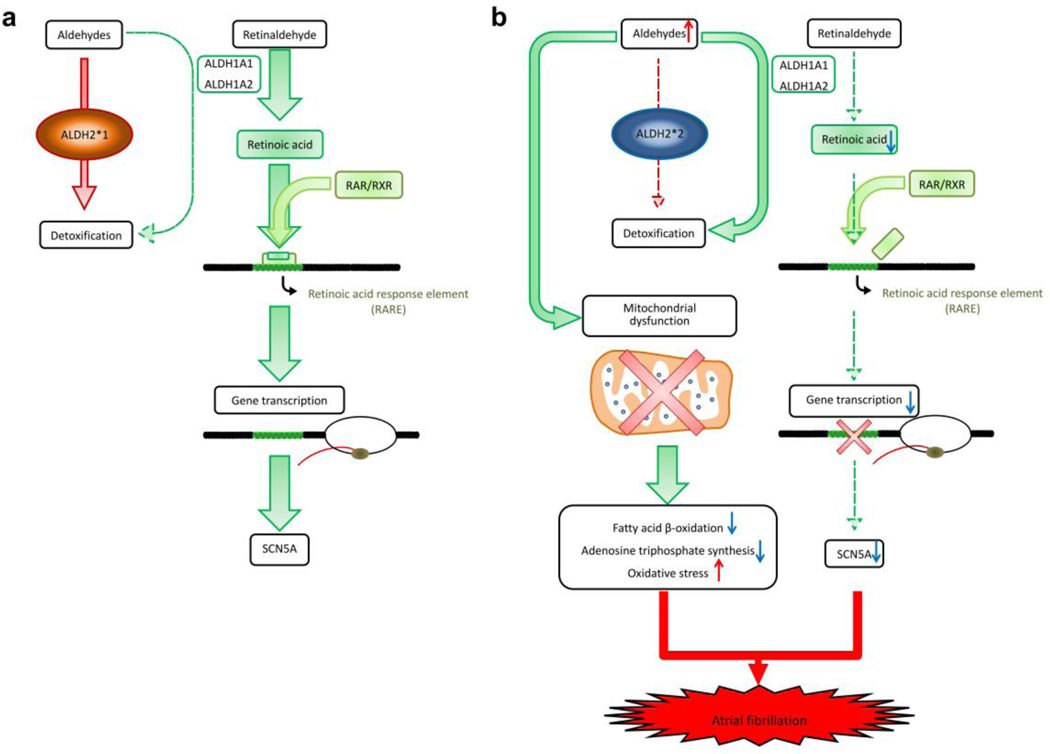
Fig. 7. The proposed mechanisms of ALDH2*2 to induce atrial fibrillation.
(a) ALDH1A1 and ALDH1A2 play a main role of retinoic acid biosynthesis and oxidize retinaldehyde to RA. These two enzymes could also metabolize aldehydes, but their contribution is minimal as ALDH2*1 genotype metabolizes aldehydes in the most efficient way. (b) ALDH2*2 is associated with decreased catalytic activity, and therefore, accumulation of toxic aldehydes would induce mitochondrial dysfunction, including dysregulated bioenergetics (reduced fatty acid β-oxidation and adenosine triphosphate synthesis), and increased oxidative stress (4HNE and MDA). In addition, an mitochondria-independent pathway induces electrical remodeling and AF through the suppression of SCN5A expressions. The accumulation of toxic aldehydes would compete retinaldehyde for catalytic activity of ALDH1A1 and ALDH1A2 (competitive inhibition). Therefore, the levels of RA and related RA/RAR pathway are further downregulated, which is accompanied by decreased transcriptional activity of SCN5A. The concurrence of dysregulated SCN5A channel and mitochondrial bioenergetics contributes to AF attack.
