Abstract
The formation of long-lived memory T cells is a critical feature of the adaptive immune response. T cells undergo metabolic reprogramming to establish a functional memory population. While initial studies characterized key metabolic pathways necessary for memory T-cell development, recent findings highlight that metabolic regulation of memory T-cell subsets is diverse. Here we describe the different requirements for metabolic programs and metabolism-related signaling pathways in memory T-cell development. We further discuss the contribution of cellular metabolism to memory T-cell functional reprogramming and stemness within acute and chronic inflammatory environments. Last, we highlight knowledge gaps and propose approaches to determine the roles of metabolites and metabolic enzymes in memory T-cell fate. Understanding how cellular metabolism regulates a functionally diverse memory population will undoubtedly provide new therapeutic insights to modulate protective T-cell immunity in human disease.
The adaptive immune response provides critical protection against infections and cancer. Upon T-cell receptor (TCR) recognition of antigen-MHC complex and costimulatory signals, activated CD8+ and CD4+ T cells proliferate and differentiate into effector cells that mediate clearance of infected and cancerous cells. T-cell expansion is balanced by programmed cell death, which mediates contraction of the effector T-cell pool. Some cells survive this phase and have the potential to develop into memory T cells, which have stem-cell-like features of self-renewal and the ability to differentiate and re-acquire effector function upon secondary antigen challenge. Within the CD8+ and CD4+ memory T-cell compartments, there is a high level of heterogeneity that is associated with diversity in tissue location, trafficking, and function. Memory T cells include, but are not limited to, the following subsets and defining markers: CD8+ effector memory (Tem; CD44hiCD62LloCCR7lo), central memory (Tcm; CD44hiCD62LhiCCR7hi), tissue resident memory (Trm; CD103+CD69+), or stem-cell memory (Tscm; CD44loCD62LhiSca-1hiCD122hiBcl-2hi) cells; and CD4+ memory Th1, memory follicular helper (Tfh), and Trm cells (Jameson and Masopust 2018; Nguyen et al. 2019). The development of a diverse memory T-cell pool is a critical feature of protective immunity.
T cells undergo metabolic reprogramming to meet bioenergetic and biosynthetic demands during times of cellular activation and quiescence. Over the past decade, studies have illuminated key metabolic programs that dictate cell fate and memory development. However, we still lack a complete understanding of how metabolites and metabolic programs regulate memory T-cell functional heterogeneity, which will have direct clinical applications for adoptive T-cell therapies targeting infectious diseases and cancers. Here, we discuss the emerging concept of T-cell metabolism as an underlying regulator of T-cell memory generation, functional diversity, and stemness.
METABOLIC REGULATION OF MEMORY T-CELL GENERATION AND DIVERSITY
Metabolic programs are critical regulators of naive T-cell activation and differentiation into effector T cells (Geltink et al. 2018; Chapman et al. 2020). However, how metabolism regulates the specific functions and fate decisions of memory T-cell subsets is still underexplored. In this section, we highlight our current understanding of the metabolic profiles of memory T cells, the metabolism-related signaling networks that regulate memory T-cell development, and how metabolism may allow for diversification of the memory T-cell pool (Fig. 1).
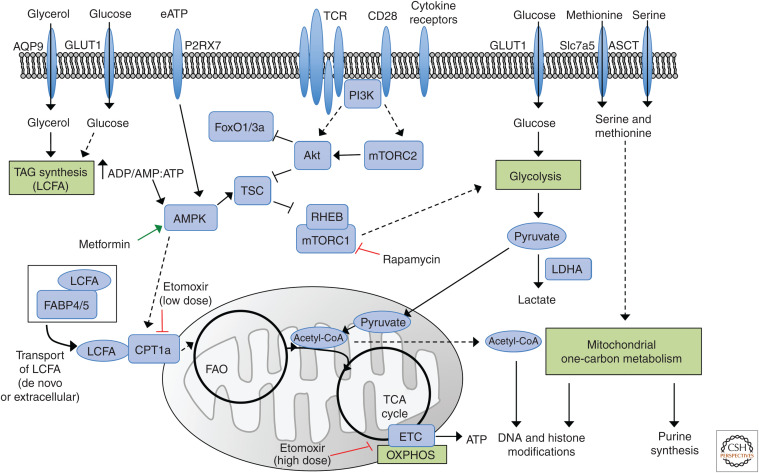
Figure 1.
Metabolic pathways and metabolism-associated signaling in memory T cells. T-cell receptor (TCR), costimulatory molecule CD28, and cytokine receptor (e.g., IL-2 receptor and IL-15 receptor) signals induce mTORC1 and mTORC2 activation via PI3K-Akt signaling (Chi 2012). mTORC1 promotes glycolysis and other anabolic programs to invoke effector T-cell responses (Zeng et al. 2016; Tan et al. 2017; Chapman et al. 2020), while low mTOR signaling favors memory T-cell development and maintenance due to increased FoxO1/3a transcriptional activity and catabolic mitochondrial metabolism (Araki et al. 2009; Rao et al. 2012; Hess Michelini et al. 2013; Pollizzi et al. 2015; Delpoux et al. 2018; Utzschneider et al. 2018). High AMPK signaling regulated by extracellular ATP (eATP) and intracellular ADP/AMP:ATP ratio also favors memory T-cell development by limiting mTORC1 signaling and promoting fatty acid oxidation (FAO) and mitochondrial oxidative phosphorylation (OXPHOS) driven by the electron transport chain (ETC) (Rolf et al. 2013; Borges da Silva et al. 2018). Triacylglyceride (TAG) synthesis from extracellular glucose or glycerol may generate long-chain fatty acids (LCFAs), or LCFAs may be transported from extracellular sources to increase the intracellular fatty acid pool. These LCFA can be transported via FABP4/5 to fuel CPT1a-dependent FAO in specific contexts (Cui et al. 2015; Pan et al. 2017). Both FAO and aerobic glycolysis allow for generation of acetyl coenzyme A (acetyl-CoA), which enters the tricarboxylic acid (TCA) cycle to support ATP production for memory T cells (Pearce et al. 2009; van der Windt et al. 2012; Maekawa et al. 2015; Phan et al. 2016) and promotes mitochondrial metabolism that poises cells for rapid secondary responses (van der Windt et al. 2012, 2013; Buck et al. 2016; Klein Geltink et al. 2017). Acetyl-CoA also likely contributes to epigenetic remodeling (Peng et al. 2016). The amino acids serine and methionine may contribute to mitochondrial one-carbon metabolism to promote memory T-cell generation or proliferation via purine synthesis and epigenetic remodeling. Red inhibitory arrows indicate suppression of mTORC1, CPT1a, and the ETC by rapamycin, low-dose etomoxir, and high-dose etomoxir, respectively. The green arrow indicates activation of AMPK by metformin. Dashed arrows indicate that parts of the pathway are omitted. (ADP) Adenosine diphosphate, (AMP) adenosine monophosphate, (AMPK) AMP-activated protein kinase, (AQP9) aquaporin 9, (ATP) adenosine triphosphate, (CPT1a) carnitine palmitoyl transferase, (FABP4/5) fatty acid–binding protein 4 and 5, (FoxO1/3a) Forkhead box O1 or 3a, (mTORC1) mechanistic target of rapamycin (mTOR) complex 1, (mTORC2) mTOR complex 2, (PI3K) phosphatidylinositol 3-kinase.
Metabolic Features of Memory T Cells
T cells undergo metabolic rewiring concomitant to memory-cell differentiation. CD8+ memory T cells have a metabolic program that is reminiscent of quiescent naive T cells, characterized by elevated mitochondrial oxidative phosphorylation (OXPHOS) relative to glycolysis (van der Windt et al. 2012). Unlike CD8+ naive T cells, CD8+ memory T cells have increased mitochondrial mass and spare respiratory capacity (SRC), an indicator of the maximum mitochondrial oxidative potential (van der Windt et al. 2012, 2013). CD28 costimulation induces mitochondrial elongation and fusion, which results in more densely packed cristae that promotes association and increases the efficiency of the electron transport chain (ETC) complexes (Cogliati et al. 2013; Buck et al. 2016; Klein Geltink et al. 2017). Together, these changes in mitochondrial quantity and functional capacity provide memory T cells with a metabolic advantage that allows for rapid recall and activation upon secondary infection (van der Windt et al. 2013), a critical feature of the memory T-cell response.
