Abstract
Functional nucleic acids, including aptamers and deoxyribozymes, have become important in a variety of applications, particularly sensors. Aptamers are useful for recognition because of their ability to bind to targets with high selectivity and affinity. They can also be paired with deoxyribozymes to form signaling aptazymes. These aptamers and aptazymes have the potential to significantly improve the detection of small molecule pollutants, such as herbicides, in the environment. One challenge when developing aptazymes is that aptamer selection conditions can vary greatly from optimal deoxyribozyme reaction conditions. Aptamer selections commonly mimic physiological conditions, while deoxyribozyme selections are conducted under a wider range of divalent metal ion conditions. Isolating aptamers under conditions that match deoxyribozyme reaction conditions should ease the development of aptazymes and facilitate the activities of both the binding and catalytic components. Therefore, we conducted in vitro selections under different divalent metal ion conditions to identify DNA aptamers for the herbicides atrazine and alachlor. Conditions were chosen based on optimal reaction conditions for commonly-used deoxyribozymes. Each set of conditions yielded aptamers that were unrelated to aptamers identified under other selection conditions. No particular set of conditions stood out as being optimal from initial binding analysis. The best aptamers bound their target with high-micromolar to low-millimolar affinity, similar to the concentrations used during the selection procedures, as well as regulatory guidelines. Our results demonstrate that different metal ion concentrations can achieve the common goal of binding to a particular target, while providing aptamers that function under alternate conditions.
Keywords: aptamer, SELEX, in vitro selection, herbicides, atrazine, alachlor
INTRODUCTION
Herbicides are useful for increasing agricultural productivity and preventing devastating crop diseases, but can have serious health and environmental consequences that are still being investigated (Lebov et al, 2016; Wan and Lin, 2016; Kim et al, 2017; Stayner et al, 2017). The ability to assess human health risks by monitoring small-molecule pollutants on-site is important. Functional nucleic acids (FNAs), such as aptamers and deoxyribozymes, can be useful components for small molecule sensors that do not require extensive sample preparation or expensive equipment (Akki and Werth, 2018; Zhang et al, 2018). Aptamers have become important recognition elements because of their ability to bind targets with high affinity and specificity, so we sought to identify aptamers for two commonly used herbicides, alachlor and atrazine, that could be joined with deoxyribozymes to form aptazymes for sensor development (McManus et al, 2012; Zhou et al, 2015). While DNA aptamers for atrazine have previously been reported (Williams et al, 2014; Abraham et al, 2018), we are unaware of any reported aptamers for alachlor.
Since the introduction of in vitro selection (Ellington and Szostak, 1990) and SELEX (Tuerk and Gold, 1990) more than 25 years ago, FNAs have been identified under a variety of reaction conditions. It is often underappreciated that the reaction conditions used to identify an aptamer can vary greatly from the reaction conditions necessary for optimal deoxyribozyme activity. Aptamers are commonly selected under near physiological conditions, including low concentrations of divalent metal ions (Huizenga and Szostak, 1995; Mann et al, 2005; Niazi et al, 2008; Sabah et al, 2018; Sadeghi et al, 2018). While some RNA-cleaving deoxyribozymes can function under these conditions ( Santoro and Joyce, 1997; Bonaccio et al, 2004), many other deoxyribozymes use higher metal ion concentrations (Purtha et al, 2005; Zelin et al, 2006; Behera et al, 2013). These differences in reaction conditions can present challenges when developing aptazymes. For example, we previously converted the RNA-ligating deoxyribozyme 10DM24 into an ATP aptazyme and found the assays had to be conducted under conditions optimal for the deoxyribozyme, as studies under the aptamer conditions led to no activity (Alila and Baum, 2011). While the aptazymes were responsive to ATP, we observed Kd values higher than previous reports (Alila and Baum, 2011), which was likely a result of sub-optimal aptamer binding conditions.
With these observations in mind, we set out to identify herbicide aptamers under conditions more amenable to deoxyribozyme activity. Specifically, we focused on different concentrations of divalent metal ion known to support deoxyribozyme activity. By testing three different conditions while targeting the same molecules, we can begin to investigate the effect of divalent metal ion concentrations on aptamer selections.
MATERIALS AND METHODS
Details concerning oligonucleotides and reagents used in these experiments are provided in Supporting Information.
