Abstract
Granulocyte colony-stimulating factor (G-CSF) is widely used in clinical settings to mobilize hematopoietic stem cells (HSC) into the circulation for HSC harvesting and transplantation. However, whether G-CSF directly stimulates HSC to change their cell cycle state and fate is controversial. HSC are a heterogeneous population consisting of different types of HSC, such as myeloid-biased HSC and lymphoid-biased HSC. We hypothesized that G-CSF has different effects on different types of HSC. To verify this, we performed serum-free single-cell culture and competitive repopulation with cultured cells. Single highly purified HSC and hematopoietic progenitor cells (HPC) were cultured with stem cell factor (SCF), SCF + G-CSF, SCF + granulocyte/macrophage (GM)-CSF, or SCF + thrombopoietin (TPO) for 7 days. Compared with SCF alone, SCF + G-CSF increased the number of divisions of cells from the lymphoid-biased HSCenriched population but not that of cells from the My-bi HSC-enriched population. SCF + G-CSF enhanced the level of reconstitution of lymphoidbiased HSC but not that of myeloid-biased HSC. Clonal transplantation assay also showed that SCF + G-CSF did not increase the frequency of myeloid-biased HSC. These data showed that G-CSF directly acted on lymphoid- biased HSC but not myeloid-biased HSC. Our study also revised the cytokine network at early stages of hematopoiesis: SCF directly acted on myeloid-biased HSC; TPO directly acted on myeloid-biased HSC and lymphoid- biased HSC; and GM-CSF acted only on HPC. Early hematopoiesis is controlled differentially and sequentially by a number of cytokines.
Introduction
Hematopoietic stem cells (HSC) are able to self-renew and differentiate into all blood lineages.1 Granulocyte colony-stimulating factor (G-CSF) is widely used in clinical settings to mobilize HSC from the bone marrow (BM) to the peripheral blood (PB) for stem cell harvesting.2 However, the effect of G-CSF on HSC is poorly understood. Several studies have reported that G-CSF drives dormant HSC into the cell cycle,3-5 whereas other studies have reported that G-CSF does not.6,7 Since in vivo G-CSF administration results in complex changes in the BM microenvironment, such as disruption of the SDF-1/CXCR4 axis,8,9 it may be difficult to determine the direct effect of G-CSF on HSC in vivo. To avoid this issue, Ogawa’s research group combined in vitro culture with transplantation assay and reported that G-CSF can induce HSC self-renewal.10
The above studies are informative but quite controversial. This study aimed to clarify whether G-CSF acts directly on HSC and drives them into cycling, thus changing their fates. HSC are a heterogeneous population. Muller-Sieburg’s research group was the first to classify HSC into myeloid-biased HSC (My-bi HSC), balanced HSC (Bala HSC), and lymphoid-biased HSC (Ly-bi HSC), based on the ratio of lymphoid to myeloid cells (the L/M ratio) in reconstituted mice.11,12 Given that G-CSF is a neutrophil-specific cytokine that is essential for granulopoiesis,13,14 we hypothesized that G-CSF acts directly on My-bi HSC but not on Ly-bi HSC.
To address this issue, we used a serum-free culture system, which enabled us to exclude the effect of unknown factors contaminated in the serum.15 To overcome the heterogeneity of HSC, we performed single-cell culture, single- cell transplantation, and single-cell reverse transcription- polymerase chain reaction (RT-PCR) on highly purified HSC and hematopoietic progenitor cells (HPC). Surprisingly, we found that G-CSF increased the number of divisions of Ly-bi HSC and maintained their repopulating activity after transplantation. SCF alone transiently activated My-bi HSC and increased their long-term reconstitution potential, but SCF + G-CSF did not show any additional effect on that potential. We conclude that GCSF acts directly on Ly-bi HSC but not on My-bi HSC. This study suggested that My-bi HSC, which are more or less equivalent to long-term HSC, remain in the quiescent state after G-CSF injection in clinical settings.
Methods
Mice
C57BL/6 (CD45.2-B6) mice were purchased from Beijing HFK Bioscience Co. (Beijing, China). C57BL/6 mice congenic for the Ly5 locus (CD45.1-B6) were bred and maintained at the State Key Laboratory of Experimental Hematology. Animal experiments were approved by the Animal Care and Use Committees, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College.
Single-cell sorting
BM cells were isolated from 8- to 10-week old female CD45.1- or CD45.2-B6 mice, and c-Kit-positive cells were enriched using anti-c-Kit antibody-conjugated MACS beads (Miltenyi Biotechnology, catalog n. 130091224). Cell surface markers used for the identification of HSC1, HSC2, HPC1, HPC2, HPC3, and HPC4 are listed in Online Supplementary Table S1. Antibodies used for flow cytometry are listed in Online Supplementary Table S2.
Single-cell culture
Single cells were cultured in serum-free medium, supplemented with 50 ng/mL recombinant mouse SCF (Peprotech, 250-03) plus 50 ng/mL recombinant mouse thrombopoietin (TPO) (Peprotech, 315-14), 10 ng/mL recombinant human G-CSF (Peprotech, 300-23), or 10 ng/mL recombinant mouse GM-CSF (Peprotech, 315-03). Cells were cultured for 7 days at 37°C with 5% CO2 in the air. Number of cells per well were counted daily under inverted microscope.
Serial competitive repopulation
Twenty HSC1 or HSC2 cells from CD45.1-B6 mice were cultured with cytokines for 7 days, and cells were transplanted into lethally irradiated CD45.2-B6 mice with 5×105 BM competitor cells from CD45.2-B6 mice. As control, 20 freshly isolated HSC1 or HSC2 cells from CD45.1-B6 mice were similarly transplanted. For secondary transplantation of HSC1 cells, 2×107 BM cells from primary recipients were transplanted into lethally irradiated CD45.2-B6 mice. PB cells were analyzed as previously described.16
Single-cell transplantation
Single HSC1 cells from CD45.1-B6 mice were cultured with SCF + TPO for 1 day, and the surviving single HSC1 cells were selected and transplanted into lethally irradiated CD45.2-B6 mice with 5×105 BM competitor cells from CD45.2-B6 mice (control). For the cultured cell group, single HSC1 cells were cultured with cytokines for 7 days, and cells of each well were transplanted into lethally irradiated CD45.2-B6 mice with 5×105 BM competitor cells from CD45.2-B6 mice. PB cells were analyzed as previously described.16
Single-cell reverse transcription polymerase chain reaction
For single-cell RT-PCR, 48 single HSC1, HSC2, HPC1, HPC2, HPC3, and HPC4 cells were sorted into each well containing RTSTA master mix. For single-cell RT-PCR for cultured cells, single HSC1 cells were cultured with SCF, SCF+G-CSF, and SCF+TPO for 7 days. Single cells were randomly picked up from 48 wells by a micromanipulator and were placed into the RT-STA master mix. Freshly isolated 48 single HSC1 cells were used as a control. PCR was performed as previously described.16 The 48 genes set used for six populations and cultured cells are listed in Online Supplementary Tables S3 and S4, respectively.
Statistical analysis
Statistical significance was assessed with the unpaired t-test and analysis of variance using GraphPad Prism 6.0 (GraphPad).