To promote mitochondrial oxidative metabolism, CD8+ memory T cells also up-regulate mitochondrial fatty acid oxidation (FAO), a key metabolic program that promotes memory T-cell generation. A seminal study that used TRAF6-deficient CD8+ effector T cells established the paradigm that links FAO with memory T-cell development (Pearce et al. 2009). Mechanistically, TRAF6 deficiency is associated with reduced adenosine monophosphate (AMP)-activated protein kinase (AMPK) signaling, which up-regulates FAO, especially in the absence of IL-2 that promotes metabolic stress; further, treatment with the AMPK agonist metformin partially restores CD8+ memory T-cell development in the absence of TRAF6 (Pearce et al. 2009). Additionally, IL-15 memory-like CD8+ T cells (term used to define peptide-activated TCR-transgenic cells differentiated with IL-15 in vitro) increase the expression of carnitine palmitoyl transferase (CPT1a), the rate-limiting enzyme for FAO (van der Windt et al. 2012). This up-regulation allows for CPT1a-mediated FAO, which promotes mitochondrial SRC and enhances CD8+ memory T-cell development and recall responses, as demonstrated by experiments that have either inhibited (via shRNA or the pharmacologic etomoxir) or promoted (by retroviral overexpression) CPT1a activity (van der Windt et al. 2012, 2013). Thus, CD8+ memory T-cell development and function are controlled by FAO. Whether FAO is the key metabolic pathway in CD4+ memory T-cell development is underexplored and will be important to determine.
Many sources of fatty acids may fuel FAO. For example, IL-15 memory-like CD8+ T cells use lysosomal lipolysis of fatty acids synthesized de novo from glucose as a cell-intrinsic source of fatty acids for FAO (O'Sullivan et al. 2014). Additionally, uptake of glycerol through aquaporin 9 promotes triacylglyceride (TAG) synthesis, which restores intracellular lipids that may be used for intrinsic lipolysis for CD8+ memory T-cell survival (Cui et al. 2015). In contrast, skin CD8+ Trm cells are dependent on uptake of extracellular free fatty acids for their survival and function (Pan et al. 2017). Indeed, T-cell-specific deletion of Fabp4 and Fabp5, two genes encoding molecules that mediate lipid uptake and transport (fatty acid-binding proteins 4 and 5 [FABP4/5]), impairs the long-term survival of CD8+ skin Trm cells, but has no effect on CD8+ Tcm cells in lymph nodes (Pan et al. 2017). Fatty acids may also be an abundant metabolite for T cells within adipose tissues, where lipolysis is required for catabolism of stored triacylglycerols upon metabolic demands (Zechner et al. 2012). In support of this view, white adipose tissue (WAT) contains diverse subsets of CD8+ and CD4+ memory T cells, including Tcm, Tem, and Trm cells, which acquire more extracellular fatty acids than memory T cells isolated from the intestinal lamina propria or spleen (Han et al. 2017), suggesting that FAO may be elevated in WAT memory T cells. These findings show that the dependence of CD8+ and CD4+ memory T cells on lipid sources for FAO is likely cell and tissue context specific, and further investigation is required to elucidate the key enzymes and transporters associated with lipid catabolism that contributes to memory T-cell development and functional recall responses in different tissues or disease contexts.
These studies provide strong evidence that at least a subset of memory T cells is reliant upon fatty acid–driven mitochondrial oxidation for long-lived persistence and secondary responses. However, a recent study that used mice with T-cell-specific deletion of Cpt1a unexpectedly demonstrated that CPT1a is largely dispensable for CD8+ T-cell effector responses and in vivo generation of functional memory T cells in response to Listeria monocytogenes-OVA bacterial infection (Raud et al. 2018). Further, high-dose etomoxir used in prior studies has off-target effects, including reducing levels of tricarboxylic acid (TCA) cycle intermediates and mitochondrial OXPHOS independently of CPT1a (Raud et al. 2018). These findings raise two possibilities: (1) there is context specificity for CPT1a-mediated FAO, such as in different tissue microenvironments as previously described; or (2) other nutrients (e.g., glucose, amino acids) may contribute to mitochondrial oxidative metabolism for memory T-cell programming. Whereas treatment of CD8+ T cells with 2-deoxyglucose (2-DG), an inhibitor of glycolysis, can enhance CD8+ memory T-cell generation in some contexts (Sukumar et al. 2013), more recent studies have also shown positive roles for glucose metabolism in memory T-cell formation. For instance, Notch signaling increases Glut1 expression to support glucose uptake and metabolism for CD4+ memory T-cell survival (Maekawa et al. 2015). Also, deletion of von Hippel–Lindau (VHL), an E3 ubiquitin ligase that inhibits glycolysis via degradation of the pro-glycolytic transcription factor hypoxia inducible factor (HIF)-1α under normoxia conditions (Phan and Goldrath 2015), does not impair memory CD8+ T-cell formation, despite elevated glycolytic metabolism and impaired mitochondrial SRC (Phan et al. 2016). Rather, elevated glycolytic metabolism allows VHL-deficient CD8+ T cells to generate normal levels of ATP (Phan et al. 2016). In summary, these results suggest that memory T cells are metabolically adaptable and can use many nutrient sources to generate sufficient ATP for cellular survival, which ultimately enables their longevity and long-term protective function.
Signaling Networks for Memory T-Cell Development
Mechanistic target of rapamycin (mTOR) is a central signaling node that integrates extracellular cues with intracellular signaling and metabolism. Several studies have revealed that the two unique mTOR complexes, mTORC1 (defined by Raptor) and mTORC2 (defined by Rictor), serve as critical regulators of CD8+ memory T-cell development and rapid recall responses (Araki et al. 2009; Gubser et al. 2013; Pollizzi et al. 2015). Inhibition of mTORC1 by rapamycin treatment during the expansion phase increases CD8+ memory precursor cell (CD127hiKLRG1lo) generation and the total memory T-cell pool, while treatment during the contraction phase accelerates memory T-cell differentiation but does not significantly impact the total number of memory T cells (Araki et al. 2009). In contrast, hyperactivation of mTORC1 signaling, mediated by deletion of tuberous sclerosis 1 (Tsc1) or Tsc2, inhibits memory T-cell differentiation and function (Shrestha et al. 2014; Pollizzi et al. 2015). mTOR signaling antagonizes CD8+ memory T-cell differentiation partly through repression of FoxO1/3a (Forkhead box O1 or 3a) activity, which promotes memory formation and persistence (Kaech and Cui 2012; Pollizzi et al. 2015; Delpoux et al. 2018; Utzschneider et al. 2018). Mechanistically, FoxO1 induces expression of memory-associated genes including Eomes and T-cell factor 1 (TCF-1), but limits T-bet expression, a transcription factor critical for effector T cells (Rao et al. 2012; Hess Michelini et al. 2013; Delpoux et al. 2017). Thus, mTOR signaling dictates memory T-cell fate and function through regulation of transcriptional programs.
mTORC1 signaling also regulates memory T-cell differentiation through metabolic reprogramming. Specifically, the switch from anabolic metabolism (e.g., aerobic glycolysis, glutaminolysis, etc.) to catabolic metabolism (e.g., FAO) likely requires the down-regulation of mTORC1 and up-regulation of AMPK signaling in response to energy stress (Kaech and Cui 2012; Buck et al. 2015), which are events associated with CD8+ memory T-cell development (Pearce et al. 2009; Rolf et al. 2013). Indeed, impaired mTORC1 function during T-cell activation is associated with increased FAO-related programs and Cpt1a expression (Pollizzi et al. 2015; Tan et al. 2017), whereas Tsc2-deficient T cells have reduced FAO programming (Pollizzi et al. 2015). In addition, antigen-experienced Tsc1-deficient CD8+ T cells have marked up-regulation of mitochondrial and glycolytic metabolism in response to IL-15 stimulation (Shrestha et al. 2014), suggesting that mTORC1 orchestrates differentiation and metabolic signals to tune memory T-cell differentiation.
Metabolites are not only critical for fueling metabolic processes, but also serve as signaling molecules that modulate critical kinase pathways, such as mTOR. For instance, the sensing of intracellular amino acids, including leucine and arginine, coordinates the recruitment and activation of mTORC1 at the lysosome (Saxton and Sabatini 2017). To date, the contribution of specific metabolites to memory T-cell development, maintenance, and effector function is underexplored. Intracellular L-arginine enhances CD8+ T-cell survival and antitumor responses, which appears to be independent of mTOR signaling but is potentially mediated by the nuclear proteins BAZ1B, PSIP1, and C3P0 (encoded by TSN) (Geiger et al. 2016). Additionally, asparagine promotes functional responses of memory CD8+ T cells (Wu et al. 2021). Beyond amino acids, the purinergic receptor P2RX7 senses extracellular ATP to promote the development of CD8+ Tcm and Trm cells by enhancing AMPK signaling and mitochondrial oxidative metabolism (Borges da Silva et al. 2018). Thus, understanding the contribution of metabolites on intracellular signaling and gene expression is an exciting and open area for future exploration and may lead to the discovery of new therapeutic targets.