In vitro selections
All selections were conducted in 50mM HEPES pH 7.5 buffer with 150mM NaCl, 1mM of the appropriate herbicide (alachlor or atrazine) and 10% (v/v) ethanol. The ethanol was included due to limited water solubility of the herbicides. Selections BC (atrazine) and BE (alachlor) also contained 5mM KCl, 40mM MgCl2, and 20mM MnCl2 (high metal conditions). Selections BG (atrazine) and BJ (alachlor) contained 5mM KCl, 1mM MgCl2, and 1mM MnCl2 (low metal conditions). Selections BL (atrazine) and BN (alachlor) contained 10mM MgCl2 and 10mM MnCl2 (cleaving conditions). Full details concerning selections are provided in Supporting Information.
Initial binding assays
Each isolated unique sequence was evaluated to determine its ability to bind the target herbicide used in its selection. Using a procedure similar to that used in selection, 200pmol of unlabeled aptamer and a tracer amount of 5′-end 32P labeled aptamer were annealed as described for selection. The annealed aptamer was then incubated with 100pmol of capture oligo on derivatized beads under the appropriate buffer conditions, followed by incubation with 1mM target herbicide in 10%(v/v) ethanol. The percent of labeled oligo eluted was determined as described for selection (Supporting Information).
Concentration-dependent binding assays
Candidate aptamers were fluorescently labeled as previously described (Baum and Silverman, 2007) and outlined in Supporting Information. Using functionalized DNA-Bind Surface plates (Supporting Information), 20pmol of labeled aptamer was annealed and aliquoted into the wells to base pair to the immobilized capture oligo. Uncaptured aptamer was collected for quantification. Varying concentrations of target herbicide in the appropriate binding buffer was added to each well and allowed to incubate with the captured aptamers for 15min. Aptamer sequences interacting with the herbicide dissociate from the capture oligo on the plate surface and are eluted in the supernatant. Fluorescence measurements were taken of both the target-bound and unbound populations using a FlexStation 3 Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA). To account for differences in fluorescence between the free and target-bound states, raw data was corrected as outlined by Samokhvalov et. al. based on a calculated correction factor, Q, to provide the fraction target-bound, Fbound (Lackowicz, 2006; Samokhvalov et al, 2018). Fbound was plotted as a function of increasing herbicide concentration. When possible, the data was fit to the equation = A0 + (Amax*Bn/Kdn+Bn) where A is fluorescence, A0 is minimum fluorescence, Amax is maximum fluorescence, B is the concentration of herbicide, Kd is the binding constant, and n is the Hill coefficient.
RESULTS AND DISCUSSION
Three different binding conditions were selected for aptamer selection. The high metal concentration selections (designated BC and BE) used concentrations of Mg2+ and Mn2+ that are known to support several ligating deoxyribozymes we have previously studied, including 10DM24 (Zelin et al, 2006). The cleaving condition selections (BL and BN) used 10mM each of Mg2+ and Mn2+, which are known to support activity by the commonly used 8–17 and 10–23 deoxyribozymes (Santoro and Joyce, 1997). The low metal concentration selections (BG and BJ) with just 1mM each of Mg2+ and Mn2+ are similar to previously used aptamer selection conditions and can possibly support activity by cleaving deoxyribozymes. By testing a series of conditions, we can begin to understand the effect of divalent metal ion concentration on aptamer selections, while also identifying aptamers that function under conditions more amenable to aptazyme conversion.
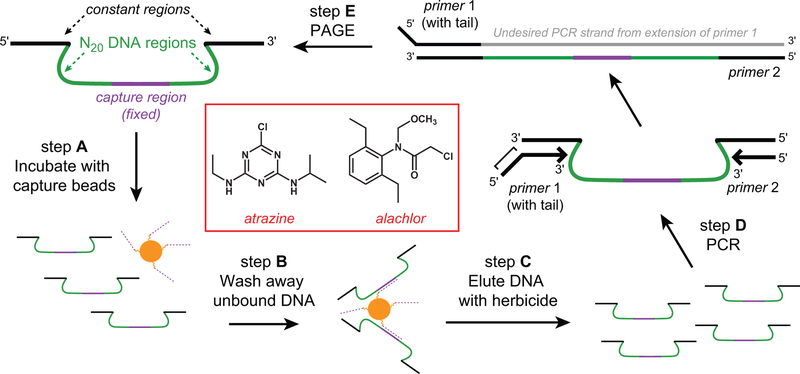
To provide access to all functional groups on the target molecules for interactions with the aptamers, we used a structure-switching (Achenbach et al, 2005; Nutiu and Li, 2005; He et al, 2011) or Capture-SELEX (Stoltenburg et al, 2012) approach in which the DNA pool sequences were immobilized via base pairing between a capture oligo attached to magnetic beads and a fixed region in the pool oligonucleotides (Figure 1). DNA sequences able to interact with the target molecule are expected to undergo a conformational change to form specific contacts with the target, which should weaken the base pairing with the capture oligo and release the binding DNA sequences into solution. The concentration of herbicide used in selections was 1mM, which is in alignment with EPA maximum contaminant levels (MCL) of 0.003 mg/l for atrazine and 0.002 mg/l for alachlor (US EPA, 2009).
