Abstract
Breast implant-associated anaplastic large cell lymphoma (BIAALCL) is a recently characterized T-cell malignancy that has raised significant patient safety concerns and led to worldwide impact on the implants used and clinical management of patients undergoing reconstructive or cosmetic breast surgery. Molecular signatures distinguishing BIA-ALCL from other anaplastic large cell lymphomas have not been fully elucidated and classification of BIA-ALCL as a World Health Organization entity remains provisional. We performed RNA sequencing and gene set enrichment analysis comparing BIA-ALCL to non-BIAALCL and identified dramatic upregulation of hypoxia signaling genes including the hypoxia-associated biomarker CA9 (carbonic anyhydrase- 9). Immunohistochemistry validated CA9 expression in all BIA-ALCL, with only minimal expression in non-BIA-ALCL. Growth induction in BIA-ALCL-derived cell lines cultured under hypoxic conditions was proportional to upregulation of CA9 expression, and RNA sequencing demonstrated induction of the same gene signature observed in BIAALCL tissue samples compared to non-BIA-ALCL. CA9 silencing blocked hypoxia-induced BIA-ALCL cell growth and cell cycle-associated gene expression, whereas CA9 overexpression in BIA-ALCL cells promoted growth in a xenograft mouse model. Furthermore, CA9 was secreted into BIA-ALCL cell line supernatants and was markedly elevated in human BIA-ALCL seroma samples. Finally, serum CA9 concentrations in mice bearing BIA-ALCL xenografts were significantly elevated compared to those in control serum. Together, these findings characterize BIA-ALCL as a hypoxia-associated neoplasm, likely attributable to the unique microenvironment in which it arises. These data support classification of BIA-ALCL as a distinct entity and uncover opportunities for investigating hypoxia-related proteins such as CA9 as novel biomarkers and therapeutic targets in this disease.
Introduction
Anaplastic large cell lymphomas (ALCL) are a heterogeneous group of CD30-positive T-cell lymphomas with varying clinical presentation, prognosis, and molecular pathogenesis.1 Breast implant-associated (BIA)-ALCL is a rare form of ALCL arising in association with breast implants placed for reconstructive or cosmetic purposes.2 It typically occurs in the peri-implant capsule and/or effusion an average of 9 years after implant placement. The cytological and immunophenotypic features of BIA-ALCL are similar to those of systemic and primary cutaneous ALK-negative ALCL, including the presence of hallmark cells, CD30 positivity, and frequent loss of T-cell markers such as CD3 and CD5. The prognosis of patients with BIA-ALCL is associated with clinical stage, particularly the presence or absence of a mass-forming lesion and/or locoregional lymph node involvement, which influence the therapeutic approach.3,4 Complete surgical excision of the peri-implant fibrous capsule is essential and sufficient in patients without a mass or lymph node involvement, whereas systemic chemotherapy is recommended in those with advanced disease.3,5 Based on these distinct clinical features, the revised World Health Organization (WHO) classification recognizes BIA-ALCL as a provisional entity.2
The molecular pathogenesis of BIA-ALCL remains incompletely understood. Recent studies have suggested a possible relationship to underlying chronic allergic reaction and bacterial biofilm infection.6,7 Rearrangements of ALK, DUSP22, and TP63 are consistently absent, referred to as the triple-negative (TN) genetic subtype.8,9 Recurrent JAK1 and STAT3 gene mutations have been identified9-12 and, like many other ALCL,13,14 BIA-ALCL shows consistent activation of the JAK-STAT3 pathway as detected by immunohistochemistry for Tyr705-phosphorylated STAT3.9,12,15 In vivo studies have demonstrated that inhibition of JAK-STAT signaling by sunitinib or ruxolitinib effectively suppresses growth of TLBR cell lines derived from BIA-ALCL,15,16 suggesting potential therapeutic utility of these drugs for patients with advanced disease. In addition to mutations affecting the JAK-STAT signaling pathway, gene alterations in epigenetic modifiers are also frequent in BIA-ALCL.17
However, these findings have not identified a molecular profile of BIA-ALCL that is unique to the peri-implant microenvironment in which it originates. Identification of unique molecular features specific for BIA-ALCL could lead to discovery of biomarkers: that improve early detection, diagnosis, and follow-up; identify candidate targets for therapy or preventive strategies; and provide justification to upgrade the WHO classification of BIA-ALCL from a provisional to a definite entity. We therefore interrogated the gene expression profile of BIA-ALCL.
Methods
Gene expression profiling
Human studies were conducted with approval of the Institutional Review Boards at Mayo Clinic and The University of Texas MD Anderson Cancer Center. We performed RNA sequencing on formalin-fixed paraffin-embedded tumor tissue from 11 patients with BIA-ALCL (Table 1). All were female and their mean age was 55 years (range, 44-73 years). All had received textured implants. As described previously,14 RNA from AllPrep extraction was used to prepare sequencing libraries (TruSeq RNA Access, Illumina) and sequenced on a HiSeq 4000 (Illumina). Reads were aligned to hg38 with MAP-RSeq18 modified to use the STAR aligner. 19 Gene-level read counts based on Ensembl version 78 were transformed into reads per kilobase per million mapped reads (RPKM). Gene expression data were compared to those of 24 previously sequenced non-BIA-ALCL of TN genetic subtype (10 primary cutaneous ALCL and 14 systemic ALK-negative ALCL14). Gene set enrichment analysis (GSEA) was performed using GSEA software (Broad Institute) as described previously.14
Immunohistochemistry
Immunohistochemistry for CA9 was carried out on formalinfixed paraffin-embedded sections of 17 BIA-ALCL and 48 non- BIA-ALCL (from patients with a mean age of 54 years). The WHO subtypes of these latter were primary cutaneous ALCL (n=13), ALK-negative ALCL (n=24), and ALK-positive ALCL (n=11). Genetic subtypes included 11 ALK-positive, ten with DUSP22 rearrangements, two with TP63 rearrangements, and 25 TN. Deparaffinized tissue sections were heated in pH 6.0 citric acid buffer in a steam cooker for 30 min. After incubation with 3% hydrogen peroxide for 10 min and 5% bovine serum albumin for 10 min, the slides were incubated with anti-CA9 rabbit monoclonal antibody (1:100 dilution, clone D47G3; Cell Signaling Technology) at 4°C overnight. Sections were then incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody Biocare Medical), developed with 3,3’-diaminobenzidine, and counterstained with hematoxylin. Stains were scored in a blind fashion by two hematopathologists (NO and ALF). For CA9, scoring was based on percentage of tumor cells with membranous staining.
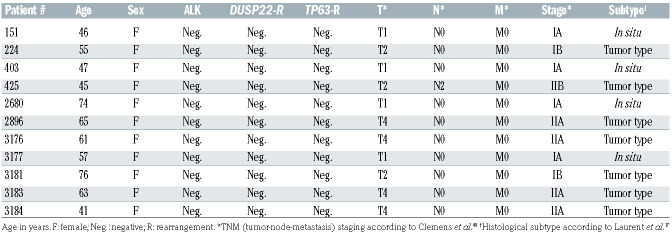
Table 1.
Clinical and pathological features of 11 patients with breast implant-associated anaplastic large cell lymphoma.