An emerging area of interest is the contribution of metabolism to T-cell epigenetic events and how this regulates cellular fate and function. Effector and memory T-cell subsets have unique chromatin states that are not always static (Philip et al. 2017; Youngblood et al. 2017; Pace et al. 2018), demonstrating that de novo changes in chromatin accessibility can program, and potentially reprogram, T-cell fate and function. These effects may also be connected to metabolism, as many enzymes involved in chromatin and nucleotide modifications (e.g., DNA methyltransferases and histone deacetylases) require substrates and cofactors (e.g., S-adenosyl methionine [SAM] and NAD+) derived from metabolic processes (Reid et al. 2017). As evidence of this concept, HIF-1α-dependent accumulation of S-2-hydroxyglutarate in CD8+ T cells promotes histone methylation associated with Tcm cells and leads to cellular persistence and antitumor responses (Tyrakis et al. 2016). In addition, metabolites derived from the methionine cycle (part of serine and glycine-dependent one-carbon metabolism), the TCA cycle, glycolysis, and FAO may also contribute to epigenetic remodeling (Reid et al. 2017). For example, lactate dehydrogenase A ([LDHA], an enzyme in the aerobic glycolysis pathway) supports aerobic glycolysis to maintain acetyl coenzyme A (acetyl-CoA) production and permissive histone acetylation of the Ifng locus in CD4+ T cells (Peng et al. 2016). Several studies have implicated important roles of one-carbon metabolism for memory T-cell development and epigenome regulation. Mitochondrial one-carbon metabolism or serine uptake promotes effector proliferation and consequently memory T-cell generation by fueling purine nucleotide synthesis (Ron-Harel et al. 2016; Ma et al. 2017). Further, inhibition of Shmt2 (a crucial one-carbon metabolism enzyme) reduces T-cell survival, possibly due to reduced redox balance mediated by glutathione that is produced via the activity of glutamate cysteine ligase (catalytic subunit encoded by Gclc) downstream of one-carbon metabolism (Ron-Harel et al. 2016; Mak et al. 2017; Lian et al. 2018). Interestingly, the accumulation of intracellular reactive oxygen species (ROS) is associated with altered H3K27me3 in Th17 cells (Johnson et al. 2018). Further, uptake of extracellular methionine by activated T cells promotes DNA histone methylation (e.g., H3K27me3 and H3K4me3) and controls T-cell effector responses (Sinclair et al. 2019; Bian et al. 2020; Roy et al. 2020). Establishing the contribution of metabolites to epigenetic-driven changes in memory T cells will be important and also challenging considering that enzymes and metabolites not only induce broad chromatin modifications, but also fuel metabolic programs that have wide influence on cellular physiology.
Metabolism as a Driver of Memory T-Cell Fate Decisions
Precisely how memory T cells arise has remained a key immunological question for decades (Ahmed et al. 2009; Jameson and Masopust 2018). One model of memory T-cell development proposes that there is an early “imprinting” of memory-cell fate that accompanies the first asymmetric cell division, which results in bipolar segregation of the proteasome and yields unequal degradation of T-bet in the two daughter cells (Chang et al. 2007, 2011). In addition, factors that regulate cellular metabolism, including mTOR and c-Myc, preferentially segregate into the proximal daughter cell near the T-cell–APC synapse (Pollizzi et al. 2016; Verbist et al. 2016). This bipolar segregation of protein signaling is associated with cell fate, as proximal daughter cells have reduced survival, while the distal daughter cells have heightened persistence, control of pathogen infections, and development into memory T cells (Chang et al. 2007; Pollizzi et al. 2016; Verbist et al. 2016). These altered cell states are associated with changes in metabolic programming, with elevated glycolytic activity in the short-lived proximal daughter cells (Pollizzi et al. 2016; Verbist et al. 2016) and increased lipid metabolism in the long-lived distal daughter cells (Pollizzi et al. 2016). Thus, early metabolic reprogramming may contribute to the fate decision of T cells into terminal effector or memory precursor cells.
A second model of linear differentiation proposes that cell fate decisions occur later in the effector T-cell stage, at which time cells either become terminally differentiated and eventually die by apoptosis, or further differentiate into persisting memory T cells (Ahmed et al. 2009; Jameson and Masopust 2018). Indeed, CD8+ effector T cells acquire de novo methylation patterns associated with suppression of naive T-cell-associated genes (e.g., Sell, encodes CD62L) that can be reversed in CD8+ memory T cells (Youngblood et al. 2017). These findings indicate that a subset of CD8+ effector T cells can undergo epigenetic reprogramming as the cells mature into long-lived memory T cells, supporting linear differentiation. Metabolic reprogramming may also dictate which effector subsets acquire memory-like signatures after activation (Fig. 2). In support of this idea, deletion of Tsc1 in antigen-experienced CD8+ T cells leads to defective memory T-cell development and function and is associated with metabolic dysregulation and defective memory precursor-cell differentiation (Shrestha et al. 2014). In contrast, rapamycin treatment at the effector stage enhances CD8+ T-cell memory development (Araki et al. 2009). Further, activated CD8+ T cells exposed to the short-chain fatty acid (SCFA) butyrate display elevated FoxO1 expression and memory potential (Bachem et al. 2019). Additionally, mitochondrial respiration supports Akt-FoxO1 signaling (Xu et al. 2021). Thus, mTOR signaling and metabolic reprogramming may contribute to memory fate differentiation at the first cell division and later at the effector stage.
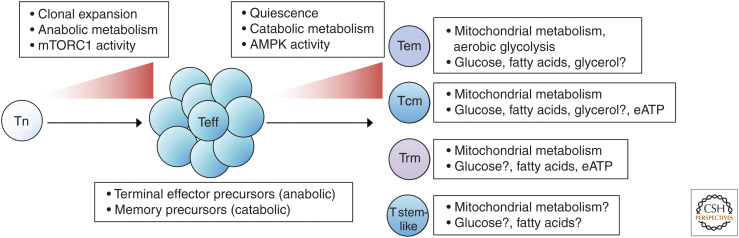
Figure 2.
Metabolic reprogramming in memory T-cell subsets. Naive T (Tn)-cell activation, proliferation, and differentiation into effector T cells (Teff) require the up-regulation of mTORC1 signaling and switch to anabolic metabolism (Chapman et al. 2020). Among Teff are terminal effector precursor cells with high anabolic programs that will undergo apoptotic-mediated death and memory precursors with catabolic programs that can survive contraction and develop into memory T cells. The development of memory T cells is associated with entering into a quiescent state with elevated catabolic metabolism, a switch that relies on up-regulated AMPK (adenosine monophosphate-activated protein kinase) signaling and down-regulated mTOR (mechanistic target of rapamycin) signaling (Araki et al. 2009; Pearce et al. 2009; Kaech and Cui 2012; Pollizzi et al. 2015). Memory T cells comprise subsets with diverse requirements for metabolic programs, nutrients, and metabolites. There are context-dependent roles for aerobic glycolysis and mitochondrial metabolism in CD8+ and CD4+ memory T-cell subsets (van der Windt et al. 2012, 2013; Maekawa et al. 2015; Buck et al. 2016; Phan et al. 2016; Klein Geltink et al. 2017). Memory T cells require fatty acid oxidation (FAO) to support mitochondrial metabolism, cell survival, and function, but the dependency on FAO and source of fatty acids, including glycerol-mediated triacylglyceride (TAG) synthesis, varies among subsets and tissue localization (Pearce et al. 2009; van der Windt et al. 2013; O'Sullivan et al. 2014; Cui et al. 2015; Pan et al. 2017; Raud et al. 2018). Extracellular ATP (eATP) enhances mitochondrial metabolism and promotes central memory (Tcm) and tissue-resident memory (Trm) development (Borges da Silva et al. 2018). mTORC1 signaling and up-regulation of glycolytic metabolism may also promote maturation and loss of cellular stemness (Karmaus et al. 2019). (Tem) Effector memory, (T stem-like) stem-cell-like memory T cell.
Metabolism also influences differentiation into different subsets of memory T cells. For instance, VHL-deficient CD8+ T cells preferentially develop into Tem cells, which have different metabolic signatures compared to other Tm cell subsets (Phan et al. 2016). Tcm cells are shown to have elevated mitochondrial SRC compared to Tem cells, suggesting Tcm cells may rely more on mitochondrial respiration (Phan et al. 2016). Tem cells may also be less dependent on FAO when glucose levels are limiting (Ecker et al. 2018). Consistent with these observations, in vivo AMPK activation by metformin treatment promotes Tcm and Trm cell generation but has little effect on Tem cells (Borges da Silva et al. 2018). This metabolic plasticity may allow for the establishment of memory T cells in the face of altered nutrient availability, but further work is needed to understand whether and how these metabolic adaptations underpin memory T-cell subset diversification.