Figure 1.
Structure-switching in vitro selections for herbicide aptamers. The structure-switching or capture-SELEX approach was used to allow candidate aptamer sequences full access to all functional groups on the target herbicides. Selections were initiated using a DNA pool containing two N20 random regions separated by a fixed 18-nt capture region. This randomized DNA pool was incubated with capture oligo-derivatized carboxyl-beads (step A). In step B, oligonucleotides that do not anneal to the capture oligo are washed away. Oligonucleotides that remain bound to the beads after step B are then eluted with herbicide-containing buffer (step C). Fractions from the elution step are collected and analyzed by Cerenkov counting. The eluted DNA is then used as a template for PCR amplification to generate the enriched pool for the next round (step D). The non-amplifiable tail of primer 1 contributes additional nucleotides to its PCR product and allows for the separation of the inactive compliment from the active pool sequence via PAGE (step E). The selection cycle is iterated until the pools are enriched with binding sequences. These active DNA pools are then cloned and sequenced to identify individual aptamer sequences.
To study individual aptamers, eluted DNA from round 7 of selections BC, BE, BG, and BJ, as well as eluted DNA from round 8 of selections BL and BN, were cloned and sequenced. We observed that the capture region for several aptamers contained mutations (often a single nucleotide difference) or was even missing, without significant changes in sequence length (Table S1). Mutations in this region are not unexpected as these could occur during PCR amplification in each selection round. Because these sequences still survived the selection process, they were included in our initial binding analysis.
Initial binding assays
Unique aptamer sequences were tested for their ability to bind atrazine or alachlor in solution under the same conditions used during selections. As shown in Table S2, each aptamer elutes with its selected herbicide, although the level of elution differs between aptamers. A particular set of conditions did not stand out as being better than other conditions in terms of binding ability.
Aptamers can be identified that are highly selective for their targets, but some aptamers can have broader binding abilities, which in our case could mean the ability to identify a class of herbicides or an herbicide and its breakdown products. We challenged the atrazine aptamers to bind to alachlor (a dissimilar herbicide) and simazine (a similar herbicide) and the alachlor aptamers to bind to atrazine (dissimilar) and metolachlor (similar). In this expanded set of studies, most of the isolated sequences had similar elution levels with each of the targets tested (parent, similar, and dissimilar), indicating that the oligos dissociated under the elution conditions regardless of the herbicide (Table S2).
However, a few sequences showed desirable aptameric properties and eluted more strongly with the herbicide used in selection and/or its related herbicide. For atrazine, there was one potential aptamer from each set of conditions. 7BC6 was identified under high divalent metal ion conditions and showed the highest elution with atrazine. 7BG1 was identified under the low divalent metal ion conditions and showed a preference for simazine, while still eluting strongly with atrazine. 8BL13 was identified under the cleaving conditions and showed a selectivity for atrazine. For alachlor, sequences isolated under the high divalent metal ion conditions did not strongly discriminate between the herbicides, but we selected one sequence, 7BE12, for further study with the other possible aptamers. The low divalent metal ion conditions produced one potential alachlor aptamer, 7BJ15, which eluted with alachlor and metolachlor. The cleaving conditions were the most promising for alachlor and three possible aptamers, 8BN15, 8BN23, and 8BN29, were carried forward for additional analysis.
Concentration dependence
To study the binding of promising aptamers under a wider range of herbicide concentrations, we used a slightly different approach. Briefly, each aptamer was labeled with a fluorophore (TAMRA for atrazine and simazine studies, and fluorescein for alachlor and metolachlor studies) and annealed to the capture oligo immobilized on a functionalized surface in 96-well trays. Varying concentrations of herbicide were then added to each well. Binding of the herbicide is expected to release the aptamer from the capture oligo, thus increasing the fluorescence observed in the supernatant. As a control, a DNA oligonucleotide of the same length as the isolated aptamers, but with a sequence not expected to form a structure or bind to the herbicides was tested, as was the starting pool (Figure S1). Fluctuations in fluorescence were observed at each concentration and there were no trends that indicated specific binding to the herbicides.