Results
Definitions of HSC1, HSC2, HPC1, HPC2, HPC3, and HPC4
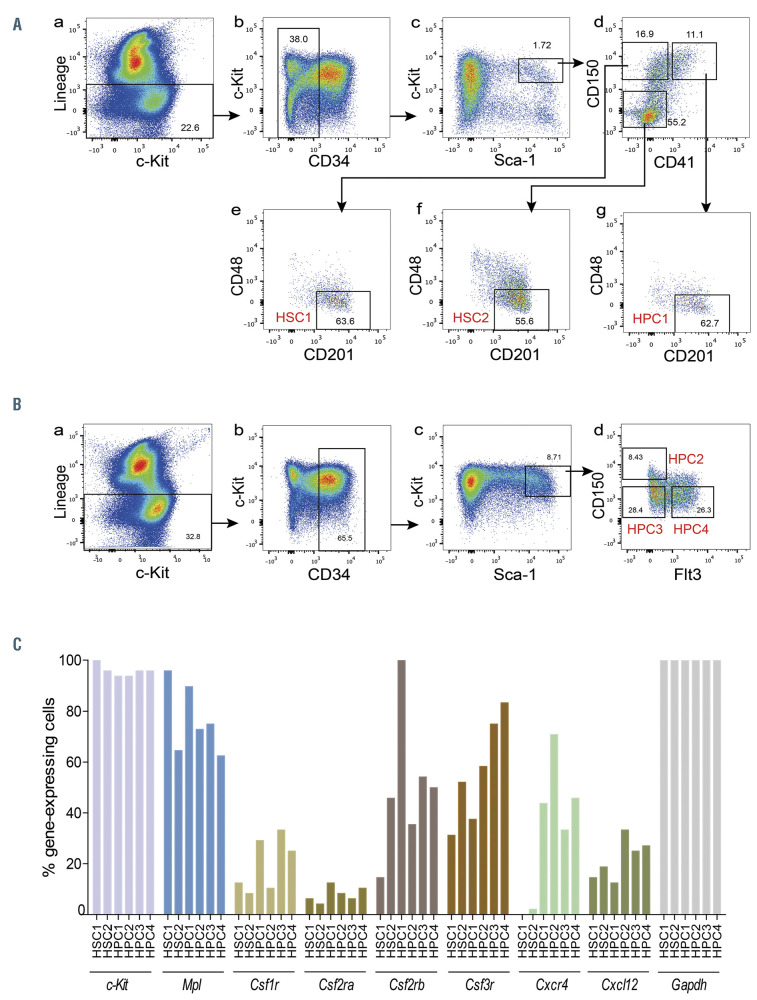
HSC1 (CD201+CD150+CD48–CD41–CD34–KSL) and HSC2 (CD201+CD150–CD48–CD41–CD34–KSL) cells were defined as shown in Figure 1A. HSC1 were separated from HSC2 by expression of CD150.17,18 We previously showed that HSC1 cells are enriched in long-term (LT, >6 months) My-bi HSC (LT-My-bi HSC), while HSC2 cells are enriched in short-term (ST, <6 months) Ly-bi HSC (ST-Ly-bi HSC).16,18-21 HPC1 (CD201+CD150+CD48– CD41+CD34–KSL), HPC2 (CD150+Flt-3–CD34+KSL), HPC3 (CD150–Flt-3–CD34+KSL), and HPC4 (CD150–Flt- 3+CD34+KSL) cells were defined as shown in Figure 1A and B. HPC1 cells are reportedly enriched in myeloidrestricted repopulating progenitors.19 HPC2, HPC3, and HPC4 cells were enriched in MPP2, MPP3, and MPP4/lymphoid- primed multipotent progenitor (LMPP), respectively (Online Supplementary Figure S1).
Cytokine receptor expression
Single-cell RT-PCR was performed on HSC1, HSC2, HPC1, HPC2, HPC3, and HPC4 cells to investigate the expression of cytokine receptors. Gene expression data are shown as heatmaps in Online Supplementary Figure S2. Figure 1C shows the percentage of gene-expressing cells. c-Kit and Mpl were detected in most cells examined. MCSF receptor, Csf1r, was not expressed in >40% of cells in any of the populations examined. GM-CSF receptor comprises Csf2ra and Csf2rb. Csf2ra was not detected in >15% of cells in any of the populations examined. Csf2rb was also expressed in <15% of HSC1 cells, but was expressed in approximately from 40 to 60% of HSC2, HPC2, HPC3, and HPC4 cells, and in 100% of HPC1. G-CSF receptor, Csf3r, was detected in approximately 30% of HSC1 and HPC1 cells, while it was detected in >50% of HSC2 and HPC2, and >70% of HPC3 and HPC4 cells. Cxcl12 was expressed in approximately from 20 to 40% of all populations. Cxcr4 was expressed in approximately from 30 to 50% of HPC1, 3, and 4 cells, and in 70% of HPC2 cells. Notably, Cxcr4 was expressed in very few HSC1 and 2 cells.
Figure 1.
Gating strategy and single-cell real-time-polymerase chain reaction (RT-PCR) of HSC1, HSC2, HPC1, HPC2, HPC3, and HPC4 cells. (A) HSC1, HSC2, and HPC1 gating. Bone marrow (BM) cells were stained with antibodies and gated as follows. (a) Gating for lineage– cells (Lin– cells). (b) Gating for Lin–CD34– cells. (c) Gating for CD34– c-Kit+ Sca-1+ Lin– cells (CD34–KSL cells). (d) Based on CD150 and CD41 expression, CD34–KSL cells were divided into CD150+CD41–, CD150–CD41–, and CD150–CD41+ cells. (e-g) CD201+CD48– cells were further selected from CD150+CD41–, CD150–CD41–, and CD150–CD41+ cells. HSC1, HSC2, and HPC1 cells were defined as CD201+CD150+CD48–CD41–CD34–KSL cells, CD201+CD150–CD48–CD41–CD34–KSL cells, and CD201+CD150–CD48–CD41+CD34–KSL cells, respectively. (B) HPC2, HPC3, and HPC4 gating. BM cells were stained with antibodies and gated as follows. (a) Gating for lineage– cells (Lin– cells). (b) Gating for Lin–CD34+ cells. (c) Gating for CD34+KSL cells. (d) CD34+KSL cells were divided into CD150+Flt-3–, CD150–Flt-3–, and CD150–Flt-3+ cells. HPC2, HPC3 and HPC4 cells were defined as CD150+Flt-3–CD34+KSL cells, CD150–Flt-3–CD34+KSL cells, and CD150–Flt-3+CD34+KSL cells, respectively. (C) Single-cell RT-PCR for six populations. The percentage of gene-expressing cells is shown. Gene-expressing cells are defined by a threshold cycle (Ct) value < 27.65.