METABOLISM AND STEMNESS IN T-CELL FUNCTION
Two critical features of CD8+ and CD4+ memory T cells are their ability to self-renew to persist long term and to differentiate upon reactivation. These two characteristics are reminiscent of hematopoietic stem cells (HSCs) (Fearon et al. 2001; Kaech et al. 2002), and CD8+ memory T cells have transcriptional profiles that overlap with HSCs (Luckey et al. 2006). These stem-cell-like qualities, also termed “stemness,” are critical for the maintenance of a functional memory T-cell pool. Here, we discuss the interplay between transcriptional and metabolic programs in dictating memory T-cell stemness in acute and chronic inflammatory environments.
Transcriptional and Metabolic Features of T-Cell Stemness
Although memory T cells are defined by their long-term persistence and ability to respond to secondary antigen challenge, not all memory T-cell subsets display equivalent stemness. Rather, several subsets with enhanced stem-cell-like qualities have been described, including CD8+ Tscm and CX3CR1int peripheral memory T (Tpm) cells (Table 1; Gattinoni et al. 2009, 2011; Turtle et al. 2009; Gerlach et al. 2016) and CD4+ Tfh and Th17 cells (Muranski et al. 2011; Karmaus et al. 2019; Nguyen et al. 2019). The stem-cell-like memory T-cell subsets have gained broad recognition in recent years due to both their longevity and enhanced function in many immune environments, including tumors, chronic infections, and autoimmune diseases. For instance, CD8+ Tscm cells, which can self-renew or differentiate into Tcm, Tem, and Trm cells, have superior antitumor activity compared to Tcm and Tem cells (Gattinoni et al. 2009). In addition, CD4+ T cells can develop into long-lived memory T cells with functional recall responses (Hale et al. 2013; Tubo et al. 2016). Using IL-21 reporter mice, Lüthje et al. (2012) found that IL-21-expressing Tfh cells develop into a CD4+ memory T-cell subset that produces effector cytokines, including IFN-γ and IL-2, and can contribute to nongerminal center responses upon secondary infections. Further, functional analysis using the lymphocytic choriomeningitis virus (LCMV) infection model shows that, while the CXCR5+ Tfh-cell subset preferentially generates a Tfh-like recall response upon secondary LCMV challenge, they can also give rise to CXCR5– Th1-cells (Hale et al. 2013). Compared to Th1 cells, the Tfh memory-cell subset retains higher functional plasticity (Lüthje et al. 2012; Hale et al. 2013; Kunzli et al. 2020). Further, Th17 cells can acquire Th1-like characteristics, including IFN-γ production, and persist in vivo (Bending et al. 2009; Lee et al. 2009; Muranski et al. 2011). A stem-cell-like CD27+ Th17 subset is also found within a chronic inflammatory environment (in the animal model of multiple sclerosis, experimental autoimmune encephalomyelitis [EAE]), which can preferentially persist and give rise to CD27– Th1-like Th17 cells (Karmaus et al. 2019). Thus, stemness is an inherent feature of memory T cells that can regulate both their persistence and function in different disease settings.
Table 1.
Stem-cell-like memory T-cell subsets
| Stem-cell-like memory subsets | Associated markers | Inflammatory context | Associated metabolism |
|---|---|---|---|
| CD4+ Tfh stem-cell-like | CXCR5+, Ly6Cint/lo, PD-1+, Bcl-6+, IL-21+, FR4+ | Acute LCMV, NP-KLH | Anabolic (glycolysis supports persistence) |
| CD4+ Th17 stem-cell-like | CD27+/lo, TCF-1+ | EAE, ocular autoimmunity | Catabolic (reduced mTORC1 activity and glycolysis-associated gene expression) |
| CD8+ Tscm | CD44lo, CD62Lhi, Sca-1hi, CD122hi | Pharmacologically induced Wnt signaling | Catabolic |
| CD8+CX3CR1int peripheral memory | CX3CR1int, CD27+, CXCR3+ | Acute LCMV | Unknown |
| CD8+CXCR5+ Tfh-like | CXCR5+, TCF-1+, Tim-3− | Chronic LCMV | Catabolic? (increased FAO-associated gene expression) |
| CD8+ progenitor-exhausted | TCF-1+, Eomeslo, T-bethi | Chronic LCMV | Unknown |
| CD8+ TME-associated stem-cell-like | TCF-1+, Tim-3−, PD-1+/−, Slamf7hi | Tumor | Catabolic (elevated autophagy) |
| Human CD8+ stem-cell-like | CD45RA+, CD62L+, CD27+, CCR7+, CD95+, CD122+, CXCR3+, LFA-1+ | Blood (expanded in immunodeficient hosts) | Unknown |
Summary of the stem-cell-like memory cells that are present in mice and humans, based on surface markers and transcription factors and their identification in different inflammatory contexts as indicated. The known associated metabolism of these stem-cell-like populations are also highlighted. CD4+ Tfh stem-cell-like (Lüthje et al. 2012; Hale et al. 2013; Kunzli et al. 2020); CD4+ Th17 stem-cell-like (Muranski et al. 2011; Karmaus et al. 2019); CD8+ Tscm (Gattinoni et al. 2009; Sukumar et al. 2016); CD8+CX3CR1int peripheral memory (Gerlach et al. 2016); CD8+CXCR5+ Tfh-like (He et al. 2016; Im et al. 2016; Leong et al. 2016; Wu et al. 2016); CD8+ progenitor-exhausted (Paley et al. 2012); CD8+ TME-associated stem-cell-like (Wu et al. 2016; Kurtulus et al. 2019; Miller et al. 2019; Siddiqui et al. 2019; Vodnala et al. 2019); human CD8+ stem-cell-like (Turtle et al. 2009; Gattinoni et al. 2011) cells. (TME) Tumor microenvironment, (LCMV) lymphocytic choriomeningitis virus, (NP-KLH) 4-hydroxy-3-nitrophenyl acetyl-keyhole limpet hemocyanin, (EAE) experimental autoimmune encephalomyelitis, (FAO) fatty acid oxidation.
TCF-1 is a critical transcriptional regulator of T-cell stemness. TCF-1 activity is regulated by the conserved Wnt-β-catenin signaling pathway, which is involved in HSC self-renewal and multipotency (Fleming et al. 2008). The induction of Wnt signaling arrests CD8+ effector T-cell development and promotes the emergence of CD8+ Tscm cells (Gattinoni et al. 2009). In addition, in vitro polarized CD4+ Th17 cells have elevated β-catenin and TCF-1 expression and can further differentiate into cells with Th1-cell features in vivo; these stem-cell-like Th17 cells can rapidly respond to secondary stimulation and persist in vivo (Muranski et al. 2011). Stem-cell-like Th17 cells also express high levels of TCF-1 in EAE (Karmaus et al. 2019). TCF-1 is likely a critical determinant of Tfh-cell plasticity during recall responses, as TCF-1 promotes Tfh-cell differentiation, and loss of TCF-1 expression allows cells to acquire a non-Tfh-cell phenotype (Choi et al. 2015; Wu et al. 2015; Xu et al. 2015). Of note, TCF-1 is a “core” transcription factor expressed in both naive and memory T cells (Gattinoni et al. 2012), so one key future direction will be to understand how TCF-1 interplays with other nuclear proteins (e.g., transcription factors, epigenetic-modifying enzymes) to influence the specific fate decisions of stem-cell-like T cells.
Whereas clear roles of metabolism have been attributed to memory T-cell survival (one facet of stemness) as previously discussed, the function of metabolism in stem-cell-like functional reprogramming is relatively understudied. Chronic LCMV infections are associated with reduced mTOR and Akt activities and increased FoxO1 activity that is essential for the persistence and function of CD8+ T cells (Staron et al. 2014), suggesting that the chronic inflammatory environment may impact cellular metabolism of CD8+ T cells. We recently showed that anabolic metabolism (e.g., glycolysis, cholesterol biosynthesis) and mTORC1 are essential for Th17 cells to differentiate into TCF-1loT-bethi IFN-γ-producing Th1-like cells, and that inhibition of either mTORC1 or glycolysis promotes acquisition of stemness-associated features (Karmaus et al. 2019). These findings are also in line with those showing that differentiation of “pathogenic” Th17 cells (i.e., those that produce IFN-γ) is dictated by the intracellular composition of lipids and cholesterol (Wang et al. 2015). Of note, mitochondrial metabolism and nutrient uptake regulate stemness of lung cancer cells (Fernandez et al. 2018), but their roles in inducing the stem-cell-like properties of memory T cells remain unclear. CD8+ T cells with low mitochondrial membrane potential are enriched in Tscm cells, further implicating mitochondrial function as a regulator of stemness (Sukumar et al. 2016). Interestingly, the uptake of lipids (e.g., the SCFA acetate) promotes increased function of reactivated CD8+ memory T cells through its interplay with glycolysis (Balmer et al. 2016). Further, sodium sensing by SGK1 may also balance stem-cell-like, nonpathogenic Th17 cells from terminal, pathogenic Th17 cells (Kleinewietfeld et al. 2013; Wu et al. 2013). In addition, methionine and SAM contribute to the regulation of cell state and epigenetics in stem cells (Shyh-Chang et al. 2013; Shiraki et al. 2014), implicating one-carbon metabolism as a possible determinant of memory T-cell stemness. The control of memory T-cell stemness by metabolism therefore remains an exciting new avenue of research that has direct therapeutic applications, such as for adoptive T-cell therapy.