BIA-ALCL xenograft models
Studies were approved by the Mayo Clinic Institutional Animal Care and Use Committee (IACUC) under protocol A00002776. Five-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from The Jackson Laboratory and maintained under standard laboratory conditions. Cell lines TLBR-1, TLBR-2, and TLBR-3 were established from BIA-ALCL by one of the authors (ALE).15,20 and were maintained in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum (Clontech), 1% penicillin/ streptomycin (Gibco), and 100 U/mL interleukin-2 (R&D 202-IL-050). TLBR-1, -2, and -3 cells were cultured at a concentration of 0.5 x 106/mL for 72 h and resuspended in phosphatebuffered saline. Mice were injected subcutaneously in the right flank with TLBR-1, -2, or -3 cells. Tumor volumes were calculated in mm3 using the formula l2×L/2, where l and L represent the shortest and longest dimensions, respectively.
Additional methods are described in the Online Supplementary Material.
Results
Increased expression of hypoxia signaling pathway genes is a hallmark of BIA-ALCL
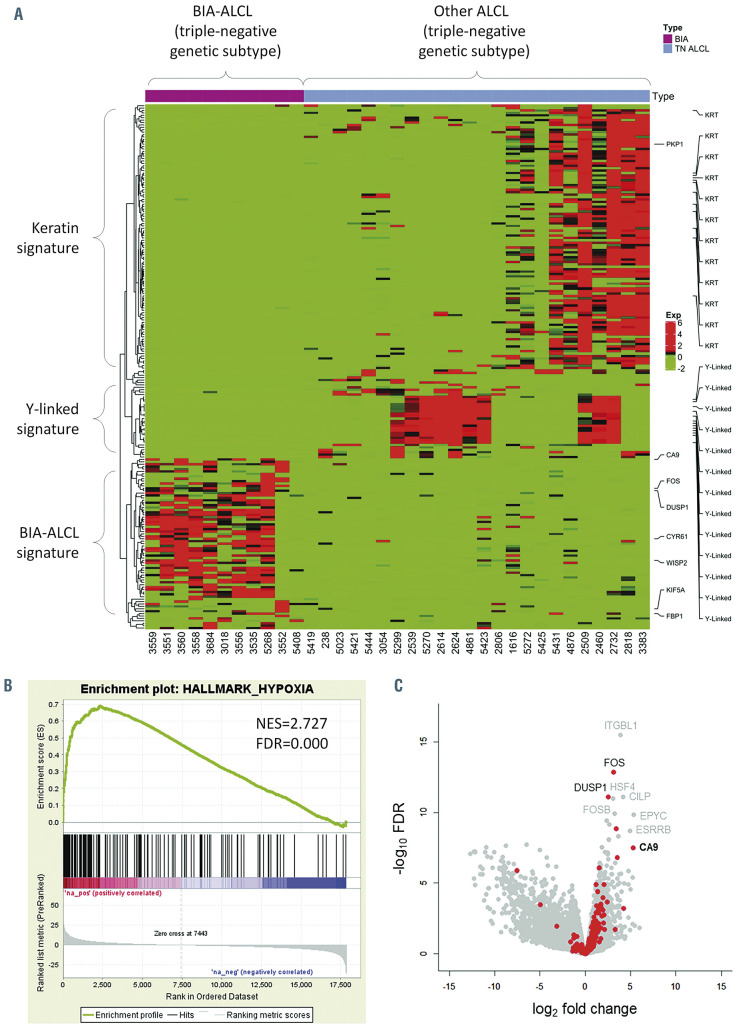
We performed RNA sequencing to identify gene expression signatures that might distinguish BIA-ALCL from other types of ALCL. Since BIA-ALCL are consistently of TN genetic subtype,9 we compared BIA-ALCL to other TN ALCL to avoid bias from the distinct expression profiles of other genetic subtypes14 (Figure 1A). A distinct cluster of genes was upregulated in BIA-ALCL, which formed the basis for subsequent analyses. Two clusters of genes were expressed only in non-BIA cases: one was enriched for keratin genes and consisted of biopsies at epithelial sites (skin and tongue) and the other contained Y-linked genes and represented male patients.
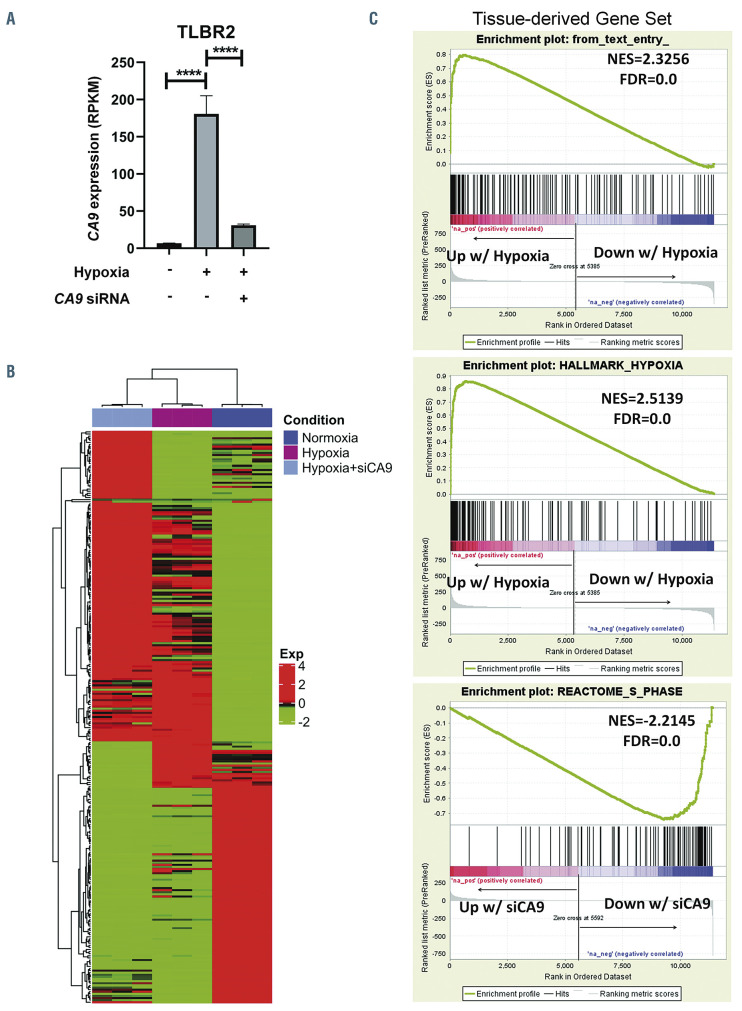
We then performed GSEA to identify candidate molecular signatures for genes upregulated in BIA-ALCL (Figure 1B, Online Supplementary Table S1). We focused on the second highest ranking gene set, HALLMARK HYPOXIA (normalized enrichment score [NES], 2.727; false discovery rate q-value [FDR], 0.000), as a candidate molecular feature distinctive of BIA-ALCL. The highest ranking gene set, HALLMARK EPITHELIAL MESENCHYMAL TRANSITION (NES, 2.963; FDR, 0.000) and other subsequent pathways mostly related to collagen formation and extracellular matrix organization, likely reflecting stromal components in the fibrous capsule surrounding the breast implant and seroma in BIA-ALCL samples.21 Supporting these GSEA results, examination of differential expression of genes and absolute RPKM values between BIA-ALCL and non-BIA-ALCL revealed significantly higher expression levels of downstream target genes of the hypoxia signaling pathway such as VEGFA, VEGFB, SLC2A3 (encoding GLUT3), and CA9 (carbonic anhydrase-9; RPKM, mean ± standard deviation: 16.5 ± 20.2 vs. 0.4 ± 0.7; P<0.001, t-test) (Online Supplementary Figure S1). Among genes associated with hypoxia, CA9 showed the highest fold-change between BIA-ALCL and non-BIA-ALCL (FC=5.296, FDR, 3.07x10-8) (Figure 1C). Collectively, these results suggest that increased expression of hypoxia signaling pathway genes is a transcriptional hallmark of BIAALCL. We did not identify a significant difference in CA9 expression between in situ and tumor-type BIA-ALCL as described by Laurent et al.17 (Table 1), or distinct gene signatures associated with clinical stage; associations between gene expression and clinicopathological features should be evaluated in future, larger studies.