For the atrazine aptamers 7BC6, 7BG1, and 8BL13, addition of increasing amount of atrazine led to increased fluorescence, but we still observed fluctuations between trials leading to large deviations (Figure S1). We estimated the Kd values for 7BC6 and 8BL13 to be around 700μM, which is slightly lower than the 1mM concentration of atrazine used in selections, but higher than previously reported atrazine aptamers (Williams et al, 2014; Abraham et al, 2018). Based on these results, we focused our analysis on the alachlor aptamers.
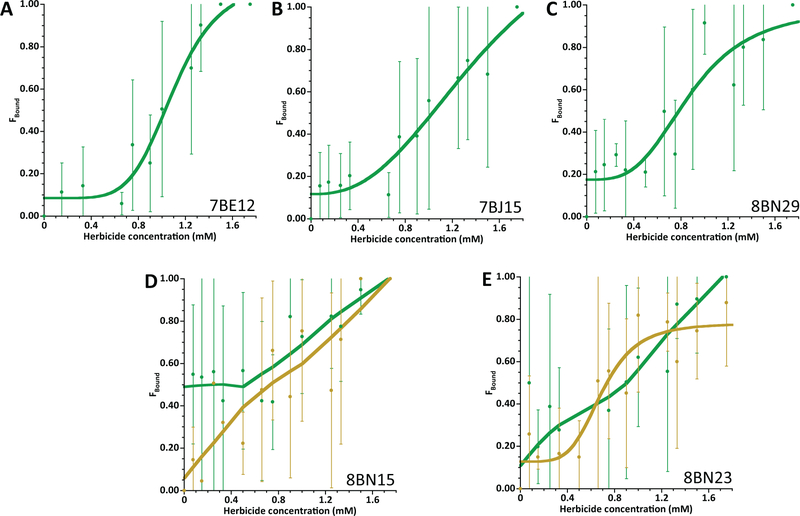
For the alachlor aptamers, estimated binding constants were close to the concentrations used in selections (1mM) or slightly above (Figure 2), which was not unexpected as there was no selection pressure towards lower target concentrations. The BN aptamers selected under the cleaving conditions had varied activity. 8BN29 (Figure 2C) showed binding similar to that of aptamers 7BE12 (Figure 2A) and 7BJ15 (Figure 2B), which were selected under high and low divalent metal ion conditions, respectively. These aptamers did not specifically bind to metolachlor (data not shown). 8BN15 showed a generally linear response to alachlor and metolachlor (Figure 2D), mirroring the results of the initial binding assays where similar elution was observed (Table S2). Interestingly, 8BN23 showed a preference for metolachlor over alachlor (Figure 2E), despite never being exposed to metolachlor during selections. As far as we are aware, these are the first DNA aptamers to be identified for the alachlor class of herbicides.
Figure 2.
Binding assays for alachlor-specific aptamers. Aptamers 7BE12 (A), 7BJ15 (B), 8BN29 (C), 8BN15 (D), and 8BN23 (E) were labeled with fluorescein and annealed to immobilized capture oligo prior to introduction of the appropriate herbicide. Fluorescence observed in the supernatant represents the aptamer released upon binding to the target. Aptamers 7BE12, 7BJ15, and 8BN29 showed distinct binding trends with alachlor (green), while 8BN15 was released with increasing concentrations of alachlor (green) and metolachlor (gold). 8BN23 showed a preference for metolachlor over alachlor.
Comparing results of different selection conditions
Aptamers for small molecules have the potential to be useful in sensor development. However, different sensor platforms may require incubation conditions different than those used in selections, which can affect the binding interactions of the aptamer and the ligand. This can often be seen in aptazyme development, which relies on the catalytic activity of deoxyribozymes to amplify the signal generated upon ligand binding to the aptamer. Thus we tested conditions known to support deoxyribozyme activity. These conditions were also attractive because they cover a wide range of divalent metal ion concentrations and they differ from those conditions commonly used with aptamers.