Metabolism Balances Stemness with T-Cell Exhaustion
Much of our understanding of stemness and memory T-cell reprogramming has been derived from studies using acute pathogen models. However, upon chronic antigen exposure in inflammatory disorders such as chronic pathogen infection or tumors, CD8+ T cells can acquire an exhausted cellular phenotype. Exhausted CD8+ T cells have high expression of inhibitory receptors (e.g., PD-1, Tim3) and impaired capacity to expand and produce cytokines (Schietinger and Greenberg 2014; Wherry and Kurachi 2015; Hashimoto et al. 2018). Within these chronic inflammatory environments, there is a subpopulation of CD8+ T cells that retains the capacity to proliferate, regenerate, and differentiate into terminally exhausted cells. This TCF-1-dependent CD8+ T-cell subset, called either “progenitor-exhausted” or “stem-cell-like” CD8+ T cells (Paley et al. 2012; He et al. 2016; Im et al. 2016; Leong et al. 2016; Utzschneider et al. 2016; Wu et al. 2016), helps sustain the immune response and may prevent collapse of the memory T-cell pool during chronic infections (Utzschneider et al. 2016).
As noted above, TCF-1 promotes CD8+ T-cell identity (Xing et al. 2016) and the generation and long-term persistence of the CD8+ memory T-cell pool during acute infections (Jeannet et al. 2010; Zhou et al. 2010). TCF-1 expression, promoted by the transcription factor c-Myb (Gautam et al. 2019), allows for increased CD8+ T-cell proliferation during chronic viral infections (Utzschneider et al. 2016). The TCF-1+ population can share transcriptional similarities to Tfh cells, identified by a Tfh-associated marker CXCR5 (He et al. 2016; Im et al. 2016; Leong et al. 2016; Wu et al. 2016). These CXCR5+TCF-1+PD-1+Tim3–CD8+ T cells predominantly reside within the T-cell zones of lymphoid tissues and promote the control of chronic LCMV infection (He et al. 2016; Im et al. 2016; Leong et al. 2016; Wu et al. 2016). The tumor microenvironment (TME) is another chronic inflammatory site that induces an exhausted-like phenotype in CD8+ T cells. Similar to the chronic LCMV environment, a stem-cell-like subset that is TCF-1+ exists within the TME (Wu et al. 2016; Kurtulus et al. 2019; Miller et al. 2019; Siddiqui et al. 2019). Further, a TCF-1+CD8+ T-cell population has been identified in human tumor samples (Wu et al. 2016; Brummelman et al. 2018; Sade-Feldman et al. 2018). What is perhaps most exciting about the identification of these CD8+ stem-cell-like T cells is that they are sensitive to anti-PD-1 and Tim3 or PD-L1 antibody checkpoint blockade therapy (He et al. 2016; Im et al. 2016; Kurtulus et al. 2019; Miller et al. 2019; Siddiqui et al. 2019), and, in melanoma patients, the increased expression of TCF-1 and PD-1 predicts better therapeutic responses to checkpoint blockade therapy and patient outcome (Sade-Feldman et al. 2018). Unlike PD-1+CD8+ exhausted T cells, TCF-1+CD8+ T cells may retain a chromatin state with the potential to reprogram in response to checkpoint blockade (Pauken et al. 2016; Scott-Browne et al. 2016; Ghoneim et al. 2017; Philip et al. 2017). The transcription factor TOX mediates CD8+ T-cell exhaustion, and the loss of TOX leads to a decline of TCF-1+ T cells (Alfei et al. 2019; Khan et al. 2019; Scott et al. 2019; Seo et al. 2019; Wang et al. 2019; Yao et al. 2019), although it remains unclear whether TOX is reciprocally regulated by TCF-1. Thus, a critical future direction for the field will be to determine whether TCF-1 acts as a molecular rheostat for cellular stemness and exhaustion and to determine the downstream targets that dictate these cellular states.
In addition to TCF-1-related networks, we propose that metabolic programs also promote fate decisions for the stem-cell-like cell populations and more dysfunctional exhausted populations in chronic inflammatory conditions, such as autoimmunity and cancer. Indeed, we found that CD27+TCF-1hi Th17 cells from EAE share transcriptional features with CXCR5+CD8+ stem-cell-like cells and CXCR5+ Tfh cells (Karmaus et al. 2019), raising the possibility that metabolic state is a critical determinate of CD8+ T-cell stemness. Indeed, several recent studies highlight the ability to condition cells ex vivo, in part by influencing cellular metabolism and ROS production, for enhanced cellular persistence and effector responses to tumors (O'Sullivan et al. 2019; Gurusamy et al. 2020; Klein Geltink et al. 2020; Vardhana et al. 2020; Verma et al. 2021). Further, enhanced BATF function by targeted deletion of Regnase-1 promotes mitochondrial fitness and programs long-lived CD8+ effector T cells that aid in antitumor immunity (Wei et al. 2019). Blocking the MAPK signaling pathway by MEK1/2 inhibitor treatment promotes FAO and the generation of Tscm-like cells, resulting in more potent antitumor responses in a model of adoptive T-cell therapy (Verma et al. 2021). Conversely, accumulation of depolarized mitochondria in tumor-infiltrating lymphocytes enforced terminal exhaustion (Yu et al. 2020). The metabolic requirements that dictate stem-cell-like reprogrammable responses from exhausted T-cell programs may be directly linked to epigenetic states (Philip et al. 2017; Reid et al. 2017; Pace et al. 2018; Khan et al. 2019; Yu et al. 2020). In addition, the level of PD-1 signaling may influence cellular stemness, as PD-1 signaling can induce FAO in activated T-cell subsets (Patsoukis et al. 2015). Whether other inhibitory receptors associated with exhaustion can promote metabolic reprogramming remains underexplored.
The stem-cell-like or exhausted states are also likely dictated by extracellular nutrients within tissue microenvironments. An excellent example is the TME, which is limited in some critical factors, such as oxygen, but elevated in others, such as cholesterol and potassium. Elevated cholesterol in the TME induces CD8+ T-cell exhaustion through the up-regulation of inhibitory receptor expression (e.g., PD-1 and 2B4) by the endoplasmic reticulum (ER) stress-XBP1 pathway (Ma et al. 2019). In addition, CD8+ T-cell proliferation is negatively regulated by liver X receptors (LXRs), nuclear receptors involved in cholesterol homeostasis (Bensinger et al. 2008). Altered cholesterol levels may also induce T-cell exhaustion through regulation of the strength or duration of TCR signaling. For example, inhibition of cholesterol esterification potentiates CD8+ T-cell antitumor responses by promoting TCR clustering and synapse formation (Yang et al. 2016), while elevated levels of cholesterol sulfate impair TCR signaling by inhibiting TCR-CD3 immunoreceptor tyrosine-based activation motif (ITAM) phosphorylation (Wang et al. 2016). In addition to changes in cholesterol, elevated potassium within the TME serves as an underlying, indirect positive regulator of stem-cell-like TCF-1+CD8+ T cells (Vodnala et al. 2019), but can also inhibit effector T-cell responses in the TME, likely through repression of glycolytic programming that is linked to effector responses (Eil et al. 2016; Peng et al. 2016). Moreover, increased extracellular potassium limits nutrient uptake (e.g., glucose), leading to starvation-induced autophagy and depletion of cytoplasmic acetyl-CoA. In turn, reduced acetyl-CoA is correlated with less histone acetylation, which limits the epigenetic imprint of T-cell exhaustion to promote a stem-cell-like state (Vodnala et al. 2019). These studies highlight that local extracellular nutrients can alter the cellular metabolic state and intracellular metabolites to influence functional and epigenetic reprogramming.