CA9 protein is consistently expressed in BIA-ALCL but not in other ALCL
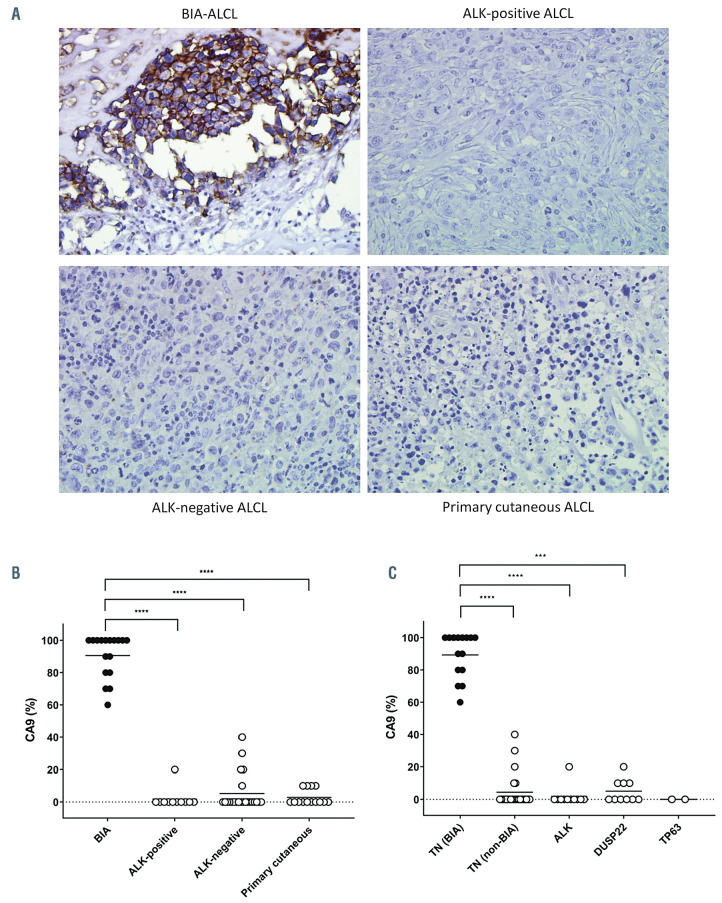
CA9 is a well-established biomarker of hypoxia and tumoral expression of CA9 is widely used in the histopathological diagnosis of hypoxia-related cancers.22 Therefore, having identified a hypoxia-associated signature and high CA9 mRNA expression in BIA-ALCL, we performed immunohistochemistry to investigate CA9 protein expression in BIA-ALCL and non-BIA-ALCL. CA9 was expressed on the cell membrane of BIA-ALCL cells (% positive staining, mean ± standard deviation, 91 ± 15%) but not in admixed inflammatory cells, validating the RNA sequencing data at the protein level (Figure 2A). A relatively narrow range of protein scores was observed by immunohistochemistry, compared to a wide range of CA9 gene expression values. The correlation between the two was not statistically significant, likely due to gene expression values reflecting contributions from non-neoplastic cells whereas immunohistochemistry was scored only in the tumor cells. Conversely, CA9 was mostly negative in non-BIA-ALCL (ALK-positive, 2 ± 6%; ALK-negative, 5 ± 11%; cutaneous, 3 ± 5%; P<0.0001, Dunn multiple comparison test) (Figure 2B). We also stratified CA9 protein expression by genetic subtype of ALCL (Figure 2C). BIA-ALCL (all TN) showed significantly more CA9 expression than ALK-positive ALCL, DUSP22-rearranged ALCL, and TN non-BIA-ALCL, suggesting that high CA9 expression in BIA-ALCL is attributable to BIA presentation rather than TN genetic subtype. Taken together, these data indicate that CA9 is specifically expressed in BIAALCL at the mRNA and protein levels.
Hypoxia-induced CA9 expression drives growth of BIA-ALCL cells
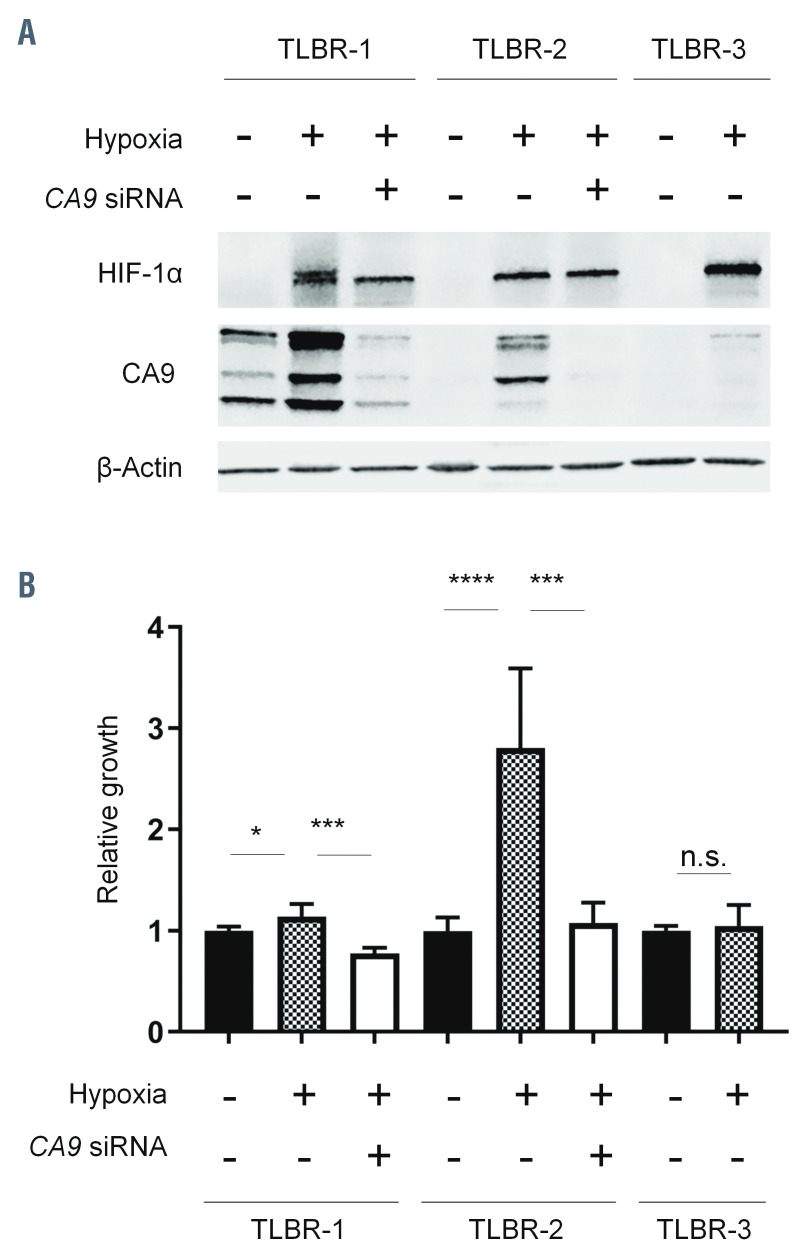
We next examined CA9 expression in BIA-ALCL cell lines under normoxic and hypoxic conditions. Expression of HIF-1α was evaluated to confirm the response to hypoxia. Western blotting of TLBR-1, -2, and -3 cells cultured under normoxic or hypoxic conditions revealed distinct patterns of CA9 expression in each cell line, providing unique models for further study (Figure 3A). In TLBR- 1, CA9 was expressed under baseline normoxic conditions, suggesting constitutive expression of the hypoxic program. CA9 expression was further induced by hypoxia. In TLBR-2, CA9 was absent under normoxic conditions but was induced under hypoxic conditions, consistent with a canonical hypoxia response. In contrast, CA9 expression in TLBR-3 was absent under normoxic conditions and only minimally induced by hypoxia.