4.1. Whole cell transcriptome analysis: the dysregulation of ion channels in ALDH2*2 mice
The results of the in vivo experiment revealed that the mutation in ALDH2 gene, ALDH2*2, imposed a significant risk for AF attack as more than 80% of the mice in the mutant group ALDH2*2 developed AF. This indicated a detrimental role of ALDH2*2 mutation in the development of AF, and established its causality. Several previous studies have reported a relevant role of ALDH2 activation in arrhythmia. In particular, ALDH2 activation has been shown to prevent reperfusion ventricular arrhythmia by inhibiting reactive aldehydes-induced release of renin in mast cells [24]. The pathogenic role of the RA/RAR pathways to regulate SCN5A identified in the present study has not been previously reported in AF or ALDH2-related signal pathways. The signaling pathways involving RA/RAR are known to be critical for the cardiac development and morphogenesis, especially for atrium, during the embryonic stage [25]. The deficiency of RA has been previously reported to cause cardiac teratogenesis [25, 26]. Besides this, it might also induce varying cardiac structural remodeling patterns in normal rats or post myocardial infarction [27, 28]. In the present study, we observed the link between RA/RAR signaling and electrical remodeling through the transcriptional regulation of voltage-gated sodium channels (SCN5A) and electrical instability. The reduction in sodium current is previously known to decrease velocity of cardiac conduction. This decrease in intra-atrial conduction could in turn initiate AF without any structural remodeling [29]. Several clinical studies have previously reported an association between the loss-of-function mutations in SCN5A and increased incidences of AF [30]. Thus, the present study provided new insights into the mechanisms involved in ALDH2*2 mutation and AF, that might be instrumental in devising a promising clinical treatment for the prevention of AF in humans carrying this mutant ALDH2*2 allele.
4.2. Multi-omics analysis for mitochondrial dysfunction
In general, ALDH2 enzyme is located in the mitochondrial matrix. ALDH2*2 enzyme deficiency is known to be associated with higher susceptibility to oxidative stress owing to the accumulation of toxic aldehydes [3]. ALDH2*2, causes an increase in the production of ROS and accumulation of toxic aldehydes during ischemia or reperfusion, which further results in cardiac injury. A similar mechanism has been also reported in case of alcoholic cardiomyopathy [31, 32]. In addition to this, ALDH2 deficiency also results in increased mitochondrial permeability, altered mitochondrial membrane potential, increased DNA damage, and reduced oxidative phosphorylation [3]. However, the alterations in the mitochondrial molecular networks in relation to ALDH2 deficiency has not been studied in detail. In addition, most of the previously reported studies have majorly focused on the analysis of ventricles, instead of atria. The genes encoding for mitochondrial proteins and those involved in the functional regulation have not been well characterized owing to lower expression in pan-transcriptome analysis. Thus, an integrated analysis of whole cell microarray in conjunction with mitochondrial proteomics analysis would be more useful in getting detailed insights into the underlying pathogenic mechanisms. In addition to comprehensively delineate mitochondria-related pathways, a critical dysregulation hub that converged at Coenzyme Q and complex III was identified. This particularly mediated a crosstalk between reduced fatty acid β-oxidation mediated ATP production from electron transport chain (oxidative phosphorylation) and increased oxidative stress. The use of Coenzyme Q10 supplemented diet in the in vivo murine model to compensate for the mitochondrial dysregulation and reduce AF attack supported the proposed mechanism. The supplement of Coenzyme Q could be used as a personalized therapeutic agent, which could effectively reduce AF in the transgenic murine model for ALDH2*2, and possibly in the patients with ALDH2*2 genotypes. The changes in the glutathione-redox cycle is an alternative pathway that might be induced by mitochondrial aldehyde stress and increase in oxidative stress [33]. However, this pathway was not found to be associated with increased oxidative stress in the present study. Future studies are warranted to further dissect the interaction between mitochondrial metabolic adaptation and other cellular responses [34].
5. Conclusions
The stepwise multi-omics analysis revealed the concurrent pathophysiology mechanisms of AF, including reduction in SCN5A and dysregulation of mitochondrial bioenergetics in ALDH2*2 genotype. The Coenzyme Q10 could be used as a personalized therapeutic agent to provide protection against AF in patients characterized by a popular ALDH2*2 genotype.
Supplementary Material
Acknowledgements
The present study was supported by Taipei Veterans General Hospital (grant nos. V109C-070, VN108-12, and VN109-03), the Ministry of Science and Technology (grant no. 108-2628-B-075-003, 109–2314-B-075–077), Szu-yuan Research Foundation of Internal Medicine (grant no. 107-041), National Health Research Institutes (grant no. NHRI-EX108-10513SC), Academia Sinica (grant no. AS-TM-109-01-05), a research grant awarded to D. Mochly-Rosen from National Institute of Health, U.S.A. (NIAAA grant no. AAA11147), and financial support provided to C.H. Chen at Stanford University.