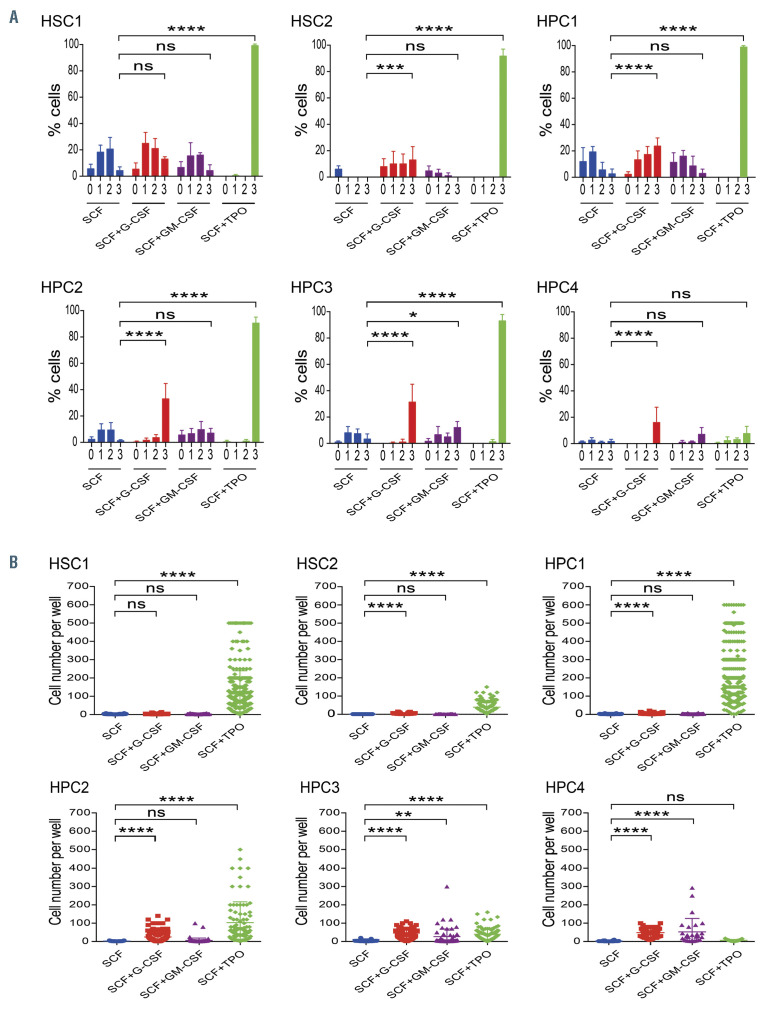
Figure 2.
Single-cell culture of six populations. (A) Percentage of cells that underwent divisions is shown as mean±standard deviation (SD). (n=5). Number of cell division was estimated from the number of cells per well at days 1-7 of culture. When the number of cells per well was 1, 2, 3-4, or ≥5, single cells were considered to have undergone 0, 1, 2, or ≥ 3 division(s), respectively. Wells that had no cells were excluded from this analysis. Two-way ANOVA with Tukey’s multiple comparisons test was used for statistical analysis. (B) Cell number per well at day 7 of culture is shown as mean±SD. (n=5). Unpaired t-test with Welch’s correction was used for statistical analysis. *P<0.05; **P<0.01; ***P<0.001; and ****P<0.0001; ns: not significant.
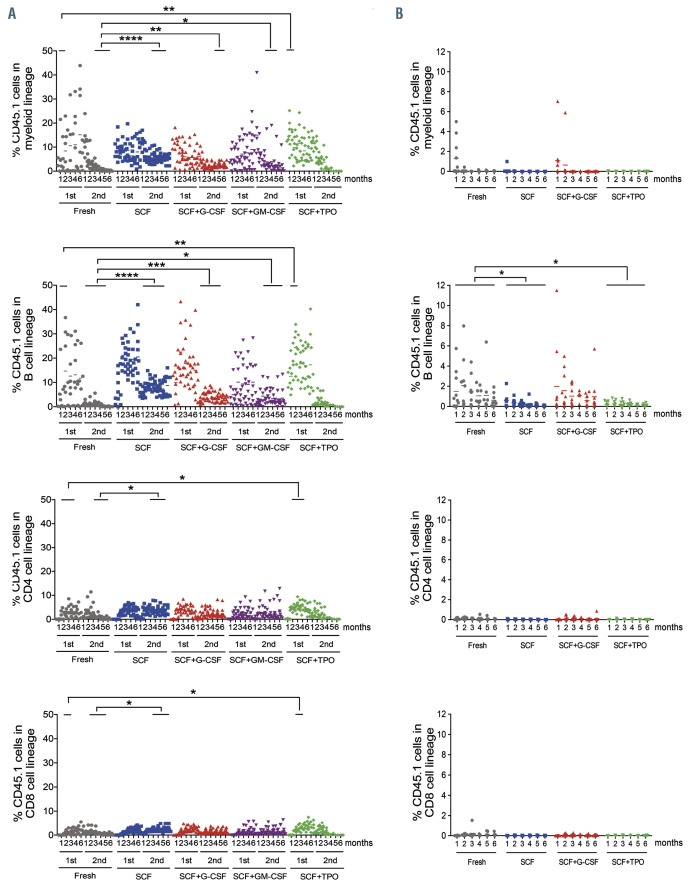
Figure 3.
Repopulating activity of cultured HSC1 and HSC2 cells. (A) Percentages of CD45.1 cells derived from HSC1 cells are shown. Percentage of CD45.1 cells in the myeloid lineage, B-cell lineage, CD4 T-cell lineage, and CD8 T-cell lineage in the culture with stem cell factor (SCF) alone were greater than those of freshly isolated cells after secondary transplantation. Percentage of CD45.1 cells in the myeloid lineage and B-cell lineage in the culture with SCF + granulocyte colony-stimulating factor (G-CSF) or SCF + granulocyte/macrophage (GM)-CSF were greater than those of freshly isolated cells after secondary transplantation. Percentage of CD45.1 cells in the myeloid lineage, B-cell lineage, CD4 T-cell lineage, and CD8 T-cell lineage in the culture with SCF+ thrombopoietin (TPO) were greater than those of freshly isolated cells at early months after primary transplantation. (B) Percentages of CD45.1 cells derived from HSC2 cells are shown. Percentage of CD45.1 cells in the B-cell lineage of the culture with SCF or SCF + TPO was significantly smaller than that of freshly isolated cells. Dots represent % chimerism in myeloid lineage, B-cell lineage, CD4 T-cell lineage and CD8 T-cell lineage from individual mice at a time point after transplantation. The lines above dots indicate the time periods compared. The unpaired t-test with Welch’s correction was used for statistical analysis between freshly isolated cells and cells in different culture conditions. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Effect of granulocyte colony-stimulating factor and granulocyte/macrophage colony-stimulating factor on the division of single hematopoietic stem cells and hematopoietic progenitor cells
We next compared the effects of SCF alone, SCF + GCSF, SCF + GM-CSF, and SCF + TPO on these six populations by single-cell culture. Figure 2A shows the percentage of cells that underwent divisions. Figure 2B shows the cell number per well at day 7 of culture. SCF supported the survival of a proportion of HSC1, HPC1, HPC2, and HPC3 cells, and induced their division 1-2 times. However, SCF alone did not support the survival of most HSC2 and HPC4 cells. SCF + G-CSF did not support division of HSC1 more than SCF alone. However, SCF + G-CSF significantly increased the number of divisions in HSC2, HPC1, HPC2, HPC3, and HPC4 cells, leading to a significant increase in the cell number per well. SCF + GM-CSF did not support the division of HSC1, HSC2, HPC1, and HPC2 cells but significantly increased the number of divisions of HPC3 and HPC4 cells, leading to an increase in the cell number per well. SCF + TPO significantly increased the number of divisions and cells per well in HSC1, HSC2, HPC1, HPC2, and HPC3 cells, but not in HPC4 cells. These data suggested that the target cells of GCSF, GM-CSF, SCF, and TPO were different among HSC and HPC: SCF acted directly on HSC1, HPC1, HPC2, and HPC3 cells, but not on HSC2 and HPC4 cells. TPO acted on HSC1, HSC2, HPC1, HPC2, and HPC3 cells, but not on HPC4 cells. G-CSF acted directly on HSC2 and HPC1-4 cells, but not on HSC1 cells. GM-CSF acted directly on HPC3 and HPC4 cells.