To explore the functional significance of these distinct patterns of CA9 expression, we evaluated the effects of hypoxia with or without siRNA-mediated silencing of CA9 on BIA-ALCL cell growth (Figure 3B). In TLBR-1, which showed evidence of a constitutive hypoxia program under normoxic conditions, hypoxia induced only a slight increase in growth (14 ± 13% above normoxic baseline, P<0.05, Mann-Whitney test). Silencing CA9 inhibited this growth to 77 ± 6% of normoxic baseline (P<0.001). In TLBR-2, a proposed model of canonical hypoxia response, hypoxia substantially increased growth by 281 ± 79% (P<0.0001) and this increase was reversed nearly to normoxic baseline by CA9 silencing (P<0.001 vs. control siRNA). In TLBR-3, which was resistant to hypoxiainduced CA9 expression, no significant increase in growth was induced by hypoxia. In summary, these data indicate that hypoxia-induced growth in BIA-ALCL cell lines follows a pattern similar to that of hypoxia-induced expression of CA9 and that CA9 drives BIA-ALCL cell growth under hypoxic conditions.
Figure 1.
Breast implant-associated anaplastic large cell lymphomas show upregulation of hypoxia-associated genes. (A) Heatmap of genes differentially expressed between breast implant-associated (BIA) anaplastic large cell lymphomas (ALCL) and other ALCL of triple-negative (TN) genetic subtype (lacking rearrangements of ALK, DUSP22 and TP63). A distinct signature of genes overexpressed in BIA-ALCL is seen. Signatures in subgroups of non-BIA ALCL include a keratin (KRT) signature seen in biopsies at epithelial sites (skin and tongue) and a Y-linked signature seen in male patients. See also Online Supplementary Figure S1. (B) Gene set enrichment analysis shows upregulation of genes associated with hypoxia in BIA-ALCL. See also Online Supplementary Table S2. NES: normalized enrichment score; FDR: false discovery rate. (C) A volcano plot indicating differentially expressed genes between BIA-ALCL and other TN ALCL. Hypoxia-related genes from HALLMARK HYPOXIA and SEMENZA HIF1 TARGETS gene sets are indicated in red.
Figure 2.
Breast implant-associated anaplastic large cell lymphomas consistently express CA9. (A) Representative microscopic images of immunohistochemistry for CA9 in breast implant-associated (BIA) anaplastic large cell lymphoma (ALCL), systemic ALK-positive ALCL, systemic ALK-negative ALCL, and primary cutaneous ALCL (40× original magnification). (B) BIA-ALCL show significantly higher CA9 expression than other forms of ALCL. (C) The increased expression of CA9 in BIA-ALCL is independent of genetic subtype. All BIA-ALCL tested have triple-negative (TN) genetics (lacking rearrangements of ALK, DUSP22, and TP63). BIA-ALCL show significantly higher CA9 expression than ALCL with any of these rearrangements, as well as TN non-BIA-ALCL. ***P<0.001; ****P<0.0001.
Hypoxia and CA9 expression drive unique gene signatures in BIA-ALCL cells
We examined the effects of hypoxia and CA9 knockdown on gene expression in BIA-ALCL cells by performing RNA sequencing in TLBR-2 cells, which showed evidence of a canonical hypoxia response in the preceding experiments. As anticipated, CA9 mRNA was markedly upregulated under hypoxic conditions and effectively downregulated by CA9 siRNA (both, P<0.0001) (Figure 4A). A heatmap of genes whose expression varied significantly showed clusters of genes with unique expression patterns as well as clusters of genes with expression patterns shared between two of the three conditions (Figure 4B). We used GSEA to explore these findings further (Figure 4C). Notably, the set of genes overexpressed in BIA-ALCL tissue samples as compared with TN non-BIAALCL (Figure 1A) was markedly enriched in TLBR-2 cells cultured under hypoxic conditions (NES=2.325; FDR=0.000), providing in vitro validation of the tissuebased finding that BIA-ALCL are characterized by a hypoxia signature. Furthermore, the HALLMARK HYPOXIA gene set identified in BIA-ALCL versus non- BIA-ALCL tissue samples (Figure 1B) was also significantly enriched in hypoxic TLBR-2 cells (NES=2.151, FDR=0.000), among other gene sets related to metabolic pathways such as HALLMARK GLYCOLYSIS, REACTOME METABOLISM OF CARBOHYDRATES, and REACTOME GLUCOSE METABOLISM (Online Supplementary Table S2). In contrast, siRNA-mediated CA9 knockdown was associated with significant depletion of cell cycle pathways, including REACTOME S PHASE (NES=-2.214; FDR=0.000) as well as MYC target gene sets and multiple other cell cycle-associated gene sets (Online Supplementary Table S3). These findings corroborate previous data showing that CA9 inhibition induced cell cycle arrest in glioblastoma cells, and specifically a marked reduction of cells in S phase.23 We also performed an exploratory analysis comparing RNA sequencing data in TLBR cell lines. Although only TLBR-1 significantly expressed CA9 at baseline, other hypoxia-related genes were relatively overexpressed in TLBR-2 or TLBR-3 (Online Supplementary Figure S5), suggesting heterogeneity that merits investigation in larger future studies.
CA9 overexpression drives BIA-ALCL growth in a xenograft model
We evaluated the effects of CA9 on BIA-ALCL cell growth further by using a lentiviral system to overexpress CA9 in TLBR-3 cells, which lack both baseline and hypoxia- inducible CA9 expression (Figure 3A). Corroborating CA9 siRNA data from TLBR-1 and -2 cells, CA9 overexpression in TLBR-3 augmented cell growth in vitro (Online Supplementary Figure S2). We then examined the effect of CA9 overexpression in a xenograft model. The median time after inoculation to establishment of palpable subcutaneous tumors was 17 days in the CA9 group and 26 days in the control (empty vector-transduced) group (P=0.004, log-rank test) (Figure 5A). At 38 days, when the first animal required euthanasia because of tumor size, tumors were 1,764 ± 1,526 mm3 in the CA9 group and 126 ± 130 mm3 in the control group (Figure 5B); differences in tumor growth were highly significant (P<0.0001, twoway repeated measure analysis of variance with the Geisser-Greenhouse correction). Using this protocoldefined euthanasia endpoint, median overall survival was 47 days in the CA9 group and 76 days in the control group (P=0.0008, log-rank test) (Figure 5C). Thus, CA9 accelerates tumor growth in the TLBR-3 BIA-ALCL xenograft model.