CONCLUDING REMARKS
In this perspective, we have highlighted that, relative to naive and effector T cells, memory T cells have altered metabolic profiles and mitochondrial morphology, which retain the cells in a metabolically quiescent-like state that enables more rapid responses upon secondary challenges. To date, the role of metabolites and metabolic programs on memory T-cell subset differentiation is highly underexplored. Further, while the contribution of metabolism on memory T-cell functional reprogramming and stemness is not well characterized, emerging evidence has identified anabolic metabolism (e.g., glycolysis) as a critical determinant of these features. This possibility is supported by studies in stem cells, which show that metabolism contributes to stem-cell pluripotency (Intlekofer and Finley 2019). Additionally, specific metabolites and nutrients (e.g., potassium) have been linked to the regulation of memory T-cell persistence and stemness, in part through epigenetics. Thus, metabolic programs likely underlie not only the generation and maintenance of memory T cells, but also the regulation of memory T-cell subsets that retain features of functional reprogramming and stemness.
Much remains unanswered in each area discussed in this perspective. Several outstanding questions include (1) How does nutrient diversity within unique microenvironments shape CD8+ and CD4+ T-cell memory formation and function? (2) Do recall responses of memory T cells rely on the same or distinct metabolic programs that promote initial effector T-cell programs? (3) How do metabolites and metabolic enzymes contribute to the programming of memory T-cell stemness? and (4) What are the molecular targets and upstream signals of TCF-1 that dictate cellular stemness or exhaustion within acute and chronic inflammatory environments? With the emergence of new technologies, including single-cell RNA sequencing and ATAC-sequencing, the field has the potential to progress rapidly in understanding how metabolism regulates specific memory T-cell subsets and developmental stages. Further, systems immunology approaches to integrate transcriptome, proteome, and metabolome analyses should allow us to better define stem-cell-like cells from terminally differentiated or exhausted cells, thus refining functional analyses of these cells. These discoveries, combined with the ability to efficiently screen and target metabolism-associated molecules and enzymes using CRISPR-Cas9 technology (Wei et al. 2019; Huang et al. 2021), may allow us to determine new therapeutic targets for improving memory T-cell generation and functional reprogramming in vaccination and immunotherapy strategies.
ACKNOWLEDGMENTS
We acknowledge all investigators whose contributions were not cited due to space constraints. We thank Dr. Y. Wang for critical reading and editing of this perspective. Funding support was provided by the National Multiple Sclerosis Society and the National Institutes of Health (NIH): AI105887, AI131703, AI140761, AI150241, AI150514, CA221290, and CA250533 (to H.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Editors: David Masopust and Rafi Ahmed
Additional Perspectives on T-Cell Memory available at www.cshperspectives.org
REFERENCES
- Ahmed R, Bevan MJ, Reiner SL, Fearon DT. 2009. The precursors of memory: models and controversies. Nat Rev Immunol 9: 662–668. 10.1038/nri2619 [DOI] [PubMed] [Google Scholar]
- Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y, et al. 2019. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571: 265–269. 10.1038/s41586-019-1326-9 [DOI] [PubMed] [Google Scholar]
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. 2009. mTOR regulates memory CD8 T-cell differentiation. Nature 460: 108–112. 10.1038/nature08155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, Whitney PG, Fernandez-Ruiz D, Dähling S, Kastenmuller W, et al. 2019. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51: 285–297. 10.1016/j.immuni.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Balmer ML, Ma EH, Bantug GR, Grählert J, Pfister S, Glatter T, Jauch A, Dimeloe S, Slack E, Dehio P, et al. 2016. Memory CD8+ T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44: 1312–1324. 10.1016/j.immuni.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Bending D, De la Peña H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. 2009. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest 119: 565–572. 10.1172/JCI37865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, et al. 2008. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 134: 97–111. 10.1016/j.cell.2008.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, Nwosu ZC, Zhang L, Czerwonka A, Pawlowska A, et al. 2020. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 585: 277–282. 10.1038/s41586-020-2682-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges da Silva H, Beura LK, Wang H, Hanse EA, Gore R, Scott MC, Walsh DA, Block KE, Fonseca R, Yan Y, et al. 2018. The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8+ T cells. Nature 559: 264–268. 10.1038/s41586-018-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelman J, Mazza EMC, Alvisi G, Colombo FS, Grilli A, Mikulak J, Mavilio D, Alloisio M, Ferrari F, Lopci E, et al. 2018. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J Exp Med 215: 2520–2535. 10.1084/jem.20180684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O'Sullivan D, Pearce EL. 2015. T cell metabolism drives immunity. J Exp Med 212: 1345–1360. 10.1084/jem.20151159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O'Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, et al. 2016. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166: 63–76. 10.1016/j.cell.2016.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. 2007. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315: 1687–1691. 10.1126/science.1139393 [DOI] [PubMed] [Google Scholar]
- Chang JT, Ciocca ML, Kinjyo I, Palanivel VR, McClurkin CE, Dejong CS, Mooney EC, Kim JS, Steinel NC, Oliaro J, et al. 2011. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity 34: 492–504. 10.1016/j.immuni.2011.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman NM, Boothby MR, Chi H. 2020. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol 20: 55–70. 10.1038/s41577-019-0203-y [DOI] [PubMed] [Google Scholar]
- Chi H. 2012. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol 12: 325–338. 10.1038/nri3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. 2015. LEF-1 and TCF-1 orchestrate TFH differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol 16: 980–990. 10.1038/ni.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, et al. 2013. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155: 160–171. 10.1016/j.cell.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, Kaech SM. 2015. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell 161: 750–761. 10.1016/j.cell.2015.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpoux A, Lai CY, Hedrick SM, Doedens AL. 2017. FOXO1 opposition of CD8+ T cell effector programming confers early memory properties and phenotypic diversity. Proc Natl Acad Sci 114: E8865–E8874. 10.1073/pnas.1618916114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpoux A, Michelini RH, Verma S, Lai CY, Omilusik KD, Utzschneider DT, Redwood AJ, Goldrath AW, Benedict CA, Hedrick SM. 2018. Continuous activity of Foxo1 is required to prevent anergy and maintain the memory state of CD8+ T cells. J Exp Med 215: 575–594. 10.1084/jem.20170697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Guo L, Voicu S, Gil-de-Gómez L, Medvec A, Cortina L, Pajda J, Andolina M, Torres-Castillo M, Donato JL, et al. 2018. Differential reliance on lipid metabolism as a salvage pathway underlies functional differences of T cell subsets in poor nutrient environments. Cell Rep 23: 741–755. 10.1016/j.celrep.2018.03.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eil R, Vodnala SK, Clever D, Klebanoff CA, Sukumar M, Pan JH, Palmer DC, Gros A, Yamamoto TN, Patel SJ, et al. 2016. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537: 539–543. 10.1038/nature19364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon DT, Manders P, Wagner SD. 2001. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science 293: 248–250. 10.1126/science.1062589 [DOI] [PubMed] [Google Scholar]
- Fernandez HR, Gadre SM, Tan M, Graham GT, Mosaoa R, Ongkeko MS, Kim KA, Riggins RB, Parasido E, Petrini I, et al. 2018. The mitochondrial citrate carrier, SLC25A1, drives stemness and therapy resistance in non-small cell lung cancer. Cell Death Differ 25: 1239–1258. 10.1038/s41418-018-0101-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming HE, Janzen V, Lo Celso C, Guo J, Leahy KM, Kronenberg HM, Scadden DT. 2008. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell Stem Cell 2: 274–283. 10.1016/j.stem.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, Wrzesinski C, Boni A, Cassard L, Garvin LM, et al. 2009. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med 15: 808–813. 10.1038/nm.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. 2011. A human memory T cell subset with stem cell-like properties. Nat Med 17: 1290–1297. 10.1038/nm.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Klebanoff CA, Restifo NP. 2012. Paths to stemness: building the ultimate antitumour T cell. Nat Rev Cancer 12: 671–684. 10.1038/nrc3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam S, Fioravanti J, Zhu W, Le Gall JB, Brohawn P, Lacey NE, Hu J, Hocker JD, Hawk NV, Kapoor V, et al. 2019. The transcription factor c-Myb regulates CD8+ T cell stemness and antitumor immunity. Nat Immunol 20: 337–349. 10.1038/s41590-018-0311-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al. 2016. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167: 829–842. 10.1016/j.cell.2016.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geltink RIK, Kyle RL, Pearce EL. 2018. Unraveling the complex interplay between T cell metabolism and function. Annu Rev Immunol 36: 461–488. 10.1146/annurev-immunol-042617-053019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, von Andrian UH. 2016. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45: 1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, et al. 2017. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170: 142–157. 10.1016/j.cell.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser PM, Bantug GR, Razik L, Fischer M, Dimeloe S, Hoenger G, Durovic B, Jauch A, Hess C. 2013. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat Immunol 14: 1064–1072. 10.1038/ni.2687 [DOI] [PubMed] [Google Scholar]