Effect of granulocyte colony-stimulating factor on reconstitution potential
To examine the effect of the cytokines on the reconstitution potential in HSC1 and HSC2 cells, we performed competitive repopulation assay. Figure 3A shows the percentage of CD45.1 cells in the myeloid, B-cell, CD4 T-cell, and CD8 T-cell lineages after transplantation with HSC1 cells. Compared with freshly isolated cells, the levels of reconstitution of each lineage in the SCF culture were significantly increased after secondary transplantation. The levels of myeloid and B-cell lineages in SCF + G-CSF or SCF + GM-CSF cultures were also significantly increased after secondary transplantation. However, when we compared the reconstitution levels among cultured cells, there was no significant difference between SCF + G-CSF or GM-CSF and SCF cultures, suggesting that SCF, but neither G-SCF nor GM-CSF, increased the long-term reconstitution potential in HSC1 cells. The levels of reconstitution of each lineage in SCF + TPO culture were significantly increased in the early months after primary transplantation, suggesting that this combination of cytokines increased the short-term reconstitution potential in HSC1 cells.
Figure 3B shows the percentage of CD45.1 cells in myeloid, B-cell, CD4 T-cell and CD8 T-cell lineages after transplantation with HSC2 cells. Freshly isolated HSC2 cells showed B-lymphoid-biased reconstitution. The level of B-cell lineage reconstitution in SCF and SCF + TPO cultures was significantly lower than that in freshly isolated cells, whereas that in SCF + G-CSF culture was comparable with that in freshly isolated cells. Taken together, these data suggested that SCF alone was sufficient to increase the long-term multilineage reconstitution potential. SCF + TPO increased the short-term multilineage reconstitution potential. SCF + G-CSF did not enhance the long-term myeloid lineage reconstitution potential but maintained the short-term lymphoid reconstitution potential.
Transplantation of clonally cultured cells
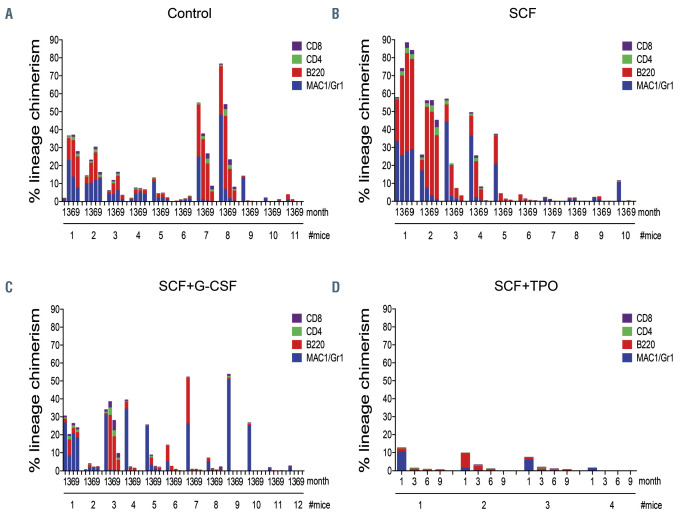
To further clarify the effect of G-CSF on HSC, we performed clonal transplantation assay. Eleven mice in the control group, 10 mice in the SCF group, 12 mice in the SCF + G-CSF group, and 4 mice in the SCF + TPO group were reconstituted (Figure 4). The percentage of chimerism and its lineage composition in single HSC1 cells varied from one another as reported.16,21 Similar to freshly isolated HSC1 cells, after one day culture single HSC1 cells showed a varying degree of reconstitution, indicating the heterogeneity of HSC.
We used the published criteria of My-bi, Bala, and Ly-bi HSC,11,12 and LT- and ST-HSC.22 In the control group of mice, 6 LT-My-bi HSC, 2 ST-Ly-bi HSC, and 3 HPC were detected (Figure 4A and Online Supplementary Table S5). After culture with SCF for 7 days, 1 LT-My-bi HSC, 1 STBala HSC, 4 ST-Ly-bi HSC, and 4 HPC were detected (Figure 4B and Online Supplementary Table S6). After culture with SCF + G-CSF, 2 LT-My-bi HSC, 1 ST-My-bi HSC, 5 ST-Ly-bi HSC, and 4 HPC were detected (Figure 4C and Online Supplementary Table S7). After culture with SCF + TPO, 3 ST-Ly-bi HSC and 1 HPC were detected (Figure 4D and Online Supplementary Table S8). These data showed no difference in reconstitution potential between SCF and SCF + G-CSF cultures, but the significant reduction in reconstitution potential after culture with SCF + TPO.
To be more precise, LT-My-bi HSC activity was detected in the mouse transplanted with three cells from SCF culture (#1 mouse), and similarly, in the mice transplanted with three cells from SCF + G-CSF culture (#1 and #2 mice) (Figure 4B and C and Online Supplementary Tables S6 and S7). ST-Ly-bi but not LT-My-bi HSC activity was detected in mice transplanted with >50 cells from SCF + TPO culture (#1, 2, and 3 mice) (Figure 4D and Online Supplementary Tables S8). These data suggested the similar effects of SCF and SCF + G-CSF on LT-My-bi HSC and the differentiation effect of SCF + TPO, associated with a number of divisions, on LT-My-bi HSC.