Candidate aptamers were isolated from each selection, which indicated that both high and low concentrations of divalent metal ions could support formation of aptamer structures that bind a small molecule ligand. Sequence comparison analysis of the aptamers isolated from each selection effort did not show any evidence of cross- contamination between the selections, as evidenced by the lack of duplicate sequences appearing in multiple selections. Thus, aptamers in each selection arose independently, demonstrating that different solutions are possible for a common problem, i.e. how to bind to the provided herbicide target. The high metal conditions provided more unique aptamers for atrazine than other conditions, but for alachlor, we saw high metal and cleaving conditions produced several unique aptamers (Table S1). The increased diversity did not mean more aptamers with useful binding properties (Table S2), but each set of conditions did result in at least one promising aptamer for further testing. In concentration-dependent binding studies, we again did not observe that a particular set of conditions led to a better binding aptamer. We also did not observe that a particular set of conditions led to more favorable specificity in terms of related herbicides, but our selections did not include pressure for selectivity. Overall, the resulting aptamers demonstrated binding affinities similar to those asked for in selection (1mM target). While this may be considered high compared to other aptamers, these concentrations are useful for herbicide detection based on EPA limits and represent the first aptamers for alachlor. From the aptamer selection point of view, exploring different conditions led to desired outcomes, the identification of binding sequences for a target of interest. Having aptamers selected under different conditions that better match deoxyribozyme conditions should facilitate aptazyme development and opens the door to exploring additional selection conditions that match with other sensor platforms.
CONCLUSIONS
We investigated three different sets of divalent metal ion concentration conditions for the selection of DNA aptamers for the herbicides atrazine and alachlor to determine what effect, if any, the choice of divalent metal ion conditions had on the selection outcomes and to identify aptamers that function optimally under deoxyribozyme reaction conditions for future aptazyme development. Aptamers were identified under each set of conditions for both alachlor and atrazine, with no set of conditions standing out as optimal for producing aptamers and the resulting binding affinities for the atrazine aptamers were higher than other reported aptamers. As far as we are aware, the resulting alachlor aptamers are the first reported aptamers for this particular family of herbicides. Using different divalent metal ion concentrations led to different aptamers with similar binding properties that can be merged more readily in terms of reaction conditions to known deoxyribozymes for aptazyme development.
Supplementary Material
ACKNOWLEDGMENTS
Work described in this manuscript was supported by National Institutes of Health (1R15GM101595-01 to DAB).
The authors thank Dr Christopher Arnatt for use of the FlexStation microplate reader and members of the Baum Lab for technical assistance.
Footnotes
COMPETING INTERESTS
None declare.
REFERENCES
- Abraham KM, Roueinfar M, Ponce AT, Lussier ME, Benson DB and Hong KL. 2018. In Vitro Selection and Characterization of a Single-Stranded DNA Aptamer Against the Herbicide Atrazine. ACS Omega 3, 13576–13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achenbach JC, Nutiu R and Li Y. 2005. Structure-switching allosteric deoxyribozymes. Anal Chim Acta 534, 41–51. [Google Scholar]
- Akki SU and Werth CJ. 2018. Critical Review: DNA Aptasensors, Are They Ready for Monitoring Organic Pollutants in Natural and Treated Water Sources? Environ Sci Technol, 52, 8989–9007. [DOI] [PubMed] [Google Scholar]
- Alila KO and Baum DA. 2011. Modulation of an RNA-branching deoxyribozyme by a small molecule. Chem Commun 47, 3227–3229. [DOI] [PubMed] [Google Scholar]
- Baum DA and Silverman SK. 2007. Deoxyribozyme-catalyzed labeling of RNA. Angew Chem Int Ed Engl 46, 3502–3504. [DOI] [PubMed] [Google Scholar]
- Behera AK, Schlund KJ, Mason AJ, Alila KO, Han M, Grout RL et al. 2013. Enhanced deoxyribozyme-catalyzed RNA ligation in the presence of organic cosolvents. Biopolymers 99, 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccio M, Credali A and Peracchi A. 2004. Kinetic and thermodynamic characterization of the RNA-cleaving 8–17 deoxyribozyme. Nucleic Acids Res 32, 916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD and Szostak JW. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822. [DOI] [PubMed] [Google Scholar]