Secreted CA9 in BIA-ALCL cell line models and patients’ samples
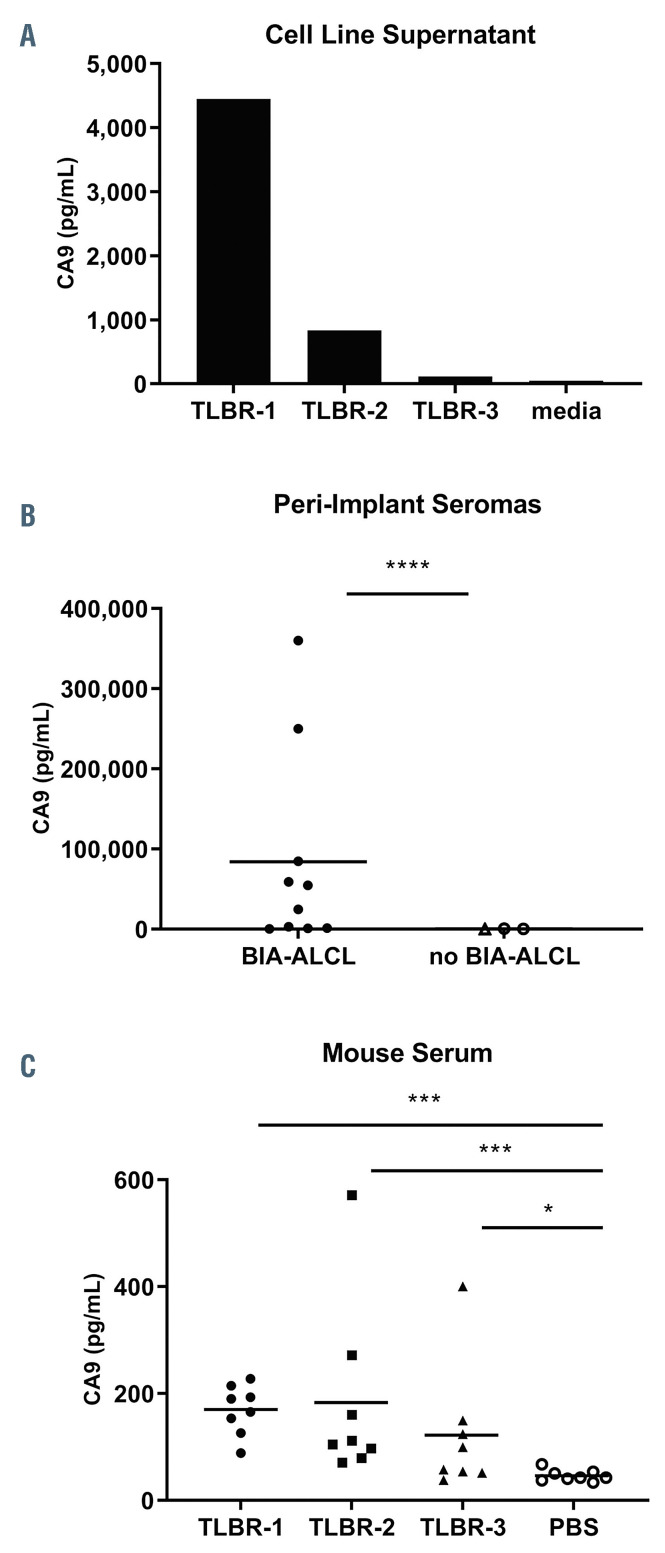
Secreted CA9 has been proposed as a biomarker for CA9-expressing malignancies.22,24-26We therefore evaluated secretion of CA9 into the supernatants of BIA-ALCL cell lines. Secreted CA9 could be detected in culture supernatants of TLBR-1, -2, and -3 cells at concentrations mirroring their cellular expression levels (Figure 6A). Since BIA-ALCL cells can secrete CA9, we next examined whether CA9 could be detected in peri-implant seroma specimens involved by BIA-ALCL. Indeed, all ten BIAALCL seroma specimens evaluated contained detectable CA9, with a mean concentration of 84,046 ± 118,695 pg/mL (range, 423-360,262 pg/mL) (Figure 6B). In contrast, seromas lacking involvement by BIA-ALCL showed a mean concentration of 502 ± 390 pg/mL (range, 9-887 pg/mL; P<0.0001, Mann-Whitney test).
Figure 3.
CA9 expression and growth of breast implant-associated anaplastic large cell lymphoma cell lines. (A) TLBR-1, -2, and -3 breast implant-associated (BIA) anaplastic large cell lymphoma (ALCL) cell lines show distinct patterns of CA9 expression under normoxic and hypoxic conditions. HIF-1α serves as a positive control for hypoxia. TLBR-1 shows constitutive CA9 expression under normoxic conditions, which is further enhanced by hypoxia. TLBR-2 lacks constitutive CA9 expression but CA9 is induced by hypoxia (canonical hypoxia response). TLBR-3 shows minimal hypoxia-induced CA9 expression. The effects of siRNAmediated CA9 silencing are shown. Representative data from three independent experiments. (B) Growth of TLBR-1, -2, and -3 cells mirrors CA9 expression. TLBR-1 cells, which constitutively express CA9, show only slight growth induction by hypoxia. Growth is inhibited by CA9 silencing. TLBR-2 cells, which show a canonical hypoxia response, have marked hypoxia-induced growth which is almost completely reversed by CA9 silencing. Hypoxia does not induce either growth or CA9 expression in TLBR-3 cells. *P<0.05; ***P<0.001; ****P<0.0001; n.s., not statistically significant.
Figure 4.
Gene signatures associated with hypoxia and CA9 expression in breast implant-associated anaplastic large cell lymphoma cells. (A) CA9 mRNA expression is induced by hypoxia and inhibited by CA9 siRNA in TLBR-2 breast implant-associated (BIA) anaplastic large cell lymphoma (ALCL) cells. RPKM: reads per kilobase per million mapped reads; ****P<0.0001. (B) Heatmap showing relative gene expression in TLBR-2 for each of the three conditions shown in panel A. RNA sequencing was performed in triplicate for each condition. (C) Top panel: TLBR-2 cells cultured under hypoxic conditions show significant enrichment for the set of genes overexpressed in BIA- versus non-BIA-ALCL tissue samples (“tissue-derived gene set,” defined as log2FC >1 and FDR ≤0.05; cf. Figure 1A). Middle panel: the HALLMARK HYPOXIA gene set identified by gene set enrichment analysis in BIA- versus non-BIA-ALCL tissue samples is also significantly enriched in hypoxic TLBR- 2 cells. See also Online Supplementary Table S2. Bottom panel: hypoxic TLBR-2 cells treated with CA9 siRNA show marked depletion of REACTOME S PHASE and other gene sets related to cell cycle and MYC targets. See also Online Supplementary Table S3. FC: fold change; NES: normalized enrichment score; FDR: false discovery rate.