Gene expression of cultured cells
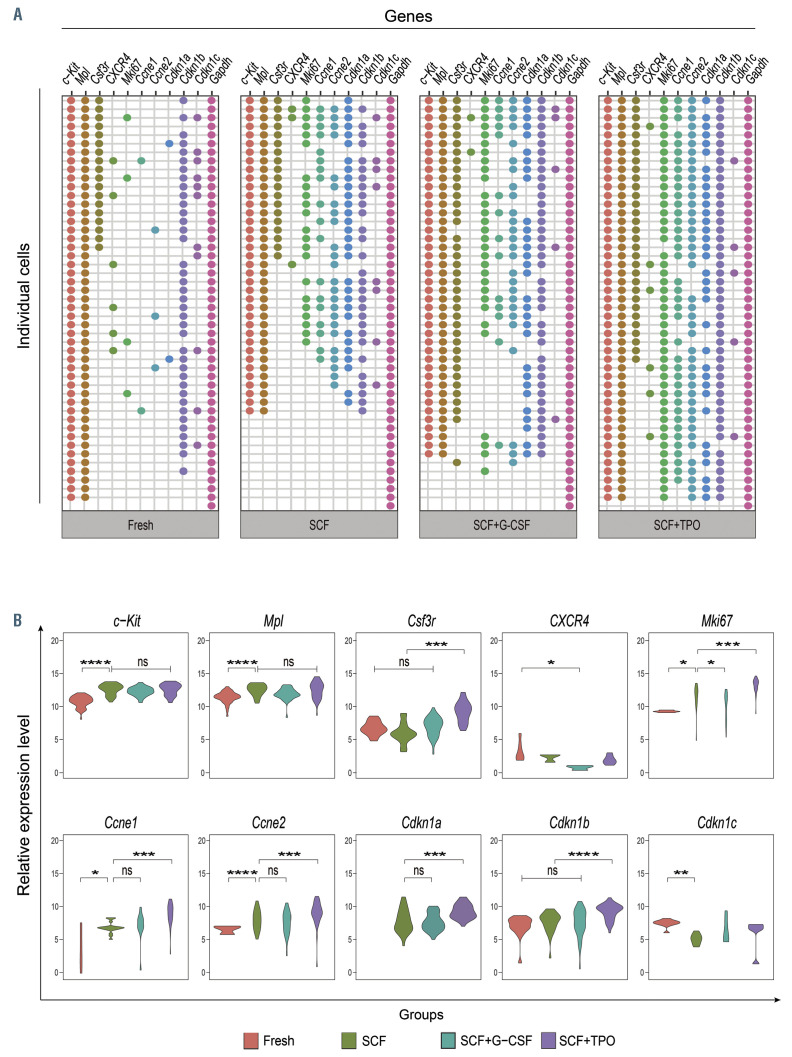
We examined the expression of cytokine receptors in day 7 cultured cells by single-cell RT-PCR. Gene expression data are shown as heatmaps in Online Supplementary Figure S3. Figure 5A depicts the gene expression in individual cells. Figure 5B depicts the relative expression level of genes. Consistent with the data in Figure 1C, both c-Kit and Mpl were expressed in the majority of freshly isolated HSC1 cells, while Csf3r was expressed in approximately 30% of the cells. After culture with SCF, the percentage of c-Kit- and Mpl-expressing cells slightly decreased but their relative expression levels significantly increased. However, neither the percentage of Csf3r-expressing cells nor the relative expression level of Csf3r changed. Interestingly, the percentage of Csf3r-expressing cells increased after culture with SCF + G-CSF or TPO. The relative expression level of Csf3r also increased after culture with SCF + TPO but not with SCF + G-CSF. Suppressor of cytokine signaling (SOCS) family are physiological regulators of several cytokine signaling.23 Less than 50% of HSC1 expressed SOCS3 while most HSC1 expressed SOCS 2-6, approximately 30% of HSC1 expressed SOCS1, and <30% of HSC1 expressed SOCS7. Interestingly, the expression of SOCS3, but not the other SOCS, increased after culture with SCF + G-CSF or TPO (Online Supplementary Figure S4A and B) suggesting a positive correlation between Csf3r and SOCS3 expression in HSC. Members of early growth response gene (Egr) family, Egr2 and Egr3, can directly induce SOCS3 expression.24 Egr3, but neither Egr1 nor Egr2, was detected in most cells (Online Supplementary Figure S4A and B). A very small number of freshly isolated HSC1 cells expressed Cxcr4, and the percentage of Cxcr4-expressing cells did not increase after culture with SCF, SCF + G-CSF, and SCF + TPO. These data suggested that these cytokines cannot directly upregulate the expression of Cxcr4.
In culture with SCF or SCF + G-CSF, most HSC1 cells divided only 1-2 times and then stopped dividing. However, the multilineage reconstitution potential was maintained in these cells. To address the question of whether these cells returned to the quiescent state, we examined their cell cycle status by single-cell RT-PCR. Mki-67 antigen is a nuclear protein exclusively expressed in proliferating cells during all phases of the cell cycle except G0.25 Mki67 was expressed in a small number of freshly isolated HSC1 cells. After culture, Mki67 was expressed in approximately 50%, approximately 60%, and >95% of cells cultured with SCF, SCF + G-CSF, and SCF + TPO, respectively, and its relative expression level was also significantly increased. Both percentage of positive cells and relative expression level expressions for Ccne1 and Ccne2 increased in Mki67+ cultured cells. These data suggested that HSC1 cells were quiescent in the steady state. After culture, most cells were continuously cycling in SCF + TPO, while approximately half of the cells were cycling in SCF or SCF + G-CSF.
Figure 4.
Transplantation of clonally cultured cells. (A) Lineage chimerism of single HSC1 cells. (B) Lineage chimerism of single-cell-derived cells in culture with stem cell factor (SCF). (C)Lineage chimerism of single-cell-derived cells in culture with SCF + granulocyte colony-stimulating factor (G-CSF). (D) Lineage chimerism of single- cell-derived cells in culture with SCF+ thrombopoietin (TPO). The percentage of the total chimerism was calculated as % (CD45.1+ cells) x 100/% (CD45.1+ cells + CD45.2+ cells). The percentage of myeloid lineage chimerism was calculated as (% CD45.1 cells) x (Mac-1/Gr-1+ cells) / (Mac-1/Gr-1+ cells + B220+ cells + CD4+ cells + CD8+ cells), in which (Mac-1/Gr-1+ cells)/(Mac-1/Gr-1+ cells + B220+ cells + CD4+ cells + CD8+ cells) was derived from CD45.1+ cells. The percentage of Bcell lineage chimerism was calculated as (% CD45.1 cells) x (B220+ cells)/(Mac-1/Gr-1+ cells + B220+ cells + CD4+ cells + CD8+ cells). The percentage of CD4 Tcell lineage chimerism was calculated as (% CD45.1 cells) x (CD4+ cells)/(Mac-1/Gr-1+ cells + B220+ cells + CD4+ cells + CD8+ cells). The percentage of CD8 T-cell lineage chimerism was calculated as (% CD45.1 cells) x (CD8+ cells)/(Mac-1/Gr-1+ cells + B220+ cells + CD4+ cells + CD8+ cells). Mice were considered to be reconstituted with donor cells when the percentage of donor-derived cells was ≥0.2%. My-bi, Bala, and Ly-bi hematopoietic stem cells (HSC) were defined by the ratio of lymphocytes to myeloid cells (L/M ratio) in the peripheral blood 6 months after transplantation. My-bi HSC were defined by the L/M ratio <3, Ly-bi HSC were defined by the L/M ratio >10, and Bala HSC were defined by 3 < L/M < 10. Long-term (LT)-HSC were defined when the percentage of myeloid cells maintained or increased by 6 months after transplantation. Short-term (ST)-HSC were defined when the percentage of myeloid cells decreased by 6 months, with myeloid, B-lymphoid, and T-lymphoid lineage reconstitution at a time after transplantation. HPC were defined when one or two lineages lacked from the definition of ST-HSC.
Figure 5.
Single-cell real-time-polymerase chain reaction (RT-PCR) of cultured HSC1 cells. (A) Gene expression in a single cell. One column represents one gene, and a row represents a single cell. Gene-expressing cells are shown as dots, which are defined by the threshold cycle (Ct) value < 27.65. (B) Violin density plots show the relative gene expression levels of gene-expressing cells. The relative expression level is defined as the (27.65-Ct) values. Statistical significance was analyzed by ANOVA with Tukey’s multiple comparisons test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; ns: not significant.