- US EPA. 2009. National Primary Drinking Water Regulations. [Google Scholar]
- He J, Liu Y, Fan M and Liu X. 2011. Isolation and identification of the DNA aptamer target to acetamiprid. J Agric Food Chem 59, 1582–1586. [DOI] [PubMed] [Google Scholar]
- Huizenga DE and Szostak JW. 1995. A DNA aptamer that binds adenosine and ATP. Biochemistry 34, 656–665. [DOI] [PubMed] [Google Scholar]
- Kim K-H, Kabir E and Jahan SA. 2017. Exposure to pesticides and the associated human health effects. Sci Total Environ 575, 525–535. [DOI] [PubMed] [Google Scholar]
- Lackowicz JR. 2006. Principles of Fluorescence Spectroscopy. Springer; US, New York. [Google Scholar]
- Lebov JF, Engel LS, Richardson D, Hogan SL, Hoppin JA and Sandler DP. 2016. Pesticide use and risk of end-stage renal disease among licensed pesticide applicators in the Agricultural Health Study. Occup Environ Med 73, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann D, Reinemann C, Stoltenburg R and Strehlitz B. 2005. In vitro selection of DNA aptamers binding ethanolamine. Biochem Biophys Res Commun 338, 1928–1934. [DOI] [PubMed] [Google Scholar]
- McManus SA, Tram K and Li Y. 2012. Making sense of catalysis: the potential of DNAzymes as biosensors. RSC Biomol Sci 26, 190–210. [Google Scholar]
- Niazi JH, Lee SJ, Kim YS and Gu MB. 2008. ssDNA aptamers that selectively bind oxytetracycline. Bioorg Med Chem 16, 1254–1261. [DOI] [PubMed] [Google Scholar]
- Nutiu R and Li Y. 2005. In vitro selection of structure-switching signaling aptamers. Angew Chem Int Ed Engl 44, 1061–1065. [DOI] [PubMed] [Google Scholar]
- Purtha WE, Coppins RL, Smalley MK and Silverman SK. 2005. General Deoxyribozyme-Catalyzed Synthesis of Native 3’–5’ RNA Linkages. J Am Chem Soc 127, 13124–13125. [DOI] [PubMed] [Google Scholar]
- Sabah JT, Zulkifli RM, Shahir S, Ahmed F, Kadir MRA and Zakaria Z. 2018. In vitro selection and characterization of single stranded DNA aptamers for luteolin: A possible recognition tool. Anal Biochem 549, 72–79. [DOI] [PubMed] [Google Scholar]
- Sadeghi AS, Mohsenzadeh M, Abnous K, Taghdisi SM and Ramezani M. 2018. Development and characterization of DNA aptamers against florfenicol: Fabrication of a sensitive fluorescent aptasensor for specific detection of florfenicol in milk. Talanta 182, 193–201 [DOI] [PubMed] [Google Scholar]
- Samokhvalov AV, Safenkova IV, Eremin SA, Zherdev AV and Dzantiev BB. 2018. Measurement of (Aptamer-Small Target) KD Using the Competition between Fluorescently Labeled and Unlabeled Targets and the Detection of Fluorescence Anisotropy. Anal Chem 90, 9189–9198. [DOI] [PubMed] [Google Scholar]
- Santoro SW and Joyce GF. 1997. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci U S A 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stayner LT, Almberg K, Jones R, Graber J, Pedersen M and Turyk M. 2017. Atrazine and nitrate in drinking water and the risk of preterm delivery and low birth weight in four Midwestern states. Environ Res 152, 294–303. [DOI] [PubMed] [Google Scholar]
- Stoltenburg R, Nikolaus N and Strehlitz B. 2012. Capture-SELEX: Selection of DNA Aptamers for Aminoglycoside Antibiotics. J Anal Methods Chem 2012, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C and Gold L 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249 505–510. [DOI] [PubMed] [Google Scholar]
- Wan N and Lin G 2016. Parkinson’s Disease and Pesticides Exposure: New Findings From a Comprehensive Study in Nebraska, USA. J Rural Health 32, 303–313.+++. [DOI] [PubMed] [Google Scholar]
- Williams R, Crihfield C, Gattu S, Holland L and Sooter L. 2014. In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against Atrazine. Int J Mol Sci 15, 14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelin E, Wang Y and Silverman SK. 2006. Adenosine is Inherently Favored as the Branch-Site RNA Nucleotide in a Structural Context that Resembles Natural RNA Splicing. Biochemistry 45, 2767–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu QX, Guo ZH and Lin JS. 2018. Practical Application of Aptamer-Based Biosensors in Detection of Low Molecular Weight Pollutants in Water Sources. Molecules 23, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Xiao L, Xiang Y, Zhou J and Tong A. 2015. A general approach for rational design of fluorescent DNA aptazyme sensors based on target-induced unfolding of DNA hairpins. Anal Chim Acta 889, 179–186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.