Four serum and/or plasma samples from BIA-ALCL were available to evaluate CA9 concentrations (Online Supplementary Figure S3). While the CA9 concentration in normal human serum or plasma is <25 pg/mL,24,26 the plasma CA9 concentration was 128 pg/mL in one BIA-ALCL patient. Because the number of human blood samples available for testing was limited, we examined whether CA9 secreted from BIA-ALCL cells might be detectable in serum samples using mouse xenograft models. We harvested subcutaneous TLBR-1, -2, and -3 tumors when each tumor reached a volume of 1,000 mm3 and obtained simultaneous serum samples. Tumor lysate CA9 concentrations from TLBR-1, -2, and -3 were 108,175±39,252 pg/mL, 231,070 ± 88,185 pg/mL, and 6,903 ± 1,871 pg/mL, respectively, based on standardized total protein concentrations of 1 mg/mL; all pairwise comparisons showed significant differences (Online Supplementary Figure S4). A similar pattern of serum CA9 concentrations was observed, with mean values for TLBR-1, -2, and -3 tumorbearing mice of 170 ± 46 pg/mL, 183 ± 170 pg/mL, and 122 ± 119 pg/mL; values in all groups were significantly higher than CA9 concentrations in serum obtained from non-tumor-bearing mice (46 ± 11 pg/mL) (Figure 6C). Taken together, these findings indicate that CA9 can be secreted from BIA-ALCL cells and is detectable in periimplant seroma fluid involved by BIA-ALCL. Serum CA9 concentrations are elevated in sera from BIA-ALCL xenograft-bearing mice and serum levels in BIA-ALCL patients should be evaluated in larger cohorts.
Discussion
In this gene expression profiling study comparing BIAALCL to their non-BIA counterparts, we found that BIAALCL demonstrate a hypoxia signature, likely attributable to the unique microenvironment in which they arise. Notably, the carbonic anhydrase CA9 was expressed consistently in BIA-ALCL and only minimally in non-BIAALCL. CA9 promoted hypoxia-induced growth in BIAALCL cell lines in vitro and in mouse xenograft models. In addition, CA9 was significantly elevated in human seroma samples involved by BIA-ALCL and in serum from BIAALCL xenograft-bearing mice. These findings identify unique biological features of BIA-ALCL, support its classification as a WHO entity distinct from other forms of ALCL, and uncover opportunities to explore hypoxia-related proteins and pathways in novel diagnostic, preventive, or therapeutic strategies for patients with this disease.
RNA sequencing with transcriptomic analysis and GSEA revealed enhanced expression of hypoxia signaling pathway genes as a hallmark of BIA-ALCL. A recent study by Di Napoli et al. also compared the transcriptome of BIA-ALCL to that of other peripheral T-cell lymphomas including non-ALCL.27 The authors identified a number of notable findings, including upregulation of genes involved in cell motility (e.g., CCR6, MET, and HGF), myeloid cell differentiation (e.g., PPARG and JAK2), and viral gene transcription (e.g., RPS10), and downregulation of T-cell receptor signaling genes. Differentially expressed genes reported by Di Napoli et al. tended to show similar changes in our dataset (Online Supplementary Figure S6). However, the gene ontology analysis of Di Napoli et al. compared BIA-ALCL to non-neoplastic T cells, whereas our study was designed to identify differences between BIA-ALCL and TN ALCL arising at other anatomic sites. Therefore, these two studies are complementary and emphasize different aspects of BIA-ALCL pathogenesis for further study. Of note, occasional non-BIA-ALCL (especially with TN genetics) showed a moderate degree of CA9 expression, highlighting the need for additional future study of hypoxia-associated pathways in T-cell neoplasms other than BIA-ALCL.28
Figure 5.
CA9 accelerates breast implant-associated anaplastic large cell lymphoma growth in a mouse xenograft model. (A) Mice inoculated with TLBR-3 breast implant-associated anaplastic large cell lymphoma cells stably transduced with a CA9 lentiviral vector develop palpable tumors faster than mice inoculated with cells transduced with vector control. (B) Mice inoculated with TLBR-3 cells overexpressing CA9 develop larger tumors than mice inoculated with control-transduced TLBR-3 cells. (C) Overall survival is shorter in mice bearing CA9-overexpressing TLBR-3 tumors than in those bearing control-transduced tumors.
BIA-ALCL arises in a unique tumor microenvironment consisting of the breast prosthesis, seroma fluid, and surrounding fibrous capsule. Local hypoxia is a well-established factor promoting the development of tissue fibrosis, 29,30 and tissues with artificial prostheses are postulated to be hypoxic.31,32 For example, Kim et al. showed that the thickness of the fibrous capsule around silicone implants in rats was reduced by stem cell-derived endothelial precursor cell conditioned medium, which promotes wound healing at least in part by reducing tissue ischemia, suggesting the peri-implant microenvironment is hypoxic even in the non-neoplastic setting.33 The ability to tolerate low oxygen tension may be critical for pre-neoplastic cells situated between the prosthesis and peri-implant fibrous capsule to survive and proliferate in the early stages of BIAALCL lymphomagenesis. Since most patients with implants do not develop BIA-ALCL, however, future studies should examine possible interplay between hypoxia and recurrent genetic events reported in this disease, such as mutations in JAK-STAT and epigenetic modifier genes.17 Furthermore, it would be of interest to compare the molecular signature of BIA-ALCL with that of other effusionassociated malignancies such as primary effusion lymphomas of B-cell origin, in which targetable hypoxic metabolic pathways have been reported previously.34,35
Among genes within the hypoxic signature, we identified CA9 as being most robustly overexpressed in BIAALCL, a finding we validated at the protein level by immunohistochemistry. CA9 is a hypoxia-inducible enzyme that catalyzes reversible hydration of carbon dioxide to bicarbonate ions and protons.22 CA9 is expressed in a variety of solid cancers and has been associated with poor prognosis.36-38 Overexpression of CA9 represents an adaptive response to hypoxia by which cancer cells control intracellular and extracellular pH, facilitating survival and growth in an acidic tumor microenvironment.22,39-44 Our data on silencing CA9 expression in hypoxia-inducible TLBR-2 cells and overexpressing CA9 in hypoxia-insensitive TLBR-3 cells indicate that CA9 promotes growth of BIA-ALCL cells. Although CA9 inhibitors have been developed, 23 the direct therapeutic implications of our findings for BIA-ALCL are unclear since disease limited to the hypoxic seroma and surrounding capsule is adequately managed by surgery alone in most cases.3We did not have adequate tissue material from disseminated BIA-ALCL to evaluate whether CA9 expression is retained outside its native microenvironment. Nevertheless, understanding the role of CA9 and other hypoxic signaling pathways in early BIA-ALCL could lead to less invasive strategies to manage localized disease and/or novel prosthetic approaches that decrease the risk of its development.