Figure 6.
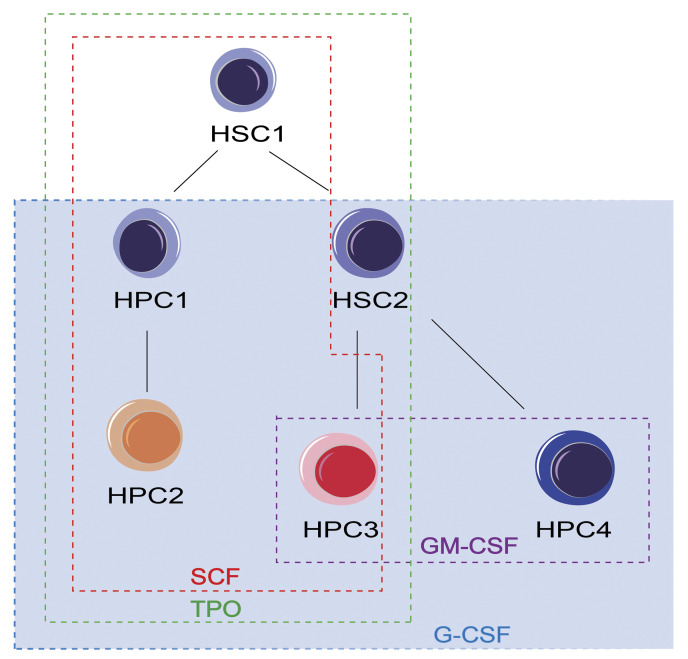
Cytokine network at the early stage of hematopoiesis. The model shows the cytokine network among HSC1, HSC2, HPC1, HPC2, HPC3, and HPC4. HSC1 cells respond to stem cell factor (SCF) and thrombopoietin (TPO). HSC2 cells respond to TPO and granulocyte colony-stimulating factor (G-CSF). HPC1 and HPC2 cells respond to SCF, TPO, and G-CSF. HPC3 cells respond to SCF, TPO, G-CSF, and GM-CSF. HPC4 cells respond to G-CSF and GM-CSF.
Cell cycle progression is regulated by Cdkn1a (p21), Cdkn1b (p27), and Cdk1c (p57).26 p21 was not expressed in the majority of freshly isolated Mki67– cells (Figure 5) but p21 was expressed in most Mki67+ cells after culture; in particular, in SCF + TPO, its relative expression level was significantly increased. Interestingly, p27 was expressed in most freshly isolated Mki67– cells as well as most Mki67+ cells after culture. p57 was expressed in some of freshly isolated Mki67– cells, and also in some Mki67+ cells after culture; however, its relative expression level was decreased after culture. These data suggested that p21 was expressed in cycling cells while p57 was expressed in some quiescent cells. p27 was expressed in both quiescent cells and cycling cells.
Discussion
Functionally distinct HSC have been classified into Mybi, Bala, and Ly-bi HSC; α, b, and γ cells; or ST- and LTHSC by different criteria. However, these classified cells overlap one another.22 Particularly, My-bi HSC overlap LTHSC, and Ly-bi HSC overlap ST-HSC. In this study, by definition, we detected LT-My-bi HSC and ST-Ly-bi HSC at the single-cell level. We used HSC1 and HSC2 cells as highly purified HSC. HSC1 cells are significantly enriched in LT-My-bi HSC, while HSC2 cells are significantly enriched in ST-Ly-bi HSC.16,18-21 As shown in clonal transplantation, however, a small proportion of HSC1 contained ST-Ly-bi HSC (Figure 4A). Therefore, ST-Ly-bi HSC co-existed with LT-My-bi HSC in HSC1. We used HPC1, HPC2, HPC3, and HPC4 as highly purified HPC. We have recently shown that LT-HSC can be similarly detected in HSC1 and HPC1 cells.21 However, this study showed that Csf2rb expression in HPC1 was significantly greater than in HSC1 (Figure 1C), and HPC1 but not HSC1 cells responded to G-CSF in single-cell culture (Figure 2A). HSC1 and HPC1 as populations remained functionally distinct.
Hematopoietic cytokines play a critical role in the regulation of hematopoiesis. In this study, we examined the effects of SCF, TPO, G-CSF, and GM-CSF on HSC1, HSC2, HPC1, HPC2, HPC3, and HPC4. SCF/c-Kit signaling plays an important role in hematopoiesis, particularly in the interaction of HSC and their niche, as shown by studies of W and Steel mutant mice.27,28 It has recently been reported that SCF is a niche factor from endothelial cells and perivascular stromal cells to maintain HSC.29 Li and Johnson were the first to report that SCF is a survival factor of HSC in culture. We now confirmed their finding by transplantation assays (Figures 3 and 4). SCF alone was sufficient to support the survival of LT-HSC. Clonal transplantation assay showed that LT-HSC activity was detected in progeny from single LT-HSC. Therefore, self-renewal division took place in SCF culture (Figure 4B and Online Supplementary Table S6). Interestingly, HSC stopped dividing after 1-2 times in SCF culture (Figure 2). Single-cell RTPCR suggested that some cells may be non-cycling, based on the expression of Mki67 (Figure 5). The role of p57 in regulating HSC quiescence has been suggested.30,31 However, we showed that p57 was not upregulated in Mki67– cells by SCF. p21 was expressed in the continuously cycling cells in Mki67+ cells from SCF + TPO culture, but not Mki67– cells from freshly isolated cells. p21 expression was upregulated in Mki67– cells after SCF culture, compared to Mki67– cells from freshly isolated cells (Figure 5). These 'stop-dividing' cells may differ from the quiescent state in vivo at the molecular level. It would be interesting to see the functional difference between G0 and pseudo-G0 HSC.
Mpl was the second highly expressed receptor in HSC and HPC (Figure 1C). SCF and TPO synergistically acted on HSC1, HSC2, HPC1, HPC2, and HPC3, but they did not exert much action on HPC4 because the percentage of division and colony sizes in HPC4 were significantly smaller than those in the others (Figure 2B). Consistent with our data, it has been reported that deletion of TPO did not affect the number of CD34+FLT3+KSL cells but significantly reduced that of CD34+/–FLT3– KSL cells.32 It also has been reported that deletion of TPO or Mpl results in the increase of cycling of HSC and subsequent reduction of the pool of quiescent HSC in mice.32,33
TPO in synergy with SCF promoted rapid division of HSC in vitro (Figures 2 and 5). SCF + TPO transiently increased the level of ST reconstitution but this was substantially decreased in LT reconstitution (Figures 3A, 4D, and Online Supplementary Table S8). In this study, SCF + TPO supported the differentiation more than self-renewal in HSC. However, HSC may behave differently, depending on different culture conditions as recently reported.34 It has also been recently reported that a low concentration of SCF and TPO can maintain HSC quiescent HSC in vitro.35 TPO may have different roles in the regulation of HSC under different conditions.