Abbreviations
- 4-HNE
4-hydroxy-2-nonenal
- AF
atrial fibrillation
- ALDH2
aldehyde dehydrogenase 2
- CC
cellular components
- CISD1
CDGSH iron-sulfur domain 1
- CYC1
Cytochrome c1
- ECG
electrocardiogram
- EP
electrophysiology
- GO
gene ontology
- LVEDD
left ventricular end-diastolic diameter
- LVESD
left ventricular end-systolic diameter
- MDA
malondialdehyde
- NRVMs
neonatal rat ventricular myocytes
- PCC
Pearson correlation coefficient
- PCR
polymerase chain reaction
- PPI
protein-protein interaction
- PVDF
polyvinylidene difluoride
- RA
retinoic acid
- RARE
retinoic acid response element
- RARs
retinoic acid receptors
- ROS
reactive oxygen species
- RXRs
retinoid X receptors
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SNP
single-nucleotide polymorphism
Footnotes
Declaration of competing interest
C. H. Chen and D. Mochly-Rosen hold patents related to ALDA-1 activation of ALDH2*1 and ALDH2*2. One of the patents is licensed to Foresee Pharmaceuticals, a company that D. Mochly-Rosen consults. However, the company did not contribute to any of the experiments included in the study. The other authors declare no conflict of interest.
References
- 1.Klyosov AA, Rashkovetsky LG, Tahir MK, Keung WM. Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry. 1996; 35: 4445–56. doi: 10.1021/bi9521093 [DOI] [PubMed] [Google Scholar]
- 2.Zambelli VO, Gross ER, Chen CH, Gutierrez VP, Cury Y, Mochly-Rosen D. Aldehyde dehydrogenase-2 regulates nociception in rodent models of acute inflammatory pain. Sci Transl Med. 2014; 6: 251ra118. doi: 10.1126/scitranslmed.3009539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014; 94: 1–34. doi: 10.1152/physrev.00017.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edenberg HJ, McClintick JN. Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcohol Clin Exp Res. 2018; 42: 2281–97. doi: 10.1111/acer.13904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang YC, Chiu YF, Lee IT, Ho LT, Hung YJ, Hsiung CA, et al. Common ALDH2 genetic variants predict development of hypertension in the SAPPHIRe prospective cohort: gene-environmental interaction with alcohol consumption. BMC Cardiovasc Disord. 2012; 12: 58. doi: 10.1186/1471-2261-12-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao Q, Weiner H, Crabb DW. The mutation in the mitochondrial aldehyde dehydrogenase (ALDH2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion. J Clin Invest. 1996; 98: 2027–32. doi: 10.1172/JCI119007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CH, Ferreira JCB, Mochly-Rosen D. ALDH2 and Cardiovascular Disease. Adv Exp Med Biol. 2019; 1193: 53–67. doi: 10.1007/978-981-13-6260-6_3 [DOI] [PubMed] [Google Scholar]
- 8.January CT, Wann LS, Alpert JS, Calkins H, Cigarroa JE, Cleveland JC Jr., , et al. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on practice guidelines and the Heart Rhythm Society. Circulation. 2014; 130: e199–267. doi: 10.1161/CIR.0000000000000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, et al. Reprogramming of the human atrial transcriptome in permanent atrial fibrillation: expression of a ventricular-like genomic signature. Circ Res. 2005; 96: 1022–9. doi: 10.1161/01.RES.0000165480.82737.33 [DOI] [PubMed] [Google Scholar]
- 10.Hsu LA, Tsai FC, Yeh YH, Chang CJ, Kuo CT, Chen WJ, et al. Aldehyde Dehydrogenase 2 Ameliorates Chronic Alcohol Consumption-Induced Atrial Fibrillation through Detoxification of 4-HNE. Int J Mol Sci. 2020; 21. doi: 10.3390/ijms21186678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano Y, Ochi H, Onohara Y, Sairaku A, Tokuyama T, Matsumura H, et al. Genetic variations of aldehyde dehydrogenase 2 and alcohol dehydrogenase 1B are associated with the etiology of atrial fibrillation in Japanese. J Biomed Sci. 2016; 23: 89. doi: 10.1186/s12929-016-0304-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CH, Ferreira JCB, Joshi AU, Stevens MC, Li SJ, Hsu JH, et al. Novel and prevalent non-East Asian ALDH2 variants; Implications for global susceptibility to aldehydes’ toxicity. EBioMedicine. 2020; 55: 102753. doi: 10.1016/j.ebiom.2020.102753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng CH, Chung FP, Chen YC, Lin SF, Huang PH, Kuo TB, et al. Pleiotropic Effects of Myocardial MMP-9 Inhibition to Prevent Ventricular Arrhythmia. Sci Rep. 2016; 6: 38894. doi: 10.1038/srep38894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ternette N, Yang M, Laroyia M, Kitagawa M, O’Flaherty L, Wolhulter K, et al. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep. 2013; 3: 689–700. doi: 10.1016/j.celrep.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi LH, Chang WM, Chang YC, Chan YC, Tai CC, Leung KW, et al. Global Proteomics-based Identification and Validation of Thymosin Beta-4 X-Linked as a Prognostic Marker for Head and Neck Squamous Cell Carcinoma. Sci Rep. 2017; 7: 9031. doi: 10.1038/s41598-017-09539-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016; 11: 2301–19. doi: 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47: D607–D13. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parasuraman S, Raveendran R, Kesavan R. Blood sample collection in small laboratory animals. J Pharmacol Pharmacother. 2010; 1: 87–93. doi: 10.4103/0976-500X.72350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4: 44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 20.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009; 25: 1091–3. doi: 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapoor N, Liang W, Marban E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013; 31: 54–62. doi: 10.1038/nbt.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das BC, Thapa P, Karki R, Das S, Mahapatra S, Liu TC, et al. Retinoic acid signaling pathways in development and diseases. Bioorg Med Chem. 2014; 22: 673–83. doi: 10.1016/j.bmc.2013.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan H, Li X, Zhang X, Kang R, Tang D. CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem Biophys Res Commun. 2016; 478: 838–44. doi: 10.1016/j.bbrc.2016.08.034 [DOI] [PubMed] [Google Scholar]
- 24.Koda K, Salazar-Rodriguez M, Corti F, Chan NY, Estephan R, Silver RB, et al. Aldehyde dehydrogenase activation prevents reperfusion arrhythmias by inhibiting local renin release from cardiac mast cells. Circulation. 2010; 122: 771–81. doi: 10.1161/CIRCULATIONAHA.110.952481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008; 134: 921–31. doi: 10.1016/j.cell.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niederreither K, Vermot J, Messaddeq N, Schuhbaur B, Chambon P, Dolle P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001; 128: 1019–31. [DOI] [PubMed] [Google Scholar]
- 27.de Paiva SA, Zornoff LA, Okoshi MP, Okoshi K, Matsubara LS, Matsubara BB, et al. Ventricular remodeling induced by retinoic acid supplementation in adult rats. Am J Physiol Heart Circ Physiol. 2003; 284: H2242–6. doi: 10.1152/ajpheart.00646.2002 [DOI] [PubMed] [Google Scholar]
- 28.Paiva SA, Matsubara LS, Matsubara BB, Minicucci MF, Azevedo PS, Campana AO, et al. Retinoic acid supplementation attenuates ventricular remodeling after myocardial infarction in rats. J Nutr. 2005; 135: 2326–8. doi: 10.1093/jn/135.10.2326 [DOI] [PubMed] [Google Scholar]
- 29.Remme CA. Cardiac sodium channelopathy associated with SCN5A mutations: electrophysiological, molecular and genetic aspects. J Physiol. 2013; 591: 4099–116. doi: 10.1113/jphysiol.2013.256461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilde AAM, Amin AS. Clinical Spectrum of SCN5A Mutations: Long QT Syndrome, Brugada Syndrome, and Cardiomyopathy. JACC Clin Electrophysiol. 2018; 4: 569–79. doi: 10.1016/j.jacep.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 31.Ebert AD, Kodo K, Liang P, Wu H, Huber BC, Riegler J, et al. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci Transl Med. 2014; 6: 255ra130. doi: 10.1126/scitranslmed.3009027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CH, Sun L, Mochly-Rosen D. Mitochondrial aldehyde dehydrogenase and cardiac diseases. Cardiovasc Res. 2010; 88: 51–7. doi: 10.1093/cvr/cvq192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endo J, Sano M, Katayama T, Hishiki T, Shinmura K, Morizane S, et al. Metabolic remodeling induced by mitochondrial aldehyde stress stimulates tolerance to oxidative stress in the heart. Circ Res. 2009; 105: 1118–27. doi: 10.1161/CIRCRESAHA.109.206607 [DOI] [PubMed] [Google Scholar]
- 34.Sivakumar A, Subbiah R, Balakrishnan R, Rajendhran J. Cardiac mitochondrial dynamics: miR-mediated regulation during cardiac injury. J Mol Cell Cardiol. 2017; 110: 26–34. doi: 10.1016/j.yjmcc.2017.07.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.