- Gurusamy D, Henning AN, Yamamoto TN, Yu Z, Zacharakis N, Krishna S, Kishton RJ, Vodnala SK, Eidizadeh A, Jia L, et al. 2020. Multi-phenotype CRISPR-Cas9 screen identifies p38 kinase as a target for adoptive immunotherapies. Cancer Cell 37: 818–833. 10.1016/j.ccell.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. 2013. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity 38: 805–817. 10.1016/j.immuni.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Glatman Zaretsky A, Andrade-Oliveira V, Collins N, Dzutsev A, Shaik J, Morais da Fonseca D, Harrison OJ, Tamoutounour S, Byrd AL, et al. 2017. White adipose tissue is a reservoir for memory T cells and promotes protective memory responses to infection. Immunity 47: 1154–1168. 10.1016/j.immuni.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, Araki K, Ahmed R. 2018. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med 69: 301–318. 10.1146/annurev-med-012017-043208 [DOI] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. 2016. Follicular CXCR5- expressing CD8+ T cells curtail chronic viral infection. Nature 537: 412–416. 10.1038/nature19317 [DOI] [PubMed] [Google Scholar]
- Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM. 2013. Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med 210: 1189–1200. 10.1084/jem.20130392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhou P, Wei J, Long L, Shi H, Dhungana Y, Chapman NM, Fu G, Saravia J, Raynor JL, et al. 2021. In vivo CRISPR screening reveals nutrient signaling processes underpinning CD8+ T cell fate decisions. Cell 10.1016/j.cell.2021.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537: 417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Finley LWS. 2019. Metabolic signatures of cancer cells and stem cells. Nat Metab 1: 177–188. 10.1038/s42255-019-0032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Masopust D. 2018. Understanding subset diversity in T cell memory. Immunity 48: 214–226. 10.1016/j.immuni.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W. 2010. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci 107: 9777–9782. 10.1073/pnas.0914127107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Maseda D, Liberti MV, Paz K, Kishton RJ, et al. 2018. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175: 1780–1795. 10.1016/j.cell.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Cui W. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12: 749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Wherry EJ, Ahmed R. 2002. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol 2: 251–262. 10.1038/nri778 [DOI] [PubMed] [Google Scholar]
- Karmaus PWF, Chen X, Lim SA, Herrada AA, Nguyen TM, Xu B, Dhungana Y, Rankin S, Chen W, Rosencrance C, et al. 2019. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 565: 101–105. 10.1038/s41586-018-0806-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE, et al. 2019. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571: 211–218. 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN, Hafler DA. 2013. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 496: 518–522. 10.1038/nature11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Geltink RI, O'Sullivan D, Corrado M, Bremser A, Buck MD, Buescher JM, Firat E, Zhu X, Niedermann G, Caputa G, et al. 2017. Mitochondrial priming by CD28. Cell 171: 385–397. 10.1016/j.cell.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Geltink RI, Edwards-Hicks J, Apostolova P, O'Sullivan D, Sanin DE, Patterson AE, Puleston DJ, Ligthart NAM, Buescher JM, Grzes KM, et al. 2020. Metabolic conditioning of CD8+ effector T cells for adoptive cell therapy. Nat Metab 2: 703–716. 10.1038/s42255-020-0256-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzli M, Schreiner D, Pereboom TC, Swarnalekha N, Litzler LC, Lotscher J, Ertuna YI, Roux J, Geier F, Jakob RP, et al. 2020. Long-lived T follicular helper cells retain plasticity and help sustain humoral immunity. Sci Immunol 5: eaay5552. 10.1126/sciimmunol.aay5552 [DOI] [PubMed] [Google Scholar]
- Kurtulus S, Madi A, Escobar G, Klapholz M, Nyman J, Christian E, Pawlak M, Dionne D, Xia J, Rozenblatt-Rosen O, et al. 2019. Checkpoint blockade immunotherapy induces dynamic changes in PD-1−CD8+ tumor-infiltrating T cells. Immunity 50: 181–194. 10.1016/j.immuni.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. 2009. Late developmental plasticity in the T helper 17 lineage. Immunity 30: 92–107. 10.1016/j.immuni.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, et al. 2016. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol 17: 1187–1196. 10.1038/ni.3543 [DOI] [PubMed] [Google Scholar]
- Lian G, Gnanaprakasam JR, Wang T, Wu R, Chen X, Liu L, Shen Y, Yang M, Yang J, Chen Y, et al. 2018. Glutathione de novo synthesis but not recycling process coordinates with glutamine catabolism to control redox homeostasis and directs murine T cell differentiation. eLife 7: e36158. 10.7554/eLife.36158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. 2006. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci 103: 3304–3309. 10.1073/pnas.0511137103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM, Nutt SL. 2012. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol 13: 491–498. 10.1038/ni.2261 [DOI] [PubMed] [Google Scholar]
- Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer ML, et al. 2017. Serine is an essential metabolite for effector T cell expansion. Cell Metab 25: 345–357. 10.1016/j.cmet.2016.12.011 [DOI] [PubMed] [Google Scholar]
- Ma X, Bi E, Lu Y, Su P, Huang C, Liu L, Wang Q, Yang M, Kalady MF, Qian J, et al. 2019. Cholesterol induces CD8+ T cell exhaustion in the tumor microenvironment. Cell Metab 30: 143–156. 10.1016/j.cmet.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y, Ishifune C, Tsukumo S, Hozumi K, Yagita H, Yasutomo K. 2015. Notch controls the survival of memory CD4+ T cells by regulating glucose uptake. Nat Med 21: 55–61. 10.1038/nm.3758 [DOI] [PubMed] [Google Scholar]
- Mak TW, Grusdat M, Duncan GS, Dostert C, Nonnenmacher Y, Cox M, Binsfeld C, Hao Z, Brüstle A, Itsumi M, et al. 2017. Glutathione primes T cell metabolism for inflammation. Immunity 46: 675–689. 10.1016/j.immuni.2017.03.019 [DOI] [PubMed] [Google Scholar]
- Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. 2019. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20: 326–336. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ, et al. 2011. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity 35: 972–985. 10.1016/j.immuni.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen QP, Deng TZ, Witherden DA, Goldrath AW. 2019. Origins of CD4+ circulating and tissue-resident memory T-cells. Immunology 157: 3–12. 10.1111/imm.13059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, et al. 2014. Memory CD8+ T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41: 75–88. 10.1016/j.immuni.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D, Sanin DE, Pearce EJ, Pearce EL. 2019. Metabolic interventions in the immune response to cancer. Nat Rev Immunol 19: 324–335. 10.1038/s41577-019-0140-9 [DOI] [PubMed] [Google Scholar]
- Pace L, Goudot C, Zueva E, Gueguen P, Burgdorf N, Waterfall JJ, Quivy JP, Almouzni G, Amigorena S. 2018. The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359: 177–186. 10.1126/science.aah6499 [DOI] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, et al. 2012. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338: 1220–1225. 10.1126/science.1229620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O'Malley JT, Gehad A, Teague JE, et al. 2017. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543: 252–256. 10.1038/nature21379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, et al. 2015. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun 6: 6692. 10.1038/ncomms7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, et al. 2016. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354: 1160–1165. 10.1126/science.aaf2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. 2009. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460: 103–107. 10.1038/nature08097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Yin N, Chhangawala S, Xu K, Leslie CS, Li MO. 2016. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354: 481–484. 10.1126/science.aaf6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Goldrath AW. 2015. Hypoxia-inducible factors regulate T cell metabolism and function. Mol Immunol 68: 527–535. 10.1016/j.molimm.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Doedens AL, Palazon A, Tyrakis PA, Cheung KP, Johnson RS, Goldrath AW. 2016. Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity 45: 1024–1037. 10.1016/j.immuni.2016.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T, et al. 2017. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545: 452–456. 10.1038/nature22367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, Delgoffe GM, Powell JD. 2015. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J Clin Invest 125: 2090–2108. 10.1172/JCI77746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi KN, Sun IH, Patel CH, Lo YC, Oh MH, Waickman AT, Tam AJ, Blosser RL, Wen J, Delgoffe GM, et al. 2016. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8+ T cell differentiation. Nat Immunol 17: 704–711. 10.1038/ni.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RR, Li Q, Gubbels Bupp MR, Shrikant PA. 2012. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity 36: 374–387. 10.1016/j.immuni.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raud B, Roy DG, Divakaruni AS, Tarasenko TN, Franke R, Ma EH, Samborska B, Hsieh WY, Wong AH, Stuve P, et al. 2018. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab 28: 504–515. 10.1016/j.cmet.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MA, Dai Z, Locasale JW. 2017. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol 19: 1298–1306. 