About 30% of HSC1 expressed Csf3r, but either G-CSF alone or G-CSF + SCF did not induce their division at all (Figure 2A and data not shown). SCF + G-CSF did not increase LT reconstitution level more than did SCF alone (Figures 3A, 4B and C, and Online Supplementary Tables S6 and S7). Csf3r expression was upregulated in SCF + G-CSF culture (Figure 5). The percentage of SOCS3+ cells increased in Csf3r+ cells after SCF + G-CSF culture (Online Supplementary Figure S4A). SOCS3 may play a negative role in G-CSF signaling in LT-HSC as reported for G-CSFdriven granulopoiesis.36,37 As a result, most Csf3r+ LTHSCs do not respond to G-CSF. Only approximately 10% of Csf3r+ cells responded to G-CSF and continuously divided more than three times and differentiated (Figure 2A), presumably escaping from the negative regulation by SOCS3. SCF + G-CSF maintained the reconstitution potential of HSC2 in vitro (Figure 3B), supporting a previous study demonstrating that SCF + G-CSF maintains Blymphoid potential in culture.38 Taken together, these data suggested that c-Kit and G-CSFR signaling in ST Ly-bi HSC is regulated differently from that in LT My-bi HSC.
Previously, HSC were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE), their division was followed by CFSE intensity, and the function of CFSElabeled HSC was examined through serial transplantation. This in vivo HSC division tracking study reported that GCSF did not induce self-renewing division of HSC.6 Similarly, the other group, using H2B-GFP label retaining system, also reported that G-CSF did not stimulate LTHSC. 7 Supporting these studies, this study showed that GCSF did not directly act on LT-My-bi HSC in vitro.
Our conclusions of the G-CSF effect may contradict previous studies.3-5,10 The levels of previous HSC purifications were not as high as ours, as they contained a mix of LTMy- bi HSC, ST-Ly-bi HSC, and HPC; serum was used in culture; some experiments lacked an appropriate control; the follow-up period after transplantation was not long enough to distinguish ST-Ly-bi HSC from LT-My-bi HSC by our definition; and experimental conditions differed. These differences may account for the discrepancies.
GM-CSF receptor is composed of two subunits, α and b. The α subunit binds GM-CSF with low affinity, while b*********subunit has no binding capacity by itself but forms a high affinity receptor with α subunit and plays a role in signal transduction.39,40 Our data showed that most HSC and HPC populations did not express Csf2ra, but most HPC1 and half of HSC2, HPC3, and HPC4 expressed Csf2rb. Only HPC2, HPC3 and HPC4 responded to GM-CSF. These data showed that target cells of GM-CSF differ from those of G-CSF, and particular progenitors can receive signals from both G-CSF and GM-CSF.
Cxcl12 is also considered to be a niche factor synthesized by bone marrow stromal cells.9,41,42 Deletion of Cxcl12 from the BM, or its receptor, Cxcr4, from hematopoietic cells reduced HSC in the BM, indicating their roles in HSC retention.9,41-43 In agreement with this, successful mobilization of HSC and HPC from BM into the circulation has been achieved by the use of G-CSF or Cxcr4 antagonists through the disruption of the Cxcr4/Cxcl12 interaction.44,45 In this study, however, we detected little expression of Cxcr4 in HSC (Figure 1C) even after stimulation with SCF, SCF + G-CSF, and SCF + TPO for 7 days (Figure 5). At least some HSC may not express Cxcr4 and be mobilized via Cxcr4-independent mechanism. As Cxcr7 has been reported as a new candidate receptor for Cxcl12.46 Nevertheless, more precise mechanisms of HSC mobilization by G-CSF should be clarified.
Hematopoiesis is a blood formation process depicted as a hierarchy with self-renewing HSC ranking at the apex. In our model (Figure 6), HSC1 cells resides at the top rank, HSC2 and HPC1 cells reside at the second rank, and HPC2, HPC3, and HPC4 cells reside at the third rank. SCF, G-CSF, GM-CSF, and TPO act on different cell types, supporting the concept that cytokines exhibit multiple functions affecting cells at different developmental stages.47 In addition, based on the cytokine responses, two major differentiation pathways from LT-HSCs are suggested: (1) the HSC1-HPC1-HPC2 pathway represents the myeloid differentiation pathway;19 and (2) the HSC1-HSC2- HPC3/4 pathway represents the lymphoid differentiation pathway. LT-HSC differentiated into either ST-HSC or myeloid progenitors like common myeloid progenitors. ST-HSC further give rise to LMPP which lose the response to SCF and TPO (Figure 2B), consistent with the decreased self-renewal ability and megakaryocyte potential in LMPP.48 G-CSF is involved in both the myeloid and lymphoid differentiation pathways, regulating the proliferation of myeloid progenitors while maintaining Ly-bi HSC. In this regard, G-CSF works as a multipotent factor in hematopoiesis. Collectively, this cytokine network model indicates that different cytokines play a role in different differentiation pathways.
This study showed that LT-My-bi HSC do not respond to G-CSF, suggesting that HSC in healthy donors are protected from the proliferation and sequential exhaustion after G-CSF administration. G-CSF enhanced the efficacy of chemotherapy for eliminating leukemia stem cells without affecting the survival of normal HSC in the mouse acute myeloid leukemia (AML) model.49 A large randomized clinical trial of G-CSF in AML patients showed that priming AML cells with G-CSF reduced the rate of relapse and improved disease-free survival without affecting hematologic recovery.50 These studies together with our own suggested that normal HSC are protected from the cytotoxic effect of chemotherapy when G-CSF is injected before chemotherapy.
Supplementary Material
Funding Statement
Funding This work was supported by grants from the National Key Research and Development Program of China Stem Cell and Translational Research (2016YFA0100600, 2017YFA0104900, and 2019YFA0110203), CAMS Initiative for Innovative Medicine (CAMS-I2M) (2016-I2M-1-017 and 2017-I2M-1-015), CAMS Fundamental Research Funds for Central Research Institutes (2019PT320017), and the National Natural Science Foundation of China (81670105, 81970119, 81670106, 81421002).