Our findings also suggest that CA9 could be useful as a biomarker for screening, detection, and/or follow-up of BIA-ALCL. CA9 expression in normal human tissues is limited to gastric, colonic, and gallbladder epithelium.22 Clear cell renal cell carcinoma is a prototypic malignancy expressing high CA9, in which recurrent VHL mutations lead to increased expression of hypoxia-associated genes including CA9; accordingly, CA9 is a widely-used immunohistochemical marker to distinguish clear cell renal cell carcinoma from other renal tumors.45 In addition, serum CA9 levels are associated with tumor size, grade, and metastatic status of clear cell renal cell carcinoma and high preoperative levels are associated with postoperative recurrence.26 We demonstrated that CA9 is readily secreted by BIA-ALCL cells in vitro and that significantly elevated CA9 concentrations are present in peri-implant seromas involved by BIA-ALCL. Various inflammatory conditions can cause peri-implant seromas,46,47 and the diagnosis of BIA-ALCL in seroma fluid can be a significant challenge if neoplastic cells are rare. Hanson et al. recently reported specificity of an enzyme-linked immunosorbent assay for CD30 in seroma specimens involved by BIA-ALCL,48 while Kadin et al. reported that BIA-ALCL cell lines secrete a unique cytokine profile that includes interleukin-13.7 A multi-analyte approach incorporating CA9 that evaluates these proteins in seroma fluid could greatly facilitate the diagnosis of BIA-ALCL when few atypical cells are present and could potentially guide the decision regarding implant removal in suspicious cases in which definite neoplastic cells cannot be identified. This prospect should encourage international collaboration and standardized seroma collection protocols to facilitate progress given the rarity of BIA-ALCL. The role of serum CA9 measurement in BIA-ALCL remains unclear because limited samples were available for analysis. However, we identified elevated CA9 concentrations in serum samples from BIA-ALCL xenograft-bearing mice, and the role of serum CA9 as a possible biomarker to predict or monitor disease activity should be evaluated in larger human studies.
Figure 6.
CA9 as a candidate biomarker in breast implant-associated anaplastic large cell lymphoma. (A) TLBR-1, -2, and -3 cell lines secrete CA9 into culture supernatant proportionally to their cellular expression, as determined by western blot (cf. Figure 3A). Data represent three replicates measured by CA9 enzyme-linked immunosorbent assay. (C) Peri-implant seroma samples involved by breast implant-associated (BIA) anaplastic large cell lymphoma (ALCL) have significantly higher CA9 concentrations than those not involved by BIA-ALCL. (C) Serum samples obtained from mice bearing 1000 mm3 subcutaneous TLBR-1, - 2, and -3 tumors have significantly higher CA9 concentrations than those from non-tumor-bearing mice. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001 (Mann-Whitney test). PBS: phosphate-buffered saline.
Supplementary Material
Funding Statement
Funding This work was supported by Award Numbers R01 CA177734 (ALF), P30 CA15083 (Mayo Clinic Cancer Center), P50 CA97274 (University of Iowa/Mayo Clinic Lymphoma SPORE), and UL1 TR002377 (Mayo Clinic Center for Clinical and Translational Science) from the National Institutes of Health; by Award Number CI-48-09 from the Damon Runyon Cancer Research Foundation (ALF); by a grant from the Plastic Surgery Foundation, American Society of Plastic Surgeons (MD Anderson Cancer Center); and by the Department of Laboratory Medicine and Pathology and the Center for Individualized Medicine, Mayo Clinic.
References
- 1.Xing X, Feldman AL. Anaplastic large cell lymphomas: ALK positive, ALK negative, and primary cutaneous. Adv Anat Pathol. 2015;22(1):29-49. [DOI] [PubMed] [Google Scholar]
- 2.Feldman AL, Harris NL, Stein H, et al. Breast implant-associated anaplastic large cell lymphoma. Swerdlow SH, Campo E, Harris NL, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th ed. Lyon: International Agency for Research on Cancer, 2017:421-422. [Google Scholar]
- 3.Miranda RN, Aladily TN, Prince HM, et al. Breast implant-associated anaplastic largecell lymphoma: long-term follow-up of 60 patients. J Clin Oncol. 2014;32(2):114-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrufino-Schmidt MC, Medeiros LJ, Liu H, et al. Clinicopathologic features and prognostic impact of lymph node involvement in patients with breast implant-associated anaplastic large cell lymphoma. Am J Surg Pathol. 2018;42(3):293-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemens MW, Medeiros LJ, Butler CE, et al. Complete surgical excision is essential for the management of patients with breast implant-associated anaplastic large-cell lymphoma. J Clin Oncol. 2016;34(2):160-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu H, Johani K, Almatroudi A, et al. Bacterial biofilm infection detected in breast implant-associated anaplastic large-cell lymphoma. Plast Reconstr Surg. 2016;137(6): 1659-1669. [DOI] [PubMed] [Google Scholar]
- 7.Kadin ME, Morgan J, Xu H, et al. IL-13 is produced by tumor cells in breast implantassociated anaplastic large cell lymphoma: implications for pathogenesis. Hum Pathol. 2018;78:54-62. [DOI] [PubMed] [Google Scholar]
- 8.Laurent C, Delas A, Gaulard P, et al. Breast implant-associated anaplastic large cell lymphoma: two distinct clinicopathological variants with different outcomes. Ann Oncol. 2016;27(2):306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oishi N, Brody GS, Ketterling RP, et al. Genetic subtyping of breast implant-associated anaplastic large cell lymphoma. Blood. 2018;132(5):544-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blombery P, Thompson E, Ryland GL, et al. Frequent activating STAT3 mutations and novel recurrent genomic abnormalities detected in breast implant-associated anaplastic large cell lymphoma. Oncotarget. 2018;9(90):36126-36136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Napoli A, Jain P, Duranti E, et al. Targeted next generation sequencing of breast implant-associated anaplastic large cell lymphoma reveals mutations in JAK/STAT signalling pathway genes, TP53 and DNMT3A. Br J Haematol. 2018;180(5):741-744. [DOI] [PubMed] [Google Scholar]
- 12.Letourneau A, Maerevoet M, Milowich D, et al. Dual JAK1 and STAT3 mutations in a breast implant-associated anaplastic large cell lymphoma. Virchows Arch. 2018;473(4): 505-511. [DOI] [PubMed] [Google Scholar]