10.1038/ncb3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolf J, Zarrouk M, Finlay DK, Foretz M, Viollet B, Cantrell DA. 2013. AMPKα1: a glucose sensor that controls CD8 T-cell memory. Eur J Immunol 43: 889–896. 10.1002/eji.201243008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Spinelli JB, et al. 2016. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T Cell activation. Cell Metab 24: 104–117. 10.1016/j.cmet.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy DG, Chen J, Mamane V, Ma EH, Muhire BM, Sheldon RD, Shorstova T, Koning R, Johnson RM, Esaulova E, et al. 2020. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab 31: 250–266. 10.1016/j.cmet.2020.01.006 [DOI] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. 2018. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175: 998–1013. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. 2017. mTOR signaling in growth, metabolism, and disease. Cell 168: 960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schietinger A, Greenberg PD. 2014. Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol 35: 51–60. 10.1016/j.it.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AC, Dündar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S, et al. 2019. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571: 270–274. 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM. 2016. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45: 1327–1340. 10.1016/j.immuni.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Chen J, Gonzállez-Avalos E, Samaniego-Castruita D, Das A, Wang YH, López-Moyado IF, Georges RO, Zhang W, Onodera A, et al. 2019. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc Natl Acad Sci 116: 12410–12415. 10.1073/pnas.1905675116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. 2014. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab 19: 780–794. 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Shrestha S, Yang K, Wei J, Karmaus PW, Neale G, Chi H. 2014. Tsc1 promotes the differentiation of memory CD8+ T cells via orchestrating the transcriptional and metabolic programs. Proc Natl Acad Sci 111: 14858–14863. 10.1073/pnas.1404264111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. 2013. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339: 222–226. 10.1126/science.1226603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. 2019. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50: 195–211. 10.1016/j.immuni.2018.12.021 [DOI] [PubMed] [Google Scholar]
- Sinclair LV, Howden AJ, Brenes A, Spinelli L, Hukelmann JL, Macintyre AN, Liu X, Thomson S, Taylor PM, Rathmell JC, et al. 2019. Antigen receptor control of methionine metabolism in T cells. eLife 8: e44210. 10.7554/eLife.44210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staron MM, Gray SM, Marshall HD, Parish IA, Chen JH, Perry CJ, Cui G, Li MO, Kaech SM. 2014. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8+ T cells during chronic infection. Immunity 41: 802–814. 10.1016/j.immuni.2014.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, et al. 2013. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest 123: 4479–4488. 10.1172/JCI69589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar M, Liu J, Mehta GU, Patel SJ, Roychoudhuri R, Crompton JG, Klebanoff CA, Ji Y, Li P, Yu Z, et al. 2016. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab 23: 63–76. 10.1016/j.cmet.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Yang K, Li Y, Shaw TI, Wang Y, Blanco DB, Wang X, Cho JH, Wang H, Rankin S, et al. 2017. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46: 488–503. 10.1016/j.immuni.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo NJ, Fife BT, Pagan AJ, Kotov DI, Goldberg MF, Jenkins MK. 2016. Most microbe-specific naive CD4+ T cells produce memory cells during infection. Science 351: 511–514. 10.1126/science.aad0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle CJ, Swanson HM, Fujii N, Estey EH, Riddell SR. 2009. A distinct subset of self-renewing human memory CD8+ T cells survives cytotoxic chemotherapy. Immunity 31: 834–844. 10.1016/j.immuni.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrakis PA, Palazon A, Macias D, Lee KL, Phan AT, Veliça P, You J, Chia GS, Sim J, Doedens A, et al. 2016. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540: 236–241. 10.1038/nature20165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. 2016. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45: 415–427. 10.1016/j.immuni.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Utzschneider DT, Delpoux A, Wieland D, Huang X, Lai CY, Hofmann M, Thimme R, Hedrick SM. 2018. Active maintenance of T cell memory in acute and chronic viral infection depends on continuous expression of FOXO1. Cell Rep 22: 3454–3467. 10.1016/j.celrep.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. 2012. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36: 68–78. 10.1016/j.immuni.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, O'Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, et al. 2013. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci 110: 14336–14341. 10.1073/pnas.1221740110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhana SA, Hwee MA, Berisa M, Wells DK, Yost KE, King B, Smith M, Herrera PS, Chang HY, Satpathy AT, et al. 2020. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat Immunol 21: 1022–1033. 10.1038/s41590-020-0725-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbist KC, Guy CS, Milasta S, Liedmann S, Kamiński MM, Wang R, Green DR. 2016. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532: 389–393. 10.1038/nature17442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V, Jafarzadeh N, Boi S, Kundu S, Jiang Z, Fan Y, Lopez J, Nandre R, Zeng P, Alolaqi F, et al. 2021. MEK inhibition reprograms CD8+ T lymphocytes into memory stem cells with potent antitumor effects. Nat Immunol 22: 53–66. 10.1038/s41590-020-00818-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodnala SK, Eil R, Kishton RJ, Sukumar M, Yamamoto TN, Ha NH, Lee PH, Shin M, Patel SJ, Yu Z, et al. 2019. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363: eaau0135. 10.1126/science.aau0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yosef N, Gaublomme J, Wu C, Lee Y, Clish CB, Kaminski J, Xiao S, Meyer Zu Horste G, Pawlak M, et al. 2015. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell 163: 1413–1427. 10.1016/j.cell.2015.10.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Beck-García K, Zorzin C, Schamel WW, Davis MM. 2016. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat Immunol 17: 844–850. 10.1038/ni.3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He Q, Shen H, Xia A, Tian W, Yu W, Sun B. 2019. TOX promotes the exhaustion of antitumor CD8+ T cells by preventing PD1 degradation in hepatocellular carcinoma. J Hepatol 71: 731–741. 10.1016/j.jhep.2019.05.015 [DOI] [PubMed] [Google Scholar]
- Wei J, Long L, Zheng W, Dhungana Y, Lim SA, Guy C, Wang Y, Wang YD, Qian C, Xu B, et al. 2019. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576: 471–476. 10.1038/s41586-019-1821-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Kurachi M. 2015. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15: 486–499. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, Kuchroo VK. 2013. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496: 513–517. 10.1038/nature11984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Shin HM, Moseman EA, Ji Y, Huang B, Harly C, Sen JM, Berg LJ, Gattinoni L, McGavern DB, et al. 2015. TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep 12: 2099–2110. 10.1016/j.celrep.2015.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. 2016. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1: eaai8593. 10.1126/sciimmunol.aai8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li G, Li L, Li D, Dong Z, Jiang P. 2021. Asparagine enhances LCK signalling to potentiate CD8+ T-cell activation and anti-tumour responses. Nat Cell Biol 23: 75–86. 10.1038/s41556-020-00615-4 [DOI] [PubMed] [Google Scholar]
- Xing S, Li F, Zeng Z, Zhao Y, Yu S, Shan Q, Li Y, Phillips FC, Maina PK, Qi HH, et al. 2016. Tcf1 and Lef1 transcription factors establish CD8+ T cell identity through intrinsic HDAC activity. Nat Immunol 17: 695–703. 10.1038/ni.3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, He R, Hao Y, Wang H, Zhao T, et al. 2015. The transcription factor TCF-1 initiates the differentiation of TFH cells during acute viral infection. Nat Immunol 16: 991–999. 10.1038/ni.3229 [DOI] [PubMed] [Google Scholar]
- Xu K, Yin N, Peng M, Stamatiades EG, Shyu A, Li P, Zhang X, Do MH, Wang Z, Capistrano KJ, et al. 2021. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 371: 405–410. 10.1126/science.abb2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Bai Y, Xiong Y, Zhang J, Chen S, Zheng X, Meng X, Li L, Wang J, Xu C, et al. 2016. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 531: 651–655. 10.1038/nature17412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Sun HW, Lacey NE, Ji Y, Moseman EA, Shih HY, Heuston EF, Kirby M, Anderson S, Cheng J, et al. 2019. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat Immunol 20: 890–901. 10.1038/s41590-019-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al. 2017. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552: 404–409. 10.1038/nature25144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YR, Imrichova H, Wang H, Chao T, Xiao Z, Gao M, Rincon-Restrepo M, Franco F, Genolet R, Cheng WC, et al. 2020. Disturbed mitochondrial dynamics in CD8+ TILs reinforce T cell exhaustion. Nat Immunol 21: 1540–1551. 10.1038/s41590-020-0793-3 [DOI] [PubMed] [Google Scholar]
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. 2012. FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling. Cell Metab 15: 279–291. 10.1016/j.cmet.2011.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Cohen S, Guy C, Shrestha S, Neale G, Brown SA, Cloer C, Kishton RJ, Gao X, Youngblood B, et al. 2016. mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity 45: 540–554. 10.1016/j.immuni.2016.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. 2010. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33: 229–240. 10.1016/j.immuni.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]