References
- 1.Till JE, McCulloch EA, Siminovitch L. A stochastic model of stem cell proliferation, based on the growth of spleen colony-forming cells. Proc Natl Acad Sci U S A. 1964; 51:29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bensinger W, Singer J, Appelbaum F, et al. Autologous transplantation with peripheral blood mononuclear cells collected after administration of recombinant granulocyte stimulating factor. Blood. 1993;81(11):3158-3163. [PubMed] [Google Scholar]
- 3.Morrison SJ, Wright DE, Weissman IL. Cyclophosphamide/granulocyte colonystimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc Natl Acad Sci U S A. 1997; 94(5):1908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright DE, Cheshier SH, Wagers AJ, Randall TD, Christensen JL, Weissman IL. Cyclophosphamide/granulocyte colonystimulating factor causes selective mobilization of bone marrow hematopoietic stem cells into the blood after M phase of the cell cycle. Blood. 2001;97(8):2278-2285. [DOI] [PubMed] [Google Scholar]
- 5.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6): 1118-1129. [DOI] [PubMed] [Google Scholar]
- 6.Kovtonyuk LV, Manz MG, Takizawa H. Enhanced thrombopoietin but not G-CSF receptor stimulation induces self-renewing hematopoietic stem cell divisions in vivo. Blood. 2016;127(25):3175-3179. [DOI] [PubMed] [Google Scholar]
- 7.Bernitz JM, Daniel MG, Fstkchyan YS, Moore K. Granulocyte colony-stimulating factor mobilizes dormant hematopoietic stem cells without proliferation in mice. Blood. 2017;129(14):1901-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petit I, Szyper-Kravitz M, Nagler A, et al. GCSF induces stem cell mobilization by decreasing bone marrow SDF-1 and up-regulating CXCR4. Nat Immunol. 2002; 3(7):687-694. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25(6):977-988. [DOI] [PubMed] [Google Scholar]
- 10.Abe T, Masuya M, Ogawa M. An efficient method for single hematopoietic stem cell engraftment in mice based on cell-cycle dormancy of hematopoietic stem cells. Exp Hematol. 2010;38(7):603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller-Sieburg CE, Cho RH, Karlsson L, Huang JF, Sieburg HB. Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood. 2004;103(11):4111-4118. [DOI] [PubMed] [Google Scholar]
- 12.Muller-Sieburg CE, Cho RH, Thoman M, Adkins B, Sieburg HB. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood. 2002;100(4): 1302-1309. [PubMed] [Google Scholar]
- 13.Lieschke GJ, Grail D, Hodgson G, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84(6):1737-1746. [PubMed] [Google Scholar]
- 14.Cohen AM, Zsebo KM, Inoue H, et al. In vivo stimulation of granulopoiesis by recombinant human granulocyte colonystimulating factor. Proc Natl Acad Sci U S A. 1987;84(8):2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ieyasu A, Ishida R, Kimura T, et al. An allrecombinant protein-based culture system specifically identifies hematopoietic stem cell maintenance factors. Stem Cell Reports. 2017;8(3):500-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Dong F, Zhang S, et al. TGF-beta1 negatively regulates the number and function of hematopoietic stem cells. Stem Cell Reports. 2018;11(1):274-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent DG, Copley MR, Benz C, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009; 13(25):6342-6350. [DOI] [PubMed] [Google Scholar]
- 18.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207(6):1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto R, Morita Y, Ooehara J, et al. Clonal analysis unveils self-renewing lineage- restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013; 154(5):1112-1126. [DOI] [PubMed] [Google Scholar]
- 20.Dong F, Bai H, Wang X, et al. Mouse acute leukemia develops independent of selfrenewal and differentiation potentials in hematopoietic stem and progenitor cells. Blood Adv. 2019;3(3):419-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Liu Z, Zhang S, et al. Lineage marker expression on mouse hematopoietic stem cells. Exp Hematol. 2019;76:13-23.e2. [DOI] [PubMed] [Google Scholar]
- 22.Ema H, Morita Y, Suda T. Heterogeneity and hierarchy of hematopoietic stem cells. Exp Hematol. 2014;42(2):74-82.e2. [DOI] [PubMed] [Google Scholar]
- 23.Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol. 2002;2(6):410-416. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Miao T, Sebastian M, et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigeninduced proliferation of B and T cells. Immunity. 2012;37(4):685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133(4):1710-1715. [PubMed] [Google Scholar]
- 26.Pietras EM, Warr MR, Passegue E. Cell cycle regulation in hematopoietic stem cells. J Cell Biol. 2011;195(5):709-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barker JE. Sl/Sld hematopoietic progenitors are deficient in situ. Exp Hematol. 1994;22(2):174-177. [PubMed] [Google Scholar]
- 28.Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335(6185):88-89. [DOI] [PubMed] [Google Scholar]
- 29.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481(7382):457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto A, Takeishi S, Kanie T, et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2011;9(3):262-271. [DOI] [PubMed] [Google Scholar]
- 31.Zou P, Yoshihara H, Hosokawa K, et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 2011;9(3):247-261. [DOI] [PubMed] [Google Scholar]
- 32.Qian H, Buza-Vidas N, Hyland CD, et al. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1(6):671-684. [DOI] [PubMed] [Google Scholar]
- 33.Kimura S, Roberts AW, Metcalf D, Alexander WS. Hematopoietic stem cell deficiencies in mice lacking c-Mpl, the receptor for thrombopoietin. Proc Natl Acad Sci U S A. 1998;95(3):1195-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson AC, Ishida R, Kikuchi M, et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature. 2019;571(7763):117-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi H, Morikawa T, Okinaga A, et al. Environmental optimization enables maintenance of quiescent hematopoietic stem cells ex vivo. Cell Rep. 2019;28(1):145-158.e9. [DOI] [PubMed] [Google Scholar]
- 36.Kimura A, Kinjyo I, Matsumura Y, et al. SOCS3 is a physiological negative regulator for granulopoiesis and granulocyte colonystimulating factor receptor signaling. J Biol Chem. 2004;279(8):6905-6910. [DOI] [PubMed] [Google Scholar]
- 37.Croker BA, Metcalf D, Robb L, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20(2):153-165. [DOI] [PubMed] [Google Scholar]
- 38.Hirayama F, Shih JP, Awgulewitsch A, Warr GW, Clark SC, Ogawa M. Clonal proliferation of murine lymphohemopoietic progenitors in culture. Proc Natl Acad Sci U S A. 1992;89(13):5907-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishinakamura R, Nakayama N, Hirabayashi Y, et al. Mice deficient for the IL-3/GM-CSF/IL-5 beta c receptor exhibit lung pathology and impaired immune response, while beta IL3 receptor-deficient mice are normal. Immunity. 1995;2(3):211-222. [DOI] [PubMed] [Google Scholar]
- 40.Hayashida K, Kitamura T, Gorman DM, Arai K, Yokota T, Miyajima A. Molecular cloning of a second subunit of the receptor for human granulocyte-macrophage colonystimulating factor (GM-CSF): reconstitution of a high-affinity GM-CSF receptor. Proc Natl Acad Sci U S A. 1990;87(24):9655-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495(7440):231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenbaum A, Hsu YM, Day RB, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495(7440):227-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzeng YS, Li H, Kang YL, Chen WC, Cheng WC, Lai DM. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood. 2011;117(2):429-439. [DOI] [PubMed] [Google Scholar]
- 44.Lévesque J-P, Hendy J, Takamatsu Y, Simmons PJ, Bendall LJ. Disruption of the CXCR4/CXCL12 chemotactic interaction during hematopoietic stem cell mobilization induced by GCSF or cyclophosphamide. J Clin Invest. 2003;111(2):187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broxmeyer HE, Orschell CM, Clapp DW, et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med. 2005;201(8):1307-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns JM, Summers BC, Wang Y, et al. A novel chemokine receptor for SDF-1 and ITAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203(9):2201-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa M. Differentiation and proliferation of hematopoietic stem cells. Blood. 1993;81(11):2844-2853. [PubMed] [Google Scholar]
- 48.Adolfsson J, Månsson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic Potential. Cell. 2005;121(2):295-306. [DOI] [PubMed] [Google Scholar]
- 49.Saito Y, Uchida N, Tanaka S, et al. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat Biotechnol. 2010;28(3):275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowenberg B, van Putten W, Theobald M, et al. Effect of priming with granulocyte colony-stimulating factor on the outcome of chemotherapy for acute myeloid leukemia. N Engl J Med. 2003;349(8):743-752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.