- 13.Crescenzo R, Abate F, Lasorsa E, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27(4):516-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luchtel RA, Dasari S, Oishi N, et al. Molecular profiling reveals immunogenic cues in anaplastic large cell lymphomas with DUSP22 rearrangements. Blood. 2018;132(13):1386-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lechner MG, Megiel C, Church CH, et al. Survival signals and targets for therapy in breast implant-associated ALK- anaplastic large cell lymphoma. Clin Cancer Res. 2012;18(17):4549-4559. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Zhang Y, Petrus MN, et al. Cytokine receptor signaling is required for the survival of ALK- anaplastic large cell lymphoma, even in the presence of JAK1/STAT3 mutations. Proc Natl Acad Sci U S A. 2017;114(15):3975-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurent C, Nicolae A, Laurent C, et al. Gene alterations in epigenetic modifiers and JAKSTAT signaling are frequent in breast implant-associated ALCL. Blood. 2020;135(5):360-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalari KR, Nair AA, Bhavsar JD, et al. MAPRSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics. 2014;15: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lechner MG, Lade S, Liebertz DJ, et al. Breast implant-associated, ALK-negative, T-cell, anaplastic, large-cell lymphoma: establishment and characterization of a model cell line (TLBR-1) for this newly emerging clinical entity. Cancer. 2011;117(7):1478-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aladily TN, Medeiros LJ, Amin MB, et al. Anaplastic large cell lymphoma associated with breast implants: a report of 13 cases. Am J Surg Pathol. 2012;36(7):1000-1008. [DOI] [PubMed] [Google Scholar]
- 22.Pastorek J, Pastorekova S. Hypoxia-induced carbonic anhydrase IX as a target for cancer therapy: from biology to clinical use. Semin Cancer Biol. 2015;31:52-64. [DOI] [PubMed] [Google Scholar]
- 23.Boyd NH, Walker K, Fried J, et al. Addition of carbonic anhydrase 9 inhibitor SLC-0111 to temozolomide treatment delays glioblastoma growth in vivo. JCI Insight. 2017;2(24):e92928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zavada J, Zavadova Z, Zat'ovicova M, Hyrsl L, Kawaciuk I. Soluble form of carbonic anhydrase IX (CA IX) in the serum and urine of renal carcinoma patients. Br J Cancer. 2003;89(6):1067-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith AD, Truong M, Bristow R, Yip P, Milosevic MF, Joshua AM. The utility of serum CA9 for prognostication in prostate cancer. Anticancer Res. 2016;36(9):4489-4492. [DOI] [PubMed] [Google Scholar]
- 26.Li G, Feng G, Gentil-Perret A, Genin C, Tostain J. Serum carbonic anhydrase 9 level is associated with postoperative recurrence of conventional renal cell cancer. J Urol. 2008;180(2):510-513; discussion 513-514. [DOI] [PubMed] [Google Scholar]
- 27.Di Napoli A, De Cecco L, Piccaluga PP, et al. Transcriptional analysis distinguishes breast implant-associated anaplastic large cell lymphoma from other peripheral T-cell lymphomas. Mod Pathol. 2019;32(2):216-230. [DOI] [PubMed] [Google Scholar]
- 28.Nasu K, Yamaguchi K, Takanashi T, et al. Crucial role of carbonic anhydrase IX in tumorigenicity of xenotransplanted adult Tcell leukemia-derived cells. Cancer Sci. 2017;108(3):435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins DF, Kimura K, Iwano M, Haase VH. Hypoxia-inducible factor signaling in the development of tissue fibrosis. Cell Cycle. 2008;7(9):1128-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lokmic Z, Musyoka J, Hewitson TD, Darby IA. Hypoxia and hypoxia signaling in tissue repair and fibrosis. Int Rev Cell Mol Biol. 2012;296(139-185). [DOI] [PubMed] [Google Scholar]
- 31.de Araujo MF, Etchebehere RM, de Melo MLR, et al. Analysis of CD15, CD57 and HIF-1alpha in biopsies of patients with periimplantitis. Pathol Res Pract. 2017;213(9): 1097-1101. [DOI] [PubMed] [Google Scholar]
- 32.Rommelt C, Munsch T, Drynda A, Lessmann V, Lohmann CH, Bertrand J. Periprosthetic hypoxia as consequence of TRPM7 mediated cobalt influx in osteoblasts. J Biomed Mater Res B Appl Biomater. 2019;107(6):1806-1813. [DOI] [PubMed] [Google Scholar]
- 33.Kim CH, Kim DH, Oh SH, Song SY. Human embryonic stem cell-derived endothelial precursor cell conditioned medium reduces the thickness of the capsule around silicone implants in rats. Ann Plast Surg. 2015;75(3): 348-352. [DOI] [PubMed] [Google Scholar]
- 34.Mediani L, Gibellini F, Bertacchini J, et al. Reversal of the glycolytic phenotype of primary effusion lymphoma cells by combined targeting of cellular metabolism and PI3K/Akt/ mTOR signaling. Oncotarget. 2016;7(5):5521-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrestha P, Davis DA, Veeranna RP, Carey RF, Viollet C, Yarchoan R. Hypoxiainducible factor-1 alpha as a therapeutic target for primary effusion lymphoma. PLoS Pathog. 2017;13(9):e1006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan DJ, Jirstrom K, Kronblad A, et al. CA IX is an independent prognostic marker in premenopausal breast cancer patients with one to three positive lymph nodes and a putative marker of radiation resistance. Clin Cancer Res. 2006;12(21):6421-6431. [DOI] [PubMed] [Google Scholar]
- 37.Hussain SA, Ganesan R, Reynolds G, et al. Hypoxia-regulated carbonic anhydrase IX expression is associated with poor survival in patients with invasive breast cancer. Br J Cancer. 2007;96(1):104-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kon-no H, Ishii G, Nagai K, et al. Carbonic anhydrase IX expression is associated with tumor progression and a poor prognosis of lung adenocarcinoma. Lung Cancer. 2006;54(3):409-418. [DOI] [PubMed] [Google Scholar]
- 39.Lee SH, McIntyre D, Honess D, et al. Carbonic anhydrase IX is a pH-stat that sets an acidic tumour extracellular pH in vivo. Br J Cancer. 2018;119(5):622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svastova E, Witarski W, Csaderova L, et al. Carbonic anhydrase IX interacts with bicarbonate transporters in lamellipodia and increases cell migration via its catalytic domain. J Biol Chem. 2012;287(5):3392-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiche J, Ilc K, Brahimi-Horn MC, Pouyssegur J. Membrane-bound carbonic anhydrases are key pH regulators controlling tumor growth and cell migration. Adv Enzyme Regul. 2010;50(1):20-33. [DOI] [PubMed] [Google Scholar]
- 42.Chiche J, Ilc K, Laferriere J, et al. Hypoxiainducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res. 2009;69(1): 358-368. [DOI] [PubMed] [Google Scholar]
- 43.Lou Y, McDonald PC, Oloumi A, et al. Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res. 2011;71(9):3364-3376. [DOI] [PubMed] [Google Scholar]
- 44.Sansone P, Piazzi G, Paterini P, et al. Cyclooxygenase-2/carbonic anhydrase-IX up-regulation promotes invasive potential and hypoxia survival in colorectal cancer cells. J Cell Mol Med. 2009;13(9B):3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuroda N, Tanaka A, Ohe C, Nagashima Y. Recent advances of immunohistochemistry for diagnosis of renal tumors. Pathol Int. 2013;63(8):381-390. [DOI] [PubMed] [Google Scholar]
- 46.Gabriel SE, Woods JE, O'Fallon WM, Beard CM, Kurland LT, Melton LJ, 3rd. Complications leading to surgery after breast implantation. N Engl J Med. 1997;336(10):677-682. [DOI] [PubMed] [Google Scholar]
- 47.Spear SL, Rottman SJ, Glicksman C, Brown M, Al-Attar A. Late seromas after breast implants: theory and practice. Plast Reconstr Surg. 2012;130(2):423-435. [DOI] [PubMed] [Google Scholar]
- 48.Hanson SE, Hassid VJ, Branch-Brooks C, et al. Validation of a CD30 enzyme-linked immunosorbant assay for the rapid detection of breast implant-associated anaplastic large cell lymphoma. Aesthet Surg J. 2019;40(2):149-153. [DOI] [PubMed] [Google Scholar]
- 49.Clemens MW, Horwitz SM. NCCN consensus guidelines for the diagnosis and management of breast implant-associated anaplastic large cell lymphoma. Aesthet Surg J. 2017;37(3):285-289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.